Abstract
Despite their strong role in human health, poor bioavailability of flavonoids limits their biological effects in vivo. Enzymatically catalyzed acylation of fatty acids to flavonoids is one of the approaches of increasing cellular permeability and hence, biological activities. In this study, six long chain fatty acid esters of quercetin-3-O-glucoside (Q3G) acylated enzymatically and were used for determining their antiproliferative action in hepatocellular carcinoma cells (HepG2) in comparison to precursor compounds and two chemotherapy drugs (Sorafenib and Cisplatin). Fatty acid esters of Q3G showed significant inhibition of HepG2 cell proliferation by 85 to 90% after 6 h and 24 h of treatment, respectively. The cell death due to these novel compounds was associated with cell-cycle arrest in S-phase and apoptosis observed by DNA fragmentation, fluorescent microscopy and elevated caspase-3 activity and strong DNA topoisomerase II inhibition. Interestingly, Q3G esters showed significantly low toxicity to normal liver cells than Sorafenib (P < 0.05), a chemotherapy drug for hepatocellular carcinoma. Among all, oleic acid ester of Q3G displayed the greatest antiproliferation action and a high potential as an anti-cancer therapeutic. Overall, the results of the study suggest strong antiproliferative action of these novel food-derived compounds in treatment of cancer.
Keywords: Quercetin-3-O-glucoside, acylation, cancer, apoptosis, cell cycle, caspase-3, topoisomerase II, HepG2 cells, hepatocellular carcinoma, chemotherapy
Introduction
Flavonoids are widely found in fruits, vegetables, and plant derived beverages such as tea, coffee, and wine. Most of the flavonoids are not only strong antioxidants but also strong therapeutics with anti-cancer activity. Epidemiology and cohort studies have suggested flavonoids in the human diet might decrease the risk of various lifestyle and age-related chronic diseases including cancer, cardiovascular disease, and diabetes.1–5
Quercetin, one of the most ubiquitous dietary flavonoid, has been shown to exhibit antiproliferative effects and induce apoptosis in different types of cancer cell lines. Quercetin induces apoptosis by activating caspase 3 cascade in human promyeloleukemic HL-60 cells.6 Quercetin-induced cytotoxicity and reduction in viability of cancer cells has been shown as result of alteration in the signal transduction pathways by affecting various kinases which includes Akt/PI3K,7,8 epidermal growth factor receptor (EGF-R),8 mitogen-activated protein kinases/extracellular signal-regulated kinases (MAPK/ERK).9 Additionally, quercetin has been shown to inhibit DNA topoisomerase II activity strongly in vitro leading to cell division malfunction thereby arresting cell proliferation.10 The naturally occurring form of quercetin in plants, quercetin-3-O-glucoside (Q3G) has been shown to exhibit strong antioxidant activity.11 Q3G was also shown to exhibit stronger antiproliferative activity than its aglycone, quercetin.12
Despite their strong health benefits and biological activity, glycosylated flavonoids possess low bioavailability13 which limits their effect in vivo. The number of hydroxyl groups, presence of a methoxy group in the B ring of flavonoids and lipophilicity has been shown to influence the bioavailability of flavonoids.14 Structural modification of flavonoids by introducing hydrophobic groups could increase the cellular uptake and therefore, bioavailability of glycosylated flavonoids.15 In previous studies, omega-3 and omega-6 polyunsaturated fatty acids have been shown to exhibit antiproliferative activity.16,17 However, their use is very limited as they are prone to autooxidation. However, combining the unstaurated fatty acids with certain flavonoids with strong antioxidant properties could protect them from oxidation and perhaps improve biological properties of the flavonoid. Salem et al.15 have shown that Q3G esters had enhanced antioxidant properties by enhancing lipophilicity.
The aim of the current study was to determine the antiproliferative action of six long chain fatty acid esters of Q3G (namely, stearic acid ester, oleic acid ester, linoleic acid ester, alpha-linolenic acid ester, eicosapentaenoic acid (EPA) ester, docosahexanoic acid (DHA) ester of Q3G) and to evaluate the mechanism of their action using a human hepatocellular carcinoma cell line, HepG2. Hepatocellular carcinoma is the most common form of liver cancer, which represents the fifth worldwide malignancy and third cause of mortality among cancer-related death. The novel compounds for this study were prepared using enzymatic acylation as previously described by Ziaullah et al.18
Materials and methods
Materials and chemicals
Quercetin, Q3G, Cisplatin, propidium iodide, fatty acids namely, oleic, stearic, linoleic, α-linolenic, EPA, DHA and two-well chambered cover slides were purchased from Sigma–Aldrich (Mississauga, ON, Canada). Sorafenib (Nexovar®) was purchased from Cayman Chemical (Ann Arbor, MI, USA). Cell Titer 96TM Aqueous One solution cell proliferation (MTS) assay and CytoTox 96® non-radioactive cytotoxicity (LDH) assay kits were purchased from Promega (Madison, WI, USA). ApoTarget™ Quick apoptotic DNA ladder detection kit from Invitrogen (Burlington, ON, Canada). Caspase 3 colorimetric assay kit was purchased from BioVision, Inc. (San Francisco, CA, USA). All cell culture vessels and plates were purchased from BD Biosciences (San Jose, CA, USA). BCA protein purification kit was purchased from Thermo Scientific (Burlington, ON, Canada).
