Abstract
Background
HMGB1 has been overexpressed in the tissues or serum of patients with non-small-cell lung cancer (NSCLC) in several studies. However, the results remain inconsistent.
Objective
The aim of this study was to perform a meta-analysis to investigate the relationship between elevated level of HMGB1 and NSCLC.
Methods
Associated studies were included, and the pooled risk difference and mean difference (MD) together with 95% confidence interval (CI) were calculated.
Results
A total of ten relevant studies on HMGB1 expression were included in this meta-analysis. The pooled results suggested that the expression of HMGB1 in NSCLC tissues was notably higher than those in corresponding nontumor normal tissues by using immu-nohistochemistry (risk difference =0.38, 95% CI: 0.28–0.48, Z=7.67, P<0.00001, I2=0%), Western blot (MD =0.27, 95% CI: 0.06–0.47, Z=2.57, P<0.01), or real-time polymerase chain reaction (MD =15.15, 95% CI: 14.8–15.5, Z=2.08, P=0.04). Serum HMGB1 levels were similarly significantly higher in patients with NSCLC than those in healthy controls. The pooled MDs of HMGB1 in patients with NSCLC compared with healthy controls were 17.54 with 95% CI: 10.99–24.09, Z=5.25, P<0.00001. Two of the included studies were fully reviewed without performing meta-analysis due to the different detection methods used. The protein level of HMGB1 in patients with NSCLC of tumor, nodes, and metastasis (TNM) stages III–IV was higher than that of TNM stages I–II (P<0.047 and P<0.001, respectively).
Conclusion
The expression levels of HMGB1 in both tissues and serum of patients with NSCLC were statistically higher than those of normal lung samples, which indicated that elevated levels of HMGB1 can reveal changes that correlated with disease progression, or even the risk of NSCLC disease progression. The elevated level of HMGB1 could also be considered as a potential biomarker for the diagnosis of patients with NSCLC.
Keywords: HMGB1, non-small-cell lung cancer, meta-analysis
Introduction
Lung cancer is one of the most serious malignant tumors, and ∼80%–85% of lung cancer is non-small-cell lung cancer (NSCLC).1 During the past few years, there have been big breakthroughs in many aspects of the diagnosis and treatment of NSCLC. However, the knowledge of pathogenesis in NSCLC is still limited, and effective indicators for early diagnosis of NSCLC are lacking. It is universally considered that early diagnosis is a crucial means to decrease the mortality of NSCLC. Therefore, looking for perfect biomarkers for early diagnosis and prevention of lung cancer is of important clinical value. Overexpression of RAGE in the lung epithelium inferred that HMGB1 may play an important role in the development of lung cancer.2–5 Furthermore, the levels of HMGB1 expression are strongly correlated with tumor progression.6 However, until now studies have shown inconsistent results regarding the levels of HMGB1 expression in patients with NSCLC.
HMGB1 is a highly conserved nuclear protein that plays a pivotal role in chromatin organization and transcriptional regulation.7,8 Previously, we reported that HMGB1 can act as an inflammatory mediator and accelerate acute allograft rejection by interleukin-17+ γδ T-cell response.9 Currently, increasing number of studies suggest that HMGB1 can also act as an extracellular signaling molecule during tumor progression.10 Elevated level of HMGB1 has been associated with unlimited replicative potential, evasion of programmed cell death, the ability to develop blood vessels, tissue invasion, and metastasis.11,12 During the past 10 years, various case–control studies were performed to investigate the expression of HMGB1 in patients with NSCLC that suggested that HMGB1 plays a pivotal role in diagnosis and prognosis of NSCLC.13–19 However, these studies reported conflicting results owing to different detection methods, small sample size, and low statistical power. For example, Shen et al18 reported that the level of HMGB1 is down-regulated in NSCLC tissues, while other studies indicated that there is a higher level of HMGB1 in NSCLC tissues compared with normal controls.14,16 Thus, whether HMGB1 was overexpressed or not remains unclear in patients with NSCLC, and the clinical diagnosis and prognosis value of HMGB1 need to be investigated further.
Therefore, to reveal the relationship between HMGB1 expression and NSCLC, we performed a meta-analysis and literature review to evaluate the expression of HMGB1 in patients with NSCLC, and we found that high expression of HMGB1 was associated with NSCLC. The abovementioned evidence suggested that high serum or tissue HMGB1 expression may play an important role in the development and progression of NSCLC.
