Abstract
DNA methylation is considered as one of the most important epigenetic mechanisms and it is catalyzed by DNA methyltransferases (DNMTs). DNMT1 abundance has been frequently seen in urogenital system tumors but the reasons for this abundance are not well understood. We aimed to look into the effects of Wnt/β-catenin signaling pathway on overexpression of DNMT1 and aberrant expression of UHRF1 and HAUSP which are responsible for stability of DNMT1 at transcriptional and protein levels in urogenital cancers. In this context, firstly, Wnt/β-catenin signaling pathway was activated by using SB216763 which is a glycogen synthase kinase-3 (GSK3) β inhibitor. Cell proliferation levels in bladder cancer cells, renal cell carcinoma, and prostate cancer cells treated with GSK3β inhibitor (SB216763) were detected by WST-1 reagent. WIF-1 gene methylation profile was determined by methylation-specific PCR (MSP); expression levels of target genes β-catenin and WIF-1 by real-time PCR; and protein levels of β-catenin, DNMT1, pGSK3β(Ser9), HAUSP, and UHRF1 by Western Blot. Our results indicated that treatment with SB216763 caused an increased cell proliferation at low dose. mRNA levels of β-catenin increased after treatment with SB216273 and protein levels of pGSK3β(Ser9), β-catenin, and DNMT1 increased in comparison to control. HAUSP and UHRF1 were either up-regulated or down-regulated at the same doses depending on the type of cancer. Also, we showed that protein levels of DNMT1, β-catenin, HAUSP, and UHRF1 decreased after re-expression of WIF-1 following treatment with DAC. In Caki-2 cells, β-catenin pathway might have accounted for the stability of DNMT1 expression, whereas such relation is not valid for T24 and PC3 cells. Our findings may offer a new approach for determination of molecular effects of Wnt/β-catenin signal pathway on DNMT1. This may allow us to identify new molecular targets for the treatment of urogenital cancers.
Keywords: DNA methyltransferase 1 stability, Wnt/β-catenin pathway, Wnt inhibitory factor-1, deubiquitinase HAUSP (USP7), E3 ubiquitin ligase UHRF1, urological cancer cell lines
Introduction
Inappropriate activation of the Wnt/β-catenin signaling pathway is a common feature of many human cancers, which entails uncontrolled cell growth and impaired differentiation. Genetic and epigenetic alterations are responsible for this process.1 DNA methylation is an important epigenetic mechanism that ensures correct gene expression and maintains genetic stability. DNA methylation is typically mediated by DNA methyltransferases (DNMTs). DNMT1, DNMT2, DNMT3a, DNMT3b, and DNMT3L have been identified in mammalian cells. Despite their genetic homology, functional differences have been ascribed to these enzymes.2
DNMT1 is a key enzyme that methylates CpG islands located near the regulatory regions of genes during replication. These regions affect transcription of specific genes involved in cancer development and progression. All protein interactions and post-translational modifications reported for DNMT1 clearly show that DNMT1 is functionally linked with several other epigenetic pathways and cellular processes. DNMT1 undergoes cell cycle-dependent changes during acetylation and ubiquitination. DNMT1 is ubiquitinated by the E3 ubiquitin ligase (UHRF1) and deubiquitinated by HAUSP (USP7). Expression levels of these proteins are directly related to the DNMT1 stability and activity.3 Song et al.4 found that HAUSP is overexpressed in prostate cancer and more importantly that high levels of HAUSP are directly correlated with tumor aggressiveness.