Synthesis of the long chain fatty acid esters of Q3G
The compounds for the study were synthesized as previously described by Ziaullah et al.18 Briefly, in a round bottom flask, defined quantities of Q3G and individual fatty acids were dissolved in acetone. Enzymatic reactions were initiated by the addition of lipase (Novozyme 435®); with an activity of 10,000 propyl laurate units). The mixture was stirred and heated at 45℃ for 12–24 h and was monitored by thin layer chromatography (TLC), followed by staining with anisaldehyde spray reagent and then heating at 110℃. After completion of reaction, it was filtered, evaporated and column chromatography (acetone/toluene; 35:75 to 50:50) was performed to get the pure fatty acid esters of Q3G. The pure compounds were then analyzed by IR, 1H NMR and 13C NMR spectroscopy to confirm the structures. The yield of the pure compounds varies between 81% and 97% and the physical characteristic was greenish yellow to light brownish yellow spongy solid.18
HepG2 cell culture system
HepG2 cells were obtained from American Type Culture Collection (ATCC, 8065) and maintained according to ATCC's instructions. Briefly, the cells were cultured in Eagle's Minimum Essential Growth Medium (EMEM) with 2 mM l-glutamine and 10% fetal bovine serum (FBS) at 37℃ and 5% CO2. T-75 tissue culture flasks with 12–15 mL of media were used for regular culturing. Sub-culturing was performed in 1:4 or 1:5 ratio every three to four days when cells reached a confluence of 70–80%. Cells were counted under Nikon Eclipse TS 100 phase contrast microscope using haemocytometer and then transferred to fresh flasks.
Measurement of cell proliferation
The assay was performed as described previously by Talib et al.19 and Shan et al.20 Commercially available Cell Titer 96TM Aqueous One solution cell proliferation (MTS) assay was employed for the assay. Manufacturer's instructions were followed for performing assay. Briefly, HepG2 cells in the exponential growth phase were collected and seeded in 96-well microplate in density of 2 × 104 cells per well, final volume being 200 μL/well with the help of multichannel pipette. The microplates were placed in culture incubator in standard conditions (37℃ with 5% CO2) and cultured for 24 h. After incubation, 100 μM of long chain fatty acid esters of Q3G and control samples in fresh media were added to each well in triplicates. The DMSO concentration for 100 μM of test compounds in all assays was less than 1%. The plates were then incubated for different time intervals (6 and 24 h) in culture incubator (37℃, 5% CO2, 90% humidity). According to manual instructions, 20 μL of MTS was added to each well (5 g/L in PBS) and again incubated for 1–4 h. Absorbance was recorded directly at 490 nm using Fluostar Optima micro plate reader (BMG Labtech, Ortenberg, Germany).
Measurement of cell cytotoxicity
Commercially available CytoTox 96® Non-Radioactive Cytotoxicity (LDH) assay kit from Promega was utilized for this analysis. The manufacturer's instructions were followed for the assay. Briefly, HepG2 (5000 cells/well) were plated in 96-well microplates, the final volume per well was kept at 100 µL. The controls included: (i) assay medium without cells, (ii) low control of cells and assay medium to see spontaneous or normal LDH activity and (iii) high control of cells treated with Triton X-100 to achieve maximum LDH release which served as positive control. The microplates were placed in culture incubator in standard conditions (37℃ with 5% CO2) and cultured for 24 h. After incubation, 100 µM of the long chain fatty acid esters of Q3G and control samples in fresh media were added to each well in triplicates. The plates were incubated for different time intervals (mainly, 6 and 24 h) in culture incubator (37℃, 5% CO2, 90% humidity). After treatment, the 96-well microplate was centrifuged and supernatant was transferred to a fresh 96-well microplate and subjected to LDH assay. Absorbance was taken at 490–492 nm using Fluostar Optima micro plate reader (BMG Labtech, Ortenberg, Germany).
DNA fragmentation
This assay was performed by utilizing commercially available ApoTarget™ Quick Apoptotic DNA Ladder Detection Kit. The manufacturer's instructions were followed for the assay. Briefly, HepG2 cells (5 × 105 cells/well) were grown in 12-well culture plate up to 75–80% confluence and then treated with 100 µM of test compounds for 24 and 48 h. Cells were collected and total DNA was isolated from each sample. Extracted DNA pellet was dissolved in 30 μL of DNA suspension buffer (provided with kit) and resolved on 1.2% agarose gel containing GelRed™ stain (used 1X) in 1X TAE buffer (pH 8.5, 20 mM Tris-acetic acid, 2 mM EDTA) in BioRad mini-gel electrophoresis kit. The DNA gel was then moved to UV sample tray for examining the bands, which were visualised and photographed by BioRad's Gel Doc™ EZ system (Mississauga, ON, Canada).