Methods
Search strategy and selection criteria
A bibliographic search was conducted on PubMed, VIP Database for Chinese Technical Periodicals and Embase, Wanfang Data Knowledge Service Platform, and China National Knowledge Infrastructure (up to December 2015) by two investigators (Quansong Xia and Juan Xu) by using the following terms: high-mobility group box 1 (HMGB1) and NSCLC. Manual searches of reference lists from potentially relevant researches were also performed to identify any additional articles that may have been missed using the computer-assisted strategy. No limits were set in terms of language used or study design. The data from the full published article instead of any meeting or abstract were taken. Studies included in this meta-analysis must be focusing on: 1) the expression of HMGB1 in patients with NSCLC was compared with normal controls and usable data can be extracted and 2) the use of real-time polymerase chain reaction (RT-PCR), Western blot, and immunohistochemistry for the detection of tissue HMGB1 expression. Enzyme-linked immunosorbent assay (ELISA) was used for serum HMGB1 detection. All studies were double checked with inclusion and exclusion criteria independently by two investigators (Quansong Xia and Juan Xu).
Data extraction
The first author, published year, country, journal, language, sample size of NSCLC, detection methods, tissue types, and confirmation of diagnosis were extracted from each included studies.13–24 These processes were performed independently by two investigators (Quansong Xia and Juan Xu), and a consensus was reached.
Statistical analysis
The pooled mean difference (MD) and risk difference (RD) together with 95% confidence intervals (CI) were calculated. Then, χ2-based Q statistics and I2 metrics were used to assess the heterogeneity between studies. When I2<50%, a fixed effect model was used to calculate pooled MD or RD, otherwise a random effect model was used. HMGB1 protein or gene expression between NSCLC and normal tissue was compared by the meta-analysis. Two studies investigated the HMGB1 expression levels in different tumor, nodes, and metastasis (TNM) stages of NSCLC, and they are listed in Table 1. All the statistical analyses were performed by using Review manager software (v.5.2, The Nordic Cochrane Centre, The Cochrane Collaboration, Copenhagen, Denmark, http://tech.cochrane.org) and STATA (v.12.0, StataCorp LP, College Station, TX, USA).
Table 1.
Expression of HMGB1 in TNM patients with NSCLC
| Study | Sample size | Stages I–II | Stages III–IV | P-value | Detection methods |
|---|---|---|---|---|---|
| Wang et al14 | 30 | 0.62±0.28 | 0.85±0.32 | 0.047 | Western blot |
| Shang et al16 | 145 | 52.9±23.8 | 102.1±29.4 | <0.001 | ELISA |
Abbreviations: ELISA, enzyme-linked immunosorbent assay; HMGB1, high-mobility group box 1; NSCLC, non-small-cell lung cancer; TNM, tumor, nodes, and metastasis.
Results
A database, including first author, published year, country, language, sample size, methods, and tissue types, was established according to the extracted information from ten studies that met the inclusion criteria (Table 2). The original study yielded a total of 125 articles related to the searched keywords. Figure 1 summarizes the selection process of this study. By screening the title, keywords, and abstracts, 110 of these articles were excluded. Full-text articles from 15 articles were reviewed, and an additional five articles were excluded (four were excluded for not presenting the usable data and one due to the same article with different languages), leaving ten studies for further review.
Table 2.
Main characteristics of included studies
| Study | Year | Country | Language | NSCLC sample size | Method | Tissue/serum |
|---|---|---|---|---|---|---|
| Shang et al16 | 2009 | People’s Republic of China | English | 145 | ELISA | Serum |
| Naumnik et al17 | 2009 | Poland | English | 40 | ELISA | Serum |
| Shen et al18 | 2009 | People’s Republic of China | English | 63 | PCR/Western blot | Tissue |
| Su et al23 | 2012 | People’s Republic of China | Chinese | 69 | Immunohistochemistry | Tissue |
| Zhang et al15 | 2013 | People’s Republic of China | English | 106 | Immunohistochemistry | Tissue |
| Yang and Yang22 | 2013 | People’s Republic of China | Chinese | 64 | Immunohistochemistry | Tissue |
| Wang et al14 | 2014 | People’s Republic of China | English | 30 | Immunohistochemistry/PCR/Western blot | Tissue |
| Yan et al24 | 2014 | People’s Republic of China | Chinese | 30 | PCR/Western blot | Tissue |
| Zhengsen et al20 | 2014 | People’s Republic of China | Chinese | 30 | ELISA | Serum |
| Jakubowska et al13 | 2015 | Poland | English | 45 | ELISA | Serum |
Abbreviations: ELISA, enzyme-linked immunosorbent assay; NSCLC, non-small-cell lung cancer; PCR, polymerase chain reaction.