DNMT1 has been reported to be a molecular target in multimodality-resistant phenotype in tumor cells.5 Dysregulation of DNMT1 activity causes various diseases including cancer.6 DNMT1 is overexpressed in several tumor types including prostate,7–9 renal,10–12 and bladder13–16 cancers. Furthermore, higher expression levels of DNMT1 are correlated with more aggressive tumor growth and treatment resistance in bladder cancer.15 Although bone morphogenetic proteins and WNTs are mediators of prostate cancer (PCa)-induced osteoblastic activity, the relation between the two in PCa bone metastases is unknown.17 Numerous studies have pointed to interactions between the tumor suppressor von Hippel–Lindau and the oncogenic Wnt/β-catenin signaling cascade; however, the mechanism of this crosstalk has remained elusive.18 Activation of the Wnt signaling pathway in the bladder of mice fails to drive urothelial cell carcinoma; however, it strongly cooperates with PTEN loss to drive tumourigenesis.19 Since APC and β-catenin mutations are rare in urological tumors, epigenetic regulators might be common traits of urological neoplasms.20
Epigenetics is one of the mechanisms that could play a major role in the activation of Wnt/β-catenin signaling pathway. β-catenin levels are normally kept low by a phosphorylation process that is mediated by glycogen synthase kinase-3 (GSK3) β, which targets β-catenin for ubiquitination and proteasomal degradation. Wnt blocks this phosphorylation process, thereby allowing β-catenin to accumulate and co-activate transcription in the nucleus. How Wnt exactly inhibits GSK3 activity towards β-catenin remains unclear despite being the focus of intensive research. The role of GSK3β in tumorigenesis and cancer progression remains controversial; it may actually function as a “tumor suppressor” for certain types of tumors, but may at the same time promote growth and development of some others. GSK3β also mediates drug sensitivity/resistance in cancer chemotherapy.21 It has been reported that SB216763 (ATP-competitive), a GSK3 inhibitor, effectively activated β-catenin-mediated transcription.22 Naito et al.23 showed that GSK3β nuclear expression was associated with high-grade tumors, metastasis, and worse survival in bladder cancer patients. Dose and time-dependent SB216763 improved the cytotoxic effect against the two human urothelial carcinoma cell lines, UMUC3 and UMUC14, suggesting potential therapeutic uses for GSK3β inhibitors.24 Bilim et al.25 showed that GSK3 inhibitors could decrease viability of renal cancer cell lines. Therefore, nuclear accumulation of GSK3β is a new promising marker of human renal cell carcinoma (RCC) cells.25 GSK3 is up-regulated in prostate cancer.26 SB216763 was able to increase β-catenin expression level in androgen receptor (AR) positive and negative cell lines.27 The proliferation rate of the AR-negative PC3 cells remained almost unaffected after SB216763 treatment. These findings are in perfect agreement with those of a previous study by Mazor et al.,28 which analyzed the effects of SB216763 on the proliferation of PCa cells.
We investigated whether abnormal Wnt/β-catenin signaling might be caused by epigenetic deregulation in urological tumors. Studies till date failed to exactly reveal the underlying mechanisms for elevated DNMT1 protein in the urogenital tract cancer. The aim of this study is to highlight the role of DNMT1 in urological cancer cell lines. In this context, firstly, Wnt/β-catenin signaling pathway was activated by using SB216763. Expression profiles of DNMT1, β-catenin, pGSK3β (Ser9), HAUSP, and UHRF1 proteins were determined after Wnt/β-catenin signaling pathway activation. Secondly, Wnt/β-catenin signaling pathway was inactivated using an unspecific DNMT1 inhibitor (DAC), and then expression profiles of DNMT1, HAUSP, and UHRF1 proteins were determined. This study is the first one to draw attention to the important role of Wnt/β-catenin signaling pathway in the expression levels of DNMT1, HAUSP, and UHRF1 proteins, all of which actively participate in the inheritance of DNA methylation in urogenital cancers.
Materials and methods
Cell culture and reagents
The prostate cancer cell line (PC3) was kindly provided by Prof. Levent Türkeri, MD (Marmara University, Urology Department, Istanbul, Turkey). Renal cell carcinoma cell line (Caki-2) and bladder cell line (T24) were purchased from ATCC (Manassas VA, USA). PC3 and Caki-2 cells were grown in RPMI-1640 containing 10% FBC, whereas T24 cells were grown in McCoy’s 5 A containing 10% FBC. DNMT inhibitor 5-aza-2′-deoxyctidine (5-Aza-dC, DAC) and GSK3β inhibitor SB216763 were obtained from Sigma (St. Louis, MO, USA). It has to be noted that our control group comprises untreated cancer cells, not healthy cells.
Cell proliferation assay
The effects of SB216763 on the viability of cells were determined by cell proliferation reagent WST-1 (Roche, Mannheim, Germany). The growing cells were treated with SB216763 (0.5 to 30 µM) for 24 h, 48 h, and 72 h. WST-1 assay was performed as previously described12,21 and absorbance was measured at 450 nm in ELISA reader.
5-Aza-dC (DAC) treatment
All three cell lines were treated with 0 to 20 µM DAC. We determined the optimal dosage of DAC first by real-time PCR and then by MSP-PCR analyses. To determine the epigenetic alteration of Wnt inhibitory factor-1 (WIF-1) gene, Caki-2 cells (1 × 106) were seeded in six-well plates and treated with 10 µM DAC after 48 h. The medium was changed 24 h after drug treatment. The cells were harvested after two days of DAC treatment.
