Abstract
The WWOX gene spans the common chromosomal fragile site FRA16D that is located within a massive (780 kb) intron. The WWOX gene is very long, at 1.1 Mb, which may contribute to the very low abundance of the full-length 1.4 kb mRNA. Alternative splicing also accounts for a variety of aberrant transcripts, most of which are devoid of C-terminal sequences required for WWOX to act as an oxidoreductase. The mouse WWOX gene also spans a chromosomal fragile site implying some sort of functional relationship that confers a selective advantage. The encoded protein domains of WWOX are conserved through evolution (between humans and Drosophila melanogaster) and include WW domains, an NAD -binding site, short-chain dehydrogenase/reductase enzyme and nuclear compartmentalization signals. This homology has enabled functional analyses in D. melanogaster that demonstrate roles for WWOX in reactive oxygen species regulation and metabolism. Indeed the human WWOX gene is also responsive to altered metabolism. Cancer cells typically exhibit altered metabolism (Warburg effect). Many cancers exhibit FRA16D DNA instability that results in aberrant WWOX expression and is associated with poor prognosis for these cancers. It is therefore thought that aberrant WWOX expression contributes to the altered metabolism in cancer. In addition, others have found that a specific (low-expression) allele of WWOX genotype contributes to cancer predisposition.
Keywords: Chromosomal fragile site, FRA16D, oxidoreductase, altered metabolism
Introduction
WWOX gene spans the common chromosomal fragile site, FRA16D
Chromosomal fragile sites are enigmatic regions of human chromosomes that are of particular interest because of their contribution to human disease (Figure 1). Fragile sites are distinguished by their frequency – rare fragile sites are found in only a minority of the population and are inherited, while common fragile sites are found in all individuals. Common fragile sites are also distinguished by the chemicals that cause their cytogenetic appearance. The inducing chemicals include agents typically found in the diet (e.g. folate, caffeine, and ethanol1,2), as well as in the environment (e.g. chemicals in cigarette smoke3). The major group of rare fragile sites are induced by folic acid, while the majority of common fragile sites are induced in vitro by aphidicolin, an inhibitor of DNA polymerase. It is this group of common, aphidicolin-inducible fragile sites that are associated with stress during replication and consequent DNA instability. Common fragile sites are also noteworthy because of their frequent colocation with regions of chromosomal instability in cancer.1,44 There is a hierarchy in the cytogenetic appearance of fragile sites in vitro in response to chemical induction and that is matched by their DNA instability observed in vivo in various cancers – indicating a direct relationship between cytogenetic appearance and DNA breakage in cancer. This DNA instability appears to be an early event in carcinogenesis occurring as a result of exposure to environmental agents such as the chemicals in cigarette smoke.3 The two fragile sites, FRA3B and FRA16D, that are the most readily induced in vitro are also those that most frequently exhibit DNA instability in vivo in cancer.2,4,5 Common fragile sites and the genes in their vicinity that are affected by their DNA instability in cancer are therefore of interest for their causal contribution to neoplasia.
Figure 1.
Summary box 1 – common chromosomal fragile sites and their genes.43
WWOX is a protein encoded by a common fragile site gene (Figure 2). The WWOX gene spans FRA16D that is located within a very large, 780 kb intron of this alternatively spliced, massive gene of 1.1 Mb in length.4,45 The time taken for transcription of such a length of DNA makes it likely that just one or a few full-length spliced transcripts are produced per cell cycle. As has been found for several common fragile site genes, the relationship between the WWOX gene and a common fragile site is conserved between mouse and human, implying a functional relationship with a selective advantage of some sort. The alternative splicing of the WWOX gene transcript leads to an abundance of aberrant transcripts incapable of encoding the full-length WWOX protein.4 While these appear to be the subject of nonsense-mediated decay, some truncated proteins are produced that may act as dominant negative competitors for the full-length protein.7 Some of these alternative splice RNA products are conserved between mouse and human, also implying functional significance.
Figure 2.