Caspase assay
The caspases activation was quantified by utilising Caspase-3/CPP32 Colorimetric Assay Kit. The assay was performed according to the manufacturer's instruction. Briefly, HepG2 cells (1 × 106 cells/well) were plated in six-well tissue culture plate. After treatment with the test compounds for 24 h, the cells were lysed with lysis buffer provided by the manufacturer and centrifuged at 13,000 r/min. After collecting the supernatant, the protein was quantified using BCA protein quantification kit and 250 μg of protein per treatment sample was used for the assay. Reaction buffer (50 μL) was added to each treatment well of microplate reader followed by addition of 5 μL DEVD-pNA (Asp-Glu-Val-Asp p-nitroanilide) caspase substrate. The microplate was incubated at 37℃ for 1–2 h. The absorbance of the samples was read at 405 nm in Fluostar Optima micro plate reader (BMG Labtech, Ortenberg, Germany). After subtracting background readings from all the samples (induced and uninduced), fold-increase in CPP32 activity was determined by comparing these results with the level of the uninduced control.
Fluorescence microscopy
GFP-Certified™ Apoptosis/Necrosis Detection Kit was purchased from Enzo Life Sciences Inc. (Farmingdale, NY, USA) for the detection of adherent apoptotic cells. Briefly, 2 × 105 HepG2 cells were seeded on two-well chambered cover slides (Sigma–Aldrich, Mississauga, ON, Canada) and grown to about 75% confluence and followed by treatment with 100 μM long chain fatty acid esters of Q3G or vehicle for 24 h. Adherent cells were stained according to the manufacturer's instructions by dual detection reagent (containing Annexin V coupled with PI). The dual-labeled cells were visualized by fluorescence microscopy with a Leica DMBL (20×/.040) fluorescent microscope (Houston, TX, USA) incorporated with Nikon Cool Pix 4500 Digital camera (Mississauga, ON, Canada). Cells with bound Annexin-V show green staining in the plasma membrane. Cells that have lost membrane integrity show red staining (PI) throughout the cytoplasm with an impression of green staining on the plasma membrane. Cells with green staining were scored as apoptotic; cells with both green and red staining were scored as late apoptotic, whereas those with only red staining were considered necrotic.
Cell cycle analysis
HepG2 cells were plated in a six-well culture plate (1 × 106 cells/well). After 24 h of incubation at 37℃, 5% CO2, the cells were treated with 100 μM long chain fatty acid esters of Q3G for another 24 h. Briefly, cells were trypsinized and centrifuged at 1200 r/min at 4℃ for 10 min followed by another PBS wash. The pellet was re-suspended in 0.3 mL of PBS. The cells were then fixed by adding 0.7 mL of ice cold ethanol for 2 h. After fixation, the cells were centrifuged again at 1200 r/min at 4℃ for 10 min and cell pellet was re-suspended in 0.25 mL of PBS with the addition of 5 μL of 10 mg/mL Rnase A (the final concentration being 0.2–0.5 mg/mL) and incubation at 37℃ for 1 h. After incubation, 10 μL of 1 mg/mL PI solution (the final concentration being 10 μg/mL) was added to the cell suspension and kept in the dark at 4℃ until analysis. The cells were then analyzed for cell cycle using flow cytometer FACS Caliber (BD Biosciences, San Jose, CA, USA) with an excitation wavelength at 488 nm and emission at 670 nm. DNA content was determined by MotFit LT™ software, version 4.0 (Topsham, ME, USA), which provided histograms to evaluate cell cycle distribution.
Topoisomerase II Assay
Commercially available topoisomerase II drug screening kit (TopoGEN, Inc., Columbus, OH, USA) was utilized and assay was performed as explained by Patra et al.21 Briefly, substrate supercoiled pHot1 DNA (0.25 µg) was incubated with four units (2 µL) of human DNA topoisomerase II, test compounds (2 µL) and assay buffer (4 µL) in 37℃ for 30 min. The reaction was terminated by the addition of 10% sodium dodecyl sulphate (2 µL) followed by digestion with proteinase K (50 µg/µL) at 37℃ for 15 min. After incubation, DNA was run on 1% agarose gel in BioRad gel electrophoresis system at 70 V for 1–2 h. The gel was stained with GelRed™ stain for 2 h and destained for 15 min with TAE buffer followed by gel imaging via BioRad's Gel Doc™ EZ system. Supercoiled DNA and linear strand DNA were incorporated in the gel as markers for DNA topology and DNA topoisomerase II poison (inhibitor). Additionally, a positive control drug VP16 (provided with the kit) and a known DNA topoisomerase II inhibitor (poison) were also incorporated in the gel for reference. The presence of a single linear band in the positive control reaction confirmed the inhibitory effect of the VP16 on DNA topoisomearse II activity. The inhibitory activity was calculated as relative activity of topoisomerase enzyme (in this case, intensity of the supercoiled band and presence/absence of single linear band) in the presence of test compounds in comparison to that in the negative control solution (DMSO control).
