Abstract
The capability to modify the genome precisely and efficiently offers an extremely useful tool for biomedical research. Recent developments in genome editing technologies such as transcription activator-like effector nuclease and the clustered regularly interspaced short palindromic repeats system have made genome modification available for a number of organisms with relative ease. Here, we introduce these genome editing techniques, compare and contrast each technical approach and discuss their potential to study the underlying mechanisms of human disease using patient-derived induced pluripotent stem cells.
Keywords: TALENs, CRISPR/Cas9, gene editing, disease modeling, gene therapy
Introduction
The development of recombinant DNA technologies has provided scientists with the fundamental tools to modify DNA sequences. These tools paved the way for the introduction of conditional alleles at specific genomic loci. To edit the genome, the introduction of a targeting construct coupled with homologous recombination (HR) was traditionally employed. Unfortunately, this approach could prove inefficient and labor intensive.1–3 Moreover, genomic targeting in eukaryotes was restricted to model organisms due to the ease of embryonic stem cell manipulation.2,4,5 Recently, the development and application of the sequence-specific endonucleases transcription activator-like effector nucleases (TALENs)6–10 and the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) system11–15 have made a revolutionary contribution to the genome editing toolbox. The advent of these technologies now enables researchers to readily manipulate any gene of interest in numerous model organisms.
TALEN and CRISPR technologies provide precise and efficient genetic modification by inducing a double-strand break (DSB) at a specific target site – an essential step for performing targeted genomic editing. The presence of a DSB activates innate cellular DNA damage repair mechanisms,16 including the dominant error-prone non-homologous end joining (NHEJ) pathway17,18 and the less frequent homology recombination-directed repair (HDR) pathway.19 DNA repair by NHEJ has the potential to lead to gene disruption by introducing deletions or mutations, while the HDR-repair pathway can be used to introduce precise changes in the presence of a donor DNA template.
In this review, we introduce TALEN and CRISPR technologies and review the extensive scientific progress made using these novel approaches. We also summarize the application of these tools in modeling human disease and discuss the future prospect of utilizing these techniques coupled with induced pluripotent stem cells (iPSCs) for future gene therapies.
TALEN-mediated genome editing
TAL (transcription activator-like) effectors or TALEs are site-specific DNA-binding proteins derived from the plant pathogen Xanthomonas sp., which uses TALE proteins to weaken host defenses by activating genes favorable to bacterial infection.6–8 Each DNA-binding module of a TALE protein typically consists of 34 amino acids arranged in tandem. These repeats are nearly identical in sequence except for two highly variable amino acids in the 12th and 13th amino acid positions (termed the repeat variable diresidue or RVD). The RVD position establishes base-recognition specificity, and thus distinct RVDs allow TALEs to recognize a specific target DNA base (NI = A, HD = C, NG = T, NH = G or NN = G/A). An array of four different repeat units has been shown to be sufficient to generate TALEs with novel DNA recognition sites.9,10,20,21
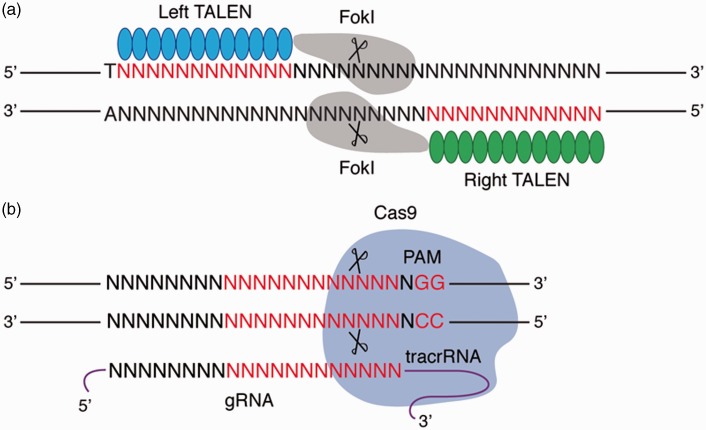
The ability to recognize and subsequently cut DNA at specific sites is accomplished by fusion of the catalytic domain of the FokI endonuclease to TALE repeats,22 which subsequently generate a TALEN protein (Figure 1(a)). Since dimerization of the catalytic domain of FokI is mandatory for nuclease activity, a pair of TALENs must be designed to recognize DNA sequences to the left and right of the intended cut site.6–10 Consequently, TALENs can be employed to generate site-specific DSBs to facilitate genome editing through NHEJ or HDR.