Summary box 2 – WWOX in cancer initiation and progression
A genetic variant of the WWOX locus, that has lower levels of WWOX, has been found to be associated with cancer predisposition. Specifically, carriers of loss variant (CNV-67048) genotypes have been found to have significantly increased risk of lung cancer, in a dose-dependant manner.8 Recently, a similar association has been found between the CNV-67048 genetic variant and risk of gliomas.9
Characteristics of the WWOX protein sequence
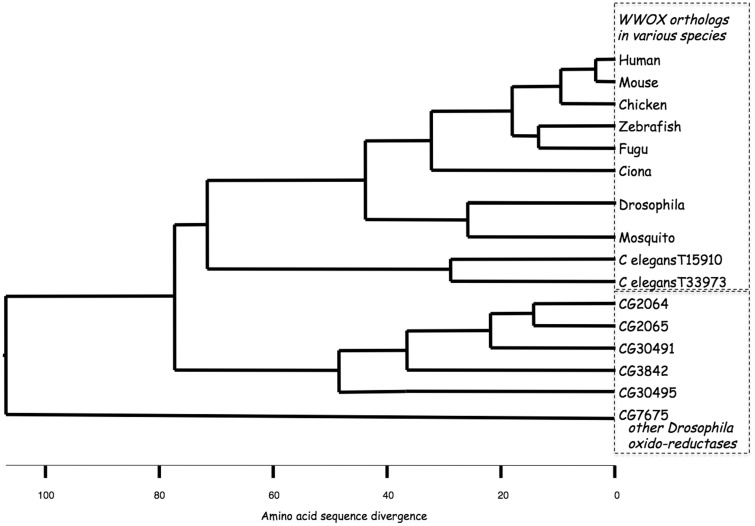
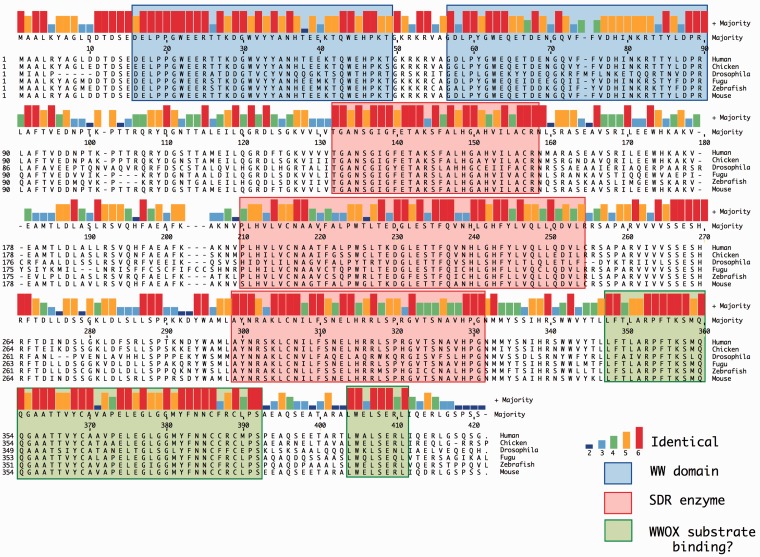
The WWOX protein itself comprises multiple distinct functional domains as well as sequences encoding a small chain dehydrogenase/reductase (SDR) enzyme. Phylogenetic analysis (Figure 3) of the WWOX amino acid sequence indicates evolutionary conservation of WWOX as a discrete ortholog, from species as diverse as humans and insects.10 The functionally important sequences in WWOX, including the WW domains and the SDR enzyme sequences, are evident as more highly conserved regions of the WWOX sequence (Figure 4). SDR enzymes typically have their specific substrate-binding sequences located C-terminal to the catalytic region. The WWOX orthologs exhibit sequence homology in this WWOX-specific domain that is as high as that for the enzyme co-factor and catalytic regions, consistent with these sequences being the region where WWOX binds to its specific (as yet unknown) substrate. Each of the SDR enzymes contains an nicotinamide adenine dinucleotide (oxidized form) (NAD+)/nicotinamide adenine dinucleotide phosphate (NADP+) co-factor-binding site as well as characteristic sequences required for the catalytic active site. The in vivo substrate and reaction product for the SDR enzyme activity of WWOX are unknown, although several in vitro targets have been identified11 and ablation of WWOX in mice leads to impaired steroidogenesis.12 Steroids are therefore likely suspects although impaired steroidogenesis is not evident in the Drosophila melanogaster WWOX mutants.
Figure 3.
Phylogenetic tree of WWOX orthologs from various species and other oxidoreductases in Drosophila melanogaster. Distinct sequence relationships through evolution of WWOX orthologs in various species are evident when compared to other oxidoreductases (in this case from Drosophila). Note: all species have a single WWOX ortholog except Caenorhabditis elegans with two, neither of which has WW domains. Figure modified from that of O’Keefe et al.10
Figure 4.
