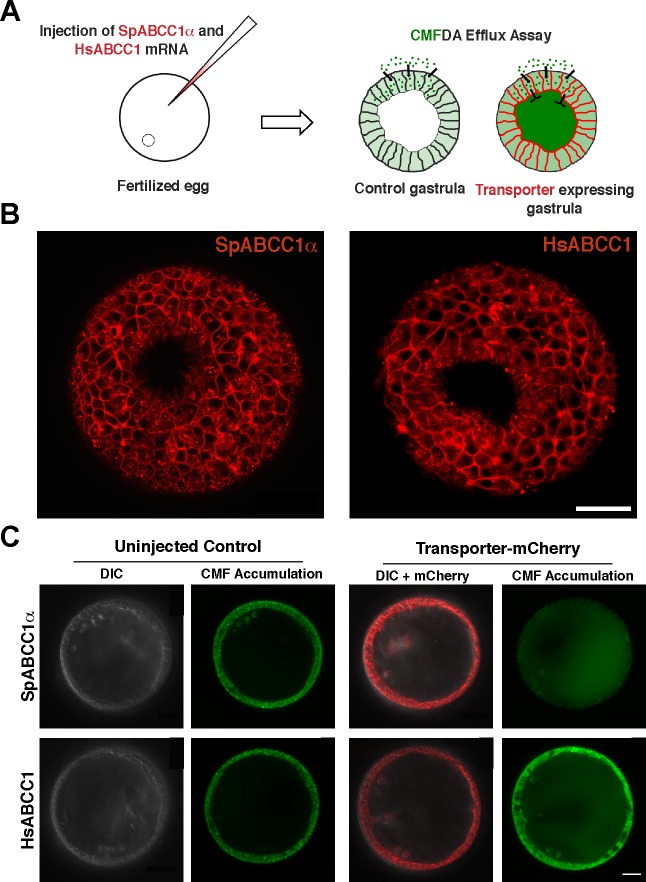
Abstract
The multidrug resistance protein (MRP) family encodes a diverse repertoire of ATP-binding cassette (ABC) transporters with multiple roles in development, disease, and homeostasis. Understanding MRP evolution is central to unraveling their roles in these diverse processes. Sea urchins occupy an important phylogenetic position for understanding the evolution of vertebrate proteins and have been an important invertebrate model system for study of ABC transporters. We used phylogenetic analyses to examine the evolution of MRP transporters and functional approaches to identify functional forms of sea urchin MRP1 (also known as SpABCC1). SpABCC1, the only MRP homolog in sea urchins, is co-orthologous to human MRP1, MRP3, and MRP6 (ABCC1, ABCC3, and ABCC6) transporters. However, efflux assays revealed that alternative splicing of exon 22, a region critical for substrate interactions, could diversify functions of sea urchin MRP1. Phylogenetic comparisons also indicate that while MRP1, MRP3, and MRP6 transporters potentially arose from a single transporter in basal deuterostomes, alternative splicing appears to have been the major mode of functional diversification in invertebrates, while duplication may have served a more important role in vertebrates. These results provide a deeper understanding of the evolutionary origins of MRP transporters and the potential mechanisms used to diversify their functions in different groups of animals.
Keywords: ATP-binding cassette transporters, multidrug resistance protein 1 (also known as ABCC1), development, sea urchin, protein evolution
multidrug resistance protein (MRP) 1 [also known as ATP-binding cassette (ABC) C1 (ABCC1)] is a conserved efflux transporter that belongs to the C subfamily of ABC transporters, which comprises 12 structurally and functionally diverse proteins (12). MRP1 was first characterized for transport of cytotoxic chemotherapeutic compounds, including doxorubicin, daunorubicin, vincristine, and etoposide (8, 18). However, in addition to chemotherapeutics, MRP1 also effluxes physiological neutral and anionic hydrophobic substrates and their glutathione (GSH), glucuronide, and sulfate conjugates, such as leukotriene C4 (LTC4), estradiol-17β-d-glucuronide, and estrone 3-sulfate (9, 33, 35, 39, 54).
The broad substrate portfolio of MRP1 enables this protein to perform diverse functions. For instance, in prostate cancer, breast cancer, and neuroblastoma (24, 61, 64), MRP1 has been implicated in drug resistance. However, even in cancer, its functions are likely not only limited to drug disposition, but also include broader roles such as control of cell motility (16). Similarly, under nonpathological conditions in mammals, MRP1 has been demonstrated to play various roles that include control of inflammatory response of mast cells (3, 37) and regulation of dendritic cell migration (57, 58). These functions are presumably related to the capacity of this protein to transport endogenous substrates that act directly or indirectly as signaling molecules.
An unresolved question that is central to understanding the basis for these diverse functions is the evolutionary origin of MRP1. In mammals, MRP1 shares homology with eight other efflux transporters [MRP2, MPR3, MRP4, MRP5, MRP6, MRP7, MRP8, and MRP9 (ABCC2, ABCC3, ABCC4, ABCC5, ABCC6, ABCC10, ABCC11, and ABCC12)] and three nonefflux membrane proteins [cystic fibrosis transmembrane conductance regulator (CFTR/ABCC7), a chloride ion channel, and 2 ATP-sensitive potassium channel regulators (SUR1-SUR2/ABCC8-ABCC9)] (48).
Although there have been very few functional studies of MRP transporters outside mammals, several observations suggest that these proteins and their endogenous substrates could be conserved. For instance, MRP transporter homologs from Saccharomyces cerevisiae and Drosophila melanogaster, YCF1 and dMRP (CG6214), can efflux LTC4 effectively when expressed in insect cells (41, 55), and both YCF1 and dMRP cluster with human MRP1, MRP2, MRP3, and MRP6. In addition, protective roles of ABCC transporters have also been proposed in diverse species (6, 36, 59, 60), suggesting that C-type transporters may have ancestral roles in cell protection.
In this study we examined the phylogenetic position and efflux functions of sea urchin ABCC1 relative to members of the C subfamily of vertebrate ABC proteins. Sea urchins, Strongylocentrotus purpuratus (Sp), along with hemichordates, are early diverging deuterostomes that occupy an important phylogenetic position for understanding the evolution of vertebrate proteins. Extensive functional studies of MDR transporters encoded in the sea urchin genome (19, 22, 23, 27, 46, 47, 58) make it one of the best-characterized invertebrate models for MDR proteins. However, until this study, the transporter(s) that could be responsible for sea urchin MRP-like transport activity had yet to be identified. Using functional and phylogenetic approaches, we characterize SpABCC1 and identify alternatively spliced regions of this transporter that could be critical for diversification of its functions.
MATERIALS AND METHODS
Phylogenetic analysis.
To assess the evolutionary positions of the sea urchin ABCC transporters, we generated a phylogenetic tree of C-type ABC transporters with homologs from Homo sapiens, Xenopus tropicalis, Danio rerio, Petromyzon marinus, Ciona intestinalis, Drosophila melanogaster, Nematostella vectensis, and Saccharomyces cerevisiae (1, 22, 40, 52). Using similarity searches at Metazome v3.0 and the Branchiostoma Joint Genome Institute (JGI) page, we also included reciprocal National Center for Biotechnology Information (NCBI) BLASTp matches for ABCC1-like proteins in the hemichordate Saccoglossus kowalevskii and the cephalochordate Branchiostoma floridae, respectively.
Sea urchin ABCC proteins were previously identified as part of the genome annotation of sea urchins (19, 49). In these studies we used reciprocal BLAST of conserved domains to identify putative ABCCs and manual analysis and validation of gene models against tiling array data. We have further refined ABCC annotations and gene models based on high-quality quantitative PCR (qPCR) analyses of ABCC gene expression and functional assays (22, 23, 46). Protein sequences were aligned with MUSCLE 3.8 (15) and manually trimmed to the region corresponding to the nucleotide-binding domains (NBDs), indicated by Pfam descriptions on NCBI conserved domains, and the intervening amino acids. Phylogenetic analysis was performed on the trimmed protein region (NBDs + intervening sequence, ∼660 amino acids) as well as NBDs only (∼415 amino acids). NBDs + intervening sequence was selected for the phylogenetic analysis, although the resulting trees from each alignment were congruent. Phylogenetic relationships for all proteins were determined using maximum-likelihood and Bayesian analysis. Maximum-likelihood analysis was conducted with RAxML version 8.2.0 (51) (http://bioinformatics.oxfordjournals.org/content/30/9/1312) and Bayesian analyses with MrBayes version 3.2.5 (42) using the LG + G model (model determined with ProtTest version 3.2) (11). In the maximum-likelihood analysis, support for individual nodes was assessed through 1,000 bootstrap replicates. For the Bayesian analysis, two independent analyses were performed with five chains run for 5 × 106 generations, with trees sampled every 500 generations. The first 1.25 × 106 generations were discarded as burn-in. Log-likelihood values were plotted and found to be asymptotic well before the burn-in fraction. The rooted likelihood tree with bootstrap and posterior probabilities was visualized and illustrated with FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Embryo preparation and reagents.
