Abstract
Diet and exercise underpin the risk of obesity-related metabolic disease. Diet alters the gut microbiota, which contributes to aspects of metabolic disease during obesity. Repeated exercise provides metabolic benefits during obesity. We assessed whether exercise could oppose changes in the taxonomic and predicted metagenomic characteristics of the gut microbiota during diet-induced obesity. We hypothesized that high-intensity interval training (HIIT) would counteract high-fat diet (HFD)-induced changes in the microbiota without altering obesity in mice. Compared with chow-fed mice, an obesity-causing HFD decreased the Bacteroidetes-to-Firmicutes ratio and decreased the genetic capacity in the fecal microbiota for metabolic pathways such as the tricarboxylic acid (TCA) cycle. After HFD-induced obesity was established, a subset of mice were HIIT for 6 wk, which increased host aerobic capacity but did not alter body or adipose tissue mass. The effects of exercise training on the microbiota were gut segment dependent and more extensive in the distal gut. HIIT increased the alpha diversity and Bacteroidetes/Firmicutes ratio of the distal gut and fecal microbiota during diet-induced obesity. Exercise training increased the predicted genetic capacity related to the TCA cycle among other aspects of metabolism. Strikingly, the same microbial metabolism indexes that were increased by exercise were all decreased in HFD-fed vs. chow diet-fed mice. Therefore, exercise training directly opposed some of the obesity-related changes in gut microbiota, including lower metagenomic indexes of metabolism. Some host and microbial pathways appeared similarly affected by exercise. These exercise- and diet-induced microbiota interactions can be captured in feces.
Keywords: gut microbiome, exercise and oxidative metabolism, obesity, insulin resistance, diabetes
the mammalian gut contains a large and diverse community of bacteria that have been implicated in infectious and chronic diseases, which can manifest in the gastrointestinal tract and systemically (9, 16, 31). Colonization of the gut is influenced by complex environmental factors that include host genetics, age, diet, lifestyle, diseases, and antibiotic use (12, 15, 53). The reciprocal relationship between the gut microbiota and host metabolism can influence disease risk. Understanding these connections may yield avenues to manipulate resident microbes to mitigate metabolic disease risk or severity including obesity and insulin resistance.
Two major factors contributing to the development of obesity and obesity-related insulin resistance are diet and exercise. Genetic models of hyperphagia-induced obesity and an obesity-causing high-fat diet (HFD) alter the gut microbiota (35, 58). The microbes in this relationship are not just a marker of obesity or metabolic disease, because transmissible components of the fecal microbial community in obese individuals can increase adiposity independently of host genetics (50, 57). Compared with diet, little is known about how exercise may influence the microbiota. However, some connections between the microbiota, exercise status, and daily activity levels are emerging. There are a limited number of studies associating exercise and gut microbiota alterations in rodent models. Studies have found that exercise increases the Bacteroidetes-to-Firmicutes ratio in the absence of obesity. For example, voluntary access to exercise (i.e., wheel running) in rats on a standard diet increased Bacteroidetes and decreased Firmicutes in the feces (49). In addition, voluntary exercise in juvenile chow-fed rats increased Bacteroidetes and decreased Firmicutes in the feces (44). In HFD-fed mice, the extent of voluntary exercise correlated with a change in Bacteroidetes/Firmicutes ratio (20). The effect of exercise on the gut microbiota depends on the mode of exercise performed, such as volitional wheel running compared with forced treadmill training (1). A targeted assessment found that low-intensity exercise can influence the cecal microbiome in obese, hyperglycemic mice (33). However, a complicating factor of all of the above studies is that these types of exercise can lower body mass and/or prevent weight gain during obesity, which may not allow microbial changes to be ascribed to exercise vs. obesity or adiposity. It is currently unknown whether exercise training can overcome changes to the microbiota that occur during diet-induced obesity independently of body or fat mass. It is also important to discern the acute effects of an exercise session vs. repeated exercise training.
Furthermore, gut segment-dependent changes in the microbiome due to exercise are ill defined. We thought it was important to investigate how the stress of exercise could alter gut segment and fecal differences in the microbial communities that include differences in anaerobic and aerobic bacteria, particularly since exercise reduces splenic and intestinal blood flow (26, 45). In fact, exercise can alter blood flow to different parts of gastrointestinal tract, which is modified by repeated exercise training. For example, an acute exercise session reduces blood flow to the large and small intestines in foxhounds, but after 8–12 wk of exercise training, blood flow was not lower in the small intestine (46). Exercise training is also linked to functional changes in the gastrointestinal tract, such as increased mouth to colon transit time without altering intestinal absorption (27). It is important to characterize exercise-induced alterations in the intestinal microbiome as a first step toward understanding whether microbial changes are responding to the stress of exercise and whether microbes are related to functional outcomes.
Endurance exercise training alters a myriad of host responses in both lean and obese individuals, including improvements in insulin sensitivity and increased cardiorespiratory fitness, effects associated with increased aerobic respiration (V̇o2) (52) and muscle mitochondrial enzymes such as those involved in the tricarboxylic acid (TCA) cycle (28, 29). High-intensity interval training (HIIT) has been shown to induce very similar metabolic effects to that elicited by endurance exercise training, including improvements in insulin sensitivity (25, 36). The mechanisms by which HIIT improves insulin sensitivity are not well understood, since obesity status is usually not altered, and recent studies in HFD-fed mice have demonstrated that HIIT improves both adipose and liver insulin sensitivity without changing body mass or adiposity (38). Remarkably, these changes in liver and adipose tissue insulin sensitivity occurred independently of liver lipid content or adipose tissue inflammation, suggesting that other factors may have been responsible for the insulin-sensitizing effects of HIIT.
Given the known connections between the gut microbiota and insulin sensitivity, we sought to determine whether HIIT alters the microbiota in mice after the establishment of obesity. We assessed whether HIIT influenced the taxonomy and predicted functional metagenomics of the murine gut and fecal microbiota during a HFD. The results show that HIIT opposed some of the effects of a HFD to reduce the predicted metabolic genetic capacity of the fecal microbiota independently of obesity. These data suggest that HIIT alters similar metabolic pathways in both the host and microbiota and raises the possibility that this type of exercise training may elicit some of its beneficial effects on metabolism through alterations in the gut microbiome.
MATERIALS AND METHODS
Mice.
All experiments were approved by the McMaster University Animal Research Ethics Board (Hamilton, ON, Canada). All mice were born at McMaster University. Littermate mice were randomly assigned to exercise trained or untrained conditions using male offspring from a given in-house breeding pair of C57 BL/6 mice. This resulted in a mix of different mothers, breeding pairs, and cages in each experimental condition, which was done to limit any environmental or inherited influence on the results obtained for microbiota analysis. Eight-week-old male mice were maintained on a 12:12-h light-dark cycle and fed a HFD (45% kcal from fat, D12451; Research Diets, New Brunswick, NJ) for 12 wk. After 6 wk of HFD feeding, mice were either exercise trained (described in detail below) or left untrained. Untrained mice were exposed to the treadmill environment for equal periods of time as the exercise trained group. After a 3-day acclimation to treadmill running (19), exercise capacity was measured using a graded exercise test during which mice began running at 8 m/min on a 5% grade and treadmill speed was gradually increased by 1 m/min every 2 min until exhaustion, as described previously (38, 54). This test was also repeated after 6 wk of exercise training, which involved treadmill running 3 days per week for 1 h each day, as previously described (38). Specifically, the treadmill running involved running for 2 min at 17 m/min at a 5% grade (100% of average pretrained maximal running speed/exercise capacity) and then resting for 2 min. The treadmill running speed was increased by 1 m/min per week, so that by the end of the training protocol mice were running at 22 m/min. After 6 wk of exercise training, and 24 h after an exercise session, a 6-h fasted insulin tolerance test (ITT) was conducted. Tail vein blood glucose was measured using a glucometer during an insulin tolerance test (1 IU/kg), and epididymal adipose tissue was removed and massed, as described (30, 51). Two additional groups of mice were included as a comparison for microbiota analysis. Littermate male mice from multiple in-house breeding pairs were randomly assigned to standard chow diet feeding (n = 7 mice) or at 6–7 wk of age a subset of mice was fed the HFD (n = 9 mice) for 12 wk. The purpose of these additional groups was to provide context to the comparison between untrained and HIIT gut microbes.