Homology between orthologs WWOX protein sequences. Alignment of WWOX protein sequences from human, chicken, Drosophila melanogaster, zebrafish, and mouse. Majority sequence is indicated with color coding for identity between 6, 5, 4, 3, and 2 sequences. The two WW domains are indicated by blue shading. Sequences comprising the SDR enzyme are indicated by red shading, and the putative “WWOX substrate binding site” is shaded in green. (A color version of this figure is available in the online journal.)
It is noteworthy that the closest organism that does not have a complete WWOX ortholog, Caenorhabditis elegans, has two genes with homology to WWOX but both are devoid of the two N-terminal WW domains (Figure 3). However, both have sequence homology to the putative “WWOX substrate binding region,” suggesting that these enzymes act on the same substrate/substrates and that this is independent of the WW domain interactions seen in, and presumably needed for, WWOX activity in other species.
The link between the WWOX substrate binding domain and SDR enzyme function appears evident in Opisthokonts (Casaspora owczarzaki) and C. elegans orthologs not having WW domains but sharing substrate-binding domain homology. The addition of WW domains to WWOX is an ancient event, albeit of unknown biological significance, with the sea sponge (Amphimedon queenslandica) WWOX protein having both WW domains and all other WWOX domains, including substrate-binding domain homology (not shown).
WW domains are known to act as protein–protein recognition and binding domains, pairing up with proteins through proline-containing motifs (PPXY or similar). WW-mediated protein interactions are thought to be a physical mechanism that causes a functional relationship between the partner proteins. The identity of the WW-binding motif partners of WWOX has therefore been sought in order to gain insight into the function of WWOX. WW domains have been classified into subgroups with specificity in their interacting motifs, e.g. the WW1 motif of WWOX is expected to interact with PPXY or LPXY motifs. Mass spectrometry and phage display techniques have been used in an effort to identify the specific PPXY-containing WWOX-interacting proteins.13 This analysis revealed that the WW1 domain of WWOX is capable of binding to many different proteins, although the significance of most of these interactions to the function of WWOX is yet to be determined.
In addition to the WW domains, the WWOX protein contains compartmentalization signals for localization to the nucleus and the mitochondria, although the latter is not well defined.14,15 The SDR-related sequences define WWOX as a member of this family of enzymes. These enzymes are NAD(P)(H)-dependent oxidoreductases that catalyze diverse small molecule reactions including those involving lipids, amino acids, carbohydrate, and steroids.16 NAD(P)(H) binding is not only for the role of this co-factor in catalysis but also as a means for SDR enzymes to act as metabolic sensors of NAD+/nicotinamide adenine dinucleotide (reduced form) (NADH)/NADP+/NADP levels and in so doing contribute to the regulation of metabolism, transcription, and signal transduction. The typical small molecule substrates and products of SDRs contribute to their designation as a “druggable” enzyme class, suitable for targeting with potential pharmaceuticals.
WWOX is able to act as a “non-classical” tumor suppressor
When cancer cells exhibit DNA instability at the FRA16D site, this is typically associated with a reduction in full-length WWOX mRNA and an increase in either normal incomplete splice forms and/or the appearance of novel aberrant transcripts. While the resultant cancer cells therefore have lower levels of WWOX, they do not exhibit second allele loss typical of Knudsen’s two-hit hypothesis and therefore retain some level of WWOX protein. Even so the transfer of intact WWOX gene back into tumor cells that are deficient in this protein, does reduce the ability of these WWOX-replenished cells to form tumours6 and therefore WWOX is regarded as a tumor suppressor, albeit a “non-classical” one – i.e. a reduction in WWOX level, rather than its absence, appears sufficient for its functional contribution to cancer. The mechanism by which WWOX might act in this capacity is unclear although some possibilities have been put forward on the basis of the proteins with which WWOX physically interacts. For example, the WW domains of WWOX may serve as negatively regulating, competitor-binding sites for proteins such as ErbB-4, thereby competing with these proteins for their ability to physically and functionally interact with other WW domain-containing proteins, in particular yes-associated protein (YAP).17 Reduction in WWOX levels below a threshold may therefore facilitate the activation by YAP of tumor-promoting pathways mediated by ErbB-4 and perhaps other proteins.
Consistent with its role as a tumor suppressor, patients with tumors who exhibit diminished levels of WWOX, have a poorer prognosis than those with normal levels of the protein. Nunez et al.18 reported that WWOX protein expression varies among ovarian carcinoma histotypes and correlates with less-favorable outcome. In addition, Pluciennik et al.19 found that WWOX expression correlated with breast cancer progression and prognosis. Furthermore, Aqeilan et al.20 and Wang et al.21 reported the prognostic significance of WWOX expression levels in patients with breast cancer and its association with the basal-like phenotype. The reduction in WWOX levels, by DNA instability at one allele, appears to be sufficient to enable a functional contribution of some sort to cancer cell biology. The normal biological function of WWOX is therefore of particular interest as it represents a target for improving the prospects of individuals with WWOX deficient cancers.