Purple sea urchin (S. purpuratus) embryos were prepared as described previously (7). The fluorescent dyes CellTracker Green [5-chloromethylfluorescein (CMF)-diacetate (CMFDA)], 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein-acetoxymethyl ester (BCECF-AM), and bodipy-verapamil (b-VER) were purchased from Invitrogen; fluorescein-diacetate (FDA) from Sigma; and calcein-AM from Biotium (Hayward, CA). All stock solutions were prepared in DMSO.
Expression profiles of sea urchin ABCC1 isoforms.
Expression profiles of SpABCC1α (GenBank ID: KT725593) and SpABCC1δ (GenBank ID: JQ354984) splice variants during sea urchin development were characterized by quantitative real-time PCR (qPCR) as described by Shipp and Hamdoun (46). Two pairs of qPCR primers were designed to differentiate the expression profiles of each splice variant. The expression profile of the SpABCC1α splice variant was characterized by the primer pair 5′-ATCTCGTATTCCACGCCTTG-3′ (forward) and 5′-TTAACTGCCGGGAGGTACAG-3′ (reverse), specific to exon 22α. The expression profile of the SpABCC1δ splice variant was determined with the forward primer 5′-CAGGGCTACCTCTCACATGC-3′, specific to exon 22δ, and the reverse primer 5′-CCGGTTCAACATCTGACCTT-3′, binding to exon 23. In addition, a generic qPCR primer pair (pan-SpABCC1), 5′-AGTCTTGGGTTGCTGCTCAT-3′ (forward) and 5′-TATACGGCTGGCAAGTCTCC-3′ (reverse), that recognizes a shared region in exon 3 present in both splice variants was designed to measure the relative levels of each splice variant.
DNA constructs, in vitro mRNA synthesis, and microinjection of SpABCC1 isoforms.
In-Fusion HD Cloning Kit (Clontech Laboratories, Mountain View, CA) was used to clone sea urchin SpABCC1α and human ABCC1 cDNA (a gift from Dr. Susan P. C. Cole) into AcsI-PacI sites of pCS2+8CmCherry plasmid (22). Cloning of SpABCC1δ into pCS2+8CmCherry was described previously (22). mRNA was synthesized and transporter mRNAs were microinjected into the fertilized eggs for protein overexpression as described previously (22, 58).
Confocal microscopy for localization and efflux assays.
A laser scanning confocal microscope (TCS SP8, Leica) equipped with a ×40/1.10 water-immersion objective was used to determine transporter localization in early blastulae and early gastrulae. Micrographs were prepared using Imaris 7.3.1 software (Bitplane, Zurich, Switzerland). Calcein-AM (250 nM) and b-VER (125 nM) assays were performed as described previously (7, 22). Other fluorescent dye efflux assays were also done in early blastula-expressing recombinant transporters. mRNA-injected and non-mRNA-injected control embryos were incubated with BCECF-AM (250 nM), CMFDA (100 nM), and FDA (100 nM) at 15°C for 1 h. Embryos were washed 10 times with filtered seawater to remove the background fluorescence and incubated for an additional 30 min before imaging. Intracellular and blastocoel fluorescent dye accumulation was imaged in the blastula (∼16 h postfertilization) and early gastrula (∼40 h postfertilization) on a laser scanning confocal microscope (LSM 700, Zeiss) equipped with a ×20/0.8 apochromatic air objective. Images were captured with the ZEN software package (Zeiss, revision 5.5). Micrographs were prepared and analyzed with ImageJ 1.47v (National Institutes of Health) as described previously (22).
RESULTS
Phylogenetic analysis of ABCC proteins.
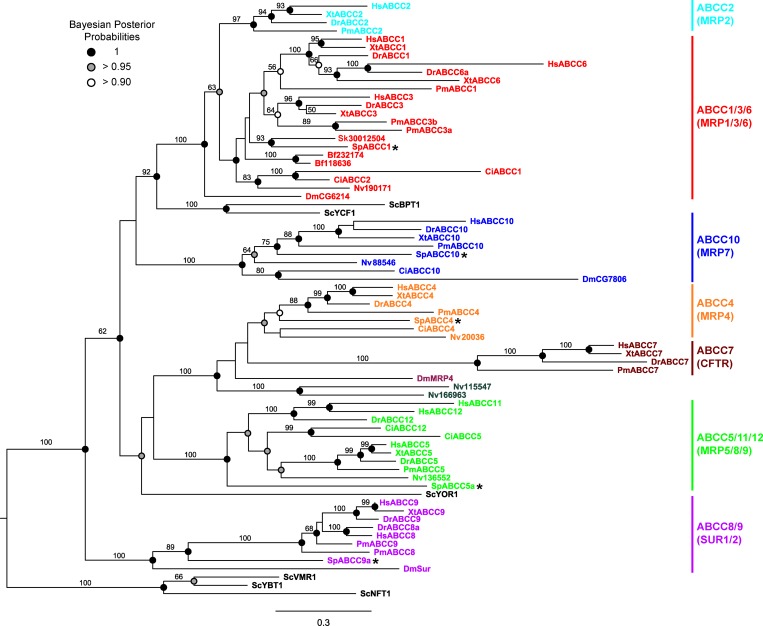
To understand the phylogenetic position of sea urchin SpABCC1, we constructed a tree of sea urchin ABCC transporters in relation to human, zebrafish, frog, lamprey, sea squirt, sand lancelet, acorn worm, fruit fly, anemone, and yeast C-type ABC transporters. SpABCC1 clusters with vertebrate ABCC1, ABCC3, and ABCC6 and is co-orthologous to these transporters. The ABCC1-ABCC3-ABCC6 clade also includes single proteins from the fly (dMRP, GC6214), the hemichordate Saccoglossus (Metazome Sakowv30012504m), the anemone (JGI190171), and inferred paralogs for the cephalochordate Branchiostoma (JGI118636 and JGI232174) and urochordates Ciona (XP_009862422.1 and XP_009859093.1), suggesting an ancient origin for this protein group to at least a cnidarian-bilaterian ancestor (>600 million years). These results also indicate that human and other vertebrate ABCC1, ABCC3, and ABCC6 share a common ancestor with SpABCC1.
Vertebrate ABCC2 transporters form a separate cluster and join to the same node with the ABCC1-ABCC3-ABCC6 clade. Finally, two vacuolar yeast transporters, YCF1 and BPT1, which are involved in cellular detoxification of GSH conjugates (28, 44), cluster together and are positioned at the root of the metazoan ABCC1-ABCC3-ABCC6 and ABCC2 clades, but not with other yeast ABCC transporters, suggesting that this group of transporters evolved from a common ancestor.
Sea urchin SpABCC5a is located at the base of the clade that includes vertebrate orthologs of the ABCC5 transporter and the closely related ABCC12 transporter and its paralog ABCC11, which is present in primates but not found in rodents (45). A similar evolutionary pattern was also observed with sea urchin SpABCC9a, which shares co-orthology with the vertebrate ABCC8 and ABCC9 potassium channel regulators and is positioned at the root of the deuterostome lineage. The sea urchin genome does not have an ABCC8 gene, but there is an expansion of the ABCC9 clade with 11 annotated genes (19), suggesting that functional diversity in the sea urchin ABCC9 group might be achieved through gene duplication.
Although phylogenetic relationships between sea urchin and vertebrate ABCC transporters follow the general pattern of co-orthologous gene pairing with vertebrate-specific gene duplications, two sea urchin transporters, SpABCC4 and SpABCC10, showed one-to-one orthology with their respective vertebrate transporters (Fig. 1). This direct orthology indicates that these transporters may have conserved substrates and functions in invertebrates and vertebrates (Fig. 1).