Statistical analysis
Data were analyzed using Minitab 16 statistical software. The assays were replicated three times using a completely randomised design (CRD) model. Data were analyzed using one way ANOVA. All treatments were checked for normality and constant variation check before running ANOVA. Tukey's test was performed for achieving significant difference between different treatment compounds. Significance level in all assays was taken at P < 0.05. All data were expressed as mean ± SD with at least three independent experiments.
Results
Antiproliferative and cytotoxic effects of long chain fatty acid esters of Q3G on HepG2 cells
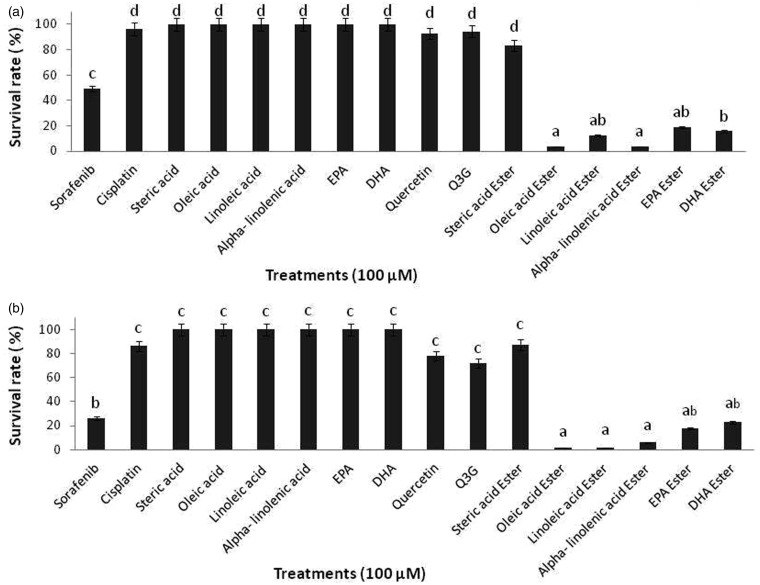
The potential effects of long chain fatty acid esters of Q3G on cell proliferation were investigated by using the MTS assay which measures the metabolically live cells based on their mitochondrial dehydrogenase activity.22 Incubation of HepG2 cells with 100 µM of long chain fatty acid esters of Q3G for 6 h and 24 h resulted in significant decrease (approximately 85–90%) in cell proliferation (P < 0.05) in comparison to the precursor compounds quercetin and Q3G, free fatty acids and prescribed drugs Sorafenib and Cisplatin (Figure 1). Within 6 h of incubation, all long chain fatty acid esters of Q3G except stearic acid ester of Q3G treated cells showed a drastic reduction in cell viability as compared to the respective controls (Figure 1(a)). After 24 h of incubation, viability of HepG2 cells further decreased significantly and dramatically in Q3G fatty acid esters treated cells as compared to the precursor compounds and control drugs treated cells (Figure 1(b)). Also, at low concentrations (mainly,10, 30 and 50 µM) of long chain fatty acid esters of Q3G, longer incubations of 48 h and 72 h were necessary to obtain a significant reduction in cell viability (data not included).
Figure 1.
Antiproliferative effects of long chain fatty acid esters of Q3G on HepG2 cells. The figure describes the percentage of viable HepG2 cells after treatment with long chain fatty acid esters of Q3G. Cells (2 × 104 cells per well; 96-well plate) were treated with 100 µM of test compounds for 6 h (a) and 24 h (b). After treatment viable cell percentage was determined by MTS assay. All long chain fatty acid esters of Q3G except stearic acid ester showed over 85% inhibition of cell proliferation as seen by the cell viability data of 6 h and 24 h. Mean separation between groups was conducted using Tukey's test (n = 6). Significance level was taken at P value < 0.05. Results are expressed relative to control (6 h and 24 h incubation with test compound-free medium)
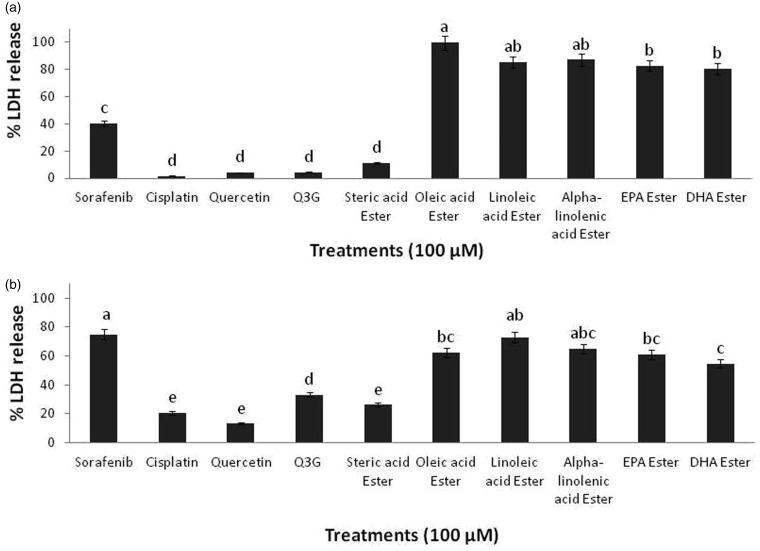