Figure 1.
Flow diagram of the search and selection process of this study.
Abbreviation: CNKI, China National Knowledge Infrastructure.
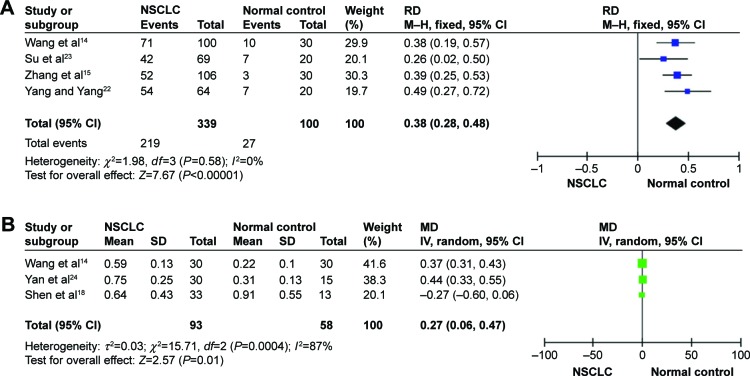
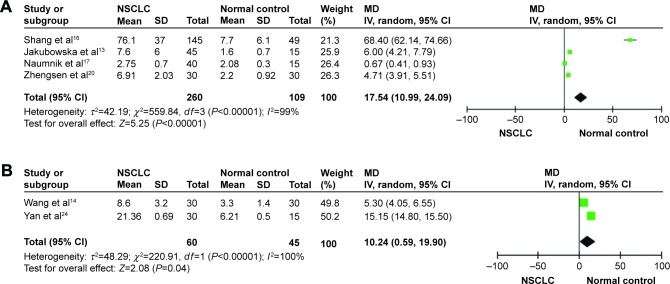
The pooled results indicated that the levels of HMGB1 in NSCLC tissues were notably higher than those in corresponding nontumor normal tissues by using immunohistochemistry (RD =0.38, 95% CI: 0.28–0.48, Z=7.67, P<0.00001, I2=0%; Figure 2A), Western blot (MD =0.27, 95% CI: 0.06–0.47, Z=2.57, P<0.01; Figure 2B). Serum HMGB1 levels were similarly significantly higher in patients with NSCLC than those in healthy controls. The pooled MDs of HMGB1 in patients with NSCLC compared with healthy controls were 17.54 with 95% CI: 10.99–24.09, Z=5.25, P<0.00001 (Figure 3A). By using RT-PCR, the pooled MDs of HMGB1 in patients with NSCLC compared with healthy controls were 15.15 with 95% CI: 14.8–15.5, Z=2.08, P=0.04 (Figure 3B).
Figure 2.
Forest plots for HMGB1 in tissue with NSCLC and normal controls detected by immunohistochemistry (A) and Western blot (B).
Abbreviations: CI, confidence interval; HMGB1, high-mobility group box 1; IV, inverse variance; MD, mean difference; M–H, Mantel–Haenszel; NSCLC, non-small-cell lung cancer; RD, risk difference; SD, standard devaition.
Figure 3.
Forest plots for HMGB1 in serum with NSCLC and normal controls detected by ELISA (A) and for HMGB1 in tissue with NSCLC and normal controls detected by PCR (B).
Abbreviations: CI, confidence interval; ELISA, enzyme-linked immunosorbent assay; HMGB1, high-mobility group box 1; IV, inverse variance; MD, mean difference; NSCLC, non-small-cell lung cancer; PCR, polymerase chain reaction; SD, standard devaition.
Two of the included studies were carefully reviewed without conducting a meta-analysis due to the different detection methods used. The protein level of HMGB1 in patients with NSCLC of TNM stages III–IV was higher than TNM stages I–II (two studies showed P<0.047 and P<0.001, respectively; Table 1).
