Abstract
Objective
To evaluate the performance of the Decipher test in predicting lymph node invasion (LNI) on radical prostatectomy (RP) specimens.
Methods
We identified 1,987 consecutive patients with RP who received the Decipher test between February and August 2015 (contemporary cohort). In the contemporary cohort, only the Decipher score from RP specimens was available for analysis. In addition, we identified a consecutive cohort of patients treated with RP between 2006 and 2012 at the University of California, San Diego, with LNI upon pathologic examination (retrospective cohort). The retrospective cohort yielded seven, 22, and 18 tissue specimens from prostate biopsy, RP, and lymph nodes (LNs) for individual patients, respectively. Univariable and multivariable logistic regression analyses were used to evaluate the performance of Decipher in the contemporary cohort with LNI as the endpoint. In the retrospective cohort, concordance of risk groups was assessed using validated cut-points for low (<0.45), intermediate (0.45–0.60), and high (>0.60) Decipher scores.
Results
In the contemporary cohort, 51 (2.6%) patients had LNI. Decipher had an odds ratio of 1.73 (95% confidence interval, 1.46–2.05) and 1.42 (95% confidence interval, 1.19–1.7) per 10% increase in score on univariable and multivariable (adjusting for pathologic Gleason score, extraprostatic extension, and seminal vesicle invasion), respectively. No significant difference in the clinical and pathologic characteristics between the LN positive patients of contemporary and retrospective cohorts was observed (all P>0.05). Accordingly, among LN-positive patients in the contemporary cohort and retrospective cohort, 80% and 77% had Decipher high risk scores (P=1). In the retrospective cohort, prostate biopsy cores with the highest Gleason grade and percentage of tumor involvement had 86% Decipher risk concordance with both RP and LN specimens.
Conclusion
Decipher scores were highly concordant between pre- and post-surgical specimens. Further, Decipher scores from RP tissue were predictive of LNI at RP. If validated in a larger cohort of prostate biopsy specimens for prediction of adverse pathology at RP, Decipher may be useful for improved pre-operative staging.
Keywords: prostate, biopsy, lymph node invasion, genomic classifier, radical prostatectomy, decipher, prognosis
Introduction
Recently, development of genomic signatures in prostate cancer has improved our understanding of disease biology.1,2 Decipher® represents one such test that was developed to predict disease progression after radical prostatectomy (RP) in a cohort of 359 patients.3 Since its conception, Decipher has been validated in multiple cohorts with varying levels of risk and patient populations.4–13 In a majority of previous studies, Decipher was performed using formalin-fixed paraffin embedded specimens from whole gland pathology.4–13 Only recently, performance and feasibility of Decipher on prostate biopsy (PBx) specimens have been studied.10 In the present study, we set to: 1) determine the agreement of Decipher among matched PBx, RP, and lymph node (LN) pathologic specimens from individual patients and 2) evaluate the performance of the Decipher genomic classifier in predicting lymph node invasion (LNI) on RP specimens.
Materials and methods
Patient cohorts
We identified 1,987 consecutive patients with RP who received the Decipher test between February 1 and August 1, 2015, from 308 centers. This contemporary cohort was deidentified, and study researchers were blinded to patient identities and had no access to personally identifiable health information (ClinicalTrials.gov number, NCT02609269). The Qurorum Institutional Review Board (record number: 31079) approved the research protocol for this study. Since all patient related data was de-identified and study researchers were blinded to patient identities and did not have access to personally identifiable health information, it was exempt from human subjects review, and members of the study population did not have to provide informed consent. In the contemporary cohort, only the Decipher score from RP specimens was available for analysis.
The retrospective cohort consisted of 25 consecutive patients treated with RP between 2006 and 2012 with pathologically determined LN metastases (LNI) and available tissue blocks and clinical data at the University of California, San Diego. For each patient, three PBx cores from their diagnostic PBx with tumor tissue were selected based on availability of sufficient tissue for genomic analysis. Likewise, RP and LN specimens positive for tumor were retrieved. For 16 patients, a matched diagnostic PBx core specimen was not available. After filtering for specimens that failed microarray quality control, retrospective cohort yielded seven, 22, and 18 tissue specimens from PBx, RP, and LNs for individual patients, respectively. All specimens were rereviewed by an expert uropathologist (A.S). The index tumor was considered as the lesion with the highest Gleason score sampled using a 1.5 mm punch tool. The “index” PBx was defined as the core with the highest Gleason score and highest percentage of tumor involvement. The University of California, San Diego, institutional review board approved the research protocol under which this study was conducted.