Fig. 1.
Phylogenetic relationships of ATP-binding cassette C subfamily (ABCC) transporters of Strongylocentrotus purpuratus (Sp) with human (Hs), Xenopus tropicalis (Xt), Danio rerio (Dr), Petromyzon marinus (Pm), Ciona intestinalis (Ci), Branchiostoma floridae (Bf), Saccoglossus kowalevskii (Sk), Drosophila melanogaster (Dm), Nematostella vectensis (Nv), and Saccharomyces cerevisiae (Sc). The sea urchin efflux transporter SpABCC1 clusters in a well-supported clade containing ABCC1-ABCC2-ABCC3-ABCC6 from vertebrates and single ABCC proteins or likely lineage-specific paralogs from other invertebrates (Ci, Bf, Sk, Nv, and Dm). SpABCC1 is co-orthologous to vertebrate ABCC1-ABCC3-ABCC6 transporters. The sea urchin also has orthologs to ABCC4 and ABCC10 and a co-ortholog to ABCC5-ABCC11-ABCC12 and ABCC8-ABCC9 clades in vertebrates. The maximum-likelihood phylogeny is presented with posterior probability (>0.9) and maximum-likelihood bootstraps (>50%) indicated for each branch.
Genomic structure of sea urchin SpABCC1 transporter and expression of its splice variants.
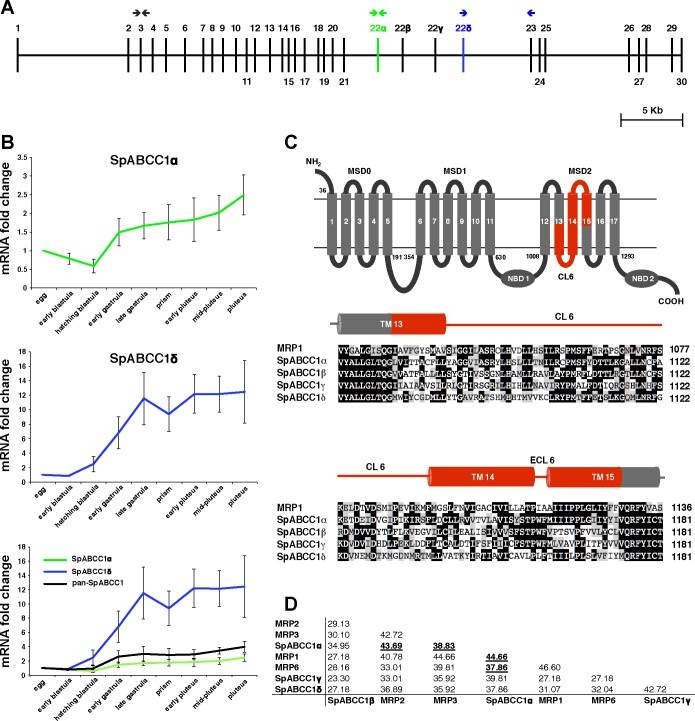
Gene duplication and alternative splicing are two common mechanisms to generate novel functions in gene families (27). Unlike vertebrates, the sea urchin genome has not undergone gene duplications in the ABCC1-ABCC3-ABCC6 clade, and SpABCC1 is the only gene found in this clade. Therefore, we examined the potential functional consequences of alternative splicing of this protein by examining the genomic structure of ABCC1 and cloning an alternative full-length ABCC1 mRNA we found to be expressed in the embryos.
The SpABCC1 gene comprises 33 exons and 32 introns and spans 54.6 kb of genomic DNA. At least four splice variants, SpABCC1α, SpABCC1β, SpABCC1γ, and SpABCC1δ, are predicted in the genome and are generated by alternative splicing of exon 22 (Fig. 2A). We detected two of these splice variants, SpABCC1α and SpABCC1δ, in embryos. qPCR profiles of SpABCC1 splice variants in the first 3 days of sea urchin development showed that SpABCC1α and SpABCC1δ have similar expression profiles with a steady increase to mid-pluteus (Fig. 2B). However, comparison of the expression profiles of the two splice variants against the generic pan-ABCC1 expression profile showed that SpABCC1α has an expression profile similar to pan-ABCC1 (Fig. 2B) and suggests that SpABCC1α might be more abundant than SpABCC1δ.
Fig. 2.
Sea urchin SpABCC1 gene has 4 forms of exon 22 that correspond to membrane-spanning domain 2 (MSD2) of SpABCC1 transporters. A: gene structure of sea urchin SpABCC1 shows that it consists of 33 exons, 4 of which encode exon 22. Arrows indicate positions of primers used to determine the quantitative PCR (qPCR) profiles of SpABCC1α (green) and SpABCC1δ (blue) splice variants and pan-SpABCC1 (black). B: qPCR expression profiles of SpABCC1α and SpABCC1δ show that both genes are expressed throughout embryonic development and that the SpABCC1α expression profile is similar to the pan-SpABCC1 expression profile. Values are means ± SD. C: variable exon 22 corresponds to a region in MSD2 marked in red. This exon encodes part of transmembrane segment 13 (TM13), cytoplasmic loop 6 (CL6), all of TM14, and part of TM15. Alignment of sea urchin SpABCC1 exon 22 splice variants with human multidrug resistance protein (MRP) 1 exon 24 indicates that some residues are conserved. D: protein sequence percent identity matrix of 4 splice variants of sea urchin exon 22 and the corresponding regions in human MRP1, MRP2, MRP3, and MRP6. Bold and underlined numbers indicate percent similarity of exon 22 of SpABCC1α in relation to the human MRP transporter sequences.
Both SpABCC1α and SpABCC1δ and two other predicted splice variants, SpABCC1β and SpABCC1γ, consist of 1,577 amino acids and have “long MRP” architecture with three membrane-spanning domains (MSD0, MSD1, and MSD2) and two NBDs (NBD1 and NBD2). Amino acid sequence alignment (Fig. 2C) of the four splice variants of sea urchin exon 22 with the corresponding region in human ABCC1 shows that exon 22 corresponds to the predicted transmembrane helices 13–15 of human ABCC1 (13) and that the variable region of SpABCC1α is more closely related, in terms of amino acid identity, to human ABCC1, ABCC2, ABCC3, and ABCC6 than to other sea urchin SpABCC1 splice variants (Fig. 2D).
Functional characterization of sea urchin SpABCC1α and SpABCC1δ.
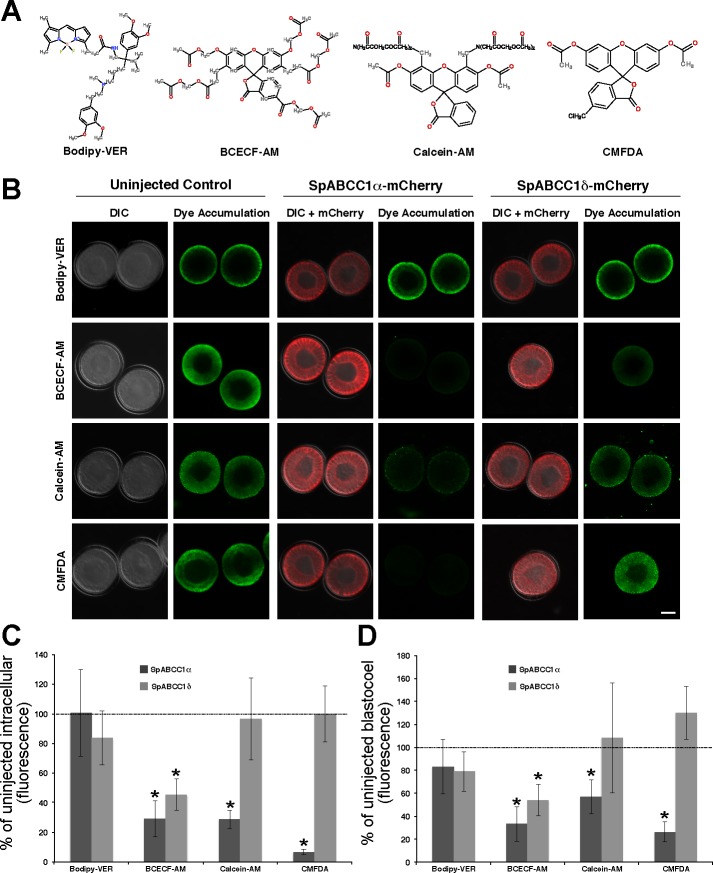
Next, we characterized the efflux activities of recombinant SpABCC1α and SpABCC1δ overexpressed at high levels in embryos. We used three fluorescent substrates, calcein-AM, BCECF-AM, and CMFDA (Fig. 3A). We also used a non-MRP substrate (Fig. 3A), b-VER, a fluorescent derivative of verapamil, which is effluxed by P-glycoprotein-type transporters, but not MRP transporters (22, 34).
