Abstract
A number of key regulatory proteins contain one or two copies of the WW domain known to mediate protein–protein interaction via proline-rich motifs, such as PPxY. The Hippo pathway components take advantage of this module to transduce tumor suppressor signaling. It is becoming evident that tyrosine phosphorylation is a critical regulator of the WW proteins. Here, we review the current knowledge on the involved tyrosine kinases and their roles in regulating the WW proteins.
Keywords: Hippo pathway, non-receptor tyrosine kinases, cell fate decision-making, protein–protein interaction, protein tyrosine phosphorylation
Introduction
The WW domain was first defined and identified by Marius Sudol in his analysis of the Yes-associated protein (Yap). Yap was isolated as an interacting protein of the Src family tyrosine kinase Yes.1–3 WW domains are of approximately 40 amino acids long with a triple stranded beta-sheet fold.4 Two tryptophan (W) residues located 20–22 amino acids apart are the hallmark of this domain (x6Wx20–22Wx5). There are at least four classes of WW domains, with each class having a preferred binding motif.5 The binding motifs are enriched for proline, with the PPxY motif being the major one. Of interest is the conditional binding motif favored by group IV WW domains, which is a phosphorylated S/T-P module, thus requiring a kinase for interaction with its binding partner.5,6 Due to the similarity of the WW domain-binding motifs to PxxP, the consensus binding motif of SH3 domains, there is the possibility for competition between WW and SH3 proteins to bind this motif. Although both the SH3 and WW domain bind their motifs independently, surrounding sequence can influence affinity and specificity.4 WW domains are found in E3 ligases, transcription coactivators, isomerases, scaffold and signaling proteins. Consistent with their involvement in many basic cell processes, they are implicated in disease processes such as neurodegenerative diseases and cancer. Protein interaction modules such as the WW domain provide convenient means to facilitate protein interaction to promote processes such as signal propagation upon modification. Here, we review the current knowledge on tyrosine kinases and their roles in regulating the WW proteins.
Tyrosine phosphorylation of the WW domain and its binding motif
The WW domain sequence consensus typically has one or more tyrosine residues in the second β-strand of the motif.5 The conservation of tyrosine residues within the consensus sequences of the WW domain and its interaction domains suggests that direct phosphorylation of these residues will affect binding. The WW protein Wwox provides a nice illustration of this concept. The Wwox (WW domain containing oxidoreductase) tumor suppressor gene is localized to one of the most common fragile sites active in cancer, FRA16D (reviewed in Del Mare et al.7). Wwox has two WW domains at its N-terminus, through which it interacts with a number of different proteins. Wwox interacts with p73 through the first WW domain of Wwox, and the 482PPPPY motif of p73.8 Interestingly, this is the same motif through which p73 interacts with both Yap9 and Itch.10 Phosphorylation of Wwox at Y33, a site within the first WW domain of Wwox, enhances Wwox binding to p73.8 This leads to cytoplasmic sequestration of p73 by Wwox. Association of Wwox with the AP-2γ transcription factor is also governed by the N-terminal WW domain of Wwox, and the PPPY motif of AP-2γ. Like p73, association of Wwox with AP-2γ leads to cytoplasmic sequestration of AP-2γ. Y33 of Wwox is also important for this interaction.11 In response to TGF-β stimulation, pY33 Wwox complexes with Hyal-2, the complex localizes to the nucleus, and controls the activation of Smad-driven promoters.12 Wwox phosphorylated at Y33 has also been shown to associate with S46-phosphorylated p53 in cells exposed to DNA damage and other stress.13,14 This interaction stabilizes pS46 p53, and promotes apoptosis. Although this association implicates the WW domain of Wwox in the interaction with p53, binding by p53 in this case is not via a classical motif. Wwox also interacts with activated JNK1, and this also requires phosphorylation of Y33 of Wwox.15 This interaction competes with Wwox binding to p53. However, the nature of this binding motif has not been completely resolved.
The Y residue of the PPxY WW-binding motif is also a potential target of tyrosine phosphorylation. In fact, tyrosine phosphorylation of this motif has been shown to regulate the interaction between several WW proteins and their interacting proteins. For example, tyrosine phosphorylation of the PPxY motif regulates the binding of the WW protein dystrophin and its cognate binding partner β-dystroglycan.16 Dystrophin forms a complex with membrane-associated proteins called the dystrophin-associated protein complex (DAPC) in the membrane of skeletal muscle cells. This complex is thought to act as a link between the actin cytoskeleton and the extracellular matrix (ECM), and is implicated in cell signaling events. β-dystroglycan is needed for the interaction between dystrophin and the DAPC. Tyrosine phosphorylation of β-dystroglycan disrupts its interaction with dystrophin, with phosphorylation of the tyrosine residue of the PPPY motif being the most crucial in mediating the disruption.16
WW proteins and PTPases
Regulation of protein–protein interactions by phosphorylation implies that in addition to the kinases providing phosphorylation, there must be phosphatases capable of reversing the effect. One example of a tyrosine-phosphorylated WW domain was identified through a screen for substrates of a phosphatase.17 The WW domain protein Magi1 (membrane-associated guanylate kinase, WW and PDZ domain containing 1) was found to be a substrate of Ptprz (Protein-tyrosine phosphatase receptor type z). Interestingly, Y373, a site within the second WW domain of Magi1, was dephosphorylated by Ptprz, in addition to Y858, within a PDZ domain.17 Although the functional significance of these phosphorylation sites remains to be explored, it is likely that they affect the binding properties of Magi1. Furthermore, this study, and information curated in PhosphositePlus®18 indicate several other sites of phosphorylation on Magi1 that are also likely to be physiologically significant.