Calculation of Decipher
The expression values for the 22 prespecified biomarkers that constitute Decipher were extracted from the normalized data matrix and entered into the locked random forest algorithm with tuning and weighting parameters defined previously.3 The Decipher readout is a continuous risk score between 0 and 1, with higher scores indicating a greater probability of disease progression or metastasis.3,7,10 Decipher was also stratified into low (<0.45), intermediate (0.45–0.60), and high (>0.60) risk groups based on previously developed and validated cut-points.9,14
Statistical analysis
Univariable and multivariable (MVA) logistic regression analyses were used to evaluate the performance of Decipher in predicting LNI in the contemporary cohort. The c-index of the pathologic and Decipher plus pathologic models was estimated by subjecting the model to bootstrapping with 10,000 resamples. Fisher’s exact test was used to study the association of clinicopathologic variables between study cohorts. Exact binomial confidence intervals (CIs) were constructed to measure the concordance of Decipher score among matched PBx, RP, and LN pathologic specimens in the retrospective cohort. Pairwise agreement of Decipher scores in the retrospective cohort was evaluated using Bland–Altman plots.15 All statistical tests involving biopsy samples were performed after index PBx sample selection unless stated otherwise. In the retrospective cohort, Kaplan–Meier method was used to estimate two endpoints: 1) biochemical recurrence rate, which was defined as a postoperative prostate-specific antigen (PSA) value >0.2 on two consecutive measurements; and 2) clinical recurrence rate, which was defined as local, and/or distant recurrence defined by imaging, and/or prostate-bed biopsy. Statistical analyses were performed using the R statistical software (R Foundation for Statistical Computing, Vienna, Austria) considering a statistical significance at P<0.05.
Results
Patient characteristics
The clinical and pathologic characteristics of patients tested with Decipher in the contemporary cohort are summarized in Table 1. Median age at surgery was 67 years (interquartile range [IQR], 61–71). Median pre-operative PSA was 6.4 ng/mL (IQR: 4.7–9.6). Overall, 75.5%, 11.1%, and 13.3% of patients had pathologic Gleason score ≤7, 8, and ≥9, respectively; 58.9% of patients harbored pT3 disease; and 55.4% had positive surgical margins. Fifty-one (2.6%) patients presented with LNI at RP.
Table 1.
Patient clinical and pathologic characteristics in the contemporary cohort
| Variables | Total (%) |
|---|---|
| No patients (%) | 1,987 (100) |
| Age, year, median (IQR) | 65 (62–69) |
| Pre-operative PSA (ng/mL), median (IQR) | 6.4 (4.7–9.6) |
| Pathologic Gleason score, n (%) | |
| ≤7 | 1,500 (75.5) |
| 8 | 220 (11.1) |
| ≥9 | 264 (13.3) |
| Unknown | 3 (0.2) |
| Pathologic stage, n (%) | |
| T2 | 772 (38.9) |
| T3a | 780 (39.3) |
| T3b | 390 (19.6) |
| T4 | 14 (0.7) |
| Unknown | 31 (1.6) |
| Positive surgical margins, n (%) | 1,100 (55.4) |
| Lymph node invasion, n (%) | 51 (2.6) |
Abbreviations: IQR, interquartile range; PSA, prostate-specific antigen.
In the retrospective cohort, 22 patients had complete clinical and pathologic characteristics and Decipher RP scores for analysis. A subset of 7 and 18 patients had tissue available from PBx and LN specimens, respectively. Retrospective cohort patients’ clinical and pathologic characteristics are summarized in Table 2. Median age at surgery was 60 years (IQR, 57–66). Median pre-operative PSA value was 6.9 ng/mL (IQR, 4.8–16.1). At biopsy, 91% had Gleason score 8–10, 41% had clinical stage T2b disease, and 90.9% were classified as high risk by National Comprehensive Cancer Network (NCCN) criteria. At RP, 90.9% of patients had Gleason score 8–10, 81.9% harbored pT3 disease, and 54.5% had positive surgical margins. The median follow-up time for this cohort was 16 months (IQR, 4–35). At 5 years post-RP, the estimated biochemical recurrence rate and clinical recurrence rate were 73% and 24%, respectively (Figure S1). A comparison of the clinical and pathologic characteristics of the LNI patients in the contemporary and retrospective cohorts did not show any significant difference (all P<0.1) between the two cohorts (Table S1).