FRA16D/WWOX damage is an early event in a population of precancerous cells
A clear illustration of the mechanism through which environmental factors are thought to trigger cancer through effects on common fragile sites, is the contribution of cigarette smoke to fragile site DNA instability in lung cancer.3 Much greater levels of cytogenetic fragile site expression are well documented in the “normal” cells of young cigarette smokers, well before the onset of lung cancer.22 There is a direct correlation between in vitro-induced cytogenetic fragile site expression and DNA instability observed as a consequence of events in vivo.2,23 The influence of tobacco smoke chemical exposure on fragile site expression is followed by chromosome breakage and/or rearrangement that, in turn, contributes to carcinogenesis. The deleterious “hit” related to small cell lung cancer may occur at any time during tobacco exposure, but, given that carcinogenesis is a stepwise process, the actual tumor may not arise for several years after the individual stops smoking.3 Such “fragile site damaged” cells will not exist in isolation but as a population of similarly “damaged” cells. Cigarette smoke chemicals preferentially affect the most readily induced common fragile sites (FRA3B and FRA16D) and, therefore, common fragile site genes (FHIT and WWOX). Any one of these FRA16D/WWOX damaged cells has the potential to act as a cancer stem cell (CSC), particularly after additional damage in genes that also contribute to cancer initiation. We have found that common fragile site damage at FRA16D is an early event in cancer cell progression.23 Two cancer cell lines developed from a primary carcinoma (KM12C) and secondary metastasis (KM12SM) have identical FRA16D deletions despite having drastic differences in their karyotype.
The normal biological function/functions of WWOX
The biological function of WWOX has been explored by genetic approaches, both by targeted mutagenesis and by the identification and characterization of spontaneous WWOX mutations in a rat, and more recently, in humans. A “non-classical” tumor suppressor function for WWOX is evident in the analysis of rodent mutants for the WWOX gene. Some (but not all) lines of loss-of-function WWOX mutant mice have higher incidence of tumors, however tumors from heterozygous mutant mice still express WWOX.24–27 A spontaneous WWOX mutant rat does not exhibit higher incidence of tumors.28,29 Rodent WWOX mutants typically exhibit metabolic disorders leading to early death (which may preclude development of some tumors) limiting the utility of rodent models in understanding WWOX protein function.24–29 While these rodent studies have provided some insight into the biological role for WWOX, there is still clearly a need to understand the molecular processes and pathways in which WWOX participates; particularly how WWOX contributes to metabolism and how this altered metabolism can contribute, at least in certain circumstances, to cancer.
Three reports detail clinical consequences in humans due to inherited mutations in WWOX. The first of these30 involves a disorder of sex development due to a deletion, in one allele of the WWOX gene, removing exons 6–8, with exon 5 being spliced on to exon 9. The resultant encoded protein would have intact WW domains, but absence of the SDR catalytic domain. Only one allele is affected and is therefore acting in a dominant manner. While haploinsufficiency is possible, another explanation is that the mutant WWOX-truncated protein is acting in a dominant negative manner. Two recent reports31,32 detail recessive epilepsy and other symptoms due to loss of function mutations in both alleles of the WWOX gene. Three different homozygous WWOX mutations were identified in three consanguineous families. The neurological symptoms in each of the affected individuals are similar to those observed for the spontaneous rat WWOX mutation indicating likely common pathogenic pathway for these symptoms. The human and rat WWOX mutations also share a lack of noted increase in spontaneous tumours28,29,31,32 as has been reported for mouse WWOX mutations.24–27
Insights into the role of WWOX in metabolism
Given the phenotypic consequences of homozygous loss-of-function WWOX mutations in mammals, it is somewhat surprising that loss-of-function WWOX mutations in the D. melanogaster ortholog of WWOX do not exhibit a phenotype.33 The D. melanogaster ortholog has 49% amino acid sequence identity with its human counterpart, with all of the functional domains and SDR enzyme-related sequences retained (Figures 3 and 4). The function of WWOX protein in D. melanogaster has become evident from the conduct of genetic and biochemical analyses in WWOX-deficient flies. Microarray and proteomic experiments identified quantitative and qualitative changes in other proteins consistent with a role for WWOX in metabolism. An advantage of the D. melanogaster system is the ability to undertake genetic analyses of function and this revealed a role for WWOX in pathways that included the proteins superoxide dismutase (SOD1) and isocitrate dehydrogenase (IDH).33 SOD1 has a role in the control of reactive oxygen species (ROS) in cells, while IDH is an integral component of the citric acid cycle. Both the viability and life span of SOD1 mutant D. melanogaster were impacted by altered WWOX levels, while both D. melanogaster and human cancer cells showed a correlation between SOD1 and WWOX mRNA levels.33 Reducing IDH levels in D. melanogaster impacts on viability and this impact is exacerbated by also reducing WWOX levels and relieved by increasing WWOX levels.33 Furthermore, WWOX and IDH mRNA levels correlate in cancer cells.33 Roles for WWOX in regulation of ROS and metabolism are both intriguing, in terms of the possible mechanism of WWOX contributing to cancer. ROS levels are lower in at least certain types of CSCs than corresponding nontumourigenic cells.34 These lower ROS levels in CSCs are associated with increased free radical scavenging and decreased sensitivity to ionizing radiation. In this setting, the lower level of ROS brought about by reduced WWOX could therefore contribute to tumor radioresistance, and therefore poorer prognosis for patients with WWOX-depleted cancers.