Microbiome sampling.
Feces were collected directly from the anus of mice into sterile tubes, which were immediately snap-frozen in liquid nitrogen before the mice were placed on the treadmill. Before repeated exercise training was initiated, the acute effects of a single exercise session were assessed by collecting feces within 1 h and 1 wk after the initial graded exercise capacity test. To test the chronic effects of repeated exercise training, fecal samples were collected 3 days after the last exercise training session following 6 wk of exercise training, with gut segments collected the following week 1 day after the final exercise session. At the completion of the 6-wk exercise training protocol, the duodenum plus jejunum, ileum, cecum, and colon were snap-frozen in liquid nitrogen and stored at −80°C. Fecal pellets were removed before processing any intestinal segment.
Bacterial profiling.
Genomic DNA was extracted from fecal and gut segment samples. PCR amplification of the variable 3 (V3) region of the 16S rRNA gene was done on each sample, which included Illumina-compatible adapter sequences and barcoding for multiplexing. DNA products of this PCR amplification were sequenced using the MiSeq platform followed by preliminary analysis by the McMaster Genome Center (McMaster University). A custom in-house pipeline was used to process the FASTQ files as described (17). Cutadapt was used to trim sequences beyond the 16S rRNA V3 region, and PANDAseq was used to align paired-end sequences (39, 40). AbundantOTU+ grouped reads into operational taxonomic units (OTUs) based on 97% similarity (41, 60). Taxonomy was assigned to OTUs Ribosomal Database Project (RDP) classifier in Quantitative Insights Into Microbial Ecology (QIIME) (6) against the 2011 version of the Greengenes reference database (18). QIIME was used to calculate the diversity within communities (alpha diversity) and between community diversity (beta diversity), as previously described (6, 17). At the genus level, OTUs were assigned to the corresponding genus and represented to the closest root of the phylogenic tree. This can result in different OTUs being assigned to the same classification. Principal coordinates analysis (PCoA) used the Bray-Curtis dissimilarity values to position the points relative to each other. Sequencing characteristics are described in Table 1. Prediction of metagenome functional content from the 16S rDNA library was developed using PICRUSt software, and PICRUSt predictions were categorized as levels 1 to 3 into KEGG pathways (34). QIIME was used to visualize the predicted functions within KEGG pathways.
Table 1.
QIIME 16S rDNA sequencing analysis
| Trained vs. Untrained Mouse Gut Segments | Trained vs. Untrained Mouse Feces | Chow- vs. HFD-Fed Mouse Feces | |
|---|---|---|---|
| Number of samples | 75 | 14 | 33 |
| Minimum sequences count | 9,881 | 92,644 | 32,649 |
| Maximum sequences count | 216,269 | 131,501 | 76,187 |
| Median sequences count | 76,756 | 100,643 | 42,021 |
| Normalized sequences* | 9,880 | 92,640 | 32,640 |
| Number of 97% phylotypes (genus level-assignable) | 335 | 187 | 165 |
Numbers of sequences in each dataset (of a given sample) were normalized by rarefaction to allow for intra-sample comparisons of the datasets.
Statistical analysis.
Results were analyzed by an unpaired, two-tailed Student's t-test where two means are compared or ANOVA (for more than two means) using GraphPad Prism 6 software. Subsequently, false discovery rate (FDR) was accounted for via implementation of the Benjamini-Hochberg multiple testing adjustment procedure using R, where FDR-corrected P values were estimated for all taxonomic data or predicted metagenomic data within a specific PICRUSt level. Statistical significance was accepted at P < 0.05 after adjustment for FDR.
RESULTS
HFD alters the gut and fecal microbiota.
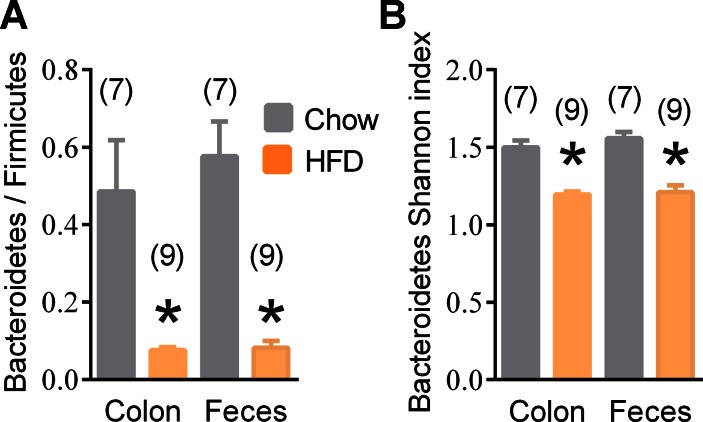
Consistent with previous findings (17, 58), we observed a lower Bacteroidetes/Firmicutes ratio in the distal gut and feces in HFD-fed mice compared with chow-fed mice (Fig. 1A). This comparison of chow-fed mice (average body mass 32 + 0.5 g) to HFD-fed mice (average body mass 50 + 0.7 g) was separate from all exercise-related experiments. Compared with chow-fed mice, HFD-fed mice were insulin and glucose intolerant (data not shown). We found no evidence that the type of diet changed the overall alpha diversity (data not shown), but we found lower alpha diversity (i.e., Shannon index) within the phylum Bacteroidetes in both the colon and feces of HFD-fed mice (Fig. 1B). Additional differences in the colonic and fecal microbiome taxonomy from chow-fed vs. HFD-fed mice are shown in Supplemental Table S1 (supplemental materials are found with the online version of this paper).
Fig. 1.
Phylum and diversity changes in chow- and high-fat diet (HFD)-fed mice. Relative Bacteroidetes-to-Firmicutes ratio (A) and alpha diversity (i.e., Shannon index) within Bacteroidetes (B) in the colon and fecal microbiota from chow-fed and HFD-fed mice. Data are means ± SE. *P < 0.05 Chow vs. HFD. Nos. of mice analyzed for each condition are shown in brackets.
Exercise training during obesity improves insulin tolerance independently of adiposity.