Table 2.
Patient clinical and pathologic characteristics in the retrospective cohort
| Variables | Total (%) |
|---|---|
| No patients (%) | 22 (100) |
| Age, year, median (IQR) | 60 (57–66) |
| Pre-operative PSA (ng/mL), median (IQR) | 6.9 (4.8–16.1) |
| Pathologic Gleason score, n (%) | |
| ≤7 | 2 (9.1) |
| 8 | 9 (40.9) |
| ≥9 | 11 (50.0) |
| Unknown | 0 (0) |
| Pathologic stage, n (%) | |
| T2 | 3 (13.6) |
| T3a | 8 (36.4) |
| T3b | 10 (45.5) |
| T4 | 1 (4.5) |
| Positive surgical margins, n (%) | 12 (54.5) |
| Lymph node invasion, n (%) | 22 (100) |
| Follow-up time (censored), mo, median (IQR) | 16 (4–35) |
Abbreviations: IQR, interquartile range; mo, months; PSA, prostate-specific antigen; yr, year.
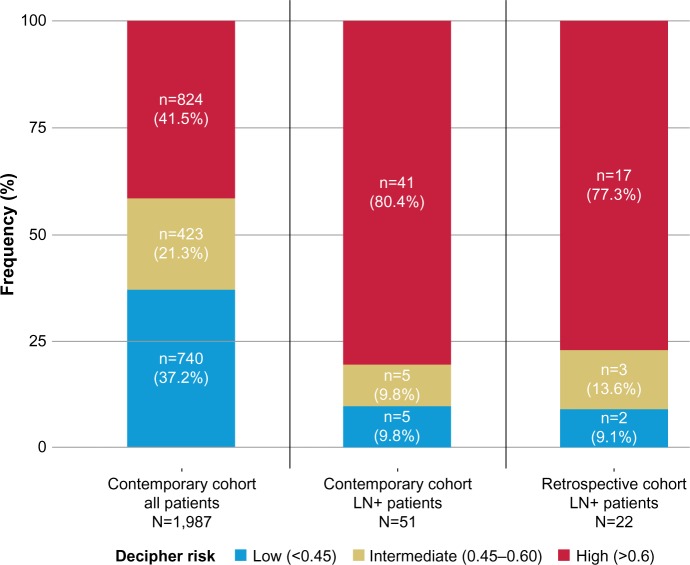
Patients were also stratified using Decipher risk groups (low, intermediate, and high). In the contemporary cohort, 9.8%, 9.8%, and 80.4% of LN positive patients were categorized as Decipher low, intermediate, and high risk, respectively (Figure 1). In the retrospective cohort, we observed a similar distribution with 9.1%, 13.6%, and 77.3% categorized into Decipher low, intermediate, and high risk, respectively. There was no significant difference with respect to Decipher distribution of LN positive patients between the two cohorts (P>0.1).
Figure 1.
Bar charts showing frequency of Decipher risk groups in (left) contemporary cohort, (center) Ln+ patients of contemporary cohort, and (right) Ln+ patients of retrospective cohort from genomic marker assessment in RP specimens.
Abbreviations: Ln, lymph node; RP, radical prostatectomy.
Evaluation of Decipher for LnI prediction in the contemporary cohort
On univariable, Decipher had an odds ratio (OR) of 1.73 (95% CI, 1.46–2.05, P<0.001) per 10% increase in score for predicting the presence of LNI (Table 3). Gleason score 8 disease had an OR of 8.77 (95% CI, 1.18–65.29, P=0.03). Extraprostatic extension (EPE) and seminal vesicle invasion (SVI) also were significant predictors of LNI with OR of 6.10 (95% CI, 2.59–14.35, P<0.001) and 5.74 (95% CI, 3.25–10.13, P<0.001), respectively. Patient age at RP and pre-operative PSA were not significant predictors of LNI (data not shown). Patient age at RP and pre-operative PSA were not modeled on MVA as these variables were missing for a significant portion of patients.
Table 3.