Altered cellular metabolism in cancer cells was first discovered over 80 years ago by Warburg,35 but has only recently been added to the list of recognized hallmarks of cancer.36 Warburg revealed that cancer cells favor adenosine triphosphate (ATP) production via rapid consumption of glucose and formation of lactic acid rather than via the more efficient mitochondrial respiration, even when oxygen is available.35 There are clear instances of altered metabolism playing causal roles in the development of cancer e.g. mitochondrial DNA mutations in cancers of the head and neck37 and the recent finding of very specific leukemia-associated IDH1 and IDH2 mutations lead to neomorphic enzyme activity, converting alpha-ketoglutarate to the “oncometabolite” 2-hydroxyglutarate.38 Although this change in metabolism has been exploited in the development of drugs for cancer therapy,39 until very recently, it has been unclear what causal cellular changes bring about this phenotype. Therefore with few exceptions, therapeutics have not yet specifically targeted the causal changes in cancer cells that are responsible for altered cellular metabolism. There is, however, growing recognition that because of this distinction between normal and cancer cells, altered metabolism has the potential for such exploitation.40,41 Thus the molecular cause/causes of altered metabolism in cancer cells need/needs to be defined in order to enable the identification of therapeutic leads.
WWOX not only contributes to metabolic regulation, its expression is also responsive to changes in metabolism.42 Altering metabolism from glycolysis to oxidative phosphorylation causes an increase in the steady-state levels of WWOX mRNA, whereas hypoxic conditions, in which cells rely on glycolysis, cause a decrease in WWOX mRNA. Both the WWOX gene and its encoded protein are therefore monitors of and contributors to the metabolic state of cells.
Perturbation of WWOX in cancer cells is therefore a clear candidate for contributing to the altered metabolism (Warburg effect35) seen in these cells and associated with poor prognosis, and therefore an ideal target for therapeutic intervention.
Conclusions/future directions
The presence of a distinct WWOX protein, in species as diverse and distant in evolution as sea sponge and humans, would suggest that it serves a fundamental function in biology. Indeed the protein has normal roles in metabolism. The level of WWOX protein is affected by DNA instability within its gene in human cancers. Reduced WWOX level appears to facilitate cancer progression, rather than act as a classical tumor suppressor. Lower levels of WWOX, due to genetic variation, contribute to the risk of carriers to increased cancer incidence. Lower levels of WWOX in cancers correlate with poorer prognosis. Clearly in both cases, mutations in other genes are required to both initiate and establish cancer.
The altered metabolism of cancer cells is an intriguing possibility for the contribution of WWOX to cancer cell biology. Identifying the role of WWOX in metabolism and targeting this role in cancers found to be deficient in WWOX may lead to better therapeutic interventions.
The WW domains of WWOX have been the focus of attention as the identity of their partner proteins can give insight into the pathways to which WWOX contributes. However, the role of WWOX in metabolism is more likely to be mediated by the enzyme function of WWOX, as SDR enzymes are known monitors of NAD+/NADP+ through their co-factor binding sites.16 Indeed the C. elegans orthologs of WWOX do not contain WW domains. The substrate and product of WWOX are therefore of particular interest. Protein homology searches indicate that the likely “substrate-binding domain” indicated by amino acid sequence homology C-terminal to the enzyme catalytic site (Figure 4) appears to be unique to WWOX orthologs (including those of C. elegans) and therefore likely to have a unique substrate. Either the accumulation of substrate or the reduced level of product of the WWOX enzyme, present potential targets to compensate for the contribution that lower WWOX levels, which lead to increased cancer risk and/or poorer cancer prognosis.