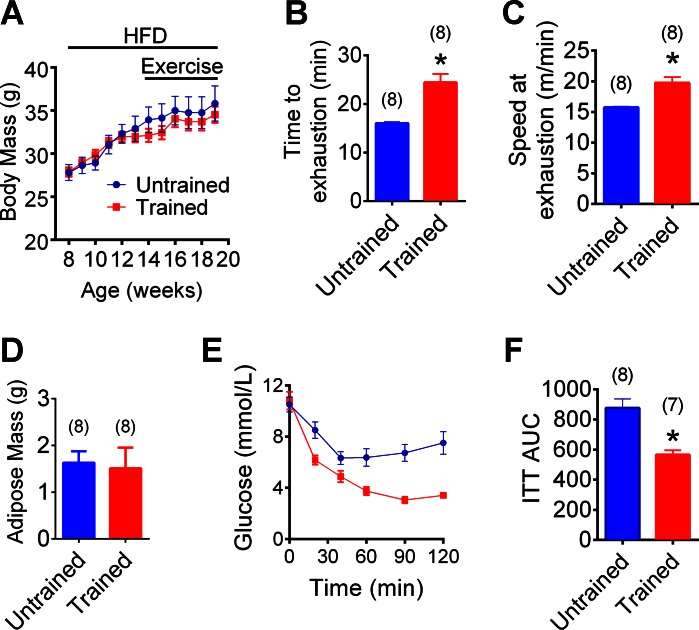
Mice were on an obesity-causing HFD for 12 wk with HIIT during the final 6 wk (Fig. 2A). HIIT was initiated after the establishment of obesity, and this type of repeated exercise training did not alter body mass in HFD-fed mice (Fig. 2A). As expected, HIIT increased the time to exhaustion and running speed at the completion of a graded exercise test (Fig. 2, B and C). Exercise training also increased insulin tolerance, but it did not alter fasting blood glucose and did not alter epididymal adipose tissue mass in HFD-fed mice (Fig. 2, D–F). We (38) recently demonstrated that this HIIT exercise protocol increased oxygen consumption (V̇o2), carbon dioxide consumption (V̇co2), respiratory exchange ratio (RER), food intake, and water intake in HFD-fed mice. We also published that this HIIT exercise protocol does not alter body mass, whole body adiposity, adipose mass, liver mass, or heart mass (38). Importantly, we made every effort to control for different environmental microbial exposures in the mice, including placing all of the untrained mice on the exercise treadmill (which was turned off) for an equal duration that corresponded to each exercise training session.
Fig. 2.
Host physiology associated with high-intensity interval training (HIIT) in HFD-fed mice. Weekly body mass in HFD-fed mice that were HIIT 3 times a week or untrained (A; n = 8 mice per group). Time to exhaustion (B) and running speed at exhaustion (C) during a graded treadmill exercise test in HFD-fed mice exercise trained for 6 wk vs. untrained mice. Epididymal adipose tissue mass in HFD-fed mice exercise trained for 6 wk or untrained (D). Blood glucose (E) and cumulative area under the curve (AUC; F) during insulin tolerance test (ITT; 1 IU/kg) in HFD-fed mice exercise trained for 6 wk or untrained. Data are means ± SE. *P < 0.05 vs. untrained. Nos. of mice analyzed for each condition are shown in brackets.
Exercise training during obesity alters the microbiota in a gut segment-dependent manner.
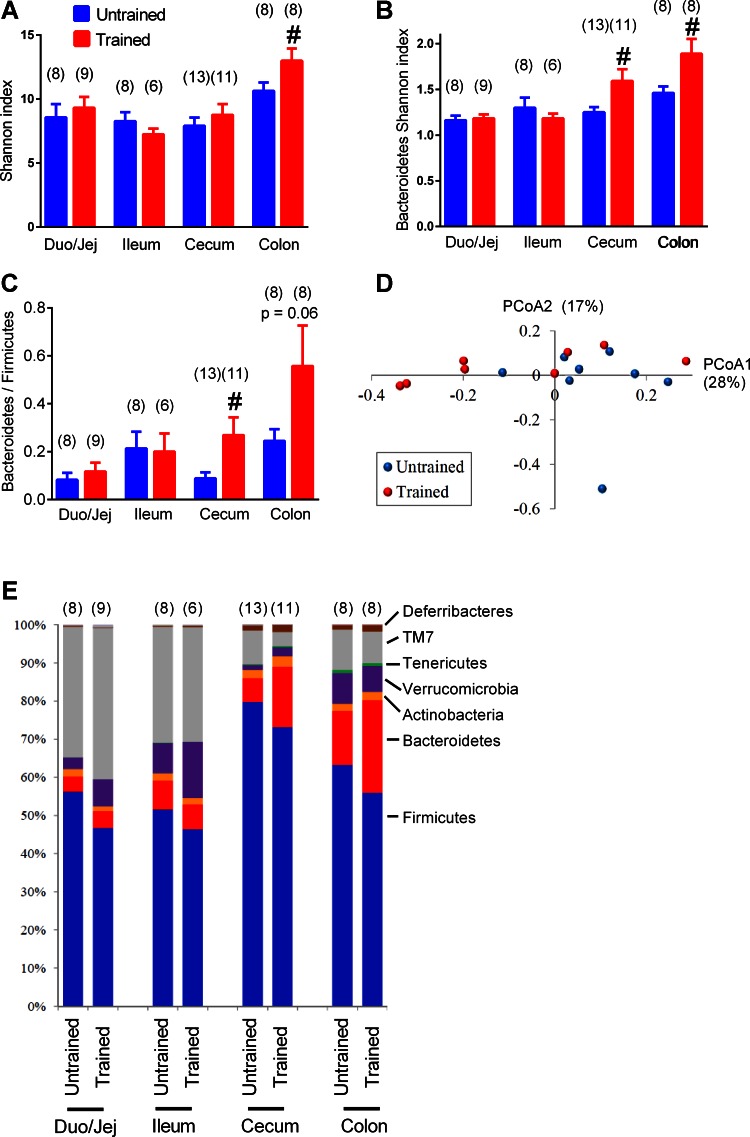
Exercise training of HFD-fed, obese mice increased the overall alpha diversity of the microbiota in the colon (Fig. 3A). Exercise training increased the alpha diversity within the phylum Bacteroidetes in both the cecum and colon (Fig. 3B). Exercise training increased the Bacteroidetes/Firmicutes ratio in the cecum, and a similar trend (P = 0.06) was seen in the colon (Fig. 3C). No clear pattern of exercise training-related changes could be seen after PCoA (Fig. 3D). Exercise training did not significantly alter any other phylum-level microbiota characteristics as depicted by relative quantity of each phylum in various gut segments (Fig. 3E). These data indicate that exercise training opposed the effects of the HFD, since a HFD decreased, but exercise training increased, the Bacteroidetes/Firmicutes ratio and diversity within Bacteroidetes in the distal gut.
Fig. 3.
Phylum and diversity changes in the gut microbiome associated with repeated exercise in HFD-fed mice. Overall alpha diversity (i.e., Shannon index) in microbiota from various gut segments (A) and alpha diversity within Bacteroidetes (B) in untrained and exercise-trained mice that were all HFD fed. Relative Bacteroidetes/Firmicutes ratio in various gut segments in HFD-fed untrained and exercise-trained mice (C). Principal coordinates analysis (PCoA) performed on Bray-Curtis distances in the colon of exercise-trained vs. untrained mice (D). Average relative abundance of the major phyla in HFD-fed untrained and exercise trained mice (E). A–C: data are means ± SE; E: data are mean values. #P < 0.05 trained vs. untrained. Nos. of mice analyzed for each condition are shown in brackets.