Fig. 3.
SpABCC1α and SpABCC1δ have different efflux activity profiles. A: chemical structures of fluorescent substrates used in efflux assays. B: micrographs showing that overexpression of mCherry-tagged SpABCC1α reduced intracellular accumulation of BCECF-AM, calcein-AM, and 5-chloromethylfluorescein (CMF)-diacetate (CMFDA), but not verapamil (bodipy-VER), while the SpABCC1δ splice variant can efflux only BCECF-AM. DIC, differential interference contrast. Scale bar = 35 μm. C and D: quantitative analysis of intracellular and blastocoel accumulation of fluorescent substrates relative to noninjected control embryos. Values are means ± SD (n = 18–20 embryos in total over 3 experiments). *P < 0.01.
Expression of mCherry-tagged SpABCC1α reduced intracellular accumulation of calcein to 28.41 ± 6.17% of the control noninjected embryos (Fig. 3, B and C). Similarly, BCECF accumulation was 28.9 ± 12.14% and CMF accumulation was 6.29 ± 1.88% of that of the control embryos (Fig. 3, B and C). As expected, b-VER was not effluxed by SpABCC1α (Fig. 3C). We also tested efflux of FDA by SpABCC1α. FDA is structurally similar to CMFDA and is a known substrate for sea urchin SpABCC5a (47). FDA accumulation of SpABCC1α-expressing embryos was only 0.53 ± 0.2% of that of the control embryos.
Interestingly, SpABCC1δ effluxed only BCECF-AM. Expression of SpABCC1δ reduced intracellular accumulation of BCECF-AM to 45.40 ± 10.67% of the control embryos (Fig. 3, B and C). Together, these data show that sea urchin SpABCC1α has a broad substrate profile similar to its mammalian co-orthologs (5, 34), whereas SpABCC1δ may have a more restricted substrate profile.
Finally, to determine whether the observed efflux activities of SpABCC1 splice variants are mediated by activity of the protein present at apical vs. basolateral cell membranes, we measured accumulation of the fluorescent substrate in the blastocoel. We expected that efflux transport from basolateral surfaces would lead to translocation of substrates into the blastocoel, whereas protein at the apical surface would prevent uptake altogether by blocking passage through the ectoderm.
The data show that overexpression of either splice variant of SpABCC1 did not cause significant fluorescent substrate accumulation in the blastocoel (Fig. 3, B and D), suggesting that the efflux activity is mediated by transporters at the apical membrane. However, activity from basolateral transporters became more apparent later in development (see below).
Membrane localization of recombinant SpABCC1α.
Membrane localization of human ABCC transporters has been extensively studied in different cell types. The human ABCC1 and closely related ABCC3 and ABCC6 localize to basolateral cell membranes of polarized epithelial cells (32, 43). In contrast, the major drug transporter ABCC2 localizes to apical membranes (38).
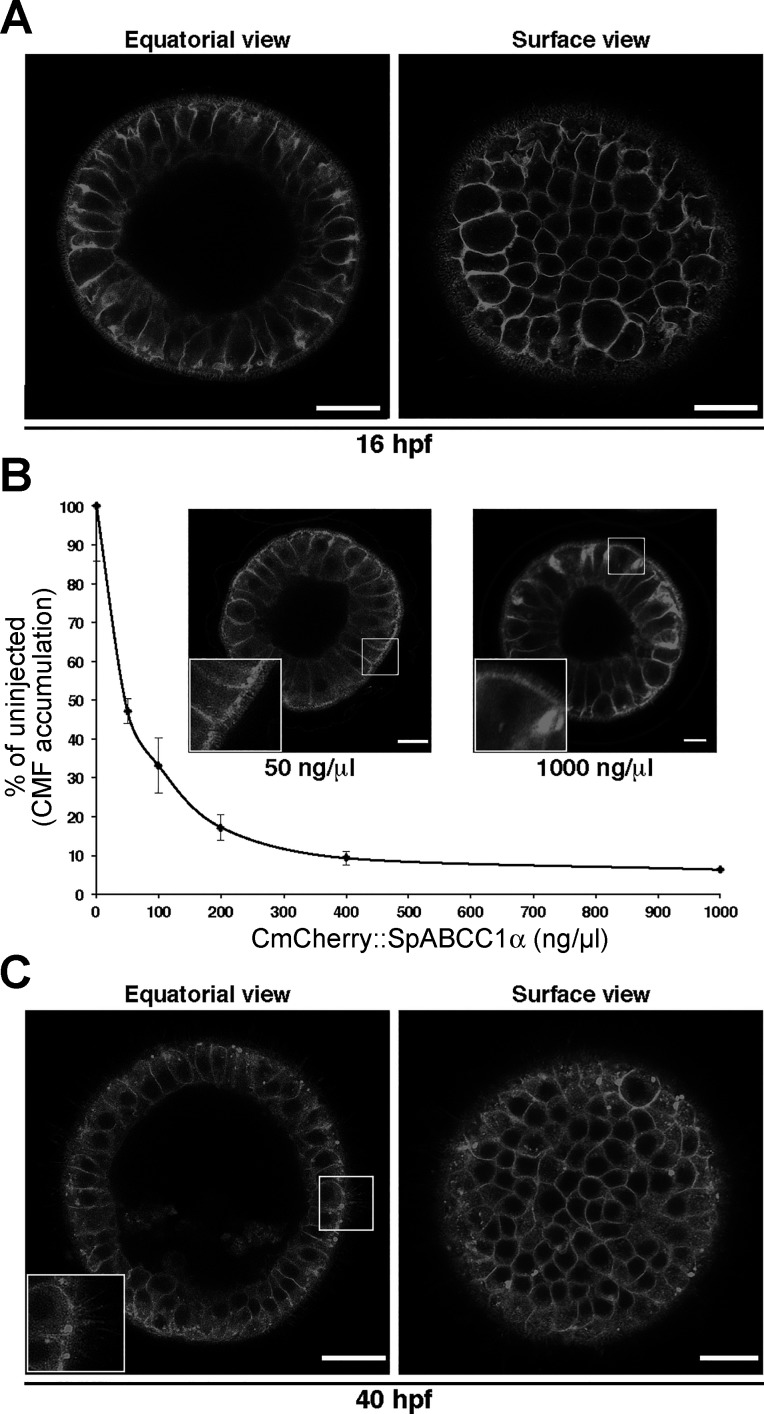
Here we determined the subcellular localization of SpABCC1α from expression of the COOH-terminal mCherry fusion from 1 μg/μl injected mRNA. SpABCC1α localized to both apical and basolateral membranes in early blastula (Fig. 4A). This result shows that subcellular localization of SpABCC1α could be different from that of its human co-orthologs. Since the amount of exogenous transporter in embryos is above the physiological level of SpABCC1α protein, we next sought to determine if this apical localization of SpABCC1α could be due to excessive amounts of exogenous protein.
Fig. 4.
Dynamic localization of SpABCC1α in sea urchin blastulae and early gastrulae. A: representative equatorial and surface images of blastula [16 high-power fields (hpf); n = 10–12 embryos in total over 2 experiments] show that mCherry-tagged SpABCC1α localizes to both apical and basolateral membranes. Scale bars = 200 μm (equatorial view) and 150 μm (surface view). B: titration of SpABCC1α mRNA with CMFDA efflux assay shows that <50 ng/μl mRNA is required to achieve physiologically relevant SpABCC1α expression. Micrographs of blastulae expressing SpABCC1α from 50 and 1,000 ng/μl mRNA show that apical localization of SpABCC1α (magnified in insets) is independent of mRNA concentration. Values are means ± SD (n = 24–26 embryos in total over 4 experiments for each mRNA concentration). Scale bar = 150 μm. C: representative equatorial and surface images of early gastrula (40 hpf; n = 10–12 embryos in total over 2 experiments) show that apical localization of SpABCC1α in this stage is significantly reduced and becomes more basolateral. Scale bars = 200 μm (equatorial view) and 150 μm (surface view).