Other tyrosine phosphatases have been shown to interact with WW proteins. The non-receptor type protein tyrosine phosphatase 14 (Ptpn14) binds to and negatively regulates Yap.19–21 Ptpn14 interacts with Yap through its PPxY motif and the WW domain of Yap. While Yap is a substrate of Ptpn14,19 the inhibition of Yap activity by Ptpn14 appears to involve several mechanisms, as inhibition of Yap was also observed independently of Ptpn14 phosphatase activity and the PPxY motif.21 In addition, Ptpn14 interacts with the Hippo pathway proteins Kibra and Lats. Ptpn14 and Kibra activate Lats both independently and cooperatively, and this activation does not require Mst kinases.22 The PTP domain of Ptpn14 is required for the interaction, but whether the phosphatase activity is required is not known. Activation of Lats by Ptpn14/Kibra then leads to the cytoplasmic sequestration and functional inhibition of Yap. Thus, Ptpn14 inhibits Yap through direct and indirect mechanisms.
Yap and its paralog Taz interact with the tyrosine phosphatase Ptpn11, also known as Shp2.23 Shp2 is involved in activation of the Ras-Erk pathway in the cytoplasm, and in the nucleus it promotes Wnt target activation. Shp2 binds to Yap and Taz at the WW- and PDZ-binding domains, and its localization is dependent on the localization of Yap/Taz.23 Thus, Shp2 localization is governed by the Hippo pathway (see further text), which regulates Yap/Taz localization. It has been shown recently that the Polyomavirus middle T-antigen (PyMT) oncogene induces cytoplasmic retention of Taz and Yap, and consequently of Shp2. In this context, Taz WW domain is required to assist PyMT-induced cellular transformation.24
Another tyrosine phosphatase, Ptpn13, was found to be in a complex comprised of Taz, Ephrin B1, and Nherf1 in bone marrow stromal cells.25 This complex, and the release of dephosphorylated Taz to the nucleus, is important for proper osteoblast differentiation.25 It is not yet known whether tyrosine phosphatase activity plays a role here in mediating the interactions.
In Drosophila, the evolutionarily conserved protein Pez interacts with the WW domain containing protein Kibra, one of the upstream regulators of the Hippo pathway.26 Pez is homologous to the mammalian non-receptor phosphatases Ptpn21 and Ptpn14. Pez has a FERM domain and a protein tyrosine phosphatase (PTP) domain in addition to a PPxY motif. Interaction with Kibra depends on the first WW domain of Kibra, but the FERM, PTP and PPxY motifs of Pez are dispensable, and instead binding depends on a central proline-rich region of Pez. Pez is required for proper regulation of intestinal stem cells, and is an upstream regulator of the Yap ortholog Yki, but this activity is independent of the PTP domain.26 These examples demonstrate that the potential exists for tyrosine phosphatase activity to affect interaction between a WW protein and its substrate, although more work is needed to confirm this hypothesis.
Tyrosine phosphorylation of WW proteins and the physiological effects
Tyrosine phosphorylation of WW proteins and transcription
The first WW protein, Yap, was isolated as a protein that binds the SH3 domain of the non-receptor tyrosine kinase Yes.1–3 Yap binds other members of this tyrosine kinase family such as Src, Nck, Crk and c-Abl SH3 domain. About a decade later, it was found that Yap is associated with both Yes and Src tyrosine kinases in osteoblasts. Based on a pharmacologic inhibitor of Src (PP2) and a Src dominant negative mutant, it was suggested that Src family tyrosine kinase signaling increases binding of Yap to transcription factor Runx2 in the process of repressing the osteocalcin gene.27 However, these studies did not demonstrate that Yap was tyrosine phosphorylated.
The ErbB-4 receptor tyrosine kinase is protealytically processed in response to stimulation, and the cleaved C-terminal fragment translocates to the cell nucleus. Yap associates with this fragment, and together the complex has transcription activation activity. The association depends upon the first WW domain of Yap, and the most carboxy-terminal PPXY of ErbB-4.28 Interestingly, phosphorylation of the PPXY motif of ErbB-4 interrupts its binding with Yap.29 Furthermore, WWOX can compete with Yap for binding to full-length ErbB-4 in the cytoplasm, via the first WW domain of WWOX.30 In this manner, WWOX antagonizes Yap/ErbB-4 transcriptional activity, through cytoplasmic sequestration of ErbB-4.
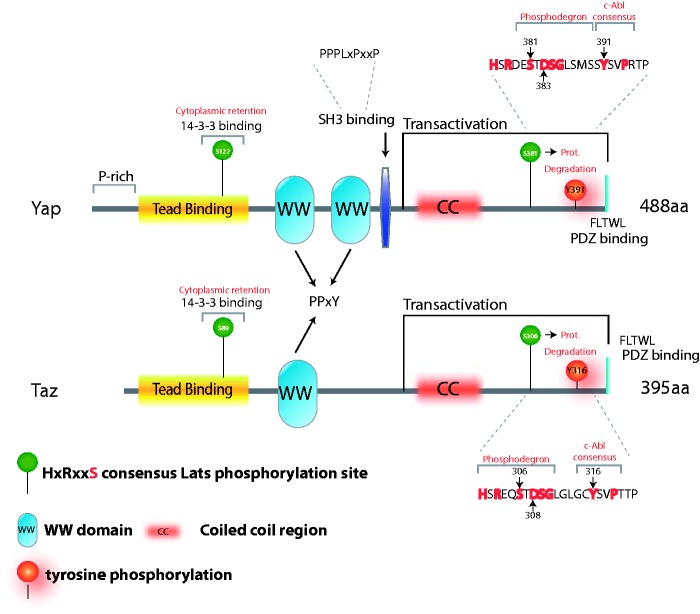
Direct Yap tyrosine phosphorylation was demonstrated in response to DNA damage.31 Under this condition, c-Abl phosphorylates Yap at Y357 (Yap isoform with one WW domain) or Y391 (Yap isoform with two WW domains) (Figure 1). This tyrosine residue is located at the Yap transcription activation domain. Whether Src phosphorylates this residue is an open question. In a cell-free system, it has been reported that Src phosphorylates Yap.19 Others failed to detect an interaction between Yap1 and Src or Fyn in colon cancer cell lines, although overexpressed Src in HEK293 cells is active in phosphorylating Yap.32 However, our direct comparison between c-Abl and Src in their capacity to phosphorylate Yap revealed that Src is a rather inefficient Yap kinase (unpublished observation).