Figure 1.
Genome targeting by TALEN and CRISPR/Cas9 systems. (a) TAL (transcription activator-like) effector (TALE) proteins (shown as blue and green spheres) bind to target sequences (shown in red) to generate a site-specific double-stranded break (DSB) upon dimerization of fused FokI endonucleases (shown in grey). TALE targeting sites are typically preceded by a thymine (T) at the 5′ end. N is any base. (b) The CRISPR/Cas9 system relies on an engineered guide RNA (gRNA) to target DNA. The gRNA is constructed from the fusion of a crRNA and a tracrRNA (transactivating CRISPR RNA) (see text). The gRNA complexes with Cas9 (shown in purple) to induce cleavage of target DNA sites complementary to 20 nucleotides of the gRNA and located adjacent (5′) to the PAM sequence. Studies have demonstrated that the 12 base pairs closest to the PAM sequence are the most important for specificity (shown in red). N is any base
To date, TALENs have been used to target genomic loci in a number of human cell lines, including embryonic stem cells (hESCs) and iPSCs.7,23–27 Furthermore, TALEN-mediated genome targeting has been shown to also function in plants,6 fruit flies28,29 worms,30 frogs,31 and zebrafish.8,32–34 Most recently, TALEN proteins have been used for rapid gene modification in mouse, rat, and rabbit by embryo microinjection.35–37
CRISPR/Cas9-mediated genome editing
CRISPR are segments of repetitive DNA sequence found in the bacterial genome.38 These segments serve to protect the organism from invading foreign nucleic acids, such as viruses or plasmids.39–41 CRISPR systems have been shown to integrate invading foreign DNA between repeats. This integration provides a novel DNA template for the transcription of hybrid RNA molecules (crRNAs) that contain sequences from both the adjacent CRISPR arrays and the invading DNA (termed the protospacer sequence). Following transcription, each crRNA hybridizes with a second RNA (known as the transactivating CRISPR RNA or tracrRNA), and together, these molecules form a complex with the Cas9 (CRISPR associated protein 9) nuclease.42,43 DNA cleavage by the Cas9 nuclease targets DNA by relying on the protospacer-encoded region of the crRNA to direct the complex to a region called the protospacer adjacent motif or protospacer adjacent motif (PAM).
The type II CRISPR/Cas9 system that has been adapted from Streptococcus pyogenes is capable of inducing sequence-specific DSBs that allow targeted genome editing (Figure 1(b)). This modified CRISPR/Cas9 system requires the interaction between the Cas9 nuclease and a newly engineered guide RNA (gRNA).11,12,43 The gRNA is a single RNA chimera that is constructed from the fusion of a crRNA and a tracrRNA, and modification of 20 nucleotides at the 5′ end of the gRNA (corresponding to the protospacer region of the crRNA) serves to guide Cas9 to putative cleave sites to generate DSBs. CRISPR/Cas9 has been demonstrated to induce targeted cleavage at predicted sites in a number of different mammalian cell lines, including stem cells, and in a number of eukaryotes.13–15,44–56 Moreover, Cas9 has also been engineered into a nicking enzyme to increase specificity and facilitate homology-directed repair with minimal off-target rates.57–60 Additional studies have demonstrated that the CRISPR/Cas9 system is additionally capable of multiplex genome engineering by simultaneous introduction of multiple gRNAs, easing programmability and exhibiting broad applicability of this system.14,15,44
Efficiency and specificity of TALENs and CRISPR/Cas9 systems
The efficiency of genome targeting has always depended on a number of technical factors, including the cell type being targeted and the location of the targeting site. Importantly, the targeting efficiencies of TALEN and CRISPR/Cas9 technologies are much higher in a number of different cell types compared to traditional genome editing methods.7,8,12,14 However, when targeting hESCs and iPSCs, studies have shown that use of the CRISPR/Cas9 system is substantially better at promoting NHEJ and HDR to generate mutant clones than TALENs.61,62
One significant concern related to the use of TALENs and/or CRISPR/Cas9 is the risk of off-target mutagenic effects that can be introduced during the genome targeting processes. In principle, TALENs only function when dimers between the FokI nuclease domains come together, thus, the specificity is determined by the combination of two TALEN DNA-binding domains. The specificity of the CRISPR/Cas9 system is instead determined by the presence of the PAM sequence and the 20 nucleotides upstream of the PAM site in the target genome (included in the gRNA). Recent studies on CRISPR/Cas9 have demonstrated that the 12 base pairs closest to the PAM sequence are the most important for specificity.11,63