Further, to evaluate the potential cytotoxic effects of long chain fatty acid esters of Q3G on HepG2 cells, LDH release assay was performed. This method estimates the cell viability by measuring the release of LDH enzyme from the cell membrane upon loss of membrane integrity. Incubation of HepG2 cells with 100 µM of long chain fatty acid esters of Q3G for 6 h and 24 h resulted in significant increase in LDH enzyme release (P < 0.05) in comparison to the precursor compounds quercetin and Q3G and control drugs, Sorafenib and Cisplatin (Figure 2). The LDH release data was consistent with the MTS data. Within 6 h of incubation, all long chain fatty acid esters of Q3G except stearic acid ester of Q3G treated cells showed a strong increase in LDH release as compared to the respective controls (Figure 2(a)). On the other hand, after 24 h of incubation, LDH release was seen relatively lower than 6 h incubation data (Figure 2(b)). This may be explained through the fact that LDH has a half-life of 8–9 h23,24 and since the compounds cause a significant amount (over 85%) of cell death within 6 h, by 24 h LDH gets degraded in the medium and hence 24 h incubation readings were seen comparatively lesser than 6 h readings. Nevertheless, the LDH release which signifies the cytotoxic extent of the long chain fatty acid esters of Q3G was significantly greater than the precursor compounds and control cancer drug, Cisplatin (P < 0.05) (Figure 2(b)). Interestingly, oleic acid ester of Q3G appeared to be the most effective compound showing the greatest inhibitory action (over 95%), while all other long chain fattyacid esters of Q3G except stearic acid ester showed relatively lesser but significant inhibitory action (over 85%).
Figure 2.
Cytotoxicity effects of long chain fatty acid esters of Q3G on HepG2 cells. The figure describes the percentage of LDH release from HepG2 cells after treatment with the test compounds. Cells (5 × 103 cells per well; 96-well plate) were treated with 100 µM of the test compounds for 6 h (a) and 24 h (b). After treatment, the cells were centrifuged and the percentage of LDH release from the cells was determined by LDH assay. All long chain fatty acid esters of Q3G except stearic acid ester showed over 80% cell death via LDH release within 6 h of treatment. Mean separation between groups was conducted using Tukey's test (n = 6). Significance level was taken at P value < 0.05
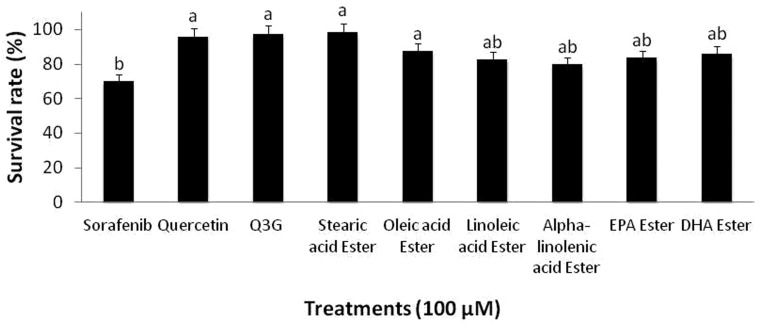
To assess the cytotoxicity effect of the long chain fatty acid esters of Q3G on normal hepatocytes, 100 µM long chain fatty acid esters of Q3G esters were incubated with normal hepatocytes and MTS assay was performed as described earlier. The results showed a significantly higher viability in long chain fatty acid esters of Q3G-treated normal hepatocytes compared to the long chain fatty acid esters of Q3G-treated transformed HepG2 cells. Additionally, the long chain fatty acid esters of Q3G-treated normal hepatocytes showed significantly higher viability than Sorafenib (Figure 3).
Figure 3.