Figure 1.
Schematic representation of the WW proteins Yap and Taz. Yap and Taz structural features, including the WW domains, Tead binding domain, coiled-coil region, and proline-rich and PDZ-binding domains are indicated. Phosphorylation sites described in the text are depicted. The Yap Y391 site is the same as Y357 in the Yap isoform with one WW domain. The sequences encompassing the phosphodegrons are shown. (A color version of this figure is available in the online journal.)
The Yes tyrosine kinase also phosphorylates Yap. Yap phosphorylation by Yes causes an increase in Yap association with TEAD, and activation of the pluripotency genes Oct-3/4 and Nanog.33 In the context of colon cancer, Yap forms a complex with β-catenin and Tbx5. Yes-mediated tyrosine phosphorylation of Yap at Y391 causes this complex to localize to promoters of antiapoptotic genes.32
The paralog of Yap, Taz, is also regulated by tyrosine phosphorylation. Upon hyperosmotic stress, Taz is phosphorylated at Y316 by c-Abl.34 Interestingly, Taz Y316 is the homologous tyrosine residue that is phosphorylated in Yap (Y391) (Figure 1). Phosphorylated Taz interacts with NFAT5 (nuclear factor of activated T cells 5), a major osmoregulatory transcription factor. This interaction suppresses DNA binding and transcriptional activation activity of NFAT5.
The WW protein FE65 is an adaptor protein that mediates transcription activation by the cleaved C-terminus of APP (amyloid precursor protein) (reviewed in Russo et al.35). c-Abl phosphorylates both APP and FE65, but it is the phosphorylation of FE65 at Y547 that is responsible for stimulating APP/FE65-mediated gene transcription.36 The phosphorylation site is located within the second phosphotyrosine binding domain of FE65, not the WW domain of FE65. In these examples, tyrosine phosphorylation affects the activity of WW proteins that are also transcription coactivators. However, in each of these cases, the phosphorylated tyrosine residue lies outside of the WW domain. The example of ErbB-4 association with Yap represents an example of phosphorylation of the PPxY motif affecting association with the WW domain protein.
Role of tyrosine phosphorylation of WW proteins in protein stability
Tyrosine phosphorylation of the E3-ubiquitin ligase WW proteins
The Nedd4 family of E3 ubiquitin ligases is characterized by several conserved domains: a Ca2+/lipid binding domain, a HECT ubiquitin ligase domain, and two to four WW domains (reviewed in Harvey and Kumar37 and An et al.38). Its family members mediate the degradation of a wide array of substrates, many of them central components of signaling pathways such as PTEN, TGF-β, and MAPK pathways (reviewed in An et al.39). The Nedd4 family member Nedd4.2 mediates the degradation of AMOTL1.40 AMOTL1 is a member of the Angiomotin (AMOT) family, membrane-associated proteins originally isolated based on binding to angiostatin and implicated in cell migration, and recently shown to be regulators of the Hippo pathway (reviewed in Moleirinho et al.41). Yap protects AMOTL1 from Nedd4.2-mediated degradation, and this depends on the first WW domain of Yap. This protection is not via competition for binding, but rather through Yap recruitment of c-Abl, which then phosphorylates Nedd4.2 at Y71 and Y457, sites outside of the WW domains, and inhibits its E3 ligase activity.40
Nedd4.1 is also regulated by tyrosine phosphorylation. FGFR1 is ubiquitinated by Nedd4.1 following FGFR activation, which leads to receptor internalization and degradation. FGFR1 activation of c-Src leads to c-Src-dependent phosphorylation of Nedd4.1 at tyrosine residues including Y43 in the C2 domain and Y585 in the HECT domain. These phosphorylations increase Nedd4.1 ubiquitin ligase activity.42
PTEN is a substrate of Nedd4.1, and the Src family kinase Rak regulates this degradation. Phosphorylation of PTEN by Rak at Y336 prevents its association with Nedd4.1, and its ubiquitination and degradation.43 Surprisingly, the Y366 site is not part of a PPxY motif, although it clearly affects the binding of PTEN to Nedd4.1.
The E3 ubiquitin ligase Itch is another member of the Nedd4 family, and has four WW domains. Itch mediates the degradation of a variety of proteins, including the Jun proteins,44 p73,10 p63,45 the Hippo pathway kinase Lats1,46 and TXNIP.47 Jun, p73, p63, Lats1, and TXNIP binding to Itch involve interaction between the PPxY motifs of these proteins and WW domain(s) of Itch. Interestingly, an intramolecular interaction between the Itch WW and HECT domains keeps it in an inactive form. Itch is activated by JNK1 phosphorylation of S199, S232, and T222, located within a proline-rich region.48 These phosphorylations induce a conformational change that enables the E3 ligase activity. In contrast, tyrosine phosphorylation negatively regulates Itch E3 ligase activity. Phosphorylation of Itch by Fyn at Y371, located between the N-terminal second and third WW domain in Itch, reduces its interaction with JunB.49 Interestingly, phosphorylation of c-Jun by c-Abl at the PPxY motif of c-Jun interrupts association of c-Jun with Itch, and thereby prevents its ubiquitination and degradation.50
Tyrosine phosphorylation of WW proteins and proteasomal degradation
WW proteins are substrates of proteasomal degradation. In a number of cases, tyrosine phosphorylation affects their proteasomal susceptibility. For example, tyrosine phosphorylation of Wwox at Y287 by Ack1 (activated Cdc42-associated kinase) leads to the dissociation of Wwox and Ack1, and the ubiquitination and proteasomal degradation of Wwox.51 This tyrosine residue is positioned outside the WW domains described above; Wwox therefore exemplifies several variations on the theme of regulation by tyrosine phosphorylation of WW domain proteins.