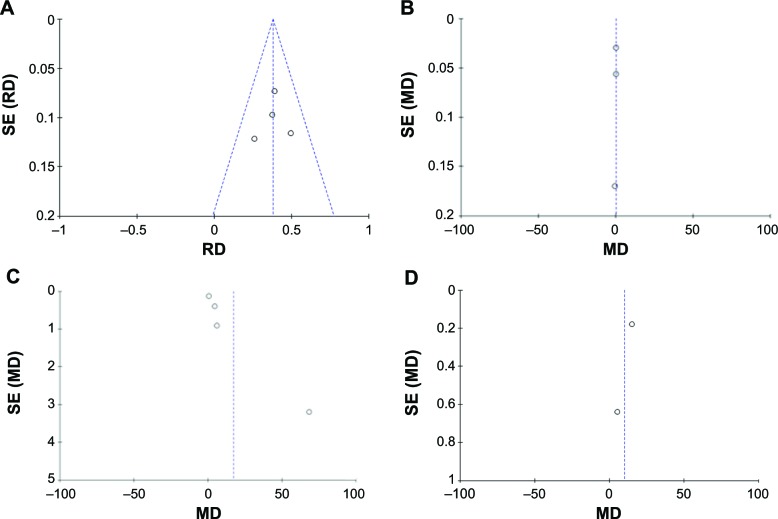
A sensitivity analysis was performed to assess the result stability by removing one study at a time. The pooled MD or RD of HMGB1 in patients with NSCLC compared with normal controls by RT-PCR, Western blot, ELISA, and immunohistochemistry was not significantly changed, suggesting the stability of our results. The funnel plots were substantially symmetric (Figure 4), indicating that no publication biases exist in the meta-analysis of HMGB1 expression. Finally, we performed Egger’s test to calculate the publication bias by using STATA software. No publication bias was detected in the Egger’s test that performed to provide statistical evidence for funnel plot symmetry (P=0.188 for immunohistochemistry, P=0.097 for Western blot, P=0.854 for ELISA, and P=0.317 for RT-PCR).
Figure 4.
Funnel plots for publication bias.
Notes: HMGB1 in tissue with NSCLC and normal controls detected by immunohistochemistry (A) and Western blot (B). HMGB1 in serum with NSCLC and healthy controls detected by ELISA (C). HMGB1 in tissue with NSCLC and normal controls detected by RT-PCR (D).
Abbreviations: ELISA, enzyme-linked immunosorbent assay; HMGB1, high-mobility group box 1; MD, mean difference; NSCLC, non-small-cell lung cancer; RD, risk difference; RT-PCR, real-time polymerase chain reaction; SE, standard error.
Discussion
Cancer development is a complicated dynamic process. Accumulated evidences indicate that HMGB1 is associated with cancer development.25 Nevertheless, the clinical value of HMGB1 in the diagnosis and prognosis of NSCLC is not fully understood at present. In this study, we performed a meta-analysis and indicated that the levels of HMGB1 in NSCLC tissues were notably higher than those in corresponding nontumor normal tissues by using immunohistochemistry, RT-PCR, and Western blot. Moreover, the serum HMGB1 levels were similarly significantly higher in patients with NSCLC than those with normal controls, suggesting that overexpression of HMGB1 can be regarded as an important molecular marker for NSCLC diagnosis. In addition, after a literature review of two included studies, it was found that the protein level of HMGB1 in patients with NSCLC of TNM stages III–IV was higher than TNM stages I–II, suggesting that HMGB1 plays a pivotal role in the progression of NSCLC.
As a late-acting proinflammatory cytokine, HMGB1 plays an important role in many crucial pathological processes including inflammation, cell migration, and tissue regeneration.26–28 Furthermore, HMGB1 may be actively involved in the development and evolution of malignant processes by promoting cell proliferation and migration, modulating the adhesive properties,29 and stimulating tumor neoangiogenesis.30,31 Currently, some reports even suggest that downregulation of HMGB1 may affect the progression of NSCLC,32,33 and interference with HMGB1 expression may increase the sensitivity to chemotherapy drugs by inhibiting HMGB1-mediated cell autophagy or cell apoptosis.34,35 In this regard, the rising degree of HMGB1 serum and tissue level can amplify the progression of lung cancer and may be closely associated with the severity of NSCLC and its development.