Univariable logistic regression analysis of Decipher and pathologic variables for prediction of lymph node invasion in the contemporary cohort
| Variables/endpoint | Lymph node invasion
|
|
|---|---|---|
| OR (95% CI) | P-value | |
| Deciphera | 1.73 (1.46–2.05) | <0.001 |
| Pathologic Gleason score 3+3 | ref | 1 |
| Pathologic Gleason score 3+4 | 0.78 (0.08–7.58) | 0.83 |
| Pathologic Gleason score 4+3 | 2 (0.24–16.78) | 0.52 |
| Pathologic Gleason score 8 | 8.77 (1.18–65.29) | 0.03 |
| Pathologic Gleason score ≥9 | 6.01 (0.79–45.89) | 0.08 |
| Extraprostatic extension | 6.1 (2.59–14.35) | <0.001 |
| Seminal vesicle invasion | 5.74 (3.25–10.13) | <0.001 |
Note:
Decipher reported per 0.1 unit increase. A statistical significance at P<0.05 was applied.
Abbreviations: CI, confidence interval; OR, odds ratio; ref, reference group.
On MVA, adjusting for pathologic Gleason score, EPE, and SVI Decipher had an OR of 1.42 (95% CI, 1.19–1.7, P<0.001) per 10% increase in score for predicting the presence of LNI (Table 4). EPE and SVI also remained significant predictors of LNI on MVA. In contrast, Gleason score 8 disease did not remain significant for predicting LNI after adjusting for Decipher and other pathologic variables. Similar results were observed in univariable and MVA for Decipher risk groups (Tables S2 and S3). On MVA analysis adjusting for pathologic Gleason score, EPE, and SVI, the Decipher high risk group (>0.6) had an OR of 3.23 (95% CI, 1.2–8.73, P=0.02), similar to the OR of EPE and SVI for predicting the presence of LNI. Finally, discrimination analysis showed that Decipher had a c-index of 0.78 (95% CI, 0.71–0.84) for prediction of LNI. Combination of Decipher with pathologic variables increased the c-index to 0.85 (95% CI, 0.73–0.86) from 0.83 (95% CI, 0.71–0.84) for pathologic model alone (Figure S2).
Table 4.
Multivariable logistic regression analysis of Decipher and pathologic variables for prediction of lymph node invasion in the contemporary cohort
| Variables/endpoint | Lymph node invasion
|
|
|---|---|---|
| OR (95% CI) | P-value | |
| Deciphera | 1.42 (1.19–1.7) | <0.001 |
| Pathologic Gleason score 3+3 | ref | 1 |
| Pathologic Gleason score 3+4 | 0.39 (0.04–3.92) | 0.43 |
| Pathologic Gleason score 4+3 | 0.56 (0.06–4.95) | 0.6 |
| Pathologic Gleason score 8 | 1.84 (0.23–14.73) | 0.57 |
| Pathologic Gleason score ≥9 | 0.85 (0.1–7.09) | 0.88 |
| Extraprostatic extension | 3.48 (1.44–8.41) | 0.01 |
| Seminal vesicle invasion | 2.73 (1.47–5.08) | <0.001 |
Note:
Decipher reported per 0.1 unit increase. A statistical significance at P<0.05 was applied.
Abbreviations: CI, confidence interval; OR, odds ratio; ref, reference group.
Concordance of Decipher in pre- and post-RP tissue specimens in the retrospective cohort
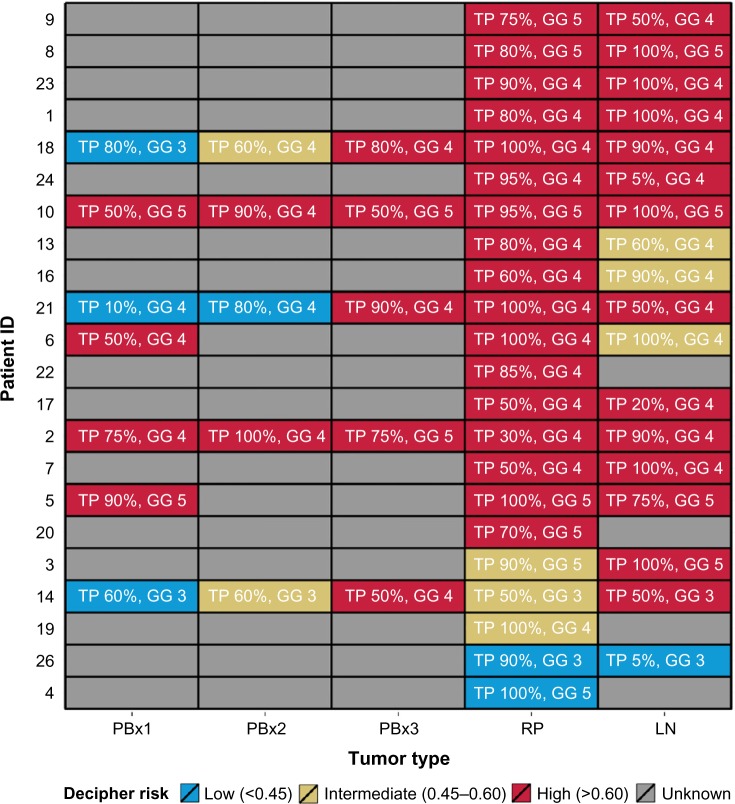
A patient-per-patient pairwise agreement among biopsy cores (≤3 cores for each patient with available Decipher scores), RP, and LN specimens is shown in Figure 2. Among all available PBx specimens, 65% were categorized as Decipher high risk, whereas 100% of selected PBx specimens (highest sampled Gleason grade and percentage of tumor involvement), 77% RP, and 78% LN specimens had high Decipher scores. Bland–Altman plots, which examined patient-per-patient agreement, showed a good overall concordance for all three compared tissue specimens (Figure S3).