Additional examples are Yap and Taz WW proteins. Phosphorylation of Yap at S381 by Lats primes further phosphorylation by CK1δ/ɛ, resulting in a phosphodegron recognized by the F-box protein β-TrCP and ubiquitination by the SCF/CRL1(β-TrCP) E3 ligase52 leading to proteasomal degradation (Figure 1). A similar mechanism regulates Taz stability.53 The Taz phosphodegron motif is phosphorylated by Lats at the S311 residue. Interestingly, the tyrosine phosphorylated sites, Y391 and Y316, of Yap and Taz respectively, are at the very same regions. Given the fact that tyrosine-phosphorylated Yap accumulates to high levels,31 it has been speculated52 that this modification downregulates β-TrCP binding to the nearby phosphodegrons, although this awaits experimental confirmation.
Accumulation of WW proteins, in turn, may protect other proteasomal substrates undergoing polyubiquitination by the WW E3 ligases. A rather well-studied case is the tumor suppressor p73, the p53 paralog. p73 is targeted by the E3 ubiquitin ligase Itch, a WW domain protein that binds to p73 through its PPxY motif.10 Yap, by competing for binding to this motif, reduces Itch-p73 interaction and ubiquitination.54,55 Thus, tyrosine phosphorylated Yap that accumulates to high levels leads to stabilization and accumulation of p73.
An interesting regulatory circuit was described that couples Itch expression to Yap and p73 tyrosine phosphorylation. The Yap-Runx complex targets the Itch gene to induce its transcription. Under DNA damage Yap is tyrosine phosphorylated by c-Abl. The modified Yap dissociates from Runx and the level of Itch is reduced. This mechanism ensures downregulation of the WW E3-ligase Itch under DNA damage, a process that supports p73 accumulation.56
Tyrosine phosphorylation of adaptor and scaffold WW proteins
Iqgap1 (IQ motif-containing GTPase activating protein 1) is a member of the Iqgap family, expressed in organisms ranging from yeast to humans (reviewed in White et al.57). Iqgap1 is a ubiquitously expressed scaffold protein, which binds or indirectly associates with over 90 different proteins, and helps to mediate receptor signaling, small GTPase function, actin dynamics, neuronal regulation, intracellular trafficking, and more. Iqgap1 has a WW domain, which is needed for Iqgap1 interaction with Erk1/2.58 Interestingly, interruption of the interaction between Erk1/2 and Iqgap1 by using a competing peptide containing the WW domain is able to inhibit RAS- and RAF-driven tumorigenesis that is dependent on Erk signaling.59 Iqgap1 is tyrosine phosphorylated in response to growth factor stimulation, such as VEGF60,61 or EGF,62 although the site(s) phosphorylated were not identified. The PhosphositePlus® site has evidence from numerous Mass Spectrometry studies that Iqgap1 is tyrosine phosphorylated on several residues, some close to the WW domain.18 The functional significance of each site is still unknown. Similarly, another GAP with a WW domain, Arhgap12, is also tyrosine phosphorylated on multiple residues of the protein according to Mass Spectrometry data curated by PhosphositePlus®.18 In addition, the WW domain-containing dystrophin-related protein Utrophin has tyrosine phosphorylated residues. The significance of these events awaits study. Table 1 summarizes the WW proteins/binding proteins for which the tyrosine kinases or the phosphorylation sites have been specifically characterized.
Table 1.
Tyrosine phosphorylation of WW domain proteins and WW binding proteins
| Protein | WW domain class | Residues modified | Kinase | Tyrosine phosphorylation in WW domain | Reference |
|---|---|---|---|---|---|
| β-dystroglycan | binds to Class I | Y892 | ND | – | 16 |
| ErbB-4 | binds to Class I | Y1056 | ligand-dependent autophosphorylation | – | 29 |
| FE65 | Class II | Y547 | c-Abl | No | 36 |
| Itch | Class I | Y371 | Fyn | No | 49 |
| c-Jun | binds to Class I | Y170 | c-Abl | – | 50 |
| Magi1 | Class I | Y373, Y858 | c-Src phosphatase:PTPrz | Yes, No | 17 |
| Nedd4.1 | Class I | Y43, Y585 | c-Src | No | 42 |
| Nedd4.2 | Class I, IV | Y71, Y457 | c-Abl | No | 40 |
| Taz (WWTR1) | Class I | Y316 | c-Abl | No | 34 |
| Wwox | Class I | Y287 | Ack | No | 51 |
| Y33 | c-Src | Yes | 8 | ||
| Yap | Class I | Y357 | c-Src/c-Yes | No | 19,32 |
| c-Abl | 31 |
Note: This table summarizes the tyrosine phosphorylated proteins described in the text whose phosphorylation sites and/or kinases have been specifically characterized.
WW proteins and Hippo pathway
The first WW protein to be discovered, Yap, is the downstream effector of the Hippo pathway. The Hippo pathway is an evolutionarily conserved pathway from flies to humans that regulates cell proliferation and organ size (reviewed in the literatures63, 64). In response to extracellular and mechanical cues indicating cell contact and crowding, a kinase cascade is initiated. At the core of the Hippo pathway cascade the S/T kinases Mst1/2, with the scaffold WW protein WW45, phosphorylate and activate S/T kinases Lats1/2. Lats in turn, together with cofactor Mob, phosphorylates Yap1/2, and its paralog Taz. Phosphorylation of the transcription coactivators Yap and Taz by Lats increases their affinity to the 14-3-3 proteins, leading to cytoplasmic retention. Lats phosphorylation of Yap and Taz also promotes their proteasomal degradation mediated by β-TrCP. These processes blunt nuclear import of Yap and Taz, preventing them from coactivating the TEAD family of transcription factors. As a result, the expression of pro-proliferative and anti-apoptotic target genes is downregulated. Thus, Hippo pathway activation arrests cell proliferation. Unfettered cell proliferation and cancer can ensue when Hippo pathway is off or dysregulated.