To gather more evidence, a literature review about the association between HMGB1 expression and TNM stage was performed. Two studies reported that the protein levels of HMGB1 in patients with NSCLC were significantly different between TNM stages I–II and III–IV (P<0.01 and P<0.0001 respectively), and the elevated HMGB1 protein expression was detected in TNM stages III–IV of patients with NSCLC. This suggests that HMGB1 may play a pivotal role in the development and progression of NSCLC. In support of this, recent studies indicated that HMGB1 was co-localized with RAGE, which was closely associated with invasive and metastatic behavior, indicating their potential ability to promote cellular migration and cancer invasion.6,36 Furthermore, the interaction between HMGB1 and RAGE leads to activation of the NF-κB, MAPK, and MMP-2/MMP-9 signaling pathways.12,37–39 What is important is that by blocking RAGE–HMGB1 complex in human and murine cancer cells, tumor growth and metastasis were suppressed.37,38 Based on the abovementioned evidence, it is suggested that HMGB1 is associated with the progression of NSCLC, besides acting as a diagnosis biomarker. However, due to the different detection methods used and lower statistical power, we did not calculate the pooled results, but we inferred that HMGB1 may act as an important mediator in the progression of NSCLC. Reports suggested that HMGB1 mainly elevated in patients with advanced NSCLC. Thus, the diagnosis value of HMGB1 is limited and HMGB1 may reveal changes in a biological pathway that correlated with disease progression, or even the risk of NSCLC progression.
On the other hand, some limitation in the current meta-analysis did exist that should be noted. First, although some reports had suggested that patients with higher level of HMGB1 expression had a poor clinical prognosis,6,40 Naumnik et al17 insisted that determination of HMGB1 concentrations has no clinical significance in the prognosis of survival time in patients with NSCLC. However, methodological assessment of these studies was not performed due to the different detection methods and lack of usable data, and more work needs to be performed in the future. Second, there is a potential publication bias in this study, since we did not take several unpublished articles and abstracts into account due to their unavailability. In addition, we picked up only eligible English or Chinese language studies in this meta-analysis, while other languages were excluded based on language criteria, which may also introduce bias and affect the findings in our study. A last potential limitation is that our meta-analysis is underpowered to acquire original data from the included studies. Despite all these mentioned limitations, to our knowledge, this is the first meta-analysis on the association of serum and tissue HMGB1 levels with the development of NSCLC.
Conclusion
The expression levels of HMGB1 in both tissues and serum of patients with NSCLC were statistically higher than those of normal lung samples, which indicated that elevated levels of HMGB1 can reveal changes associated with disease progression, or even the risk of NSCLC disease progression. The early detection of HMGB1 may help clinicians to determine valuable therapeutic strategies for patients with NSCLC.
Acknowledgments
There was no support or funding for this study.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Salgia R. Prognostic significance of angiogenesis and angiogenic growth factors in nonsmall cell lung cancer. Cancer. 2011;117(17):3889–3899. doi: 10.1002/cncr.25935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hori O, Brett J, Slattery T, et al. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin. Mediation of neurite outgrowth and co-expression of rage and amphoterin in the developing nervous system. J Biol Chem. 1995;270(43):25752–25761. doi: 10.1074/jbc.270.43.25752. [DOI] [PubMed] [Google Scholar]