Figure 2.
Heat map of Decipher risk groups for all patients in the retrospective cohort.
Abbreviations: GG, highest Gleason grade; Ln, lymph node; PBx, prostate biopsy; RP, radical prostatectomy; TP, tumor percentage.
Finally, we examined Decipher agreement in pairwise comparisons between the following 1) PBx and RP specimens, 2) PBx and LN specimens, and 3) RP and LN specimens when PBx selection was performed (Table 5). Without selection of PBx specimens (ie, taking the average Decipher score for all cores), the overall concordance with RP Decipher risk groups was 71%, which increased to 86% concordance when the selected PBx core (ie, with the highest sampled Gleason grade and percentage of tumor involvement) was compared to RP Decipher. The concordance between PBx and LN was 43% for the average of all PBx specimens in each patient and similarly increased to 86% when comparing only the PBx with highest sampled Gleason grade and percentage of tumor involvement. The concordance of RP and LN Decipher scores was 72%.
Table 5.
Concordance of Decipher scores across tumor types using various selection methods for biopsy samples with multiple tumors in the retrospective cohort
| Pairwise comparison | Overall concordance | Concordance/PBx selected based on highest TP and dominant GG |
|---|---|---|
| PBx-RP (%) | 71 (29–96)a | 86 (42–100) |
| PBx-Ln (%) | 43 (10–82)a | 86 (42–100) |
| RP-Ln (%) | 72 (47–90) | 72 (47–90) |
Note:
In case of a tie, sample with highest tumor percent and dominant Gleason grade was selected.
Abbreviations: GG, highest Gleason grade; Ln, lymph node; PBx, prostate biopsy; RP, radical prostatectomy; TP, tumor percentage.
Discussion
Here, we evaluated the performance of Decipher scores on RP specimens for prediction of LNI in a contemporary cohort and then in an exploratory analysis of a small retrospective cohort determined the agreement of Decipher scores among pre- and post-operative tissue specimens, including LN with tumor involvement. Our results show that Decipher is a strong predictor of LNI in a contemporary cohort of nearly 2,000 patients. In a retrospective cohort, with similar characteris tics to the LNI patients of a contemporary RP cohort, we observed high concordance between Decipher scores in PBx and both RP and LN specimens of individual patients. The caveat is that this is only true when the PBx sampled with highest Gleason grade and percentage of tumor involvement is selected.
Our study has several limitations. In the contemporary cohort: the lack of information on pre-operative variables, extended pelvic lymph node dissection (ePLND) status, and number of LN retrieved for all patients. In the retrospective cohort: the small sample size of the matched PBx specimens and the lack of a matched set of control patients (ie, without LNI) to determine the discrimination performance of the genomic marker expression in PBx for predicting LNI. We aimed to address these issues in part by considering a large contemporary cohort that included both LN negative and positive patients in order to show the discrimination of the genomic marker for predicting the presence of LNI. However, further validation of Decipher on larger PBx cohorts is warranted to confirm the results of the current study.