Transformed human cell lines, such as embryonic kidney 293T cells and erythromyeloblastoid leukemia K562 cells, have been used to study the off-target effects.64–66 Higher levels of off-target mutagenesis suggested that the CRISPR/Cas9 system is susceptible to cleavage at off-target loci due to mismatches in the gRNA. However, controlling the concentration of the Cas9 mRNA appears to diminish these off-target effects. The inducing single-stranded DNA breaks (nicks) generated by the CRISPR/Cas9-D10A nickase nuclease have been shown to reduce the off-target rate by more than 1500 fold.57,64 Notably, whole-genome sequencing of TALEN and CRISPR/Cas9 gene targeted cells has also shown that off-target mutations are extremely rare in hESCs and iPSCs.67–69
Generation of disease models using TALENs and CRISPR/Ca9 targeting
On-going genome targeting in human cells using TALENs and/or CRISPR/Cas9 will help establish disease models that more accurately reflect the pathogenesis observed in patients and provide proof-of-principle approaches for future gene therapies. To that end, iPSCs derived from the somatic cells of patients can provide a useful tool for studying the underlying mechanism of human disease and may serve as a promising source for cell replacement therapies.70,71
Due to the unpredictable genetic variations of patient-derived iPSCs, researchers have been challenged to distinguish minor, disease-related phenotypic changes from otherwise normal variations in different genetic backgrounds. Therefore, one of the best ways to study the function of disease-related gene mutations is to utilize TALENs or CRISPR/Cas9 technologies to introduce mutations in a more controlled environment. Recently, mutations in 15 genes related to metabolic diseases were introduced using TALENs to provide isogenic control cell lines and to demonstrate cell-autonomous phenotypes.25 The generation of additional isogenic iPSCs that differ exclusively at disease causing genomic loci will provide invaluable tools towards finding a solution to this problem.
Conversely, other laboratories have utilized TALENs and CRISPR/Cas9 technologies to repair mutations in patient-derived iPS cells. TALENs have been used to correct a mutation found in patients suffering from alpha-1 antitrypsin (AAT) deficiency, a genetic liver disorder that predisposes patients to liver cirrhosis and hepatocellular carcinoma, cystic fibrosis, and Gaucher’s disease.72,73 Moreover, CRISPR/Cas9 has been used to study cystic fibrosis by correcting a mutation found in the transmembrane conductor receptor (CFTR) gene in primary adult stem cells.74 TALENs and the CRISPR/Cas9 system have also been used to target mutations found in iPS cells derived from Duchenne muscular dystrophy (DMD) patients. This correction leads to the generation of skeletal muscle cells that express a wild type version of dystrophin, a protein that is fundamental in supporting muscle fiber strength. Together, these reports demonstrate that genome-modifying systems provide an invaluable tool for generating and investigating models of human disease.
Generation of mouse models using TALENs and CRISPR/Cas9
TALENs and CRISPR/Cas9 are being employed to quickly and directly generate mouse models of human disease. The direct microinjection of TALENs and CRISPR/Cas9 into zygotes enables instant germline modifications and accelerates the generation of mouse models.75–77 In addition to germline manipulation, a Cre recombinase-(Cre) dependent Cas9 knock-in mouse model was recently shown to enable the introduction of genetic alterations via the expression of gRNAs in specific tissues (brain, bone marrow, and lung) through adeno-associated virus (AAV), lentivirus or particle-mediated delivery.78 This technology allows genetic modifications to be introduced in the genome of somatic cells (in vivo or ex vivo) for immediate analysis of phenotypes associated with disease-causing mutations. Moreover, another study has shown viral-mediated delivery of the CRISPR/Cas9 system to alter the somatic cells of mice to introduce the fusion of the EML4-ALK oncogene. This fusion is detected in a subset of human non-small cell lung cancers (NSCLC). The fusion of EML4 (echinoderm microtubule-associated protein like 4) and ALK (anaplastic lymphoma kinase) results from an inversion of the short arm of chromosome 2, and modeling of this particular fusion event in the mouse (via traditional methods) had been previously problematic.