To determine a more physiologically relevant level, we titrated SpABCC1α mRNA and conducted efflux assays to determine the relative increase in efflux activity with increasing transporter expression (Fig. 4B). We found that embryos expressing SpABCC1α from 50 ng/μl mRNA accumulated roughly half (47.18 ± 14.14%) of the CMF of uninjected control embryos, suggesting that this mRNA concentration might produce recombinant protein levels closer to the physiological level (Fig. 4B).
Importantly, we also found that, even when expressed from 50 ng/μl mRNA, SpABCC1α localized into both apical and basolateral membranes in blastulae (Fig. 4B), indicating that this localization is not simply due to high levels of protein expression. In addition, we investigated the localization of SpABCC1α expressed from 50 ng/μl mRNA in early gastrula (∼40 h postfertilization). At this stage, the protein can still be detected on the apical surface, but the amount is reduced and SpABCC1α is more strongly basolateral (Fig. 4C), suggesting that localization could be stage/cell type-dependent.
Comparative analysis of sea urchin and human ABCC1 transporters in localization and efflux assays.
Finally, to examine species differences in ABCC1 localization, we determined heterologous expression of the COOH-terminal mCherry-tagged fusion of human HsABCC1 (MRP1) in sea urchin embryos (Fig. 5A). MRP1 was slower to be translated and trafficked to plasma membranes than was the sea urchin protein but, ultimately, localized strongly to basolateral membranes in early gastrula (Fig. 5B), consistent with its localization in polarized human epithelial cells. In gastrulae, the sea urchin SpABCC1α is predominantly basolateral, with a small, but detectable, level of apical localization (Fig. 5B).
Fig. 5.
SpABCC1α and human HsABCC1 transporters exhibit basolateral efflux activity in early gastrulae. A: schematic representation of basolateral efflux assay. Fluorescent substrate CMF (green) is effluxed into the blastocoel by MRP-type basolateral transporters (red). B: blastopore view of sea urchin early gastrulae expressing basolaterally localized SpABCC1α and HsABCC1 transporters. Scale bar = 200 μm. C: representative micrographs showing that overexpression of mCherry-tagged SpABCC1α and HsABCC1 increases blastocoel accumulation of CMF (n = 12–13 embryos in total over 2 experiments). Scale bar = 20 μm.
Given that, later in development, localization of both proteins was more strongly basolateral, we next investigated whether sea urchin and human ABCC1 transporters pump substrates into the blastocoel. Both SpABCC1α and HsABCC1 were functional in sea urchin gastrulae and increased blastocoel accumulation of CMF or CMF-GSH conjugate compared with control embryos (Fig. 5C). However, unlike HsABCC1, ectodermal CMF accumulation in gastrulae was also reduced by SpABCC1α. Thus although sea urchin MRP1 is predominantly basolateral, this result suggests its potential for dual localization and function on both apical and basolateral membranes.
DISCUSSION
MRP1 is an ancient protein with homologs in all known metazoans and is expressed in a wide range of cell types (2). Given its diversity of substrates, distinguishing its roles is a challenging task. Comparative studies in diverse model systems are an important tool to dissect the origins and potential functions of these proteins. In this study we identified a functional MRP1 homolog in sea urchin embryos and analyzed its phylogenetic position in relation to vertebrate, invertebrate, and yeast C-subfamily ABC transporters. Our results show that the members of the MRP1 clade (ABCC1, ABCC3, and ABCC6 transporters) have one-to-one orthology with the corresponding vertebrate transporters (zebrafish and Xenopus), while sea urchin SpABCC1, as well as ABCC proteins from various other invertebrates [e.g., Drosophila dMRP (CG6214) and Nematostella JGI190171], share co-orthology with the MRP1 clade (Fig. 1).
The MRP1 clade within the ABCC subfamily is ancient, emerging before the cnidarian-bilaterian split (>600 million years) and likely to be the opisthokonta (fungus and animal) ancestor. Sequencing of the genomes of the sea urchin S. purpuratus, an echinoderm, and the sea squirt C. intestinalis, a nonvertebrate chordate, revealed that ABCC2, ABCC3, and ABCC6 genes are not found in these genomes (1, 14, 19, 49), suggesting that ABCC2, ABCC3, and ABCC6 arose through gene duplication events in the vertebrate lineage.
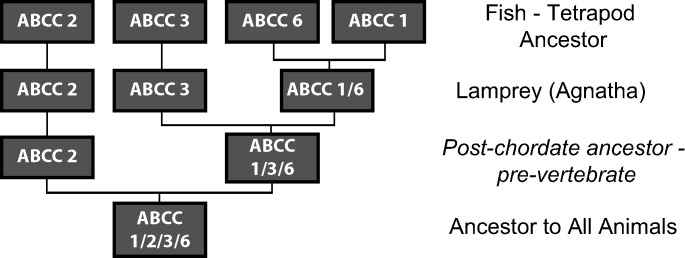
In agreement with this assessment, synteny analysis of the human ABCC1 gene with the related, but functionally different, ABCC6 transporter gene shows that they both reside on chromosome 16p13 in opposite orientations with their 3′ ends only 8–9 kb apart (30). The vertebrate ABCC2 genes are sisters to the ABCC1-ABCC3-ABCC6 clade. Our hypothesis for this positioning is that ABCC2 underwent a diversification in the vertebrate line prior to the fish-tetrapod ancestor and, hence, falls outside the ABCC1-ABCC3-ABCC6 clade as a sister group. ABCC2 could have been the first duplication event of the ancestral gene that resembled ABCC1-ABCC3-ABCC6 from invertebrates and has retained this divergent sequence since the duplication event. The ABCC1-ABCC3-ABCC6 gene then underwent additional duplication events. In fact, ABC transporters from lamprey (an intermediate taxonomic group) revealed that it has an ABCC1, an ABCC2 ortholog, and two ABCC3 genes (PmABCC3a and PmABCC3b), which are co-orthologous to vertebrate ABCC3 proteins, supporting this hypothesis, as well as the order of ABCC1-ABCC2-ABCC3-ABCC6 (40) (Fig. 6). Other gene duplication events have also apparently occurred in the urochordates and cephalochordate lineages due to paralogs in Ciona and Branchiostoma.
Fig. 6.
Diversification of the ABCC1-ABCC2-ABCC3-ABCC6 clade across the animal kingdom. Schematic representation shows the most likely duplication history of ABCC1-ABCC2-ABCC3-ABCC6 based on available evidence.
In addition to gene duplication, generation of novel proteins through alternative splicing is also a mechanism for increasing the functional diversity in most eukaryotic protein families. In the C-subfamily of human ABC transporters, numerous splice variants have been reported (50). Most of these variants are generated, however, by splice site mutations that cause removal of one or multiple exons or insertion of introns into the mature mRNA, most of which cause premature protein truncation by nonsense mutations (17, 21) or by alternate transcription start sites (4, 53). For instance, in human ovarian cancer cells, multiple splice variants of human ABCC1 were created by exon skipping and/or intron inclusion (26).
Alternative-splicing events could be critical for diversification of transporter function in invertebrates lacking the duplicated MRP members found in vertebrates. Indeed, in Drosophila, 14 different full-length splice variants of dMRP were detected. These isoforms are generated from a single gene by exchange of two variant copies of exon 4 and seven variant copies of exon 8 (20). Considering that Drosophila has only one co-orthologous gene in the MRP1 clade, generation of protein diversity by exchange of internal exons could be a way to compensate the lack of paralogs created by gene duplication and innovate novel protein diversity while maintaining a single gene. In support of this hypothesis, Kopelman et al. (31) showed an inverse correlation between alternative splicing and gene duplication. Singletons (gene families of one) are more inclined to undergo exon splicing than their orthologs that have undergone gene duplication (31).