Conclusion
In summary, in our study, Decipher was highly predictive of LNI from analysis of RP specimens in a large contemporary cohort. Improved post-operative LNI staging may be useful for selection of patients for adjuvant hormonal or whole pelvic irradiation. Decipher scores were also highly concordant between pre- and post-operative specimens. If validated in a larger cohort of PBx specimens, Decipher may improve pre-operative staging, which is useful for optimal selection of patients for neoadjuvant therapy, primary radiotherapy as well as RP with ePLND.
Supplementary materials
Kaplan–Meier estimates of survival-free (A) BCR and (B) CR in retrospective cohort.
Abbreviations: BCR, biochemical recurrence; CR, clinical recurrence; RP, radical prostatectomy.
ROC curves of Decipher and pathologic variables in the contemporary cohort.
Abbreviation: CI, confidence interval.
Pairwise Bland–Altman plots to evaluate agreement of Decipher scores between (A) PBx and RP, (B) PBx and Lns, and (C) RP and Lns in retrospective cohort.
Abbreviations: PBx, prostate biopsy; RP, radical prostatectomy; Ln, lymph node.
Table S1.
Comparison of LnI patient characteristics from retrospective and contemporary cohorts
| Variables | Retrospective cohort N=22 | Contemporary cohort LN N=51 | P-value* |
|---|---|---|---|
| Patient characteristics | |||
| Age, yr, median (IQR) | 60 (57–66) | 65 (64–68) | |
| Pre-operative PSA (ng/mL), median (IQR) | 6.9 (4.8–16.1) | 7.58 (5.1–11.8) | |
| Pathologic Gleason score, n (%) | >0.1 | ||
| ≥7 | 2 (9.1) | 10 (19.6) | |
| 8 | 9 (40.9) | 25 (49.0) | |
| ≥9 | 11 (50.0) | 15 (29.4) | |
| Unknown | 0 (0.0) | 1 (2.0) | |
| Pathologic stage, n (%) | >0.1 | ||
| T2 | 3 (13.6) | 5 (9.8) | |
| T3a | 8 (36.4) | 16 (31.4) | |
| T3b | 10 (45.5) | 29 (56.9) | |
| T4 | 1 (4.5) | 1 (2.0) | |
Note:
Fisher’s exact test. A statistical significance at P<0.05 was applied.
Abbreviations: IQR, interquartile range; Ln, lymph node; LnI, lymph node invasion; PSA, prostate-specific antigen; yr, year.
Table S2.
Univariable logistic regression analysis of categorical Decipher and pathologic variables for prediction of lymph node invasion in the contemporary cohort
| Variables/endpoint | Lymph node invasion
|
|
|---|---|---|
| OR (95% CI) | P-value | |
| Decipher low risk | ref | 1 |
| Decipher intermediate risk | 1.76 (0.51–6.11) | 0.37 |
| Decipher high risk | 7.7 (3.03–19.58) | <0.001 |
| Pathologic Gleason score 3+3 | ref | 1 |
| Pathologic Gleason score 3+4 | 0.78 (0.08–7.58) | 0.83 |
| Pathologic Gleason score 4+3 | 2 (0.24–16.78) | 0.52 |
| Pathologic Gleason score 8 | 8.77 (1.18–65.29) | 0.03 |
| Pathologic Gleason score ≥9 | 6.01 (0.79–45.89) | 0.08 |
| Extraprostatic extension | 6.1 (2.59–14.35) | <0.001 |
| Seminal vesicle invasion | 5.74 (3.25–10.13) | <0.001 |
Abbreviations: CI, confidence interval; OR, odds ratio; ref, reference group.
Note: A statistical significance at P<0.05 was applied.
Table S3.
Multivariable logistic regression analysis of categorical Decipher and pathologic variables for prediction of lymph node invasion in the contemporary cohort
| Variables/endpoint | Lymph node invasion
|
|
|---|---|---|
| OR (95% CI) | P-value | |
| Decipher low risk | ref | 1 |
| Decipher intermediate risk | 1.23 (0.35–4.38) | 0.75 |
| Decipher high risk | 3.23 (1.2–8.73) | 0.02 |
| Pathologic Gleason score 3+3 | ref | 1 |
| Pathologic Gleason score 3+4 | 0.41 (0.04–4.12) | 0.45 |
| Pathologic Gleason score 4+3 | 0.64 (0.07–5.72) | 0.69 |
| Pathologic Gleason score 8 | 2.46 (0.31–19.57) | 0.4 |
| Pathologic Gleason score ≥9 | 1.1 (0.13–9.25) | 0.93 |
| Extraprostatic extension | 3.93 (1.63–9.46) | <0.001 |
| Seminal vesicle invasion | 3.09 (1.67–5.73) | <0.001 |
Abbreviations: CI, confidence interval; OR, odds ratio; ref, reference group.