Nucleic acid extraction
After treatment with specified drug concentrations for specific periods, genomic DNA was extracted using High Pure PCR Template Preparation kit (Roche). Total RNA was extracted by Tripure reagent (Roche). DNA and RNA isolations were performed according to manufacturer’s instructions. DNA and RNA specimens were stored at −20°C and −80°C, respectively. DNA and RNA concentrations were measured by NanoDrop (ND-1000; Montchanin, DE) spectrophotometer.
Methylation-specific PCR (MSP)
The methylation status in WIF-1 was determined by chemical treatment with sodium bisulfite and subsequent MSP analysis. Bisulfite modification of genomic DNA was performed by using an EZ DNA methylation kit (Zymo Research). Bisulfite-treated genomic DNA was amplified using either a methylation-specific or an unmethylation-specific primer set. Primer sets and MSP have been described in our previous study.29
Quantitative real-time PCR
Total RNA (1 µg) was reverse-transcribed in a 20 µL reaction mixture using random hexamers and a transcript high fidelity cDNA synthesis kit (Roche, Germany) according to manufacturer’s instructions. β-catenin (CTNNB1) and WIF-1 mRNA expression levels were measured by using a real-time PCR method. Probes and primer sets for each gene were designed at the Probe Finger Design Assay Center website. The primer sets and probe numbers have been described in our previous studies.12,29 Equal amounts of cDNA were used for real-time PCR with Light Cycler 480 Taqman probe master mix (Roche) on the 480 LightCycler II System according to the manufacturer’s instructions. Each sample was tested in duplicate. PCR efficiency for each gene was tested by serial dilutions of all genes. Gene expression levels were normalized to glyceraldehyde 3-phosphate dehydrogenase.
Western blotting
Western blot for DNMT1, β-catenin, pGSK3β(ser9), HAUSP, and UHRF1 proteins have been previously described.12 Briefly, following treatment with specified drug concentrations for specific periods, cells were lysed in lysis buffer (CST, USA) containing 1 mM PMSF (Sigma, St. Louis, MO, USA) and PhosStop phosphatase inhibitors (Roche) for phospho protein. Equal amounts of protein were loaded on an SDS-PAGE and transferred to polyvinylidene difluoride membrane. After blocking 5% w/v non-fat milk or 5% w/v bovine serum albumin in tris-buffered saline with Tween 20, the membranes were incubated with β-actin (loading control) (CST, USA), DNMT1, β-catenin, HAUSP, UHRF1 (Thermo Fisher Scientific), and pGSK3β(ser9). This process was followed by horseradish peroxidase (HRP)-conjugated secondary antibodies. Proteins were visualized by Kodak Gel Logic 2200 using Lumina Crescendo Western HRP substrate (Millipore, MA, USA).
Statistical analysis
Each test point was measured in quadruplicate in three successive experiments for WST assay. mRNA and protein results were replicated twice. Statistical significance levels in mRNA expressions were analyzed by using the pairwise fixed reallocation randomization test. The REST software (2009) was used for group-wise comparison and statistical analysis of expression levels. Cell viability was analyzed by using the one-way analysis of variance. Multiple comparison analysis was performed by using SPSS software (v.15.0) (SPSS, Inc.). A P value less than 0.01 represented a statistical significance. The results were expressed as the mean ± standard deviation.
Results
Effects of GSK3β inhibitor (SB216763) on cell viability and proliferation
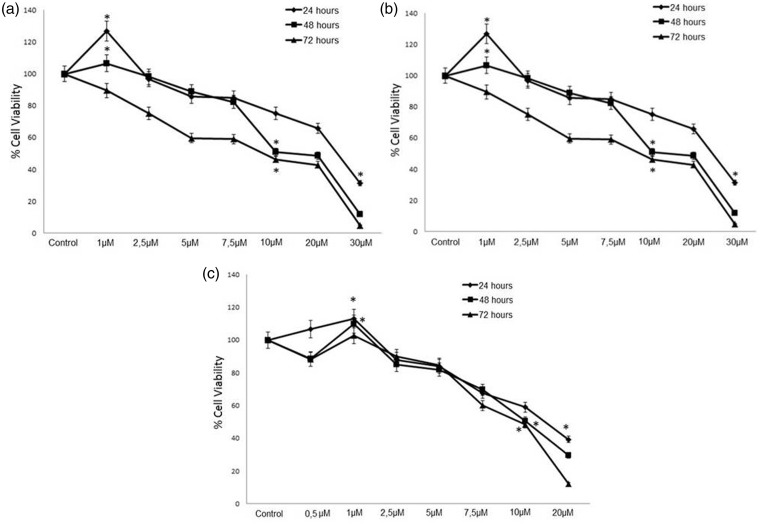
T24, Caki-2, and PC3 cell lines were exposed to different concentrations of SB216763 (0.5–30 µM) for the determination of IC50 values for all incubation periods (24 h, 48 h, and 72 h). Percentages of cell viability in all three urological cancer cells increased at low concentrations (0.5–1 µM) and decreased at high concentrations (10–30 µM) when treated with SB216763 (Figure 1). We then chose the optimal concentration and timing for SB216763 treatment for activating the Wnt/β-catenin signaling pathway in each cell line. Treatment with 1 µM SB216763 for 24 h was selected as the optimal dose–time combination in terms of least toxicity and highest cell viability for all three cell lines.