Conclusions and future perspectives
TALENs and CRISPR/Cas9 genome editing technologies have dramatically boosted the ability to manipulate a diverse set of genomes. These novel approaches are aggressively being applied to study a wider set of biological questions, including several human disorders. Naturally, limitations exist for each system, including the requirement of generating TALEN pairs to target one site and slightly higher off-target rates for CRISPR/Cas9. Targeting efficiencies between TALENs and/or CRISPR/Cas9 in certain cell types is an additional problem that will need to be resolved by further study. Moreover, the application of these technologies in the clinical setting has yet to be established, and it will be imperative to understand potential complications after genetic correction. In summary, TALENs and CRISPR/Cas9 systems are invaluable genetic manipulating tools that will continue to be improved and applied to countless future studies. These systems hold the key to revolutionize biological research and facilitate the promise of personalized medicine in the future.
Acknowledgments
The authors have no conflict of interest to declare. The work was supported by NYSTEM contracts C028129, C029556 and C026714, NIH grant NS061856 and Department of Veterans Affairs Merit Award I01BX002452 to JF. JP is a Natural Science Foundation of China (NSFC) grant awardee (81400933).
Authors’ contributions
All authors participated in the conception of the manuscript; BZ and JF provided editorial guidance; JP and DF wrote the manuscript.
References
- 1.Melton DW. Gene targeting in the mouse. Bioessays 1994; 16: 633–8. [DOI] [PubMed] [Google Scholar]
- 2.Golic KG, Golic MM. Engineering the Drosophila genome: chromosome rearrangements by design. Genetics 1996; 144: 1693–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet 2005; 6: 507–12. [DOI] [PubMed] [Google Scholar]
- 4.Koller BH, Smithies O. Inactivating the beta 2-microglobulin locus in mouse embryonic stem cells by homologous recombination. Proc Natl Acad Sci USA 1989; 86: 8932–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 1987; 51: 503–12. [DOI] [PubMed] [Google Scholar]
- 6.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic acids Res 2011; 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 2011; 29: 731–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, 2nd, Tan W, Penheiter SG, Ma AC, Leung AY, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC. In vivo genome editing using a high-efficiency TALEN system. Nature 2012; 491: 114–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009; 326: 1509–12. [DOI] [PubMed] [Google Scholar]
- 10.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science 2009; 326: 1501. [DOI] [PubMed] [Google Scholar]
- 11.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339: 819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science 2013; 339: 823–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 2013; 31: 230–2. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013; 153: 910–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 2013; 154: 1370–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyman C, Kanaar R. DNA double-strand break repair: all's well that ends well. Ann Rev Genet 2006; 40: 363–83. [DOI] [PubMed] [Google Scholar]
- 17.Barnes DE. Non-homologous end joining as a mechanism of DNA repair. Curr Biol 2001; 11: R455–7. [DOI] [PubMed] [Google Scholar]
- 18.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Ann Rev Biochem 2010; 79: 181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Bosch M, Lohman PH, Pastink A. DNA double-strand break repair by homologous recombination. Biol Chem 2002; 383: 873–92. [DOI] [PubMed] [Google Scholar]
- 20.Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat Commun 2012; 3: 968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Streubel J, Blucher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nat Biotechnol 2012; 30: 593–5. [DOI] [PubMed] [Google Scholar]
- 22.Mussolino C, Morbitzer R, Lutge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res 2011; 39: 928–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holkers M, Maggio I, Liu J, Janssen JM, Miselli F, Mussolino C, Recchia A, Cathomen T, Goncalves MA. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res 2013; 41: e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osborn MJ, Starker CG, McElroy AN, Webber BR, Riddle MJ, Xia L, DeFeo AP, Gabriel R, Schmidt M, von Kalle C, Carlson DF, Maeder ML, Joung JK, Wagner JE, Voytas DF, Blazar BR, Tolar J. TALEN-based gene correction for epidermolysis bullosa. Mol Ther 2013; 21: 1151–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, Kuperwasser N, Motola DL, Meissner TB, Hendriks WT, Trevisan M, Gupta RM, Moisan A, Banks E, Friesen M, Schinzel RT, Xia F, Tang A, Xia Y, Figueroa E, Wann A, Ahfeldt T, Daheron L, Zhang F, Rubin LL, Peng LF, Chung RT, Musunuru K, Cowan CA. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell 2013; 12: 238–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piganeau M, Ghezraoui H, De Cian A, Guittat L, Tomishima M, Perrouault L, Rene O, Katibah GE, Zhang L, Holmes MC, Doyon Y, Concordet JP, Giovannangeli C, Jasin M, Brunet E. Cancer translocations in human cells induced by zinc finger and TALE nucleases. Genome Res 2013; 23: 1182–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabir S, Hockemeyer D, de Lange T. TALEN Gene Knockouts Reveal No requirement for the conserved human shelterin protein Rap1 in telomere protection and length regulation. Cell Rep 2014; 9: 1273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katsuyama T, Akmammedov A, Seimiya M, Hess SC, Sievers C, Paro R. An efficient strategy for TALEN-mediated genome engineering in Drosophila. Nucleic Acids Res 2013; 41: e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Li C, Yu Z, Huang P, Wu H, Wei C, Zhu N, Shen Y, Chen Y, Zhang B, Deng WM, Jiao R. Efficient and specific modifications of the Drosophila genome by means of an easy TALEN strategy. J Genet Genomics 2012; 39: 209–15. [DOI] [PubMed] [Google Scholar]
- 30.Cheng Z, Yi P, Wang X, Chai Y, Feng G, Yang Y, Liang X, Zhu Z, Li W, Ou G. Conditional targeted genome editing using somatically expressed TALENs in C. elegans. Nat Biotechnol 2013; 31: 934–7. [DOI] [PubMed] [Google Scholar]
- 31.Lei Y, Guo X, Liu Y, Cao Y, Deng Y, Chen X, Cheng CH, Dawid IB, Chen Y, Zhao H. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs). Proc Natl Acad Sci USA 2012; 109: 17484–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, Khayter C, Joung JK, Sander JD, Peterson RT, Yeh JR. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res 2012; 40: 8001–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol 2011; 29: 699–700. [DOI] [PubMed] [Google Scholar]
- 34.Zu Y, Tong X, Wang Z, Liu D, Pan R, Li Z, Hu Y, Luo Z, Huang P, Wu Q, Zhu Z, Zhang B, Lin S. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat Methods 2013; 10: 329–31. [DOI] [PubMed] [Google Scholar]
- 35.Qiu Z, Liu M, Chen Z, Shao Y, Pan H, Wei G, Yu C, Zhang L, Li X, Wang P, Fan HY, Du B, Liu B, Li D. High-efficiency and heritable gene targeting in mouse by transcription activator-like effector nucleases. Nucleic Acids Res 2013; 41: e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song J, Zhong J, Guo X, Chen Y, Zou Q, Huang J, Li X, Zhang Q, Jiang Z, Tang C, Yang H, Liu T, Li P, Pei D, Lai L. Generation of RAG 1- and 2-deficient rabbits by embryo microinjection of TALENs. Cell Res 2013; 23: 1059–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tesson L, Usal C, Menoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, Gregory PD, Anegon I, Cost GJ. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol 2011; 29: 695–6. [DOI] [PubMed] [Google Scholar]
- 38.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014; 346: 1258096. [DOI] [PubMed] [Google Scholar]
- 39.Fineran PC, Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology 2012; 434: 202–9. [DOI] [PubMed] [Google Scholar]
- 40.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010; 327: 167–70. [DOI] [PubMed] [Google Scholar]
- 41.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature 2012; 482: 331–8. [DOI] [PubMed] [Google Scholar]
- 42.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011; 471: 602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012; 337: 816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li W, Teng F, Li T, Zhou Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol 2013; 31: 684–6. [DOI] [PubMed] [Google Scholar]
- 45.Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, Zhao Y. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol 2013; 31: 681–3. [DOI] [PubMed] [Google Scholar]
- 46.Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O'Connor-Giles KM. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 2013; 194: 1029–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep 2013; 4: 220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu Z, Ren M, Wang Z, Zhang B, Rong YS, Jiao R, Gao G. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics 2013; 195: 289–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods 2013; 10: 1028–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 2013; 10: 741–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Belhaj K, Chaparro-Garcia A, Kamoun S, Patron NJ, Nekrasov V. Editing plant genomes with CRISPR/Cas9. Curr Opin Biotechnol 2014; 32C: 76–84. [DOI] [PubMed] [Google Scholar]