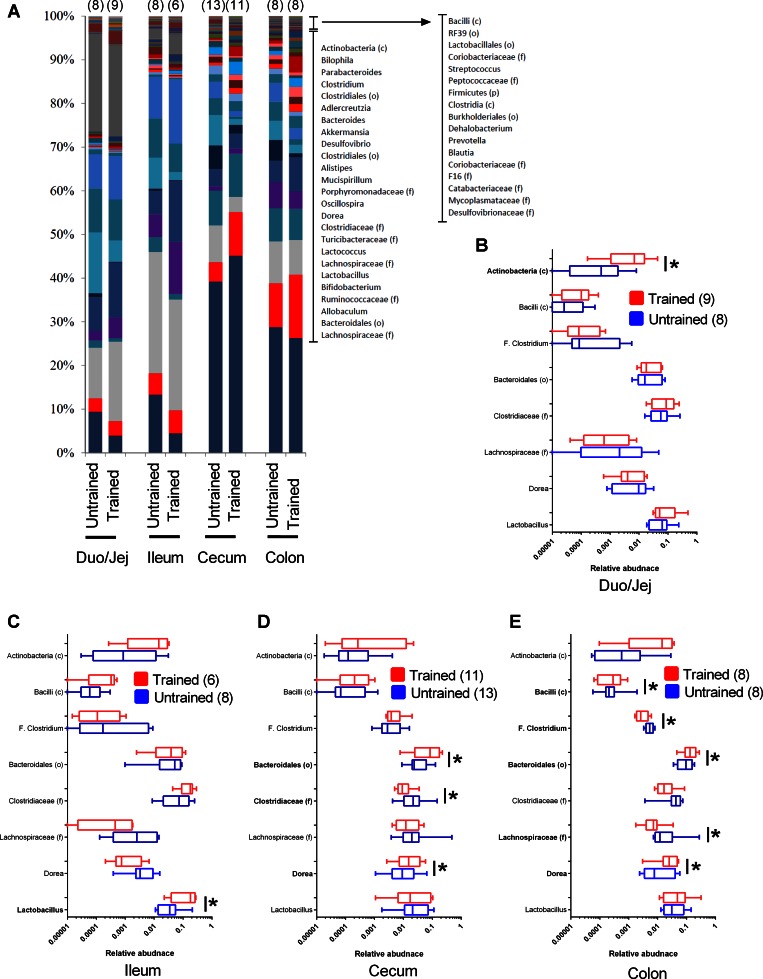
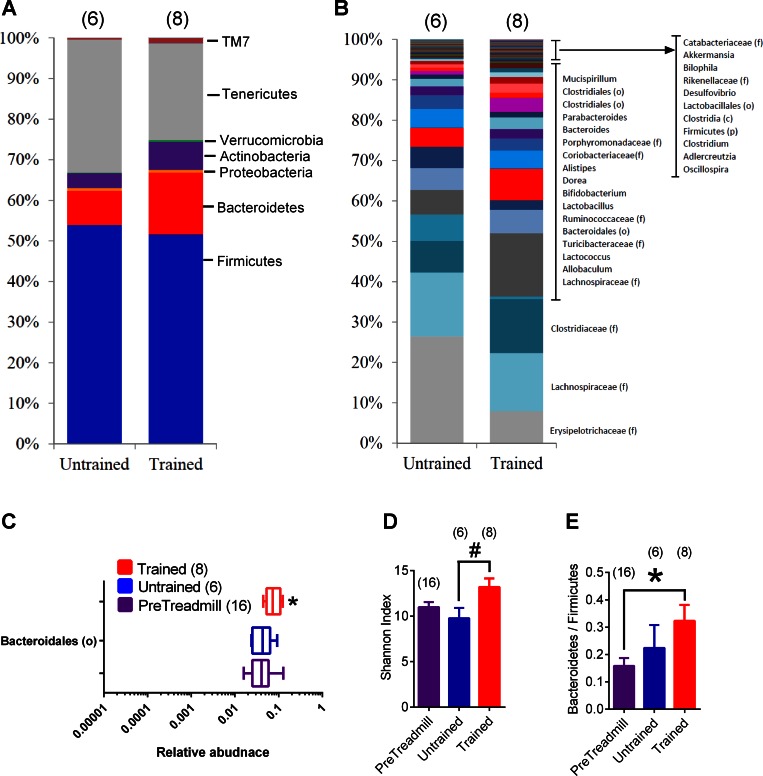
At the OTU level, exercise training during the final 6 wk of the HFD had more profound effects in the distal gut. Figure 4A depicts the average relative abundance of genera in various gut segments of HFD-fed mice that that were exercise trained or untrained. Figure 4,B–E, depicts all of the statistically significant genus-level changes associated with exercise that occurred in at least one of the gut segments after correction for FDR. Based on the relative quantity of each OTU that was detected, exercise training was associated with a significant difference in 1 OTU in the duodenum plus jejunum [Actinobacteria (c)] and 1 OTU in the ileum (Lactobacillus) compared with 3–5 significantly different OTUs in the cecum or colon (Fig. 4, B–E). Also at the OTU level, exercise training increased Bacteroidales (o) (∼1% of the relative abundance) in both the cecum and colon (Fig. 4, D and E). Overall, this data shows that repeated exercise training altered the constituents of the microbiome more profoundly in the distal gut during HFD-induced obesity and that HIIT consistently increased Bacteroidales (o) in both the cecum and colon of obese mice.
Fig. 4.
Genus level changes of gut microbiota associated with repeated exercise. Average relative abundance of genera in various gut segments of HFD-fed untrained and exercise-trained mice (A). OTUs significantly different in microbiota from any gut segment are shown from exercise-trained vs. untrained HFD-fed mice (B–E). Significant differences are noted in boldface for taxonomic classification in duodenum plus jejunum (A), ileum (B), cecum (C), and colon (D). A: data are mean values; B–E: data are box-and-whisker plots. *P < 0.05 trained vs. untrained. Nos. of mice analyzed for each condition are shown in brackets.
Exercise training during obesity alters the fecal microbiota.
Feces are easier to obtain for biomarker assessments, and use of feces allows comparison within the same individual/mouse across time. Hence, we analyzed the feces of all HFD-fed mice before exercise (i.e., PreTreadmill) compared with samples collected after 6 additional weeks from HFD-fed mice that were exercise trained and age-matched HFD-fed untrained mice. Analysis of the fecal samples collected before mice were allocated to trained or untrained groups (i.e., direct comparison within the PreTreadmill groups) showed that the exercise training-induced differences in microbiota characteristics were not derived from an existing difference in these microbiome measurements before mice initiated exercise training (data not shown). Within-subjects analysis at the phylum and genus levels (Fig. 5, A and B) revealed that 6 wk of exercise training significantly increased only 1 OTU in the feces. Bacteroidales (o) was higher in exercise-trained vs. pre-treadmill fecal samples (Fig. 5C). This is consistent with exercise training-induced effects in the cecum and colon. Exercise training increased the alpha diversity, as measured by a higher Shannon index in trained vs. untrained fecal samples (Fig. 5D), but it did not change the Shannon index within Bacteroidetes (data not shown). Exercise training also increased the Bacteroidetes/Firmicutes ratio compared with pre-treadmill fecal samples (Fig. 5E). Again, there was no discernable pattern based on PCoA of exercise-trained, untrained, and PreTreadmill fecal samples (data not shown). These data indicate that exercise training increased Bacteriodales (o) and diversity in the feces, which is reflective of exercise-induced changes in distal gut of HFD-fed mice.
Fig. 5.
Taxonomic changes in the fecal microbiome associated with repeated exercise. Average relative abundance of the major phyla (A) and genera (B) in feces of HFD-fed untrained and exercise-trained mice. Depiction of the only significantly different genus-level change [Bacteriodales (o)] in the feces (C). Alpha diversity (Shannon index) in exercise-trained vs. untrained vs. PreTreadmill conditions in feces from HFD-fed mice (D). Relative Bacteroidetes/Firmicutes ratio in exercise-trained vs. untrained vs. PreTreadmill conditions in feces from HFD-fed mice (E). A and B: data are mean values; C: data are box-and-whisker plots; D and E: data are means ± SE. #P < 0.05 Trained vs. Untrained. *P < 0.05 Trained vs. PreTreadmill. Ttaxonomic data for the PreTreadmill condition are presented in Fig. 6. Nos. of mice analyzed for each condition are shown in brackets.