Figure 1.
The effects of GSK3β inhibitor SB216763 on cell viability of (a) T24, (b) Caki-2 and (c) PC3 cells for 24 h, 48 h, and 72 h. The results were expressed as the mean ± standard deviation of at least three independent experiments. Differences were considered significant in all experiments when *P < 0.05
Expression levels of DNMT1, β-catenin, pGSK3β(ser9) in T24, Caki-2, and PC3 cell lines
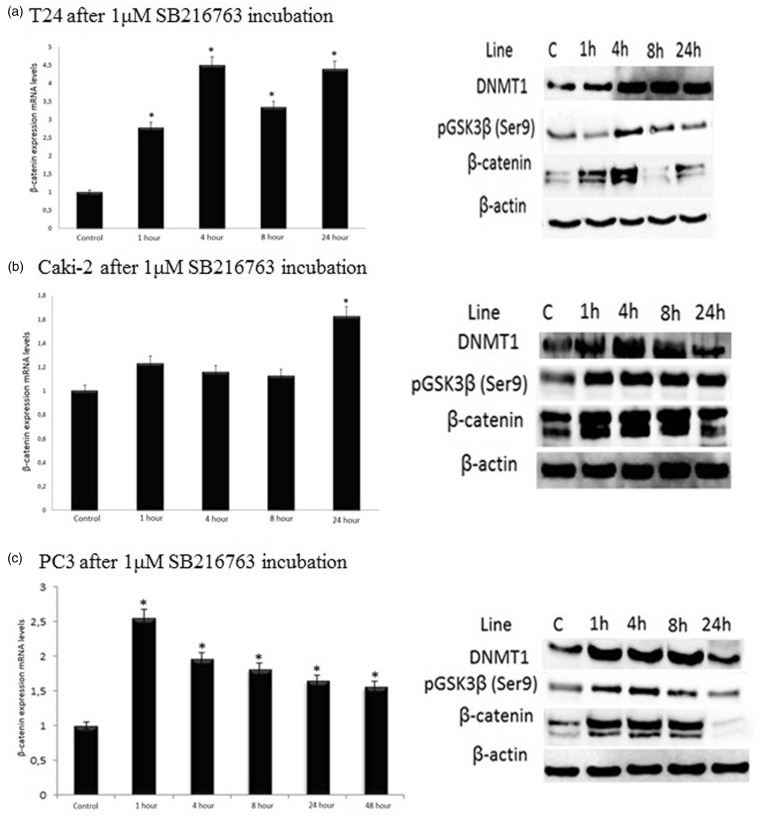
Expression levels of β-catenin in all three cell lines were examined by using both mRNA and protein analyses. As far as the T24 cells are concerned, mRNA expression levels of β-catenin in comparison to control increased 2.8 folds at 1 h, 4.5 folds at 4 h, 3.3 folds at 8 h, and 4.4 folds at 24 h (P < 0.01) in T24 cells. We observed a parallel increase in the protein levels of β-catenin and pGSK3β (ser9) after treating the cells with 1 µM SB216763 for 4 h. mRNA expression levels of β-catenin increased at 4 h and its protein levels accompanied this increase. Increased DNMT1 expression was present in T24 cells compared with control for 1 h to 24 h (Figure 2(a)).
Figure 2.
Distributions of β-catenin mRNA expression levels and DNMT1, pGSK3β(ser9), β-catenin protein levels in urological cancer cell lines: (a) bladder (T24), (b) renal (Caki-2) and (c) prostate (PC3). The results were expressed as the mean ± standard deviation of at least three independent experiments. Differences were considered significant in all experiments when *P < 0.05. C: control
As for the Caki-2 cells, after incubation with 1 µM SB216763, protein levels of β-catenin and DNMT1 as well as those of pGSK3β(ser9) increased for 1 h to 8 h. Although mRNA expression levels of β-catenin increased only at 24 h (P < 0.01), its protein levels did not accompany this increase at the same period. A slight increase in DNMT1 expression was found at 24 h in comparison to the control (Figure 2(b)).