- 52.Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 2014; 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res 2013; 23: 465–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 2013; 31: 227–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hwang WY, Fu Y, Reyon D, Maeder ML, Kaini P, Sander JD, Joung JK, Peterson RT, Yeh JR. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PloS One 2013; 8: e68708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA 2013; 110: 13904–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013; 154: 1380–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, Wang L, Hodgkins A, Iyer V, Huang X, Skarnes WC. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods 2014; 11: 399–402. [DOI] [PubMed] [Google Scholar]
- 59.Rong Z, Zhu S, Xu Y, Fu X. Homologous recombination in human embryonic stem cells using CRISPR/Cas9 nickase and a long DNA donor template. Protein Cell 2014; 5: 258–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duda K, Lonowski LA, Kofoed-Nielsen M, Ibarra A, Delay CM, Kang Q, Yang Z, Pruett-Miller SM, Bennett EP, Wandall HH, Davis GD, Hansen SH, Frodin M. High-efficiency genome editing via 2A-coupled co-expression of fluorescent proteins and zinc finger nucleases or CRISPR/Cas9 nickase pairs. Nucleic Acids Res 2014; 42: e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ding Q, Regan SN, Xia Y, Oostrom LA, Cowan CA, Musunuru K. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell stem Cell 2013; 12: 393–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang L, Guell M, Byrne S, Yang JL, De Los Angeles A, Mali P, Aach J, Kim-Kiselak C, Briggs AW, Rios X, Huang PY, Daley G, Church G. Optimization of scarless human stem cell genome editing. Nucleic Acids Res 2013; 41: 9049–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 2013; 31: 233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 2013; 31: 833–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 2013; 31: 827–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 2013; 31: 839–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Veres A, Gosis BS, Ding Q, Collins R, Ragavendran A, Brand H, Erdin S, Talkowski ME, Musunuru K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 2014; 15: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith C, Gore A, Yan W, Abalde-Atristain L, Li Z, He C, Wang Y, Brodsky RA, Zhang K, Cheng L, Ye Z. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell 2014; 15: 12–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suzuki K, Yu C, Qu J, Li M, Yao X, Yuan T, Goebl A, Tang S, Ren R, Aizawa E, Zhang F, Xu X, Soligalla RD, Chen F, Kim J, Kim NY, Liao HK, Benner C, Esteban CR, Jin Y, Liu GH, Li Y, Izpisua Belmonte JC. Targeted gene correction minimally impacts whole-genome mutational load in human-disease-specific induced pluripotent stem cell clones. Cell Stem Cell 2014; 15: 31–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131: 861–72. [DOI] [PubMed] [Google Scholar]
- 71.Jiang H, Ren Y, Yuen EY, Zhong P, Ghaedi M, Hu Z, Azabdaftari G, Nakaso K, Yan Z, Feng J. Parkin controls dopamine utilization in human midbrain dopaminergic neurons derived from induced pluripotent stem cells. Nat Commun 2012; 3: 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Choi SM, Kim Y, Shim JS, Park JT, Wang RH, Leach SD, Liu JO, Deng C, Ye Z, Jang YY. Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology 2013; 57: 2458–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramalingam S, Annaluru N, Kandavelou K, Chandrasegaran S. TALEN-Mediated generation and genetic correction of disease-specific human induced pluripotent stem cells. Curr Gene Ther 2014; 14: 461–72. [DOI] [PubMed] [Google Scholar]
- 74.Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK, Nieuwenhuis EE, Beekman JM, Clevers H. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 2013; 13: 653–8. [DOI] [PubMed] [Google Scholar]
- 75.Wefers B, Meyer M, Ortiz O, Hrabe de Angelis M, Hansen J, Wurst W, Kuhn R. Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. Proc Natl Acad Sci USA 2013; 110: 3782–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee AY, Lloyd KC. Conditional targeting of Ispd using paired Cas9 nickase and a single DNA template in mice. FEBS Open Bio 2014; 4: 637–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kasparek P, Krausova M, Haneckova R, Kriz V, Zbodakova O, Korinek V, Sedlacek R. Efficient gene targeting of the Rosa26 locus in mouse zygotes using TALE nucleases. FEBS Lett 2014; 588: 3982–8. [DOI] [PubMed] [Google Scholar]
- 78.Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014; 159: 440–55. [DOI] [PMC free article] [PubMed] [Google Scholar]