Acknowledgements
The research in the authors’ laboratory, that forms the basis for some of this review, was funded by an NHMRC Project Grant (519125) to R.I.R. and L.O’K and an ARC–NHMRC Research Network Grant (RN0457079) to R.I.R.
Authors’ contribution
R.I.R. held initial discussions with coauthors regarding the content and order of the review. He prepared the first and subsequent drafts, initial, and revised submissions. He drafted the responses to reviewers’ criticisms. A.C. provided input to the structure and content of the review and specific discussion regarding the role of WWOX in metabolism of Drosophila. Contributed revisions to multiple drafts. C.S.L. provided input to the whole structure and content of the review and specific insight into the pathways by which WWOX contributes to normal metabolism in Drosophila and altered metabolism in cancer cells. Contributed revisions to multiple drafts. S.D. helped formulate and finalize the discussion regarding the role of WWOX in cancer cells and its regulation by metabolism as well as its regulation of metabolism in both normal cells and cancer cells with reduced WWOX. Contributed revisions to multiple drafts. L.O’K provided input into the order and content of the review, particularly the Drosophila experiments that identified a role for WWOX in metabolism and specific functional interactions with SOD1 and IDH. Contributed revisions to multiple drafts.
References
- 1.Yunis JJ, Soreng AL. Constitutive fragile sites and cancer. Science 1984; 6: 1199–204. [DOI] [PubMed] [Google Scholar]
- 2.Glover TW, Arlt MF, Casper AM, Durkin SG. Mechanisms of common fragile site instability. Hum Mol Genet 2005; 14: R197–205. [DOI] [PubMed] [Google Scholar]
- 3.Stein CK, Glover TW, Palmer JL, Glisson BS. Direct correlation between FRA3B and cigarette smoking. Genes Chrom Cancer 2002; 34: 333–40. [DOI] [PubMed] [Google Scholar]
- 4.Ried K, Finnis M, Hobson L, Mangelsdorf M, Dayan S, Nancarrow J, Venter D, Richards RI. Common chromosomal fragile site FRA16D DNA sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum Mol Genet 2000; 9: 1651–63. [DOI] [PubMed] [Google Scholar]
- 5.Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C, Widaa S, Hinton J, Fahey C, Fu B, Swamy S, Dalgliesh GL, Teh BT, Deloukas P, Yang F, Campbell PJ, Futreal PA, Stratton MR. Signatures of mutation and selection in cancer genome. Nature 2010; 463: 893–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bednarek A, Keck-Waggoner CL, Daniel RL, Laflin KJ, Bergsagel PL, Kiguchi K, Brenner AJ, Aldaz CM. WWOX FRA16D gene behaves as a suppressor of tumour growth. Cancer Res 2001; 61: 8068–73. [PubMed] [Google Scholar]
- 7.Driouch K, Prydz H, Monese R, Johansen H, Lidereau R, Frengen E. Alternative transcripts of the candidate tumor suppressor gene, WWOX, are expressed at high levels in human breast tumors. Oncogene 2002; 21: 1832–40. [DOI] [PubMed] [Google Scholar]
- 8.Yang L, Liu B, Huang B, Deng J, Li H, Yu B, Qiu F, Cheng M, Wang H, Yang R, Yang X, Zhou Y, Lu J. A functional copy number variation in the WWOX gene is associated with lung cancer risk in Chinese. Hum Mol Genet 2013; 22: 1886–94. [DOI] [PubMed] [Google Scholar]
- 9.Yu K, Fan J, Ding X, Li C, Wang J, Xiang Y, Wang QS. Association study of a functional copy number variation in the WWOX gene with risk of gliomas among Chinese people. Int J Cancer 2014; 135: 1687–91. [DOI] [PubMed] [Google Scholar]
- 10.O’Keefe L, Liu Y-H, Perkins A, Dayan S, Saint RB, Richards RI. FRA16D common chromosomal fragile site oxido-reductase (FOR/WWOX) protects against the effects of ionising radiation in Drosophila melanogaster. Oncogene 2005;24:6590–6 Corrigendum: Oncogene 2006;25:7662. [DOI] [PubMed]
- 11.Saluda-Gorgui A, Seta K, Nowakowska M, Bednarek AK. WWOX oxidoreductase – substrate and enzymatic characterization. Z Naturforsch C 2011; 66: 73–82. [PubMed] [Google Scholar]