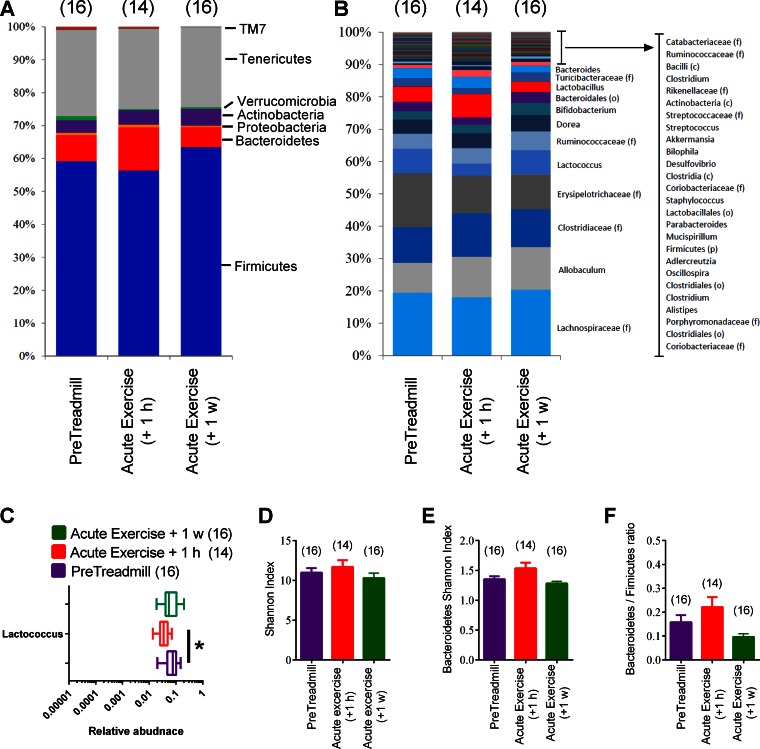
We next sought to determine whether these changes in the taxonomy of the microbiota during repeated exercise training were independent of an acute bout of exercise. In feces collected within 1 h and 1 wk after a single acute bout of exercise, we found no phylum-level changes (Fig. 6, A and B). Only one genus (Lactococcus) was decreased 1 h after acute exercise (Fig. 6C), and this change did not persist 1 wk after acute exercise. Finally, there was no change in the overall alpha diversity (Shannon index) or alpha diversity within Bacteroidetes or Bacteroidetes/Firmicutes ratio of the feces at 1 h or 1 wk after a single exercise session compared with pre-treadmill values (Fig. 6, D–F). Overall, these results show that the changes observed in repeated exercise training could not be explained by the effect of a single exercise session.
Fig. 6.
Taxonomic changes in the fecal microbiome after acute exercise. Average relative abundance of major phyla (A) and genera (B) in feces of HFD-fed (untrained) mice before exercise (PreTreadmill) and 1 h (+1 h) and 1 wk (+1 w) after a single acute exercise session. Depiction of the only significantly different genus-level change (C). Alpha diversity the feces represented by the Shannon index (D) and Shannon index within Bacteroidetes (E) in feces of HFD-fed mice before exercise (PreTreadmill) and 1 h or 1 wk after an acute exercise session. Relative Bacteroidetes/Firmicutes ratio in feces of HFD-fed mice before exercise (PreTreadmill) and 1 h or 1 wk after an acute exercise session (F). A and B: data are mean values; C: data are box-and-whisker plots;. D–F: data are means ± SE. *P < 0.05 Acute Exercise vs. PreTreadmill. Nos. of mice analyzed for each condition are shown in brackets.
Exercise training during obesity alters the predicted function of the fecal microbiota.
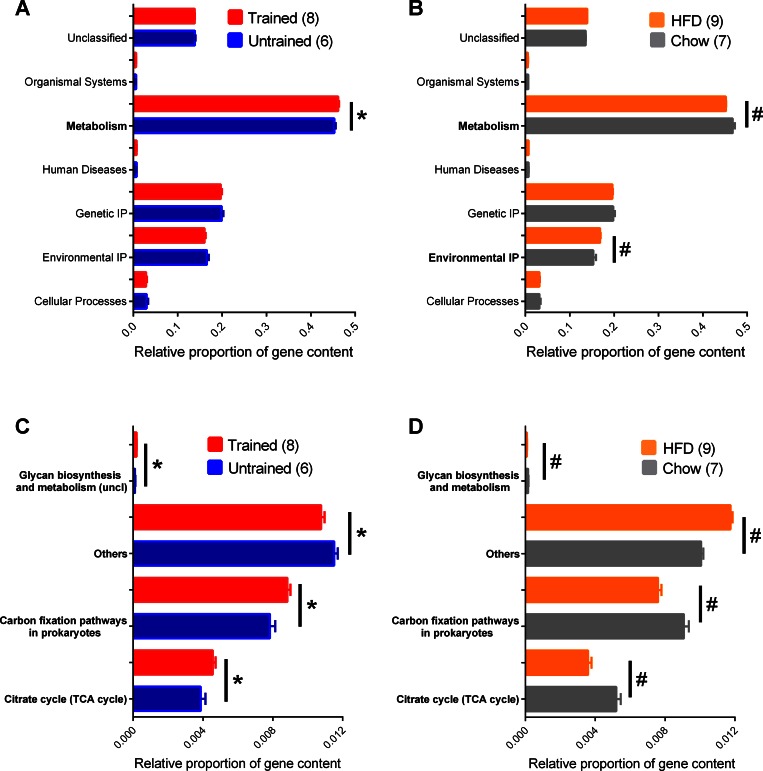
We next sought to determine whether exercise training was associated with predicted changes in the genetic capacity for microbial functions in the feces. Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) analysis showed a significant increase in the microbial genetic material that is KEGG-annotated to metabolism after exercise training (Fig. 7A). This directly opposes a decrease in the genetic capacity related to metabolism during a HFD compared with chow diet (Fig. 7B). HFD also increased the predicted genetic capacity related to Environmental IP (Fig. 7B). Further analysis revealed that exercise training altered predicted pathways only within the KEGG pathways assigned to metabolism. Exercise training increased the fecal microbial genes predicted to be involved in glycan biosynthesis and metabolism, carbon fixation, and the TCA/citrate cycle (Fig. 7C). Again, this directly opposes a subset of the changes during a HFD where all of these KEGG-assigned metabolic pathways were decreased compared with chow fed mice (Fig. 7D). A single acute exercise session did not alter these or any predicted metagenomic characteristics (data not shown).
Fig. 7.
Predicted functional metagenomic changes in the fecal microbiome associated with diet and exercise. Phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) was used to calculate and the relative proportion of genes predicted to be present in various KEGG pathways in feces from exercise-trained and untrained mice all fed a HFD (A). This was compared with the relative proportion of genes predicted to be present in various KEGG pathways in feces from chow-fed and HFD-fed mice e not exercise trained (B). Depicted are significant differences in feces from exercise-trained and untrained mice (C) and side-by-side comparison of a subset of those significant differences within the metabolic pathways in feces from chow-fed and HFD-fed mice (D). Data are means ± SE. *P < 0.05 Trained vs. Untrained; #P < 0.05 Chow vs. HFD. Nos. of mice analyzed for each condition are shown in brackets.
The effects of HFD (vs. chow diet) included over 30 changes within the metabolism annotation (Supplemental Table S1). Therefore, the effects of diet on the microbiota were more extensive compared with exercise. Nevertheless, our results show that exercise and an obesity-causing diet shift the predicted metagenomic characteristics of the fecal microbiota in opposite directions. Furthermore, our results show that exercise during diet-induced obesity directly opposes some of the obesity-related functional characteristics of the microbiota despite no change in body mass.
DISCUSSION
Diet and exercise are two factors involved in obesity. Obesity is associated with altered gut microbiota. We show that HIIT can oppose some of the microbiota changes characteristic of diet-induced obesity even though there is no change in body mass or adipose tissue mass during this type of exercise training. These exercise-related changes in the microbiota include expansion of the predicted genetic capacity related to various pathways in metabolism.