Effect of long chain fatty acid esters of Q3G on viability of normal hepatocytes. Cells (1 × 104 cells per well; 96-well plate) were treated with 100 µM of the test compounds for 24 h. After treatment, viable cell percentage was determined by MTS assay. Results are expressed relative to the control (24 h incubation without test compounds). Mean separation between groups was conducted using Tukey's test (n = 6). Significance level was taken at P value < 0.05
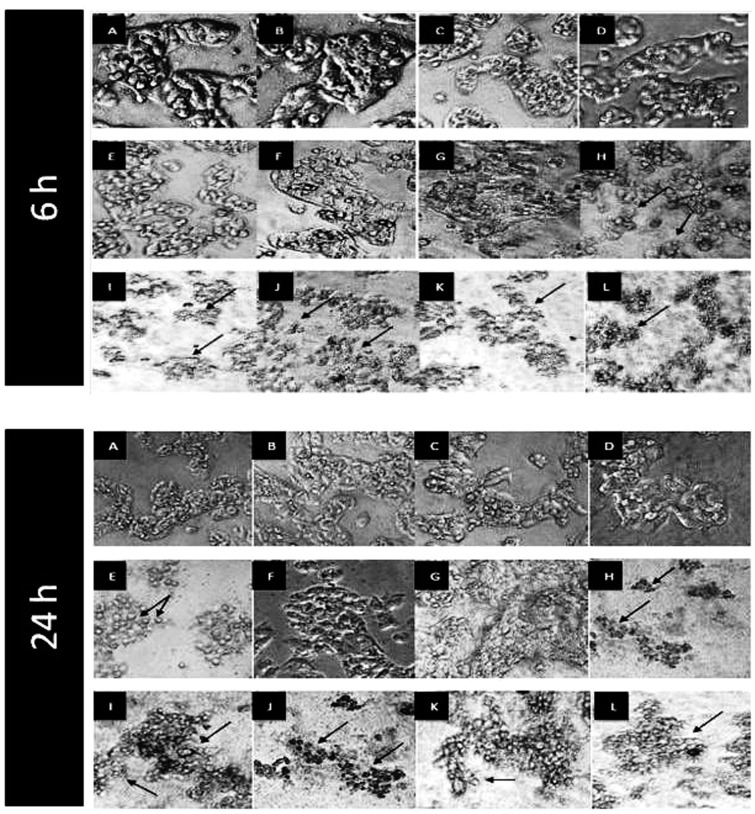
Long chain fatty acid esters of Q3G cause a drastic change in cellular morphology of HepG2 cells with decreased cell number
HepG2 cells were cultured in squared chamber slides and incubated with 100 µM of long chain fatty acid esters ofQ3G, precursor compounds (quercetin and Q3G), Sorafenib and Cisplatin for 6 h and 24 h. After incubation, cells were observed and photographed using phase contrast microscope and observation at 6 h showed a dramatic decrease in cell number of the cells treated with long chain fatty acid esters of Q3G as compared to the control cells (Figure 4). Interestingly, the long chain fatty acid esters of Q3G-treated HepG2 cells appeared to be undergoing an excessive morphology change in comparison to the precursor compounds, the drugs and the control. This was accompanied by a very low cell number. Cell membrane shrinkage, blebbing, clustering and lysis were some prominent visible features (Figure 4). Similarly, severe changes in cell number and morphology were observed after 24 h of treatment as well (Figure 4). However, consistent with above results, stearic acid ester of Q3G failed to show any major effect on cell number and morphology.
Figure 4.
Morphological changes in HepG2 cells after 6 h and 24 h of treatment with long chain fatty acid esters of Q3G. Cells (1 × 104 cells per well; 6-well plate) were treated with 100 µM of test compounds for 6 h and 24 h. After incubation, cells were observed and photographed using Nikon eclipse TS 100 phase contrast microscope equipped with Infinity 1 camera at 10 × magnification. The arrows in the pictures show the change in the morphology of cells upon treatments. As compared to the control, the long chain fatty acid esters of Q3G treated HepG2 cells showed a great decrease in cell number and complete loss of morphology. (a) No treatment control, (b) DMSO control (0.1%), (c) quercetin, (d) Q3G, (e) Sorafenib, (f) Cisplatin, (g) stearic acid ester of Q3G, (h) oleic acid ester of Q3G, (i) linoleic acid ester of Q3G, (j) alpha-linolenic acid ester of Q3G, (k) EPA ester of Q3G, and (l) DHA ester of Q3G
Long chain fatty acid esters of Q3G induce apoptosis in HepG2 cells via activation of caspase-3 family
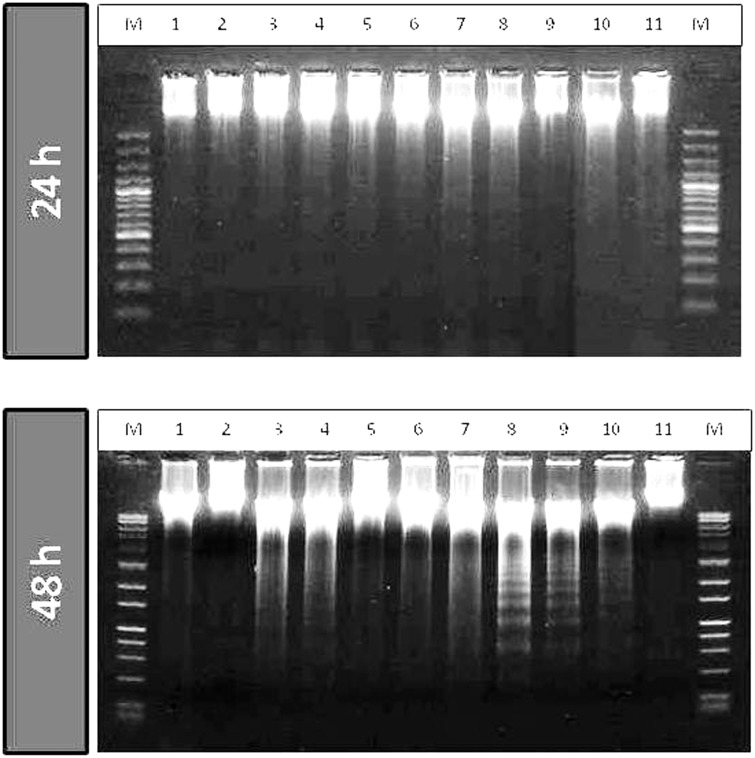
Intense morphological changes were observed under the phase contrast microscope. Further assessments were carried out to examine whether the cells were getting growth arrested upon treatment with the long chain fatty acid esters of Q3G with a possible induction of apoptosis. DNA fragmentation assay was performed as explained in materials and methods. After incubation of cells with 100 µM long chain fatty acid esters of Q3G, precursor compounds and control drugs for 24 h and 48 h, DNA was extracted and resolved on 1.2% agarose gel. The long chain fatty acid esters of Q3G treated cells showed DNA fragments within 24 h as seen on the gel image. The intensity of fragmentation increased at 48 h incubation (Figure 5). This result confirmed one of the basic hallmarks of apoptosis by the novel esters of Q3G.