In PC3 cells, mRNA expression levels of β-catenin in comparison to control increased 2.55 folds at 1 h, 1.96 folds at 4 h, 1.8 folds at 8 h, and 1.65 folds at 24 h (P < 0.01). After incubation of PC3 cells with 1 µM SB216763, protein levels of β-catenin and those of DNMT1 and pGSK3β (ser9) increased for 1 h to 8 h. The highest mRNA expression levels of β-catenin as well as the highest protein levels of DNMT1 and pGSK3β (ser9) were found at 1 h. Although mRNA expression levels of β-catenin increased at 24 h (P < 0.01), no increase was observed in its protein levels at the same period. Also, protein levels of pGSK3β (ser9) and those of DNMT1 decreased at 24 h (Figure 2(c)).
Expression levels of HAUSP and UHRF1 proteins in T24, Caki-2, and PC3 cell lines
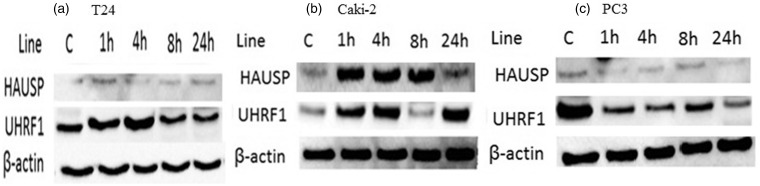
To study the role of the HAUSP deubiquitinase activity with respect to the regulation of DNMT1 and UHRF1 protein stability, expression profiles of HAUSP were examined in all three cell lines treated with 1 µM SB216763 for specified periods (Figure 3).
Figure 3.
Protein levels of HAUSP and UHRF1 in urological cancer cell lines: (a) bladder (T24), (b) renal (Caki-2) and (c) prostate (PC3). C: control
In T24 cells, there was no up-regulation in protein levels of HAUSP in comparison to control. Furthermore, there was no change in UHRF1 protein expression levels in any periods (Figure 3(a)). In Caki-2 cells, HAUSP and UHRF1 protein levels in comparison to control were up-regulated starting from hour 1, whereas HAUSP protein levels were down-regulated at 24 h (Figure 3(b)). We found that in PC3 cells, UHRF1 protein levels in comparison to control were down-regulated for 1 h to 8 h, then up-regulated at 24 h (Figure 3(c)). There were no changes in the expression profiles of HAUSP proteins throughout the 24 h period (Figure 3(c)).
Methylation status of the WIF-1 promoter region and restoration of WIF-1 mRNA expression after DAC treatment
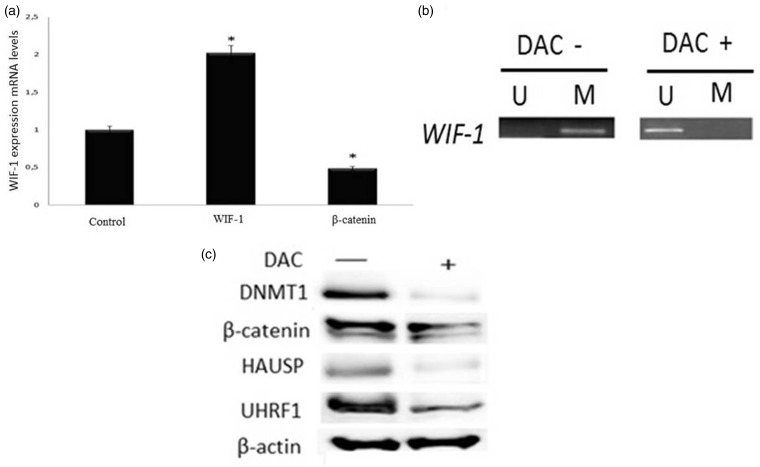
We continued only with Caki-2 cells in this part of the experiment as UHRF1 protein only increased in this cell line. We investigated whether the UHRF1-mediated proteasomal degradation could be one of the mechanisms by which DNMT1 abundance decreased. In this context, firstly, the methylation status of the WIF-1 CpG islands was characterized in the Caki-2 cell line to determine whether the loss of WIF-1 expression resulted from promoter hypermethylation. An optimal dose of DAC to remove methylation was previously determined by MSP in Caki-2 human renal cancer cells.12 The results of MSP analyses were confirmed by quantitative real-time PCR (Figure 4(a)). Caki-2 cell line indicated a high level of WIF-1 gene expression. The result displayed de-methylation in the WIF-1 promoter. After treatment with 10 µM DAC for 72 h, we found unmethylated WIF-1 with significantly increased mRNA expression levels (P < 0.01) (Figure 4(a) and (b)). In parallel, mRNA expression levels of β-catenin significantly decreased (P < 0.01). Changes in DNMT1, β-catenin, UHRF1 and HAUSP protein levels before and after DAC treatments are shown in Figure 4(c). After DAC treatments, β-catenin, DNMT1, UHRF1 and HAUSP protein levels decreased.