Interestingly, many of the Hippo pathway components are WW proteins (reviewed in Salah and Aqeilan65) and can be grouped by their possession of WW domains or WW binding motifs, particularly the PPxY (PY) motif. This modularity of the Hippo pathway with its extensive use of WW domain interactions may indicate its evolutionary origins.66,67 In addition, it permits non-linear interactions within the pathway.66
Hippo pathway and tyrosine phosphorylation
Tyrosine phosphorylation of the Hippo pathway effectors, Yap and Taz, is important in guiding specific functional output and instructing cell fate. The Yap transcription coactivator targets several different transcription factors, including the TEAD family for activation of pro-proliferative genes; Runx, for activation of differentiation and other targets, such as Itch; and p73, for induction of pro-apoptotic genes. Tyrosine phosphorylation regulates Yap binding to its transcription factor partners and to specific target genes. An example of this is Yap phosphorylation by c-Abl in response to DNA damage. Under this condition, the modified Yap binds to and coactivates p73 in targeting proapoptotic genes. Yap phosphorylation also influences the choice of p73 target genes, with phosphorylated Yap preferentially associating with pro-apoptotic genes such as Bax, rather than the cell cycle arrest gene p21.31 Phosphorylation of Yap also switches target gene expression via increasing Yap association with a distinct group of transcription factors. For example, the modified Yap preferentially binds and transactivates p73, whereas its association with Runx is compromised.31 Furthermore, phospho Y391 Yap or its phosphomimetic mutant coactivate the TEAD transcription factors less efficiently than unphosphorylated Yap.68 In this case, Yap Y391 phosphorylation does not interfere with Yap binding to TEAD, but it does inhibit transactivation of TEAD. This could mean that the Yap phosphorylation inhibits the recruitment of other factors at TEAD target genes. Thus, in response to DNA damage, c-Abl phosphorylation of Yap at Y391 switches the cell fate program away from proliferation or differentiation, by reducing Yap targeting of TEAD and Runx, and instead induces apoptosis by increasing Yap coactivation of p73.
Perspective
Our analysis of tyrosine phosphorylation of WW domain proteins and their binding partners has revealed three groups: examples where WW domains were phosphorylated, and this affected binding to substrate proteins, as in the case of Wwox; cases where the PPxY motif is targeted, affecting binding to a WW domain, as in the case of β-dystroglycan; and examples where the phosphorylation falls outside the WW domain, but still affects binding involving the WW domain, as in the case of Yap (Figure 2). Although the documented cases are still relatively few in number, as noted above there are many phosphorylations identified by mass spectrometry whose function has not been characterized. In addition, there may yet be others that exist that have not yet been detected experimentally. Nevertheless, the available data do offer some insight with mechanistic implications.
Figure 2.
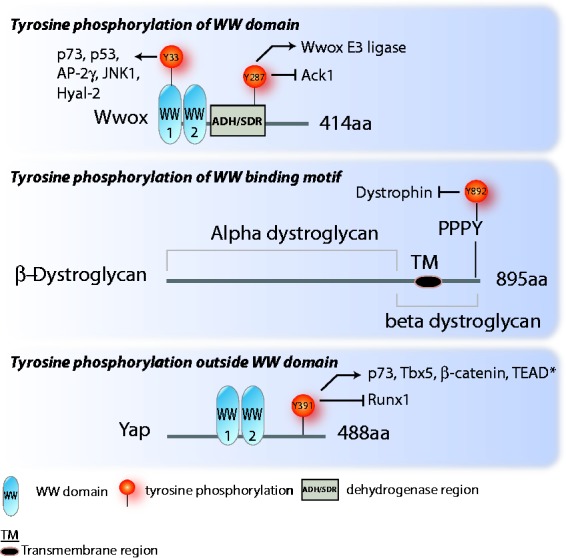
Schematic representation of WWox, β-dystroglycan, and Yap. These three proteins serve as examples for three ways tyrosine phosphorylation can affect WW domain interaction: (1) phosphorylation of the WW domain itself, as seen in Wwox; (2) phosphorylation of the WW binding domain, as seen in β-dystroglycan; and (3) phosphorylation outside of the WW domain that affects WW domain interactions, as seen with Yap. The Y391 site is the same as Y357 in Yap1, the isoform with one WW domain. The protein interactions affected by the phosphorylation are indicated by an arrow for increased interaction due to the phosphorylation, and a blunted arrow for decreased interaction. *The effect of Yap Y391 phosphorylation on Yap/TEAD interaction and activity may be different in different contexts. Tamm et al.33 showed that Yes phosphorylation of Yap increased association with and activation of TEAD. It should be noted, however, that in this work the Yap phosphorylation site was not specifically characterized. Keshet et al.68 showed that Yap Y391 phosphorylation reduced Yap coactivation of TEAD, but it did not reduce Yap-TEAD binding. (A color version of this figure is available in the online journal.)
Most of the documented cases of tyrosine phosphorylation of WW proteins involve tyrosine phosphorylation of regions of the proteins outside of the WW domains, yet these phosphorylations affect interactions involving the WW domain. For example, phosphorylation of Yap by c-Abl at Y391 reduces its interaction with Runx,31 despite the fact that Runx does interact with Yap through the Yap WW domain, and Y391 is not within the WW domain. Although the WW domain binds its motif independently,4 there are cases where regions outside the WW domain can affect this interaction, as with the interaction between dystrophin and β-dystroglycan, which depends upon the EF hands and N-terminus of dystrophin.69 We can infer that other WW domain interactions might be affected by phosphorylations outside of the interaction domain that lead to allosteric effects. Furthermore, certain WW proteins might display auto-inhibitory interactions where the WW domain interacts with another site on the protein, as in the case of Itch, whose WW domain interacts with the HECT domain, despite the non-canonical motif used for the interaction.48 In the case of Itch, the autoinhibition is relieved by JNK phosphorylation. It is logical that tyrosine phosphorylation may perform this role in other settings.
As noted earlier, the Hippo pathway employs many WW proteins. Tyrosine phosphorylation events on members of this pathway, such as Yap and Taz, demonstrate crosstalk between Hippo and other signaling pathways in the cell. Interestingly, tyrosine phosphorylation of the WW proteins reported to date most often involves non-receptor tyrosine kinases, such as Abl, Src, and Yes. These kinases possess SH2 and SH3 domains for binding to phosphotyrosine and polyproline sites, respectively, and thus may also serve a scaffolding function in bringing together proteins of different pathways. In addition, these kinases are involved in modulating actin dynamics. Recent studies have demonstrated an interplay between mechanical cues, the actin cytoskeleton, and the Hippo pathway (reviewed in the literatures70–72). Thus, involvement of these tyrosine kinases in modulating WW protein activity may be relevant to this interplay. Furthermore, several of the WW domain proteins described earlier are themselves involved in actin dynamics, such as Iqgap1, Arhgap12, and Utrophin. This common use of WW domains and tyrosine phosphorylation could help to bridge mechanosensing and Hippo pathway signaling.