Figure 5.
DNA fragmentation in HepG2 cells after 24 h and 48 h of treatment with long chain fatty acid esters of Q3G. Cells (5 × 105 cells; 12-well plate) were treated with 100 µM of the test compounds for 24 h and 48 h. Cells were collected, lysed and DNA was extracted and run on agarose gel containing GelRed™ DNA staining solution for fragmentation analysis. Lane M, DNA marker; lane 1, Cisplatin; lane 2, stearic acid ester of Q3G; lane 3, oleic acid ester of Q3G; lane 4, linoleic acid ester of Q3G; Lane 5, alpha-linolenic acid ester of Q3G; lane 6, EPA ester of Q3G; lane 7, DHA ester of Q3G; lane 8, Q3G; lane 9, quercetin; lane 10, Sorafenib; lane 11, control (no treatment). After 48 h of treatment all long chain fatty acid esters of Q3G except stearic acid ester treatment showed substantial amount of apoptosis as seen by the fragmented DNA pattern
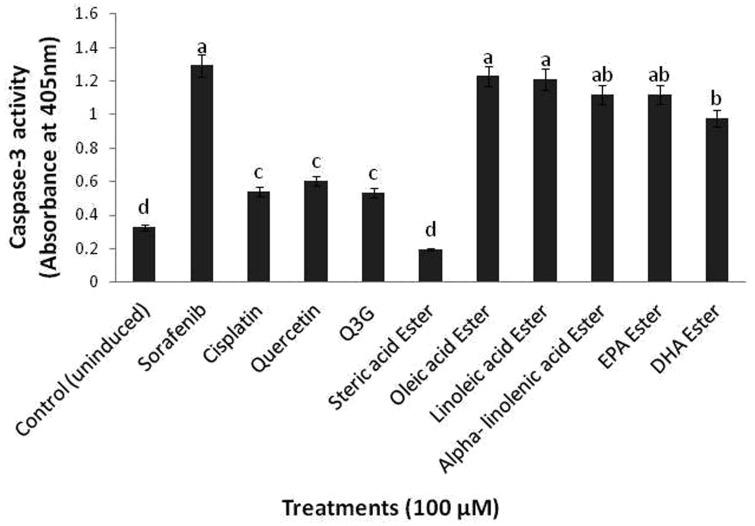
DNA fragmentation data further prompted us to examine the mechanism of apoptosis induction caused by the long chain fatty acid esters of Q3G. As caspase-3 of proteases is the principle effector caspases which leads cells to apoptosis,25 we examined its activity and level of expression upon long chain fatty acid esters of Q3G treatment. After 24 h of treatment of HepG2 cells with 100 µM of long chain fatty acid esters of Q3G, a significant increase in the caspase-3 activity was observed as compared to the control untreated cells (Figure 6). This relative change in caspase-3 activity was consistent with the MTS and LDH release assays. Consistent with above results oleic ester of Q3G showed the greatest caspase-3 activity among the other esters of Q3G.
Figure 6.