Figure 4.
(a) mRNA expression levels of WIF-1 and β-catenin in Caki-2 cells. (b) Methylation status of the WIF-1 promoter region and restoration of WIF-1 expression after 10 µM DAC treatment for 72 h. (c) protein levels of DNMT1, β-catenin, UHRF1 and HAUSP before and after 10 µM DAC treatment for 72 h. *P < 0.05. U: unmethylated; M: methylated
Discussion
In our study, percentages of cell viability in all three urological cancer cells increased at low concentrations and decreased at high concentrations of SB216763 treatment. We then picked the optimal dose–time combination of SB216763 to activate the Wnt/β-catenin signaling pathway in all three cell lines.
We observed a parallel increase in the protein levels of β-catenin and pGSK3β (ser9) after treating the T24 cells with 1 µM SB216763 for 4 h. mRNA expression levels of β-catenin increased at 4 h and its protein levels accompanied this increase. Although mRNA expression levels of β-catenin in comparison to control increased in T24 cells at 24 h, its protein levels did not accompany this increase this time. Increased DNMT1 expression was also present in T24 cells. After treating the Caki-2 cells and PC3 cells with 1 µM SB216763, protein levels of β-catenin and DNMT1 as well as those of pGSK3β(ser9) increased for 1 h–8 h. Although mRNA expression levels of β-catenin increased at 24 h (P < 0.01) in all three cell lines, protein levels of β-catenin did not accompany this increase at the end of same period. This situation may be caused by post-transcriptional editing, mRNA degradation or transcriptional regulators of β-catenin with non-coding RNAs. We also suggest that Wnt/β-catenin-independent pathways or regulatory proteins may also be involved in the stability of β-catenin protein. In Caki-2 and PC3 cells, only a faint DNMT1 protein expression was found at 24 h. This suggests that some tumor cells might maintain DNA hypermethylation in the absence of, or severely reduced levels of DNMT1. Furthermore, increased protein expression levels of DNMT1 could be caused more by increased stability of DNMT1 rather than the increase in mRNA. DNMT1 stability is regulated by post-transcriptional and translational modifications as well as DNMT1-associated proteins.30 DNMT1 stability is also regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination.31
In our study, there were no changes in the expression profiles of HAUSP proteins in T24 and PC3 cells throughout the 24 h period. However, significant overexpression of HAUSP was observed in Caki-2 cells. High expression of UHRF1 in these cells was also observed. Our results are consistent with those of Unoki et al.32 Throughout the 24 h period, expression levels of β-catenin and DNMT1 genes also increased. Therefore, we continued the experiment with Caki-2 cells only. We investigated whether the UHRF1-mediated proteasomal degradation could be the major mechanism in this cell line. In our previous study, we showed that constitutive activation of the Wnt signaling pathway as a result of epigenetic alterations in Wnt antagonist (WIF-1) provoked the accumulation of nuclear β-catenin, resulted in a functional transcription factor complex and expression of downstream target genes in Caki-2 cells.12 In the present study, we found unmethylated WIF-1 with significantly increased mRNA expression levels after treatment with an unspecific DNMT inhibitor -DAC-. In parallel, mRNA expression levels of β-catenin decreased significantly. Our results are consistent with those of Costa et al.1 After DAC treatments, β-catenin, DNMT1, UHRF1 and HAUSP protein levels decreased. Consequently, in Caki-2 cells, β-catenin pathway might have accounted for the stability of DNMT1 expression, whereas such relation is not valid for T24 and PC3 cells. It is important to keep in mind that DNMT inhibitors have also been shown to induce or enhance tumorigenesis via DNA hypomethylation-induced oncogene activation and chromosomal instability. Therefore, there is a need to develop specific DNMT inhibitors, such as procainamide, for efficient cancer therapy.
Conclusion
UHRF1 and HAUSP proteins which are responsible for the stability of DNMT1 protein levels might be cancer specific. Our findings suggest that the methylation status of Wnt antagonist (WIF-1) and that of DNMT1 may serve as surrogate biomarkers for Caki-2 cells. To examine the prognostic value of DNMT1 in RCC cells, further research on tissue-based molecular mechanisms for renal cell carcinoma is needed.