- 3.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5(4):331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 4.Klune JR, Dhupar R, Cardinal J, Billiar TR, Tsung A. HMGB1: endogenous danger signaling. Mol Med. 2008;14(7–8):476–484. doi: 10.2119/2008-00034.Klune. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lotze MT, Zeh HJ, Rubartelli A, et al. The grateful dead: damage-associated molecular pattern molecules and reduction/oxidation regulate immunity. Immunol Rev. 2007;220:60–81. doi: 10.1111/j.1600-065X.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- 6.Sasahira T, Akama Y, Fujii K, Kuniyasu H. Expression of receptor for advanced glycation end products and HMGB1/amphoterin in colorectal adenomas. Virchows Arch. 2005;446(4):411–415. doi: 10.1007/s00428-005-1210-x. [DOI] [PubMed] [Google Scholar]
- 7.Bianchi ME. HMGB1 loves company. J Leukoc Biol. 2009;86(3):573–576. doi: 10.1189/jlb.1008585. [DOI] [PubMed] [Google Scholar]
- 8.Agresti A, Bianchi ME. HMGB proteins and gene expression. Curr Opin Genet Dev. 2003;13(2):170–178. doi: 10.1016/s0959-437x(03)00023-6. [DOI] [PubMed] [Google Scholar]
- 9.Xia Q, Duan L, Shi L, Zheng F, Gong F, Fang M. High-mobility group box 1 accelerates early acute allograft rejection via enhancing IL-17+ gammadelta T-cell response. Transpl Int. 2014;27(4):399–407. doi: 10.1111/tri.12264. [DOI] [PubMed] [Google Scholar]
- 10.Kang R, Tang D, Livesey KM, Schapiro NE, Lotze MT, Zeh HJ. The receptor for advanced glycation end-products (RAGE) protects pancreatic tumor cells against oxidative injury. Antioxid Redox Signal. 2011;15(8):2175–2184. doi: 10.1089/ars.2010.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito I, Fukazawa J, Yoshida M. Post-translational methylation of high mobility group box 1 (HMGB1) causes its cytoplasmic localization in neutrophils. J Biol Chem. 2007;282(22):16336–16344. doi: 10.1074/jbc.M608467200. [DOI] [PubMed] [Google Scholar]
- 12.Taguchi A, Blood DC, del Toro G, et al. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature. 2000;405(6784):354–360. doi: 10.1038/35012626. [DOI] [PubMed] [Google Scholar]
- 13.Jakubowska K, Naumnik W, Niklinska W, Chyczewska E. Clinical significance of HMGB-1 and TGF-beta level in serum and BALF of advanced non-small cell lung cancer. Adv Exp Med Biol. 2015;852:49–58. doi: 10.1007/5584_2015_115. [DOI] [PubMed] [Google Scholar]
- 14.Wang JL, Wu DW, Cheng ZZ, Han WZ, Xu SW, Sun NN. Expression of high mobility group box-B1 (HMGB-1) and matrix metalloproteinase-9 (MMP-9) in non-small cell lung cancer (NSCLC) Asian Pac J Cancer Prev. 2014;15(12):4865–4869. doi: 10.7314/apjcp.2014.15.12.4865. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Wang H, Wang J. Expression of HMGB1 and NF-kappaB p65 and its significance in non-small cell lung cancer. Contemp Oncol (Pozn) 2013;17(4):350–355. doi: 10.5114/wo.2013.35291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang GH, Jia CQ, Tian H, et al. Serum high mobility group box protein 1 as a clinical marker for non-small cell lung cancer. Respir Med. 2009;103(12):1949–1953. doi: 10.1016/j.rmed.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 17.Naumnik W, Nilklinska W, Ossolinska M, Chyczewska E. Serum levels of HMGB1, survivin, and VEGF in patients with advanced non-small cell lung cancer during chemotherapy. Folia Histochem Cytobiol. 2009;47(4):703–709. doi: 10.2478/v10042-009-0025-z. [DOI] [PubMed] [Google Scholar]
- 18.Shen X, Hong L, Sun H, Shi M, Song Y. The expression of high-mobility group protein box 1 correlates with the progression of non-small cell lung cancer. Oncol Rep. 2009;22(3):535–539. doi: 10.3892/or_00000468. [DOI] [PubMed] [Google Scholar]
- 19.Sun KK, Ji C, Li X, et al. Overexpression of high mobility group protein B1 correlates with the proliferation and metastasis of lung adenocarcinoma cells. Mol Med Rep. 2013;7(5):1678–1682. doi: 10.3892/mmr.2013.1362. [DOI] [PubMed] [Google Scholar]
- 20.Zhengsen C, Li H, Jiang B, Zhang Z. The elevated level of HMGB1 in serum and its clinical role in patients with COPD, pneumonia and lung cancer. Acta Universitatis Medicinalis Anhui. 2014;49(6):812–815. [Google Scholar]
- 21.Deping H. Clinical pathological study of HMGB1 and β-actenin expression in non-small cell lung cancer. J Hainan Med Univ. 2014;20(6):732–735. [Google Scholar]