We found a similar mode of alternative splicing in sea urchin SpABCC1, an MRP-type transporter that shares co-orthology with human ABCC1, ABCC3, and ABCC6 transporters (Fig. 1), and characterized the consequences of the splicing event on efflux activity. The new splice variant described here, SpABCC1α, was generated by alternative splicing of exon 22 (Fig. 2). The boundaries of exon 22 are conserved in human ABCC1, ABCC2, and ABCC3, and it encodes part of TM13, CL6, TM14, and part of TM15 of MSD2 (Fig. 2). Interestingly, the frequently spliced exon 8 of Drosophila dMRP also corresponds to the 5′ end of exon 22 of sea urchin SpABCC1, suggesting that this protein region in MRP homologs is an evolutionarily active region for generating functional diversity.
Previous mutagenesis studies in human ABCC1 showed that proline, polar and charged, residues in this region are important for substrate recognition (10, 29, 62, 63). Considering that residues in MSDs are important for the substrate interactions, organisms with singleton MRP homologs may use alternative splicing of exons corresponding to the regions where substrate binding occurs. In support of this hypothesis, we showed that, by alternative splicing of exon 22, SpABCC1α can efflux calcein-AM, BCECF-AM, CMFDA, and FDA, all known fluorescent substrates for human ABCC1, ABCC2, and ABCC3, while SpABCC1δ effluxes only BCECF-AM with a lower capacity (Fig. 3). Protein alignment of exons 22α, 22β, 22γ, and 22δ with the corresponding exon sequences of human ABCC1, ABCC2, and ABCC3 proteins showed that SpABCC1α is more similar to human proteins in this region than SpABCC1δ, which may explain why SpABCC1α can efflux canonical human MRP substrates, while SpABCC1δ can efflux only BCECF-AM. We previously showed that mCherry-tagged SpABCC1δ localizes to both apical and basolateral membranes in sea urchin blastulae (23). Identical subcellular localization of SpABCC1δ also indicates that distinctive efflux activity profiles of each splice variant are determined by the changes in the substrate recognition region corresponding to exon 22, but not by the subcellular localizations.
The results of this study extend and validate the heterologous expression approach we have adapted for analysis of sea urchin ABC transporters (22, 23, 46, 47). These assays are important, because they reveal the transporters that could be responsible for observed activities and fingerprint the relevant suspects for analysis by more laborious immunochemical methods. Large overexpression has been used to ensure that the vast majority of the observed efflux activity comes from the recombinant protein (22), which is essential for characterization of substrate selectivity.
Here, we showed that for localization studies the protein can be reproducibly titrated to levels that increase corresponding transport activity only twofold and still be functionally imaged by routine confocal microscopy (Fig. 4B), albeit with use of high-sensitivity detectors. More importantly, these experiments indicated that localization of both forms of sea urchin ABCC1 could be both apical and bilateral, depending on developmental stage and independent of expression level. This dual location could be an additional mechanism for diversification of function in both protection and signaling.
Finally, an important step was to demonstrate the expression of functional human MRP1. An interesting observation was that while sea urchin protein exhibited both apical and basolateral efflux activity, the human protein only appeared to act at the basolateral membrane, possibly indicating that the differences in localization of sea urchin and human proteins are likely to be inherent features of the transporters themselves. Collectively, the results indicate that the sea urchin is an important system in which to understand the evolution of MRP proteins.
GRANTS
This research was supported, in whole or in part, by National Institutes of Health Grants HD-058070 and ES-021985, National Science Foundation Grant OCE1314480, a grant from the University of California Cancer Research Coordinating Committee, and University of California San Diego Academic Senate Grant RN141S.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
T.G. and A.H. developed the concept and designed the research; T.G., J.P.C., A.M.R., L.E.S., and G.W.M. performed the experiments; T.G., J.P.C., A.M.R., L.E.S., G.W.M., and A.H. analyzed the data; T.G., J.P.C., A.M.R., L.E.S., G.W.M., and A.H. interpreted the results of the experiments; T.G. and A.M.R. prepared the figures; T.G. drafted the manuscript; T.G., J.P.C., A.M.R., L.E.S., G.W.M., and A.H. approved the final version of the manuscript; A.H. edited and revised the manuscript.
ACKNOWLEDGMENTS
We thank Dr. Susan Cole for human ABCC1 cDNA construct and comments on the manuscript and Dr. Victor D. Vacquier for valuable scientific discussions. We also thank Dr. Sascha C. T. Nicklisch and Lisa Mesrop for discussion and assistance.
REFERENCES
- 1.Annilo T, Chen ZQ, Shulenin S, Costantino J, Thomas L, Lou H, Stefanov S, Dean M. Evolution of the vertebrate ABC gene family: analysis of gene birth and death. Genomics 88: 1–11, 2006. [DOI] [PubMed] [Google Scholar]
- 2.Bakos E, Homolya L. Portrait of multifaceted transporter, the multidrug resistance-associated protein 1 (MRP1/ABCC1). Pflügers Arch 453: 621–641, 2007. [DOI] [PubMed] [Google Scholar]
- 3.Bartosz G, König J, Keppler D, Hagmann W. Human mast cells secreting leukotriene C4 express the MRP1 gene-encoded conjugate export pump. Biol Chem 379: 1121–1126, 1998. [DOI] [PubMed] [Google Scholar]
- 4.Bera TK, Lee S, Salvatore G, Lee B, Pastan I. MRP8, a new member of ABC transporter superfamily, identified by EST database mining and gene prediction program, is highly expressed in breast cancer. Mol Med 7: 509–516, 2001. [PMC free article] [PubMed] [Google Scholar]
- 5.Boraldi F, Quaglino D, Croce MA, Garcia Fernandez MI, Tiozzo R, Gheduzzi D, Bacchelli B, Pasquali Ronchetti I. Multidrug resistance protein-6 (MRP6) in human dermal fibroblasts. Comparison between cells from normal subjects and from Pseudoxanthoma elasticum patients. Matrix Biol 22: 491–500, 2003. [DOI] [PubMed] [Google Scholar]
- 6.Bosnjak I, Uhlinger KR, Heim W, Smital T, Franekić-Colić J, Coale K, Epel D, Hamdoun A. Multidrug efflux transporters limit accumulation of inorganic, but not organic, mercury in sea urchin embryos. Environ Sci Technol 43: 8374–8380, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campanale JP, Hamdoun A. Programmed reduction of ABC transporter activity in sea urchin germline progenitors. Development 139: 783–792, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole SP. Multidrug resistance protein 1 (MRP1, ABCC1), a “multitasking” ATP-binding cassette (ABC) transporter. J Biol Chem 289: 30880–30888, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole SP, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, Almquist KC, Stewart AJ, Kurz EU, Duncan AM, Deeley RG. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science 258: 1650–1654, 1992. [DOI] [PubMed] [Google Scholar]
- 10.Conseil G, Deeley RG, Cole SP. Functional importance of three basic residues clustered at the cytosolic interface of transmembrane helix 15 in the multidrug and organic anion transporter MRP1 (ABCC1). J Biol Chem 281: 43–50, 2006. [DOI] [PubMed] [Google Scholar]