Acknowledgments
This study has been supported by the Gazi University Research Fund and assigned the project code number 01/2013-06.
Authors’ contribution
EK, NV and CYB made contributions to design this study. EK and NV performed experiments. EK, NV and CYB analyzed output data. EK and CYB prepared the manuscript.
References
- 1.Costa VL, Henrique R, Ribeiro FR, Carvalho JR, Oliveira J, Lobo F, Teixeira MR, Jerónimo C. Epigenetic regulation of Wnt signaling pathway in urological cancer. Epigenetics 2010; 5: 343–51. [DOI] [PubMed] [Google Scholar]
- 2.Turek-Plewa J, Jagodziński PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett 2005; 10: 631–47. [PubMed] [Google Scholar]
- 3.Qin W, Leonhardt H, Pichler G. Regulation of DNA methyltransferase 1 by interactions and modifications. Nucleus 2011; 2: 392–402. [DOI] [PubMed] [Google Scholar]
- 4.Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 2008; 455: 813–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mishra MV, Bisht KS, Sun L, Muldoon-Jacobs K, Awwad R, Kaushal A, Nguyen P, Huang L, Pennington JD, Markovina S, Bradbury CM, Gius D. DNMT1 as a molecular target in a multimodality-resistant phenotype in tumor cells. Mol Cancer Res 2008; 6: 243–9. [DOI] [PubMed] [Google Scholar]
- 6.Scott A, Song J, Ewing R, Wang Z. Regulation of protein stability of DNA methyltransferase 1 by post-translational modifications. Acta Biochim Biophys Sin 2014; 46: 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morey Kinney SR, Smiraglia DJ, James SR, Moser MT, Foster BA, Karpf AR. Stage-specific alterations of DNA methyltransferase expression, DNA hypermethylation, and DNA hypomethylation during prostate cancer progression in the transgenic adenocarcinoma of mouse prostate model. Mol Cancer Res 2008; 6: 1365–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gravina GL, Ranieri G, Muzi P, Marampon F, Mancini A, Di Pasquale B, Di Clemente L, Dolo V, D'Alessandro AM, Festuccia C. Increased levels of DNA methyltransferases are associated with the tumorigenic capacity of prostate cancer cells. Oncol Rep 2013; 29: 1189–95. [DOI] [PubMed] [Google Scholar]
- 9.Lin HY, Kuo YC, Weng YI, Lai IL, Huang TH, Lin SP, Niu DM, Chen CS. Activation of silenced tumor suppressor genes in prostate cancer cells by a novel energy restriction-mimetic agent. Prostate 2012; 72: 1767–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avissar-Whiting M, Koestler DC, Houseman EA, Christensen BC, Kelsey KT, Marsit CJ. Polycomb group genes are targets of aberrant DNA methylation in renal cell carcinoma. Epigenetics 2011; 6: 703–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Negrotto S, Hu Z, Alcazar O, Ng KP, Triozzi P, Lindner D, Rini B, Saunthararajah Y. Noncytotoxic differentiation treatment of renal cell cancer. Cancer Res 2011; 71: 1431–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konac E, Varol N, Yilmaz A, Menevse S, Sozen S. DNA methyltransferase inhibitor-mediated apoptosis in the Wnt/β-catenin signal pathway in a renal cell carcinoma cell line. Exp Biol Med 2013; 238: 1009–16. [DOI] [PubMed] [Google Scholar]
- 13.Nakagawa T, Kanai Y, Saito Y, Kitamura T, Kakizoe T, Hirohashi S. Increased DNA methyltransferase 1 protein expression in human transitional cell carcinoma of the bladder. J Urol 2003; 170(6 Pt 1): 2463–6. [DOI] [PubMed] [Google Scholar]
- 14.Nakagawa T, Kanai Y, Ushijima S, Kitamura T, Kakizoe T, Hirohashi S. DNA hypermethylation on multiple CpG islands associated with increased DNA methyltransferase DNMT1 protein expression during multistage urothelial carcinogenesis. J Urol 2005; 173: 1767–71. [DOI] [PubMed] [Google Scholar]
- 15.Wu CT, Wu CF, Lu CH, Lin CC, Chen WC, Lin PY, Chen MF. Expression and function role of DNA methyltransferase 1 in human bladder cancer. Cancer 2011; 117: 5221–33. [DOI] [PubMed] [Google Scholar]