Diet-induced changes in the microbiota have emerged as a contributor to host metabolism relevant to obesity-related disease. Gut-derived microbial factors can influence metabolic disease characteristics (4, 10). Multiple diet-related factors influence obesity, including caloric intake and macronutrient composition. Diets high in fat promote obesity and alter the taxonomic and metagenomic characteristics of the gut microbiome (58). The characteristic changes in the gut microbiota during an obesity-causing HFD include a lower relative level of Bacteroidetes and a relative expansion of Firmicutes (35, 59). A high-fat/sugar diet is the dominant factor influencing the gut microbiota compared with host genetics (7). Furthermore, the fecal microbiota from lean and obese twins can influence fat mass when transferred to germ-free mice (50). Therefore, diet and/or obesity appear to be a powerful factor influencing the metabolic effects of the microbiota. Our results confirm that diet (i.e., HFD vs. chow diet) is a dominant factor in shaping the microbiota, including its predicted functional genetic characteristics. Diet interacts with many host and environmental factors that can potentially influence the microbiota and obesity. We sought to determine whether exercise was a factor that could influence or overcome dietary shifts in the microbiota. Exercise can counterbalance many host metabolic processes during obesity, but little is known about how exercise influences the microbiota during diet-induced obesity.
We found that exercise training (i.e., HIIT) during obesity promoted changes in the distal gut and fecal microbiota that were opposite to those characteristic of obesity and/or a HFD. For example, exercise training increased the Bacteroidetes/Firmicutes ratio and also increased the alpha diversity (within the Bacteroidetes phylum) of the microbiota. HIIT could overcome the influence of a HFD in the distal gut and feces, since exercise training opposed some of the taxonomic and predicted metagenomic changes caused by diet-induced obesity. Altered host obesity/adiposity appears not to be a major driver of these exercise-induced changes in the microbiota, since the type of exercise used in our study did not alter body mass or adipose tissue mass during the HFD. Adaptations to repeated exercise sessions appear to be required for diversity and taxonomic and predicted metagenomic microbiota characteristics, since acute exercise did not alter these indexes. A limitation of our study was the inability to segregate improved insulin tolerance caused by repeated exercise as a factor that could contribute to alterations in the microbiota.
There is evidence mounting that sustainable perturbations to the microbiota can alter obesity-related metabolic disease characteristics. For example, a low dose of antibiotics given during an early-life window can promote increased adiposity (11). The transient early-life dysbiosis caused by certain antibiotics appears to interact with dietary stress, since it magnifies the obesity-causing effect of a HFD (15). Antibiotics can also promote intestinal dysbiosis that is sufficient to accelerate diabetes (3). Immune signals are positioned to connect microbial dysbiosis to disease characteristics (2). Diet is a powerful factor influencing the microbiota, which can influence immune underpinnings of disease (37). Unraveling the compartmentalized responses and connections among diet, dysbiosis, metabolism, and immunity is a complex challenge (43). We (8) have recently shown that diet-induced changes in intestinal immunity do not necessarily parallel immune responses in adipose tissue. It is beyond the scope of this work to determine the underlying immune or endocrine signals that are a potential cause or consequence of exercise and diet-induced changes in the microbiota. Future goals of this type of work include assessments of immune cell populations and inflammatory or endocrine mediators in the gut and metabolic tissues. This will be important, since there is a reciprocal relationship between inflammatory mediators and cellular energy sensors involved in exercise responses (22, 55). These connections could also engage adipokines and myokines and changes in bile acid metabolism and could involve increased food/water intake coincident with exercise training. Intriguingly, different dietary lipids can promote dysbiosis during aging and infectious colitis (23, 24). Therefore, a key future goal is to expand the model of a HFD and assess whether specific dietary lipids interact with the effects of exercise on microbiota and host responses. It is also important to extend this work to humans. Professional rugby players have increased diversity of the fecal microbiota (12). However, it appears very difficult to separate exercise and dietary influences on the human microbiome (48).
The evidence is mounting that exercise is a perturbation that can influence the microbiota. Our most consistent finding regarding taxonomy was an exercise training-related increase in the levels of Bacteriodales (o) in distal gut (cecum, colon) and feces. In HFD-fed mice, we also found an increase in Dorea in the cecum and colon, which is consistent with other groups that employed forced treadmill running and found increased Dorea in the cecum and feces in chow-fed mice (1). Another previous study found by investigating the interaction of exercise and nonexercised mice that were chow fed vs. HFD-fed that exercise and diet each altered the microbiota independently (32). This type of exercise caused decreased body mass in HFD-mice, which is different from the HIIT protocol in the current study. In fact, the mode of exercise could be very important in dictating changes in microbiome characteristics. As expertly reviewed, rodent models using different types of exercise have shown different gut microbiome characteristics (13). This may be related to differential immune responses in the gut. For example, macrophage number is higher in the colon of mice after forced treadmill exercise, an effect that does not occur with voluntary wheel running (13). This is an intriguing gut immune response that may relate to the exercise-related expansion of lactobacilli in the distal colon during voluntary running, but not forced treadmill running (13). These gut immune and microbe changes correspond to functional outcomes such as increased inflammatory score and mortality due to forced treadmill running if there is an additional stress such as a model of colitis in mice (14). It remains to be determined what aspects of metabolic disease, such as obesity, hyperglycemia, or even specific dietary components interact with different modes of exercise to influence the microbiome. This seems worthwhile, since it has already been shown that 6 wk of exercise increased Bifidobacterium in the cecum of control mice but not in hyperglycemic leptin receptor-deficient (db/db) littermate mice (33).
The effects of exercise training on the microbiota during a HFD diet in our study were relatively small compared with changes in diet or other stimuli that can cause dysbiosis such as antibiotics. This was not that surprising, and we chose to characterize exercise because of the overwhelming evidence of the host metabolic changes (and health benefits) induced by exercise. This descriptive information may set the stage for understanding how to assess whether microbes play a role in the metabolic health benefits of exercise during obesity. Furthermore, the altered microbial signatures may be able to be used as a biomarker of exercise status or responsiveness. Our data support investigation of exercise-induced metagenomic characteristics in the feces.
Exercise is well known to increase oxidative capacity, mitochondria, and proteins involved in the TCA cycle (28, 29, 47). Our results show that exercise training increased the predicted metagenomic capacity for metabolism and the TCA cycle in the fecal microbiota. Our data only provide a prediction of the genetic capacity of the microbial community for these KEGG pathways. Caution is warranted in assuming that these genetic indexes influence the same host pathways by producing chemical messengers or altering the metabolism of substrates. It is not yet clear which of the many host effects of exercise relate to microbiota changes. Nevertheless, it is enticing to speculate about the exercise-induced functional changes in the microbiota, which occurred only in the metabolism annotation. The observed changes in poorly classified glycan biosynthesis could be linked to the regulation of mucin O-glycan or biosynthesis of lipopolysaccharide or peptidoglycan. All of these pathways have been implicated in aspects of host metabolic disease (4, 10, 17, 21, 51, 56). The observed changes in carbon fixation and the TCA cycle pathways could be related to acetyl-CoA and short-chain fatty acid regulation, which have been implicated as microbial ligands/metabolites that influence host metabolism (5). For example, exercise has been shown to increase cecal butyrate levels in rats (42).
HFD had a more profound effect on the predicted metagenomic characteristics of the fecal microbiota, which corresponds to diet dominating other factors (7). Intriguingly, all of the exercise-induced changes in predicted microbial function occurred in the opposite direction compared with those associated with a HFD. Our data suggest that exercise counterbalances a subset of the changes in microbial function caused by obesity or obesity-causing diets. It is not clear how repeated exercise training elicits a change in the microbiota of the distal gut, but intestinal motility should be considered. Moreover, exercise reduces blood flow to the colon to a much greater degree than other parts of the intestine. This reduced blood flow might equate to a hypoxic state in the resident microbiota, which could dictate expansion of microbes with a greater metabolic or suitable respiratory capacity. It is probable that the exercise-induced changes include bacteria where the respiratory chain proteins do not necessarily require oxygen as the terminal electron acceptor. Nevertheless, future studies testing different exercise regimens (low- and high-intensity training), altered intestinal blood flow, and hypoxia will be important to delineate whether these environmental cues alter the gut microbiome.