Authors' contributions
NR and YS wrote the review, MS contributed to background research and prepared the figures.
Funding
This work was supported by the Israel Science Foundation (grant No. 551/11).
References
- 1.Sudol M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene 1994; 9: 2145–52. [PubMed] [Google Scholar]
- 2.Bork P, Sudol M. The WW domain: a signalling site in dystrophin. Trends Biochem Sci 1994; 19: 531–3. [DOI] [PubMed] [Google Scholar]
- 3.Sudol M, Bork P, Einbond A, Kastury K, Druck T, Negrini M, Huebner K, Lehman D. Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J Biol Chem 1995; 270: 14733–41. [DOI] [PubMed] [Google Scholar]
- 4.Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett 2002; 513: 30–7. [DOI] [PubMed] [Google Scholar]
- 5.Hu H, Columbus J, Zhang Y, Wu D, Lian L, Yang S, Goodwin J, Luczak C, Carter M, Chen L, James M, Davis R, Sudol M, Rodwell J, Herrero JJ. A map of WW domain family interactions. Proteomics 2004; 4: 643–55. [DOI] [PubMed] [Google Scholar]
- 6.Otte L, Wiedemann U, Schlegel B, Pires JR, Beyermann M, Schmieder P, Krause G, Volkmer-Engert R, Schneider-Mergener J, Oschkinat H. WW domain sequence activity relationships identified using ligand recognition propensities of 42 WW domains. Protein Sci: Publication Protein Soc 2003; 12: 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Del Mare S, Salah Z, Aqeilan RI. WWOX: its genomics, partners, and functions. J Cell Biochem 2009; 108: 737–45. [DOI] [PubMed] [Google Scholar]
- 8.Aqeilan RI, Pekarsky Y, Herrero JJ, Palamarchuk A, Letofsky J, Druck T, Trapasso F, Han SY, Melino G, Huebner K, Croce CM. Functional association between Wwox tumor suppressor protein and p73, a p53 homolog. Proceedings Natl Acad Sci USA 2004; 101: 4401–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, Oren M, Sudol M, Cesareni G, Blandino G. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem 2001; 276: 15164–73. [DOI] [PubMed] [Google Scholar]
- 10.Rossi M, De Laurenzi V, Munarriz E, Green DR, Liu YC, Vousden KH, Cesareni G, Melino G. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J 2005; 24: 836–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aqeilan RI, Palamarchuk A, Weigel RJ, Herrero JJ, Pekarsky Y, Croce CM. Physical and functional interactions between the Wwox tumor suppressor protein and the AP-2gamma transcription factor. Cancer Res 2004; 64: 8256–61. [DOI] [PubMed] [Google Scholar]
- 12.Hsu LJ, Schultz L, Hong Q, Van Moer K, Heath J, Li MY, Lai FJ, Lin SR, Lee MH, Lo CP, Lin YS, Chen ST, Chang NS. Transforming growth factor beta1 signaling via interaction with cell surface Hyal-2 and recruitment of WWOX/WOX1. J Biol Chem 2009; 284: 16049–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang NS, Doherty J, Ensign A, Lewis J, Heath J, Schultz L, Chen ST, Oppermann U. Molecular mechanisms underlying WOX1 activation during apoptotic and stress responses. Biochem Pharmacol 2003; 66: 1347–54. [DOI] [PubMed] [Google Scholar]
- 14.Chang NS, Doherty J, Ensign A, Schultz L, Hsu LJ, Hong Q. WOX1 is essential for tumor necrosis factor-, UV light-, staurosporine-, and p53-mediated cell death, and its tyrosine 33-phosphorylated form binds and stabilizes serine 46-phosphorylated p53. J Biol Chem 2005; 280: 43100–8. [DOI] [PubMed] [Google Scholar]
- 15.Chang NS, Doherty J, Ensign A. JNK1 physically interacts with WW domain-containing oxidoreductase (WOX1) and inhibits WOX1-mediated apoptosis. J Biol Chem 2003; 278: 9195–202. [DOI] [PubMed] [Google Scholar]
- 16.Ilsley JL, Sudol M, Winder SJ. The interaction of dystrophin with beta-dystroglycan is regulated by tyrosine phosphorylation. Cell Signal 2001; 13: 625–32. [DOI] [PubMed] [Google Scholar]
- 17.Fujikawa A, Fukada M, Makioka Y, Suzuki R, Chow JP, Matsumoto M, Noda M. Consensus substrate sequence for protein-tyrosine phosphatase receptor type Z. J Biol Chem 2011; 286: 37137–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res 2012; 40: D261–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, Yang N, Figel SA, Wilson KE, Morrison CD, Gelman IH, Zhang J. PTPN14 interacts with and negatively regulates the oncogenic function of YAP. Oncogene 2013; 32: 1266–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang JM, Nagatomo I, Suzuki E, Mizuno T, Kumagai T, Berezov A, Zhang H, Karlan B, Greene MI, Wang Q. YAP modifies cancer cell sensitivity to EGFR and survivin inhibitors and is negatively regulated by the non-receptor type protein tyrosine phosphatase 14. Oncogene 2013; 32: 2220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michaloglou C, Lehmann W, Martin T, Delaunay C, Hueber A, Barys L, Niu H, Billy E, Wartmann M, Ito M, Wilson CJ, Digan ME, Bauer A, Voshol H, Christofori G, Sellers WR, Hofmann F, Schmelzle T. The tyrosine phosphatase PTPN14 is a negative regulator of YAP activity. PloS ONE 2013; 8: e61916–e61916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson KE, Li YW, Yang N, Shen H, Orillion AR, Zhang J. PTPN14 Forms a Complex with Kibra and LATS1 proteins and negatively regulates the YAP oncogenic function. J Biol Chem 2014; 289: 23693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsutsumi R, Masoudi M, Takahashi A, Fujii Y, Hayashi T, Kikuchi I, Satou Y, Taira M, Hatakeyama M. YAP and TAZ, Hippo signaling targets, act as a rheostat for nuclear SHP2 function. Developmen Cell 2013; 26: 658–65. [DOI] [PubMed] [Google Scholar]