- 11.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27: 1164–1165, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res 11: 1156–1166, 2001. [DOI] [PubMed] [Google Scholar]
- 13.Deeley RG, Westlake C, Cole SP. Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiol Rev 86: 849–899, 2006. [DOI] [PubMed] [Google Scholar]
- 14.Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, Harafuji N, Hastings KE, Ho I, Hotta K, Huang W, Kawashima T, Lemaire P, Martinez D, Meinertzhagen IA, Necula S, Nonaka M, Putnam N, Rash S, Saiga H, Satake M, Terry A, Yamada L, Wang HG, Awazu S, Azumi K, Boore J, Branno M, Chin-Bow S, DeSantis R, Doyle S, Francino P, Keys DN, Haga S, Hayashi H, Hino K, Imai KS, Inaba K, Kano S, Kobayashi K, Kobayashi M, Lee BI, Makabe KW, Manohar C, Matassi G, Medina M, Mochizuki Y, Mount S, Morishita T, Miura S, Nakayama A, Nishizaka S, Nomoto H, Ohta F, Oishi K, Rigoutsos I, Sano M, Sasaki A, Sasakura Y, Shoguchi E, Shin-i T, Spagnuolo A, Stainier D, Suzuki MM, Tassy O, Takatori N, Tokuoka M, Yagi K, Yoshizaki F, Wada S, Zhang C, Hyatt PD, Larimer F, Detter C, Doggett N, Glavina T, Hawkins T, Richardson P, Lucas S, Kohara Y, Levine M, Satoh N, Rokhsar DS. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298: 2157–2167, 2002. [DOI] [PubMed] [Google Scholar]
- 15.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fletcher JI, Haber M, Henderson MJ, Norris MD. ABC transporters in cancer: more than just drug efflux pumps. Nat Rev Cancer 10: 147–156, 2010. [DOI] [PubMed] [Google Scholar]
- 17.Fromm MF, Leake B, Roden DM, Wilkinson GR, Kim RB. Human MRP3 transporter: identification of the 5′-flanking region, genomic organization and alternative splice variants. Biochim Biophys Acta 1415: 369–374, 1999. [DOI] [PubMed] [Google Scholar]
- 18.Gadsby DC, Vergani P, Csanády L. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature 440: 477–483, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstone JV, Hamdoun A, Cole BJ, Howard-Ashby M, Nebert DW, Scally M, Dean M, Epel D, Hahn ME, Stegeman JJ. The chemical defensome: environmental sensing and response genes in the Strongylocentrotus purpuratus genome. Dev Biol 300: 366–384, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grailles M, Brey PT, Roth CW. The Drosophila melanogaster multidrug-resistance protein 1 (MRP1) homolog has a novel gene structure containing two variable internal exons. Gene 307: 41–50, 2003. [DOI] [PubMed] [Google Scholar]
- 21.Grant CE, Kurz EU, Cole SP, Deeley RG. Analysis of the intron-exon organization of the human multidrug-resistance protein gene (MRP) and alternative splicing of its mRNA. Genomics 45: 368–378, 1997. [DOI] [PubMed] [Google Scholar]
- 22.Gökirmak T, Campanale JP, Shipp LE, Moy GW, Tao H, Hamdoun A. Localization and substrate selectivity of sea urchin multidrug (MDR) efflux transporters. J Biol Chem 287: 43876–43883, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gökirmak T, Shipp LE, Campanale JP, Nicklisch SC, Hamdoun A. Transport in technicolor: mapping ATP-binding cassette transporters in sea urchin embryos. Mol Reprod Dev 81: 778–793, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haber M, Smith J, Bordow SB, Flemming C, Cohn SL, London WB, Marshall GM, Norris MD. Association of high-level MRP1 expression with poor clinical outcome in a large prospective study of primary neuroblastoma. J Clin Oncol 24: 1546–1553, 2006. [DOI] [PubMed] [Google Scholar]
- 25.Hamdoun AM, Cherr GN, Roepke TA, Epel D. Activation of multidrug efflux transporter activity at fertilization in sea urchin embryos (Strongylocentrotus purpuratus). Dev Biol 276: 452–462, 2004. [DOI] [PubMed] [Google Scholar]
- 26.He X, Ee PL, Coon JS, Beck WT. Alternative splicing of the multidrug resistance protein 1/ATP binding cassette transporter subfamily gene in ovarian cancer creates functional splice variants and is associated with increased expression of the splicing factors PTB and SRp20. Clin Cancer Res 10: 4652–4660, 2004. [DOI] [PubMed] [Google Scholar]
- 27.Jin L, Kryukov K, Clemente JC, Komiyama T, Suzuki Y, Imanishi T, Ikeo K, Gojobori T. The evolutionary relationship between gene duplication and alternative splicing. Gene 427: 19–31, 2008. [DOI] [PubMed] [Google Scholar]
- 28.Klein M, Mamnun YM, Eggmann T, Schüller C, Wolfger H, Martinoia E, Kuchler K. The ATP-binding cassette (ABC) transporter Bpt1p mediates vacuolar sequestration of glutathione conjugates in yeast. FEBS Lett 520: 63–67, 2002. [DOI] [PubMed] [Google Scholar]
- 29.Koike K, Conseil G, Leslie EM, Deeley RG, Cole SP. Identification of proline residues in the core cytoplasmic and transmembrane regions of multidrug resistance protein 1 (MRP1/ABCC1) important for transport function, substrate specificity, and nucleotide interactions. J Biol Chem 279: 12325–12336, 2004. [DOI] [PubMed] [Google Scholar]
- 30.Kool M, van der Linden M, de Haas M, Baas F, Borst P. Expression of human MRP6, a homologue of the multidrug resistance protein gene MRP1, in tissues and cancer cells. Cancer Res 59: 175–182, 1999. [PubMed] [Google Scholar]
- 31.Kopelman NM, Lancet D, Yanai I. Alternative splicing and gene duplication are inversely correlated evolutionary mechanisms. Nat Genet 37: 588–589, 2005. [DOI] [PubMed] [Google Scholar]
- 32.Kruh GD, Belinsky MG. The MRP family of drug efflux pumps. Oncogene 22: 7537–7552, 2003. [DOI] [PubMed] [Google Scholar]
- 33.Leier I, Jedlitschky G, Buchholz U, Cole SP, Deeley RG, Keppler D. The MRP gene encodes an ATP-dependent export pump for leukotriene C4 and structurally related conjugates. J Biol Chem 269: 27807–27810, 1994. [PubMed] [Google Scholar]
- 34.Litman T, Brangi M, Hudson E, Fetsch P, Abati A, Ross DD, Miyake K, Resau JH, Bates SE. The multidrug-resistant phenotype associated with overexpression of the new ABC half-transporter, MXR (ABCG2). J Cell Sci 113: 2011–2021, 2000. [DOI] [PubMed] [Google Scholar]
- 35.Loe DW, Almquist KC, Cole SP, Deeley RG. ATP-dependent 17β-estradiol 17-(β-d-glucuronide) transport by multidrug resistance protein (MRP). Inhibition by cholestatic steroids. J Biol Chem 271: 9683–9689, 1996. [DOI] [PubMed] [Google Scholar]
- 36.Long Y, Li Q, Zhong S, Wang Y, Cui Z. Molecular characterization and functions of zebrafish ABCC2 in cellular efflux of heavy metals. Comp Biochem Physiol C Toxicol Pharmacol 153: 381–391, 2011. [DOI] [PubMed] [Google Scholar]
- 37.Mitra P, Oskeritzian CA, Payne SG, Beaven MA, Milstien S, Spiegel S. Role of ABCC1 in export of sphingosine-1-phosphate from mast cells. Proc Natl Acad Sci USA 103: 16394–16399, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nies AT, Keppler D. The apical conjugate efflux pump ABCC2 (MRP2). Pflügers Arch 453: 643–659, 2007. [DOI] [PubMed] [Google Scholar]
- 39.Qian YM, Song WC, Cui H, Cole SP, Deeley RG. Glutathione stimulates sulfated estrogen transport by multidrug resistance protein 1. J Biol Chem 276: 6404–6411, 2001. [DOI] [PubMed] [Google Scholar]
- 40.Ren J, Chung-Davidson YW, Yeh CY, Scott C, Brown T, Li W. Genome-wide analysis of the ATP-binding cassette (ABC) transporter gene family in sea lamprey and Japanese lamprey. BMC Genomics 16: 436, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren XQ, Furukawa T, Chen ZS, Okumura H, Aoki S, Sumizawa T, Tani A, Komatsu M, Mei XD, Akiyama S. Functional comparison between YCF1 and MRP1 expressed in Sf21 insect cells. Biochem Biophys Res Commun 270: 608–615, 2000. [DOI] [PubMed] [Google Scholar]
- 42.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574, 2003. [DOI] [PubMed] [Google Scholar]