In summary, our results show that repeated exercise training can overcome a distinct subset of the changes in the distal gut and fecal microbiota caused by HFD-induced obesity, independently of changes in body mass or fat mass. In the fecal microbiota, an obesity-causing diet decreased, whereas repeated exercise training increased, several predicted metagenomic traits involved in metabolism, including the TCA/citric acid cycle. Exercise training is well known to regulate these host metabolic pathways, and it is enticing to speculate that the physiological response to exercise also includes changes in analogous microbial pathways.
GRANTS
This study was supported by an operating grant to J. D. Schertzer from the Natural Sciences and Engineering Research Council (NSERC) and an operating GRS from NSERC. J. D. Schertzer holds CDA Scholar (SC-5-12-3891-JS) and CIHR New Investigator awards (MSH-136665). M. G. Surette is a Canada Research Chair in Interdisciplinary Microbiome Research; G. R. Steinberg is a Canada Research Chair in Metabolism and Obesity and the J. Bruce Duncan Chair in Metabolic Diseases.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
E.D., K.M., and J.D.S. performed experiments; E.D., K.M., M.G.S., G.R.S., and J.D.S. analyzed data; E.D., K.M., M.G.S., G.R.S., and J.D.S. interpreted results of experiments; E.D., K.M., and J.D.S. prepared figures; E.D., M.G.S., G.R.S., and J.D.S. edited and revised manuscript; E.D., K.M., M.G.S., G.R.S., and J.D.S. approved final version of manuscript; G.R.S. and J.D.S. conception and design of research; J.D.S. drafted manuscript.
Supplementary Material
REFERENCES
- 1.Allen JM, Berg Miller ME, Pence BD, Whitlock K, Nehra V, Gaskins HR, White BA, Fryer JD, Woods JA. Voluntary and forced exercise differentially alter the gut microbiome in C57BL/6J mice. J Appl Physiol 118: 1059–1066, 2015. [DOI] [PubMed] [Google Scholar]
- 2.Brown K, DeCoffe D, Molcan E, Gibson DL. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients 4: 1095–1119, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown K, Godovannyi A, Ma C, Zhang Y, Ahmadi-Vand Z, Dai C, Gorzelak MA, Chan Y, Chan JM, Lochner A, Dutz JP, Vallance BA, Gibson DL. Prolonged antibiotic treatment induces a diabetogenic intestinal microbiome that accelerates diabetes in NOD mice. ISME J 10: 321–332, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, Burcelin R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 57: 1470–1481, 2008. [DOI] [PubMed] [Google Scholar]
- 5.Cani PD, Everard A, Duparc T. Gut microbiota, enteroendocrine functions and metabolism. Curr Opin Pharmacol 13: 935–940, 2013. [DOI] [PubMed] [Google Scholar]
- 6.Caporaso J, Kuczynski J, Stombaugh J, Bittinger K, Bushman F, Costello E, Fierer N, Pena A, Goodrich J, Gordon J, Huttley G, Kelley S, Knights D, Koenig J, Ley R, Lozupone C, McDonald D, Muegge B, Pirrung M, Reeder J, Sevinsky J, Turnbaugh P, Walters W, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmody Rachel N, Gerber Georg K, Luevano Jesus M Jr, Gatti Daniel M, Somes L, Svenson Karen L, Turnbaugh Peter J. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17: 72–84, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavallari JF, Denou E, Foley KP, Khan WI, Schertzer JD. Different Th17 immunity in gut, liver, and adipose tissues during obesity: the role of diet, genetics, and microbes. Gut Microbes 7: 82–89, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan YK, Estaki M, Gibson DL. Clinical consequences of diet-induced dysbiosis. Ann Nutr Metab 63, Suppl 2: 28–40, 2013. [DOI] [PubMed] [Google Scholar]
- 10.Chi W, Dao D, Lau TC, Henriksbo BD, Cavallari JF, Foley KP, Schertzer JD. Bacterial peptidoglycan stimulates adipocyte lipolysis via NOD1. PLoS One 9: e97675, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, Li H, Alekseyenko AV, Blaser MJ. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488: 621–626, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke SF, Murphy EF, O'Sullivan O, Lucey AJ, Humphreys M, Hogan A, Hayes P, O'Reilly M, Jeffery IB, Wood-Martin R, Kerins DM, Quigley E, Ross RP, O'Toole PW, Molloy MG, Falvey E, Shanahan F, Cotter PD. Exercise and associated dietary extremes impact on gut microbial diversity. Gut 63: 1913–1920, 2014. [DOI] [PubMed] [Google Scholar]
- 13.Cook MD, Allen JM, Pence BD, Wallig MA, Gaskins HR, White BA, Woods JA. Exercise and gut immune function: evidence of alterations in colon immune cell homeostasis and microbiome characteristics with exercise training. Immunol Cell Biol 94: 158–163, 2016. [DOI] [PubMed] [Google Scholar]
- 14.Cook MD, Martin SA, Williams C, Whitlock K, Wallig MA, Pence BD, Woods JA. Forced treadmill exercise training exacerbates inflammation and causes mortality while voluntary wheel training is protective in a mouse model of colitis. Brain Behav Immun 33: 46–56, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox Laura M, Yamanishi S, Sohn J, Alekseyenko Alexander V, Leung Jacqueline M, Cho I, Kim Sungheon G, Li H, Gao Z, Mahana D, Zárate Rodriguez Jorge G, Rogers Arlin B, Robine N, Loke Pn, and Blaser Martin J. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158: 705–721, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delzenne NM, Neyrinck AM, Backhed F, Cani PD. Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nat Rev Endocrinol 7: 639–646, 2011. [DOI] [PubMed] [Google Scholar]
- 17.Denou E, Lolmède K, Garidou L, Pomie C, Chabo C, Lau TC, Fullerton MD, Nigro G, Zakaroff-Girard A, Luche E, Garret C, Serino M, Amar J, Courtney M, Cavallari JF, Henriksbo BD, Barra NG, Foley KP, McPhee JB, Duggan BM, O'Neill HM, Lee AJ, Sansonetti P, Ashkar AA, Khan WI, Surette MG, Bouloumié A, Steinberg GR, Burcelin R, Schertzer JD. Defective NOD2 peptidoglycan sensing promotes diet-induced inflammation, dysbiosis, and insulin resistance. EMBO Mol Med 7: 259–274, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeSantis T, Hugenholtz P, Larsen N, Rojas M, Brodie E, Keller K, Huber T, Dalevi D, Hu P, Andersen G. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dzamko N, Schertzer JD, Ryall JG, Steel R, Macaulay SL, Wee S, Chen ZP, Michell BJ, Oakhill JS, Watt MJ, Jørgensen SB, Lynch GS, Kemp BE, Steinberg GR. AMPK-independent pathways regulate skeletal muscle fatty acid oxidation. J Physiol 586: 5819–5831, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans C, LePard K, Kwak J, Stancukas M, Laskowski S, Dougherty J, Moulton L, Glawe A, Wang Y, Leone V, Antonopoulos D, Smith D, Chang E, Ciancio M. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS One 9: e92193, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA 110: 9066–9071, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fullerton MD, Steinberg GR, Schertzer JD. Immunometabolism of AMPK in insulin resistance and atherosclerosis. Mol Cell Endocrinol 366: 224–234, 2013. [DOI] [PubMed] [Google Scholar]
- 23.Ghosh S, DeCoffe D, Brown K, Rajendiran E, Estaki M, Dai C, Yip A, Gibson DL. Fish oil attenuates omega-6 polyunsaturated fatty acid-induced dysbiosis and infectious colitis but impairs lps dephosphorylation activity causing sepsis. PLoS One 8: e55468, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh S, Molcan E, DeCoffe D, Dai C, Gibson DL. Diets rich in n-6 PUFA induce intestinal microbial dysbiosis in aged mice. Br J Nutr 110: 515–523, 2013. [DOI] [PubMed] [Google Scholar]