- 24.Shanzer M, Ricardo-Lax I, Keshet R, Reuven N, Shaul Y. The Polyomavirus Middle T-antigen oncogene activates the Hippo-pathway tumor suppressor Lats in a Src-dependent manner. Oncogene 2014. Nov 3. doi: 10.1038/onc.2014.347. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 25.Xing W, Kim J, Wergedal J, Chen ST, Mohan S. Ephrin B1 regulates bone marrow stromal cell differentiation and bone formation by influencing TAZ transactivation via complex formation with NHERF1. Molecular Cellular Biol 2010; 30: 711–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poernbacher I, Baumgartner R, Marada SK, Edwards K, Stocker H. Drosophila Pez acts in Hippo signaling to restrict intestinal stem cell proliferation. Curr Biol 2012; 22: 389–96. [DOI] [PubMed] [Google Scholar]
- 27.Zaidi SK, Sullivan AJ, Medina R, Ito Y, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J 2004; 23: 790–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem 2003; 278: 33334–41. [DOI] [PubMed] [Google Scholar]
- 29.Omerovic J, Puggioni EM, Napoletano S, Visco V, Fraioli R, Frati L, Gulino A, Alimandi M. Ligand-regulated association of ErbB-4 to the transcriptional co-activator YAP65 controls transcription at the nuclear level. Experiment Cell Res 2004; 294: 469–79. [DOI] [PubMed] [Google Scholar]
- 30.Aqeilan RI, Donati V, Palamarchuk A, Trapasso F, Kaou M, Pekarsky Y, Sudol M, Croce CM. WW domain-containing proteins, WWOX and YAP, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res 2005; 65: 6764–72. [DOI] [PubMed] [Google Scholar]
- 31.Levy D, Adamovich Y, Reuven N, Shaul Y. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol Cell 2008; 29: 350–61. [DOI] [PubMed] [Google Scholar]
- 32.Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, Zack TI, Wang X, Tsherniak A, Schinzel AC, Shao DD, Schumacher SE, Weir BA, Vazquez F, Cowley GS, Root DE, Mesirov JP, Beroukhim R, Kuo CJ, Goessling W, Hahn WC. beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell 2012; 151: 1457–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamm C, Bower N, Anneren C. Regulation of mouse embryonic stem cell self-renewal by a Yes-YAP-TEAD2 signaling pathway downstream of LIF. J Cell Sci 2011; 124: 1136–44. [DOI] [PubMed] [Google Scholar]
- 34.Jang EJ, Jeong H, Han KH, Kwon HM, Hong JH, Hwang ES. TAZ suppresses NFAT5 activity through tyrosine phosphorylation. Mol Cell Biol 2012; 32: 4925–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russo T, Faraonio R, Minopoli G, De Candia P, De Renzis S, Zambrano N. Fe65 and the protein network centered around the cytosolic domain of the Alzheimer's beta-amyloid precursor protein. FEBS Lett 1998; 434: 1–7. [DOI] [PubMed] [Google Scholar]
- 36.Perkinton MS, Standen CL, Lau KF, Kesavapany S, Byers HL, Ward M, McLoughlin DM, Miller CC. The c-Abl tyrosine kinase phosphorylates the Fe65 adaptor protein to stimulate Fe65/amyloid precursor protein nuclear signaling. J Biol Chem 2004; 279: 22084–91. [DOI] [PubMed] [Google Scholar]
- 37.Harvey KF, Kumar S. Nedd4-like proteins: an emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol 1999; 9: 166–9. [DOI] [PubMed] [Google Scholar]
- 38.An H, Krist DT, Statsyuk AV. Crosstalk between kinases and Nedd4 family ubiquitin ligases. Mol Biosyst 2014; 10: 1643–57. [DOI] [PubMed] [Google Scholar]
- 39.An H, Krist DT, Statsyuk AV. Crosstalk between kinases and Nedd4 family ubiquitin ligases. Mol Biosyst 2014; 10: 1643–57. [DOI] [PubMed] [Google Scholar]
- 40.Skouloudaki K, Walz G. YAP1 recruits c-Abl to protect angiomotin-like 1 from Nedd4-mediated degradation. PloS ONE 2012; 7: e35735–e35735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moleirinho S, Guerrant W, Kissil JL. The Angiomotins – From discovery to function. FEBS letters. 2014. [DOI] [PMC free article] [PubMed]
- 42.Persaud A, Alberts P, Mari S, Tong J, Murchie R, Maspero E, Safi F, Moran M, Polo S, Rotin D. Tyrosine phosphorylation of NEDD4 activates its ubiquitin ligase activity. Sci Signal 2014; 7: ra95–ra95. [DOI] [PubMed] [Google Scholar]
- 43.Yim EK, Peng G, Dai H, Hu R, Li K, Lu Y, Mills GB, Meric-Bernstam F, Hennessy BT, Craven RJ, Lin SY. Rak functions as a tumor suppressor by regulating PTEN protein stability and function. Cancer Cell 2009; 15: 304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao M, Labuda T, Xia Y, Gallagher E, Fang D, Liu YC, Karin M. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science 2004; 306: 271–5. [DOI] [PubMed] [Google Scholar]
- 45.Rossi M, Aqeilan RI, Neale M, Candi E, Salomoni P, Knight RA, Croce CM, Melino G. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc Natl Acad Sci USA 2006; 103: 12753–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salah Z, Melino G, Aqeilan RI. Negative regulation of the Hippo pathway by E3 ubiquitin ligase ITCH is sufficient to promote tumorigenicity. Cancer Res 2011; 71: 2010–20. [DOI] [PubMed] [Google Scholar]