- 22.Yang XM, Yang H. Expression of high mobility group box-1 in non-small cell lung cancer and the correlation with clinical pathology. Acta Academiae Medicinae Zunyi. 2013;36(4):343–345. [Google Scholar]
- 23.Su WM, Bi MH. The expression of HMGB1/MMP9 and its clinical significance in non-small cell lung cancer. J Clin Pulm Med. 2012;17(8):1463–1465. [Google Scholar]
- 24.Yan W, Wang L, Liu Q, et al. Expression and significance of serum high mobility group box protein 1 in non-small cell lung cancer. Mod Oncol. 2014;22(08):1834–1836. [Google Scholar]
- 25.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Harris HE, Andersson U, Pisetsky DS. HMGB1: a multifunctional alarmin driving autoimmune and inflammatory disease. Nat Rev Rheumatol. 2012;8(4):195–202. doi: 10.1038/nrrheum.2011.222. [DOI] [PubMed] [Google Scholar]
- 27.Schlueter C, Weber H, Meyer B, et al. Angiogenetic signaling through hypoxia: HMGB1: an angiogenetic switch molecule. Am J Pathol. 2005;166(4):1259–1263. doi: 10.1016/S0002-9440(10)62344-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venereau E, Schiraldi M, Uguccioni M, Bianchi ME. HMGB1 and leukocyte migration during trauma and sterile inflammation. Mol Immunol. 2013;55(1):76–82. doi: 10.1016/j.molimm.2012.10.037. [DOI] [PubMed] [Google Scholar]
- 29.Zuo Z, Che X, Wang Y, et al. High mobility group Box-1 inhibits cancer cell motility and metastasis by suppressing activation of transcription factor CREB and nWASP expression. Oncotarget. 2014;5(17):7458–7470. doi: 10.18632/oncotarget.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28:367–388. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- 31.Zhang CL, Shu MG, Qi HW, Li LW. Inhibition of tumor angiogenesis by HMGB1 A box peptide. Med Hypotheses. 2008;70(2):343–345. doi: 10.1016/j.mehy.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 32.Xiao P, Liu WL. MiR-142-3p functions as a potential tumor suppressor directly targeting HMGB1 in non-small-cell lung carcinoma. Int J Clin Exp Pathol. 2015;8(9):10800–10807. [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Zhang H, Sun M, Yin Z, Qian J. High mobility group box 1- mediated autophagy promotes neuroblastoma cell chemoresistance. Oncol Rep. 2015;34(6):2969–2976. doi: 10.3892/or.2015.4278. [DOI] [PubMed] [Google Scholar]
- 34.Zhang R, Li Y, Wang Z, Chen L, Dong X, Nie X. Interference with HMGB1 increases the sensitivity to chemotherapy drugs by inhibiting HMGB1-mediated cell autophagy and inducing cell apoptosis. Tumour Biol. 2015;36(11):8585–8592. doi: 10.1007/s13277-015-3617-6. [DOI] [PubMed] [Google Scholar]
- 35.Pan B, Chen D, Huang J, et al. HMGB1-mediated autophagy promotes docetaxel resistance in human lung adenocarcinoma. Mol Cancer. 2014;13:165. doi: 10.1186/1476-4598-13-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuniyasu H, Oue N, Wakikawa A, et al. Expression of receptors for advanced glycation end-products (RAGE) is closely associated with the invasive and metastatic activity of gastric cancer. J Pathol. 2002;196(2):163–170. doi: 10.1002/path.1031. [DOI] [PubMed] [Google Scholar]
- 37.Dumitriu IE, Baruah P, Manfredi AA, Bianchi ME, Rovere-Querini P. HMGB1: guiding immunity from within. Trends Immunol. 2005;26(7):381–387. doi: 10.1016/j.it.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 38.Huttunen HJ, Fages C, Kuja-Panula J, Ridley AJ, Rauvala H. Receptor for advanced glycation end products-binding COOH-terminal motif of amphoterin inhibits invasive migration and metastasis. Cancer Res. 2002;62(16):4805–4811. [PubMed] [Google Scholar]
- 39.Chang YH, Chen CM, Chen HY, Yang PC. Pathway-based gene signatures predicting clinical outcome of lung adenocarcinoma. Sci Rep. 2015;5:10979. doi: 10.1038/srep10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu PL, Tsai JR, Hwang JJ, et al. High-mobility group box 1-mediated matrix metalloproteinase-9 expression in non-small cell lung cancer contributes to tumor cell invasiveness. Am J Respir Cell Mol Biol. 2010;43(5):530–538. doi: 10.1165/rcmb.2009-0269OC. [DOI] [PubMed] [Google Scholar]