- 25.Gillen JB, Percival ME, Ludzki A, Tarnopolsky MA, Gibala MJ. Interval training in the fed or fasted state improves body composition and muscle oxidative capacity in overweight women. Obesity 21: 2249–2255, 2013. [DOI] [PubMed] [Google Scholar]
- 26.Gisolfi CV. Is the GI system built for exercise? Physiology 15: 114–119, 2000. [DOI] [PubMed] [Google Scholar]
- 27.Harris A, Lindeman AK, Martin BJ. Rapid orocecal transit in chronically active persons with high energy intake. J Appl Physiol 70: 1550–1553, 1991. [DOI] [PubMed] [Google Scholar]
- 28.Holloszy JO, Coyle EF. Adaptations of skeletal muscle to endurance exercise and their metabolic consequences. J Appl Physiol 56: 831–838, 1984. [DOI] [PubMed] [Google Scholar]
- 29.Holloszy JO, Oscai LB, Don IJ, Molé PA. Mitochondrial citric acid cycle and related enzymes: Adaptive response to exercise. Biochem Biophys Res Commun 40: 1368–1373, 1970. [DOI] [PubMed] [Google Scholar]
- 30.Jorgensen SB, O'Neill HM, Sylow L, Honeyman J, Hewitt KA, Palanivel R, Fullerton MD, Öberg L, Balendran A, Galic S, van der Poel C, Trounce IA, Lynch GS, Schertzer JD, Steinberg GR. Deletion of skeletal muscle SOCS3 prevents insulin resistance in obesity. Diabetes 62: 56–64, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 14: 685–690, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang S, Jeraldo P, Kurti A, Miller ME, Cook M, Whitlock K, Goldenfeld N, Woods J, White B, Chia N, Fryer J. Diet and exercise orthogonally alter the gut microbiome and reveal independent associations with anxiety and cognition. Mol Neurodegen 9: 36, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lambert JE, Myslicki JP, Bomhof MR, Belke DD, Shearer J, Reimer RA. Exercise training modifies gut microbiota in normal and diabetic mice. Appl Physiol Nutr Metab 40: 749–752, 2015. [DOI] [PubMed] [Google Scholar]
- 34.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31: 814–821, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 102: 11070–11075, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Little JP, Gillen JB, Percival ME, Safdar A, Tarnopolsky MA, Punthakee Z, Jung ME, Gibala MJ. Low-volume high-intensity interval training reduces hyperglycemia and increases muscle mitochondrial capacity in patients with type 2 diabetes. J Appl Physiol 111: 1554–1560, 2011. [DOI] [PubMed] [Google Scholar]
- 37.Lukens JR, Gurung P, Vogel P, Johnson GR, Carter RA, McGoldrick DJ, Bandi SR, Calabrese CR, Walle LV, Lamkanfi M, Kanneganti TD. Dietary modulation of the microbiome affects autoinflammatory disease. Nature 516: 246–249, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcinko K, Sikkema SR, Samaan MC, Kemp BE, Fullerton MD, Steinberg GR. High intensity interval training improves liver and adipose tissue insulin sensitivity. Mol Metab 4: 903–915, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal 17.1, 2011. [Google Scholar]
- 41.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics 13: 1–7, 31, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsumoto M, Inoue R, Tsukahara T, Ushida K, Chiji H, Matsubara N, Hara H. Voluntary running exercise alters microbiota composition and increases n-butyrate concentration in the rat cecum. Biosci Biotechnol Biochem 72: 572–576, 2008. [DOI] [PubMed] [Google Scholar]
- 43.McPhee Joseph B, Schertzer Jonathan D. Immunometabolism of obesity and diabetes: microbiota link compartmentalized immunity in the gut to metabolic tissue inflammation. Clin Sci 129: 1083–1096, 2015. [DOI] [PubMed] [Google Scholar]
- 44.Mika A, Van Treuren W, González A, Herrera JJ, Knight R, Fleshner M. Exercise is more effective at altering gut microbial composition and producing stable changes in lean mass in juvenile versus adult male F344 rats. PLoS One 10: e0125889, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Musch TI, Eklund KE, Hageman KS, Poole DC. Altered regional blood flow responses to submaximal exercise in older rats. J Appl Physiol 96: 81–88, 2004. [DOI] [PubMed] [Google Scholar]
- 46.Musch TI, Haidet GC, Ordway GA, Longhurst JC, Mitchell JH. Training effects on regional blood flow response to maximal exercise in foxhounds. J Appl Physiol 62: 1724–1732, 1987. [DOI] [PubMed] [Google Scholar]
- 47.Nishida Y, Tanaka H, Tobina T, Murakami K, Shono N, Shindo M, Ogawa W, Yoshioka M, St-Amand J. Regulation of muscle genes by moderate exercise. Int J Sports Med 31: 656–670, 2010. [DOI] [PubMed] [Google Scholar]
- 48.O'Sullivan O, Cronin O, Clarke SF, Murphy EF, Molloy MG, Shanahan F, Cotter PD. Exercise and the microbiota. Gut Microbes: 1–6, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Queipo-Ortuno M, Seoane L, Murri M, Pardo M, Gomez-Zumaquero J, Cardona F, Casanueva F, Tinahones F. Gut microbiota composition in male rat models under different nutritional status and physical activity and its association with serum leptin and ghrelin levels. PLoS One 8: e65465, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341 (6150): 1241214, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schertzer JD, Tamrakar AK, Magalhães JG, Pereira S, Bilan PJ, Fullerton MD, Liu Z, Steinberg GR, Giacca A, Philpott DJ, Klip A. NOD1 activators link innate immunity to insulin resistance. Diabetes 60: 2206–2215, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Segal KR, Edano A, Abalos A, Albu J, Blando L, Tomas MB, Pi-Sunyer FX. Effect of exercise training on insulin sensitivity and glucose metabolism in lean, obese, and diabetic men. J Appl Physiol (1985) 71: 2402–24011, 1991. [DOI] [PubMed] [Google Scholar]
- 53.Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, Finlay BB. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun 76: 4726–4736, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steinberg GR, O'Neill HM, Dzamko NL, Galic S, Naim T, Koopman R, Jørgensen SB, Honeyman J, Hewitt K, Chen ZP, Schertzer JD, Scott JW, Koentgen F, Lynch GS, Watt MJ, van Denderen BJW, Campbell DJ, Kemp BE. Whole body deletion of AMP-activated protein kinase β2 reduces muscle AMPK activity and exercise capacity. J Biol Chem 285: 37198–37209, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinberg GR, Schertzer JD. AMPK promotes macrophage fatty acid oxidative metabolism to mitigate inflammation: implications for diabetes and cardiovascular disease. Immunol Cell Biol 92: 340–345, 2014. [DOI] [PubMed] [Google Scholar]
- 56.Tamrakar AK, Schertzer JD, Chiu TT, Foley KP, Bilan PJ, Philpott DJ, Klip A. NOD2 activation induces muscle cell-autonomous innate immune responses and insulin resistance. Endocrinology 151: 5624–5637, 2010. [DOI] [PubMed] [Google Scholar]
- 57.Turnbaugh P, Ley R, Mahowald M, Magrini V, Mardis E, Gordon J. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031, 2006. [DOI] [PubMed] [Google Scholar]
- 58.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host microbe 3: 213–223, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature 457: 480–484, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye Y. Identification and quantification of abundant species from pyrosequences of 16S rRNA by consensus alignment. Proc IEEE Internl Conf Bioinformatics Biomed 2010: 153–157, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.