- 43.Scheffer GL, Hu X, Pijnenborg AC, Wijnholds J, Bergen AA, Scheper RJ. MRP6 (ABCC6) detection in normal human tissues and tumors. Lab Invest 82: 515–518, 2002. [DOI] [PubMed] [Google Scholar]
- 44.Sharma KG, Kaur R, Bachhawat AK. The glutathione-mediated detoxification pathway in yeast: an analysis using the red pigment that accumulates in certain adenine biosynthetic mutants of yeasts reveals the involvement of novel genes. Arch Microbiol 180: 108–117, 2003. [DOI] [PubMed] [Google Scholar]
- 45.Shimizu H, Taniguchi H, Hippo Y, Hayashizaki Y, Aburatani H, Ishikawa T. Characterization of the mouse Abcc12 gene and its transcript encoding an ATP-binding cassette transporter, an orthologue of human ABCC12. Gene 310: 17–28, 2003. [DOI] [PubMed] [Google Scholar]
- 46.Shipp LE, Hamdoun A. ATP-binding cassette (ABC) transporter expression and localization in sea urchin development. Dev Dyn 241: 1111–1124, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shipp LE, Hill RZ, Moy GW, Gökırmak T, Hamdoun A. ABCC5 is required for cAMP-mediated hindgut invagination in sea urchin embryos. Development 142: 3537–3548, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slot AJ, Molinski SV, Cole SP. Mammalian multidrug-resistance proteins (MRPs). Essays Biochem 50: 179–207, 2011. [DOI] [PubMed] [Google Scholar]
- 49.Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC, Angerer LM, Arnone MI, Burgess DR, Burke RD, Coffman JA, Dean M, Elphick MR, Ettensohn CA, Foltz KR, Hamdoun A, Hynes RO, Klein WH, Marzluff W, McClay DR, Morris RL, Mushegian A, Rast JP, Smith LC, Thorndyke MC, Vacquier VD, Wessel GM, Wray G, Zhang L, Elsik CG, Ermolaeva O, Hlavina W, Hofmann G, Kitts P, Landrum MJ, Mackey AJ, Maglott D, Panopoulou G, Poustka AJ, Pruitt K, Sapojnikov V, Song X, Souvorov A, Solovyev V, Wei Z, Whittaker CA, Worley K, Durbin KJ, Shen Y, Fedrigo O, Garfield D, Haygood R, Primus A, Satija R, Severson T, Gonzalez-Garay ML, Jackson AR, Milosavljevic A, Tong M, Killian CE, Livingston BT, Wilt FH, Adams N, Bellé R, Carbonneau S, Cheung R, Cormier P, Cosson B, Croce J, Fernandez-Guerra A, Genevière AM, Goel M, Kelkar H, Morales J, Muln er-Lorillon O, Robertson AJ, Goldstone JV, Cole B, Epel D, Gold B, Hahn ME, Howard-Ashby M, Scally M, Stegeman JJ, Allgood EL, Cool J, Judkins KM, McCafferty SS, Musante AM, Obar RA, Rawson AP, Rossetti BJ, Gibbons IR, Hoffman MP, Leone A, Istrail S, Materna SC, Samanta MP, Stolc V, Tongprasit W, Tu Q, Bergeron KF, Brandhorst BP, Whittle J, Berney K, Bottjer DJ, Calestani C, Peterson K, Chow E, Yuan QA, Elhaik E, Graur D, Reese JT, Bosdet I, Heesun S, Marra MA, Schein J, Anderson MK, Brockton V, Buckley KM, Cohen AH, Fugmann SD, Hibino T, Loza-Coll M, Majeske AJ, Messier C, Nair SV, Pancer Z, Terwilliger DP, Agca C, Arboleda E, Chen N, Churcher AM, Hallböök F, Humphrey GW, Idris MM, Kiyama T, Liang S, Mellott D, Mu X, Murray G, Olinski RP, Raible F, Rowe M, Taylor JS, Tessmar-Raible K, Wang D, Wilson KH, Yaguchi S, Gaasterland T, Galindo BE, Gunaratne HJ, Juliano C, Kinukawa M, Moy GW, Neill AT, Nomura M, Raisch M, Reade A, Roux MM, Song JL, Su YH, Townley IK, Voronina E, Wong JL, Amore G, Branno M, Brown ER, Cavalieri V, Duboc V, Duloquin L, Flytzanis C, Gache C, Lapraz F, Lepage T, Locascio A, Martinez P, Matassi G, Matranga V, Range R, Rizzo F, Röttinger E, Beane W, Bradham C, Byrum C, Glenn T, Hussain S, Manning G, Miranda E, Thomason R, Walton K, Wikramanayke A, Wu SY, Xu R, Brown CT, Chen L, Gray RF, Lee PY, Nam J, Oliveri P, Smith J, Muzny D, Bell S, Chacko J, Cree A, Curry S, Davis C, Dinh H, Dugan-Rocha S, Fowler J, Gill R, Hamilton C, Hernandez J, Hines S, Hume J, Jackson L, Jolivet A, Kovar C, Lee S, Lewis L, Miner G, Morgan M, Nazareth LV, Okwuonu G, Parker D, Pu LL, Thorn R, Wright R, Sea Urchin Genome Sequencing Consortium. The genome of the sea urchin Strongylocentrotus purpuratus. Science 314: 941–952, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Srinivasan S, Bingham JL, Johnson D. The ABCs of human alternative splicing: a review of ATP-binding cassette transporter splicing. Curr Opin Drug Discov Dev 12: 149–158, 2009. [PubMed] [Google Scholar]
- 51.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sturm A, Cunningham P, Dean M. The ABC transporter gene family of Daphnia pulex. BMC Genomics 10: 170, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki T, Sasaki H, Kuh HJ, Agui M, Tatsumi Y, Tanabe S, Terada M, Saijo N, Nishio K. Detailed structural analysis on both human MRP5 and mouse mrp5 transcripts. Gene 242: 167–173, 2000. [DOI] [PubMed] [Google Scholar]
- 54.Tamaki A, Ierano C, Szakacs G, Robey RW, Bates SE. The controversial role of ABC transporters in clinical oncology. Essays Biochem 50: 209–232, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tarnay JN, Szeri F, Iliás A, Annilo T, Sung C, Le Saux O, Váradi A, Dean M, Boyd CD, Robinow S. The dMRP/CG6214 gene of Drosophila is evolutionarily and functionally related to the human multidrug resistance-associated protein family. Insect Mol Biol 13: 539–548, 2004. [DOI] [PubMed] [Google Scholar]
- 56.van de Ven R, Scheffer GL, Scheper RJ, de Gruijl TD. The ABC of dendritic cell development and function. Trends Immunol 30: 421–429, 2009. [DOI] [PubMed] [Google Scholar]
- 57.Weekes MP, Tan SY, Poole E, Talbot S, Antrobus R, Smith DL, Montag C, Gygi SP, Sinclair JH, Lehner PJ. Latency-associated degradation of the MRP1 drug transporter during latent human cytomegalovirus infection. Science 340: 199–202, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whalen K, Reitzel AM, Hamdoun A. Actin polymerization controls the activation of multidrug efflux at fertilization by translocation and fine-scale positioning of ABCB1 on microvilli. Mol Biol Cell 23: 3663–3672, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu X, Fu J, Wang H, Zhang B, Wang X, Wang Y. Influence of P-glycoprotein on embryotoxicity of the antifouling biocides to sea urchin (Strongylocentrotus intermedius). Ecotoxicology 20: 419–428, 2011. [DOI] [PubMed] [Google Scholar]
- 60.Zaja R, Munić V, Klobucar RS, Ambriović-Ristov A, Smital T. Cloning and molecular characterization of apical efflux transporters (ABCB1, ABCB11 and ABCC2) in rainbow trout (Oncorhynchus mykiss) hepatocytes. Aquat Toxicol (Amst) 90: 322–332, 2008. [DOI] [PubMed] [Google Scholar]
- 61.Zalcberg J, Hu XF, Slater A, Parisot J, El-Osta S, Kantharidis P, Chou ST, Parkin JD. MRP1 not MDR1 gene expression is the predominant mechanism of acquired multidrug resistance in two prostate carcinoma cell lines. Prostate Cancer Prostatic Dis 3: 66–75, 2000. [DOI] [PubMed] [Google Scholar]
- 62.Zhang DW, Cole SP, Deeley RG. Identification of an amino acid residue in multidrug resistance protein 1 critical for conferring resistance to anthracyclines. J Biol Chem 276: 13231–13239, 2001. [DOI] [PubMed] [Google Scholar]
- 63.Zhang DW, Gu HM, Situ D, Haimeur A, Cole SP, Deeley RG. Functional importance of polar and charged amino acid residues in transmembrane helix 14 of multidrug resistance protein 1 (MRP1/ABCC1): identification of an aspartate residue critical for conversion from a high to low affinity substrate binding state. J Biol Chem 278: 46052–46063, 2003. [DOI] [PubMed] [Google Scholar]
- 64.Zöchbauer-Müller S, Filipits M, Rudas M, Brunner R, Krajnik G, Suchomel R, Schmid K, Pirker R. P-glycoprotein and MRP1 expression in axillary lymph node metastases of breast cancer patients. Anticancer Res 21: 119–124, 2001. [PubMed] [Google Scholar]