- 47.Zhang P, Wang C, Gao K, Wang D, Mao J, An J, Xu C, Wu D, Yu H, Liu JO, Yu L. The ubiquitin ligase itch regulates apoptosis by targeting thioredoxin-interacting protein for ubiquitin-dependent degradation. J Biol Chem 2010; 285: 8869–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gallagher E, Gao M, Liu YC, Karin M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc Natl Acad Sci USA 2006; 103: 1717–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang C, Zhou W, Jeon MS, Demydenko D, Harada Y, Zhou H, Liu YC. Negative regulation of the E3 ubiquitin ligase itch via Fyn-mediated tyrosine phosphorylation. Mol Cell 2006; 21: 135–41. [DOI] [PubMed] [Google Scholar]
- 50.Gao B, Lee SM, Fang D. The tyrosine kinase c-Abl protects c-Jun from ubiquitination-mediated degradation in T cells. J Biol Chem 2006; 281: 29711–8. [DOI] [PubMed] [Google Scholar]
- 51.Mahajan NP, Whang YE, Mohler JL, Earp HS. Activated tyrosine kinase Ack1 promotes prostate tumorigenesis: role of Ack1 in polyubiquitination of tumor suppressor Wwox. Cancer Res 2005; 65: 10514–23. [DOI] [PubMed] [Google Scholar]
- 52.Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Develop 2010; 24: 72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu CY, Zha ZY, Zhou X, Zhang H, Huang W, Zhao D, Li T, Chan SW, Lim CJ, Hong W, Zhao S, Xiong Y, Lei QY, Guan KL. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCF{beta}-TrCP E3 ligase. J Biol Chem 2010; 285: 37159–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levy D, Adamovich Y, Reuven N, Shaul Y. The Yes-associated protein 1 stabilizes p73 by preventing Itch-mediated ubiquitination of p73. Cell death Differentiation 2007; 14: 743–51. [DOI] [PubMed] [Google Scholar]
- 55.Danovi SA, Rossi M, Gudmundsdottir K, Yuan M, Melino G, Basu S. Yes-associated protein (YAP) is a critical mediator of c-Jun-dependent apoptosis. Cell Death Differentiation 2008; 15: 217–9. [DOI] [PubMed] [Google Scholar]
- 56.Levy D, Reuven N, Shaul Y. A regulatory circuit controlling Itch-mediated p73 degradation by Runx. J Biol Chem 2008; 283: 27462–8. [DOI] [PubMed] [Google Scholar]
- 57.White CD, Erdemir HH, Sacks DB. IQGAP1 and its binding proteins control diverse biological functions. Cell Signal 2012; 24: 826–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roy M, Li Z, Sacks DB. IQGAP1 binds ERK2 and modulates its activity. J Biol Chem 2004; 279: 17329–37. [DOI] [PubMed] [Google Scholar]
- 59.Jameson KL, Mazur PK, Zehnder AM, Zhang J, Zarnegar B, Sage J, Khavari PA. IQGAP1 scaffold-kinase interaction blockade selectively targets RAS-MAP kinase-driven tumors. Nat Med 2013; 19: 626–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamaoka-Tojo M, Ushio-Fukai M, Hilenski L, Dikalov SI, Chen YE, Tojo T, Fukai T, Fujimoto M, Patrushev NA, Wang N, Kontos CD, Bloom GS, Alexander RW. IQGAP1, a novel vascular endothelial growth factor receptor binding protein, is involved in reactive oxygen species–dependent endothelial migration and proliferation. Circ Res 2004; 95: 276–83. [DOI] [PubMed] [Google Scholar]
- 61.Meyer RD, Sacks DB, Rahimi N. IQGAP1-dependent signaling pathway regulates endothelial cell proliferation and angiogenesis. PloS ONE 2008; 3: e3848–e3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heil A, Nazmi AR, Koltzscher M, Poeter M, Austermann J, Assard N, Baudier J, Kaibuchi K, Gerke V. S100P is a novel interaction partner and regulator of IQGAP1. J Biol Chem 2011; 286: 7227–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mo JS, Park HW, Guan KL. The Hippo signaling pathway in stem cell biology and cancer. EMBO reports 2014;15:642–56. [DOI] [PMC free article] [PubMed]
- 64.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Develop 2010; 24: 862–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salah Z, Aqeilan RI. WW domain interactions regulate the Hippo tumor suppressor pathway. Cell Death Dis 2011; 2: e172–e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sudol M, Harvey KF. Modularity in the Hippo signaling pathway. Trends Biochem Sci 2010; 35: 627–33. [DOI] [PubMed] [Google Scholar]
- 67.Jin J, Xie X, Chen C, Park JG, Stark C, James DA, Olhovsky M, Linding R, Mao Y, Pawson T. Eukaryotic protein domains as functional units of cellular evolution. Sci Signal 2009; 2: ra76–ra76. [DOI] [PubMed] [Google Scholar]
- 68.Keshet R, Adler J, Ricardo-Lax I, Shanzer M, Porat Z, Reuven N, Shaul Y. c-Abl antagonizes the Yap oncogenic function. Cell Death Differ 2014. Oct 31. doi: 10.1038/cdd.2014.182. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang X, Poy F, Zhang R, Joachimiak A, Sudol M, Eck MJ. Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan. Nat Struct Biol 2000; 7: 634–8. [DOI] [PubMed] [Google Scholar]
- 70.Matsui Y, Lai ZC. Mutual regulation between Hippo signaling and actin cytoskeleton. Protein Cell 2013; 4: 904–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Low BC, Pan CQ, Shivashankar GV, Bershadsky A, Sudol M, Sheetz M. YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Lett 2014; 588: 2663–70. [DOI] [PubMed] [Google Scholar]
- 72.Gaspar P, Tapon N. Sensing the local environment: actin architecture and Hippo signalling. Curr Opin Cell Biol 2014; 31C: 74–83. [DOI] [PubMed] [Google Scholar]