- 12.Aqeilan RI, Hagan JP, de Bruin A, Rawahneh M, Salah Z, Gaudio E, Siddiqui H, Volinia S, Alder H, Lian JB, Stein GS, Croce CM. Targeted ablation of the WW domain-containing oxidoreductase tumor suppressor leads to impaired steroidogenesis. Endocrinology 2009; 150: 153–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abu-Odeh M, Bar-Mag T, Huang H, Kim T, Salah Z, Abdeen SK, Sudol M, Reichmann D, Sidhu S, Kim PM, Aqeilan RI. Characterizing WW domain interactions of tumor suppressor WWOX reveals its association with multi-protein networks. J Biol Chem 2014; 289: 8865–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang NS, Pratt N, Heath J, Schultz L, Sleve D, Carey GB, Zevotek N. Hyaluronidase induction of a WW domain-containing oxidoreductase that enhances tumor necrosis factor cytotoxicity. J Biol Chem 2001; 276: 3361–70. [DOI] [PubMed] [Google Scholar]
- 15.Chang NS, Doherty J, Ensign A, Lewis J, Heath J, Schultz L, Chen ST, Oppermann U. Molecular mechanisms underlying WOX1 activation during apoptotic and stress responses. Biochem Pharmacol 2003; 66: 1347–54. [DOI] [PubMed] [Google Scholar]
- 16.Kavanagh KL, Jornvall H, Persson B, Oppermann U. The SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci 2008; 65: 3895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aqeilan RI, Donati V, Palamarchuk A, Trapasso F, Kaou M, Pekarsky Y, Sudol M, Croce CM. WW domain-containing proteins, WWOX and YAP, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res 2005; 65: 6764–72. [DOI] [PubMed] [Google Scholar]
- 18.Nunez MI, Rosen DG, Ludes-Meyers JH, Abba MC, Kil H, Page R, Klein-Szanto AJ, Godwin AK, Liu J, Mills GB, Aldaz CM. WWOX protein expression varies among ovarian carcinoma histotypes and correlates with less favorable outcome. BMC Cancer 2005; 5: 64–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pluciennik E, Kusinska R, Potemski P, Kubiak R, Kordek R, Bednarek AK. WWOX – the FRA16D cancer gene: expression correlation with breast cancer progression and prognosis. Eur J Surg Oncol 2006; 32: 153–7. [DOI] [PubMed] [Google Scholar]
- 20.Aqeilan RI, Donati V, Gaudio E, Nicoloso MS, Sundvall M, Korhonen A, Lundin J, Isola J, Sudol M, Joensuu H, Croce CM, Elenius K. Association of Wwox with ErbB4 in breast cancer. Cancer Res 2007; 67: 9330–6. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Chao L, Ma G, Chen L, Zang Y, Sun J. The prognostic significance of WWOX expression in patients with breast cancer and its association with the basal-like phenotype. J Cancer Res Clin Oncol 2011; 137: 271–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao-Shan CS, Fine RL, Whang-Peng J, Lee EC, Chabner BA. Increased fragile sites and sister chromatid exchanges in bone marrow and peripheral blood of young cigarette smokers. Cancer Res 1987; 47: 6278–82. [PubMed] [Google Scholar]
- 23.Finnis M, Dayan S, Hobson L, Chenevix-Trench G, Friend K, Ried K, Venter D, Woollatt E, Baker E, Richards RI. Common chromosomal fragile site FRA16D mutation in cancer cells. Hum Mol Genet 2005; 14: 1341–9. [DOI] [PubMed] [Google Scholar]
- 24.Aqeilan RI, Trapasso F, Hussain S, Costinean S, Marshall D, Pekarsky Y, Hagan JP, Zanesi N, Kaou M, Stein GS, Lian JB, Croce CM. Targeted deletion of Wwox reveals a tumor suppressor function. Proc Natl Acad Sci USA 2007a; 104: 3949–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aqeilan RI, Hagan JP, Aqeilan HA, Pichiorri F, Fong LY, Croce CM. Inactivation of the Wwox gene accelerates forestomach tumor progression in vivo. Cancer Res 2007b; 67: 5606–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludes-Meyers JH, Kil H, Nuñez MI, Conti CJ, Parker-Thornburg J, Bedford MT, Aldaz CM. WWOX hypomorphic mice display a higher incidence of B-cell lymphomas and develop testicular atrophy. Genes Chrom Cancer 2007; 46: 1129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludes-Meyers JH, Kil H, Parker-Thornburg J, Kusewitt DF, Bedford MT, Aldaz CM. Generation and characterization of mice carrying a conditional allele of the Wwox tumor suppressor gene. PLoS ONE 2009; 4: e7775–e7775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki H, Takenaka M, Suzuki K. Phenotypic characterization of spontaneously mutated rats showing lethal dwarfism and epilepsy. Comp Med 2007; 57: 360–9. [PubMed] [Google Scholar]