Note: A statistical significance at P<0.05 was applied.
Acknowledgments
CJK received an unrestricted research grant from GenomeDx to support the costs of sample acquisition and data generation. FA is a consultant and KY, ZH, LLCL, HS, MA, and ED are employees of GenomeDx Biosciences.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Boström PJ, Bjartell AS, Catto JWF, et al. Genomic predictors of outcome in prostate cancer. Eur Urol. 2015;68(6):1033–1044. doi: 10.1016/j.eururo.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Ross AE, D’Amico AV, Freedland SJ. Which, when and why? Rational use of tissue-based molecular testing in localized prostate cancer. Prostate Cancer Prostatic Dis. 2015 Jun 30; doi: 10.1038/pcan.2015.31. Epub. [DOI] [PubMed] [Google Scholar]
- 3.Erho N, Crisan A, Vergara IA, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8(6):e66855. doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karnes RJ, Bergstralh EJ, Davicioni E, et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol. 2013;190(6):2047–2053. doi: 10.1016/j.juro.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Den RB, Feng FY, Showalter TN, et al. Genomic prostate cancer classifier predicts biochemical failure and metastases in patients after postoperative radiation therapy. Int J Radiat Oncol Biol Phys. 2014;89(5):1038–1046. doi: 10.1016/j.ijrobp.2014.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross AE, Feng FY, Ghadessi M, et al. A genomic classifier predicting metastatic disease progression in men with biochemical recurrence after prostatectomy. Prostate Cancer Prostatic Dis. 2014;17(1):64–69. doi: 10.1038/pcan.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein EA, Yousefi K, Haddad Z, et al. A genomic classifier improves prediction of metastatic disease within 5 years after surgery in node-negative high-risk prostate cancer patients managed by radical pros-tatectomy without adjuvant therapy. Eur Urol. 2015;67(4):778–786. doi: 10.1016/j.eururo.2014.10.036. [DOI] [PubMed] [Google Scholar]
- 8.Den RB, Yousefi K, Trabulsi EJ, et al. Genomic classifier identifies men with adverse pathology after radical prostatectomy who benefit from adjuvant radiation therapy. J Clin Oncol. 2015;33(8):944–951. doi: 10.1200/JCO.2014.59.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross AE, Johnson MH, Yousefi K, et al. Tissue-based genomics augments post-prostatectomy risk stratification in a natural history cohort of intermediate- and high-risk men. Eur Urol. 2016;69(1):157–165. doi: 10.1016/j.eururo.2015.05.042. [DOI] [PubMed] [Google Scholar]
- 10.Klein EA, Haddad Z, Yousefi K, et al. Decipher genomic classifier measured on prostate biopsy predicts metastasis risk. Urology. 2016 Jan 22; doi: 10.1016/j.urology.2016.01.012. Epub. [DOI] [PubMed] [Google Scholar]
- 11.Freedland SJ, Choeurng V, Howard L, et al. Utilization of a genomic classifier for prediction of metastasis following salvage radiation therapy after radical prostatectomy. Eur Urol. 2016 Jan 21; doi: 10.1016/j.eururo.2016.01.008. Epub. [DOI] [PubMed] [Google Scholar]
- 12.Yamoah K, Johnson MH, Choeurng V, et al. Novel biomarker signature that may predict aggressive disease in African American men with prostate cancer. J Clin Oncol. 2015;33(25):2789–2796. doi: 10.1200/JCO.2014.59.8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glass AG, Leo MC, Haddad Z, et al. Validation of a genomic classifier for predicting post-prostatectomy recurrence in a community-based healthcare setting. J Urol. 2015 Nov 25; doi: 10.1016/j.juro.2015.11.044. Epub. [DOI] [PubMed] [Google Scholar]
- 14.Choeurng V, Luo B, Yousefi K, et al. Re-calibration of genomic risk prediction models in prostate cancer to improve individual-level predictions; Joint Statistical Meeting; August 8–13, 2015; Seattle, Washington. [Google Scholar]
- 15.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1(8476):307–310. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Kaplan–Meier estimates of survival-free (A) BCR and (B) CR in retrospective cohort.