- 16.Dhawan D, Ramos-Vara JA, Hahn NM, Waddell J, Olbricht GR, Zheng R, Stewart JC, Knapp DW. DNMT1: an emerging target in the treatment of invasive urinary bladder cancer. Urol Oncol 2013; 31: 1761–9. [DOI] [PubMed] [Google Scholar]
- 17.Dai J, Hall CL, Escara-Wilke J, Mizokami A, Keller JM, Keller ET. Prostate cancer induces bone metastasis through Wnt-induced bone morphogenetic protein-dependent and independent mechanisms. Cancer Res 2008; 68: 5785–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berndt JD, Moon RT, Major MB. Beta-catenin gets jaded and von Hippel-Lindau is to blame. Trends Biochem Sci 2009; 34: 101–4. [DOI] [PubMed] [Google Scholar]
- 19.Ahmad I, Morton JP, Singh LB, Radulescu SM, Ridgway RA, Patel S, Woodgett J, Winton DJ, Taketo MM, Wu XR, Leung HY, Sansom OJ. Beta-catenin activation synergizes with PTEN loss to cause bladder cancer formation. Oncogene 2011; 30: 178–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verras M, Sun Z. Roles and regulation of Wnt signaling and beta-catenin in prostate cancer. Cancer Lett 2006; 237: 22–32. [DOI] [PubMed] [Google Scholar]
- 21.Luo J. Glycogen synthase kinase 3beta (GSK3beta) in tumorigenesis and cancer chemotherapy. Cancer Lett 2009; 273: 194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirby LA, Schott JT, Noble BL, Mendez DC, Caseley PS, Peterson SC, Routledge TJ, Patel NV. Glycogen synthase kinase 3 (GSK3) inhibitor, SB-216763, promotes pluripotency in mouse embryonic stem cells. PLoS One 2012; 7: e39329–e39329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naito S, Bilim V, Yuuki K, Ugolkov A, Motoyama T, Nagaoka A, Kato T, Tomita Y. Glycogen synthase kinase-3beta: a prognostic marker and a potential therapeutic target in human bladder cancer. Clin Cancer Res 2010; 16: 5124–32. [DOI] [PubMed] [Google Scholar]
- 24.Yohn NL, Bingaman CN, DuMont AL, Yoo LI. Phosphatidylinositol 3'-kinase, mTOR, and glycogen synthase kinase-3β mediated regulation of p21 in human urothelial carcinoma cells. BMC Urol 2011; 11: 19–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bilim V, Ougolkov A, Yuuki K, Naito S, Kawazoe H, Muto A, Oya M, Billadeau D, Motoyama T, Tomita Y. Glycogen synthase kinase-3: a new therapeutic target in renal cell carcinoma. Br J Cancer 2009; 101: 2005–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darrington RS, Campa VM, Walker MM, Bengoa-Vergniory N, Gorrono-Etxebarria I, Uysal-Onganer P, Kawano Y, Waxman J, Kypta RM. Distinct expression and activity of GSK-3α and GSK-3β in prostate cancer. Int J Cancer 2012; 131: E872–83. [DOI] [PubMed] [Google Scholar]
- 27.Rinnab L, Schütz SV, Diesch J, Schmid E, Küfer R, Hautmann RE, Spindler KD, Cronauer MV. Inhibition of glycogen synthase kinase-3 in androgen-responsive prostate cancer cell lines: are GSK inhibitors therapeutically useful? Neoplasia 2008; 10: 624–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazor M, Kawano Y, Zhu H, Waxman J, Kypta RM. Inhibition of glycogen synthase kinase-3 represses androgen receptor activity and prostate cancer cell growth. Oncogene 2004; 23: 7882–92. [DOI] [PubMed] [Google Scholar]
- 29.Varol N, Konac E, Onen IH, Gurocak S, Alp E, Yilmaz A, Menevse S, Sozen S. The epigenetically regulated effects of Wnt antagonists on the expression of genes in the apoptosis pathway in human bladder cancer cell line (T24). DNA Cell Biol 2014; 33: 408–17. [DOI] [PubMed] [Google Scholar]
- 30.Estève PO, Chang Y, Samaranayake M, Upadhyay AK, Horton JR, Feehery GR, Cheng X, Pradhan S. A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat Struct Mol Biol 2011; 18: 42–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, Kao HY, Xu Y, Willis J, Markowitz SD, Sedwick D, Ewing RM, Wang Z. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal 2010; 3: ra80–ra80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unoki M, Kelly JD, Neal DE, Ponder BA, Nakamura Y, Hamamoto R. UHRF1 is a novel molecular marker for diagnosis and the prognosis of bladder cancer. Br J Cancer 2009; 101: 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]