- 29.Suzuki H, Katayama K, Takenaka M, Amakasu K, Saito K, Suzuki K. A spontaneous mutation of the Wwox gene and audiogenic seizures in rats with lethal dwarfism and epilepsy. Genes Brain Behav 2009; 8: 650–60. [DOI] [PubMed] [Google Scholar]
- 30.White S, Hewitt J, Turbitt E, van der Zwan Y, Hersmus R, Drop S, Koopman P, Harley V, Cools M, Looijenga L, Sinclair A. A multi-exon deletion within WWOX is associated with a 46,XY disorder of sex development. Eur J Hum Genet 2012; 20: 348–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallaret M1, Synofzik M, Lee J, Sagum CA, Mahajnah M, Sharkia R, Drouot N, Renaud M, Klein FA, Anheim M, Tranchant C, Mignot C, Mandel JL, Bedford M, Bauer P, Salih MA, Schüle R, Schöls L, Aldaz CM, Koenig M. The tumour suppressor gene WWOX is mutated in autosomal recessive cerebellar ataxia with epilepsy and mental retardation. Brain 2014; 137: 411–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abdel-Salam G, Thoenes M, Afifi HH, Körber F, Swan D, Bolz HJ. The supposed tumor suppressor gene WWOX is mutated in an early lethal microcephaly syndrome with epilepsy, growth retardation and retinal degeneration. Orphanet J Rare Dis 2014; 9: 12–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Keefe LV, Colella C, Dayan S, Chen Q, Choo A, Jacob R, Price G, Venter D, Richards RI. Drosophila orthologue of WWOX, the chromosomal fragile site FRA16D tumour suppressor gene, functions in aerobic metabolism and regulates reactive oxygen species. Hum Mol Genet 2011; 20: 497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, Joshua B, Kaplan MJ, Wapnir I, Dirbas FM, Somlo G, Garberoglio C, Paz B, Shen J, Lau SK, Quake SR, Brown JM, Weissman IL, Clarke MF. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 2009; 458: 780–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol 1927; 8: 519–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–74. [DOI] [PubMed] [Google Scholar]
- 37.Zhou S, Kachhap S, Sun W, Wu G, Chuang A, Poeta L, Grumbine L, Mithani SK, Chatterjee A, Koch W, Westra WH, Maitra A, Glazer C, Carducci M, Sidransky D, McFate T, Verma A, Califano JA. Frequency and phenotypic implications of mitochondrial DNA mutations in human squamous cell cancers of the head and neck. Proc Natl Acad Sci USA 2007; 104: 7540–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, Rabinowitz JD, Carroll M, Su SM, Sharp KA, Levine RL, Thompson CB. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 2010; 17: 225–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porporato PE, Dhup S, Dadhich RK, Copetti T, Sonveaux P. Anticancer targets in the glycolytic metabolism of tumors: a comprehensive review. Front Pharmacol 2011; 2: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell 2008; 13: 472–82. [DOI] [PubMed] [Google Scholar]
- 41.Kaelin WG, Thompson CB. Q&A: cancer: clues from cell metabolism. Nature 2010; 465: 562–4. [DOI] [PubMed] [Google Scholar]
- 42.Dayan S, O’Keefe LV, Choo A, Richards RI. Common chromosomal fragile site FRA16D tumor suppressor WWOX gene expression and metabolic reprograming in cells. Genes Chrom Cancer 2013; 52: 823–31. [DOI] [PubMed] [Google Scholar]
- 43.Richards RI. Fragile & unstable chromosomes in cancer: causes & consequences. Trends Genet 2001; 17: 339–45. [DOI] [PubMed] [Google Scholar]
- 44.Mangelsdorf M, Ried K, Woollatt E, Dayan S, Eyre H, Finnis M, Hobson L, Nancarrow J, Venter D, Baker E, Richards RI. Chromosomal fragile site FRA16D and DNA instability in cancer. Cancer Res 2000; 60: 1683–9. [PubMed] [Google Scholar]
- 45.Bednarek AK, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3-24.1, a region frequently affected in breast cancer. Cancer Res 2000;60:2140–5. [PubMed]