Abbreviations: BCR, biochemical recurrence; CR, clinical recurrence; RP, radical prostatectomy.
ROC curves of Decipher and pathologic variables in the contemporary cohort.
Abbreviation: CI, confidence interval.
Pairwise Bland–Altman plots to evaluate agreement of Decipher scores between (A) PBx and RP, (B) PBx and Lns, and (C) RP and Lns in retrospective cohort.
Abbreviations: PBx, prostate biopsy; RP, radical prostatectomy; Ln, lymph node.
Table S1.
Comparison of LnI patient characteristics from retrospective and contemporary cohorts
| Variables | Retrospective cohort N=22 | Contemporary cohort LN N=51 | P-value* |
|---|---|---|---|
| Patient characteristics | |||
| Age, yr, median (IQR) | 60 (57–66) | 65 (64–68) | |
| Pre-operative PSA (ng/mL), median (IQR) | 6.9 (4.8–16.1) | 7.58 (5.1–11.8) | |
| Pathologic Gleason score, n (%) | >0.1 | ||
| ≥7 | 2 (9.1) | 10 (19.6) | |
| 8 | 9 (40.9) | 25 (49.0) | |
| ≥9 | 11 (50.0) | 15 (29.4) | |
| Unknown | 0 (0.0) | 1 (2.0) | |
| Pathologic stage, n (%) | >0.1 | ||
| T2 | 3 (13.6) | 5 (9.8) | |
| T3a | 8 (36.4) | 16 (31.4) | |
| T3b | 10 (45.5) | 29 (56.9) | |
| T4 | 1 (4.5) | 1 (2.0) | |
Note:
Fisher’s exact test. A statistical significance at P<0.05 was applied.
Abbreviations: IQR, interquartile range; Ln, lymph node; LnI, lymph node invasion; PSA, prostate-specific antigen; yr, year.
Table S2.
Univariable logistic regression analysis of categorical Decipher and pathologic variables for prediction of lymph node invasion in the contemporary cohort
| Variables/endpoint | Lymph node invasion
|
|
|---|---|---|
| OR (95% CI) | P-value | |
| Decipher low risk | ref | 1 |
| Decipher intermediate risk | 1.76 (0.51–6.11) | 0.37 |
| Decipher high risk | 7.7 (3.03–19.58) | <0.001 |
| Pathologic Gleason score 3+3 | ref | 1 |
| Pathologic Gleason score 3+4 | 0.78 (0.08–7.58) | 0.83 |
| Pathologic Gleason score 4+3 | 2 (0.24–16.78) | 0.52 |
| Pathologic Gleason score 8 | 8.77 (1.18–65.29) | 0.03 |
| Pathologic Gleason score ≥9 | 6.01 (0.79–45.89) | 0.08 |
| Extraprostatic extension | 6.1 (2.59–14.35) | <0.001 |
| Seminal vesicle invasion | 5.74 (3.25–10.13) | <0.001 |
Abbreviations: CI, confidence interval; OR, odds ratio; ref, reference group.
Note: A statistical significance at P<0.05 was applied.
Table S3.
Multivariable logistic regression analysis of categorical Decipher and pathologic variables for prediction of lymph node invasion in the contemporary cohort
| Variables/endpoint | Lymph node invasion
|
|
|---|---|---|
| OR (95% CI) | P-value | |
| Decipher low risk | ref | 1 |
| Decipher intermediate risk | 1.23 (0.35–4.38) | 0.75 |
| Decipher high risk | 3.23 (1.2–8.73) | 0.02 |
| Pathologic Gleason score 3+3 | ref | 1 |
| Pathologic Gleason score 3+4 | 0.41 (0.04–4.12) | 0.45 |
| Pathologic Gleason score 4+3 | 0.64 (0.07–5.72) | 0.69 |
| Pathologic Gleason score 8 | 2.46 (0.31–19.57) | 0.4 |
| Pathologic Gleason score ≥9 | 1.1 (0.13–9.25) | 0.93 |
| Extraprostatic extension | 3.93 (1.63–9.46) | <0.001 |
| Seminal vesicle invasion | 3.09 (1.67–5.73) | <0.001 |
Abbreviations: CI, confidence interval; OR, odds ratio; ref, reference group.
Note: A statistical significance at P<0.05 was applied.