Caspase-3 activation by long chain fatty acid esters of Q3G. Cells (1 × 106 cells/well) were incubated with the test compounds in a six well plate for 24 h. Cells were lysed and protein was quantified. After quantification, 250 µg of protein was used for detection of Caspase-3 activity. Absorbance was taken at 405 nm. The long chain fatty acid esters of Q3G except stearic acid ester of Q3G showed significantly high caspase activity as compared to the parent compounds and the cancer drug Cisplatin (P value < 0.05). Mean separation between groups was conducted using Tukey's test (n = 3). Significance level was taken at P value < 0.05
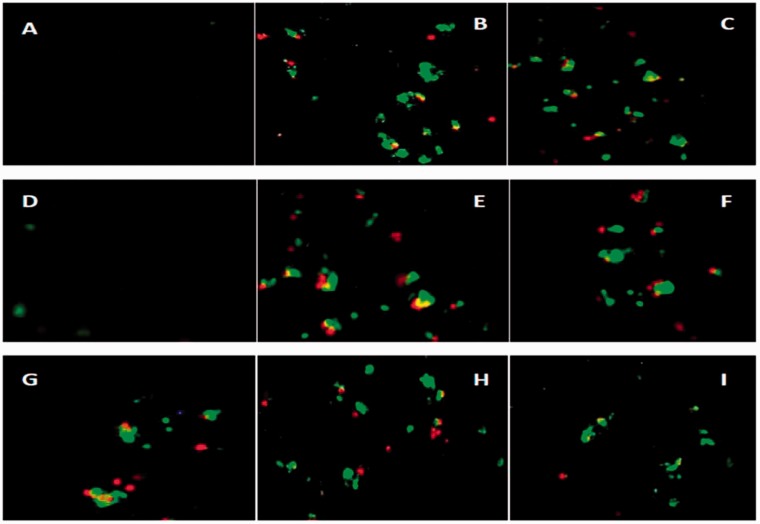
Further to analyze whether the long chain fatty acid esters of Q3G-induced inhibitory effects on cell growth and the morphological changes observed were due to apoptosis or necrosis (or both), the cells were examined by Annexin V FITC and PI staining after treatment for 24 h with 100 μM long chain fatty acid esters of Q3G via fluorescent microscopy (Figure 7). The untreated cells did not show any staining, suggesting that these cells did not undergo significant apoptosis or necrosis. Long chain fatty acid esters of Q3G treated cells displayed some staining for both Annexin V (green) and PI (yellow) which signifies late apoptotic cells and for PI only (red) which signifies necrotic cells and more staining for Annexin V only (green) which signifies apoptotic cells except for stearic acid ester of Q3G which did not show apparent staining. This data suggests that the inhibitory effects of the long chain fatty acid esters of Q3G are in fact due to the induction of apoptosis further leading to late apoptosis.
Figure 7.
Apoptosis detection through fluorescence microscopy. Cells were treated for 24 h with 100 μM long chain fatty acid esters of Q3G (d, e, f, g, h, and i) and 100 μM Sorafenib (c) in complete medium. After staining with Annexin V and PI, necrotic and apoptotic cells were detected by fluorescence microscopy (20×). (a) control (no treatment), (b) positive control, (c) Sorafenib, (d) stearic acid ester, (e) oleic acid ester, (f) linoleic acid ester, (g) alpha-linolenic acid ester, (h) EPA ester and (i) DHA ester. (A color version of this figure is available in the online journal.)
Long chain fatty acid esters of Q3G cause alterations in HepG2 cell cycle progression via induction of S-phase arrest
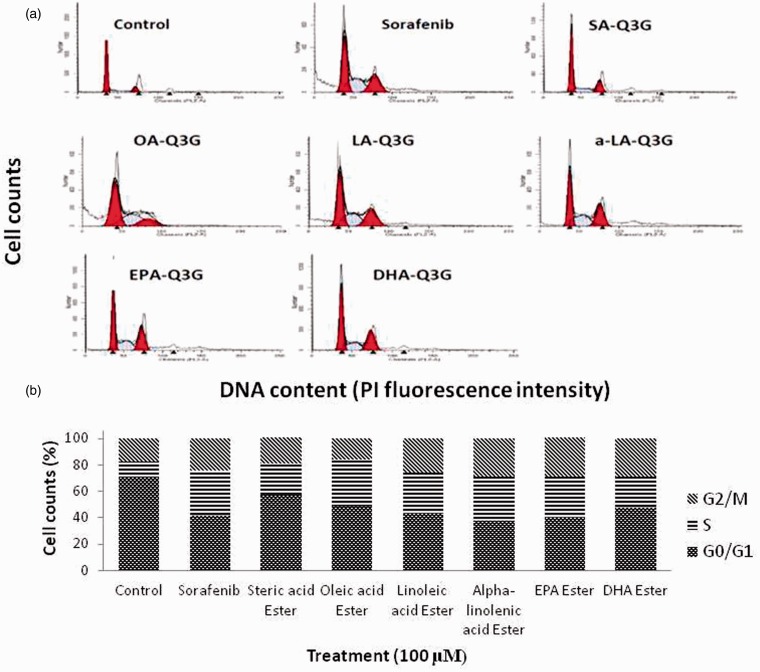
To assess whether long chain fatty acid esters of Q3G esters-induced cell growth inhibition is mediated through changes in cell cycle progression, asynchronous cells were incubated with 100 μM of long chain fatty acid esters of Q3G and control drug Sorafenib for 24 h. The effect of long chain fatty acid esters of Q3G on cell cycle phase distribution was determined by flow cytometry analysis. A representative histogram is shown in Figure 8(a), and the data are summarized in Figure 8(b). Consistent with their growth inhibitory effects, the long chain fatty acid esters of Q3G increased the population in the S phase with a corresponding decrease of cells in the G1 phase after 24 h of treatment, implying that the DNA synthesis was retarded. In addition to increasing the population of cells in the S phase from 11.1% (control) to 35.4% (Oleic acid ester of Q3G), long chain fatty acid esters of Q3G also appeared to increase the cell population in the G2-M phase, implying the cell mitosis stage was inhibited for the cells that managed to move from S phase to G2-M phase. These data support the potent inhibitory effect of long chain fatty acid esters of Q3G on DNA synthesis and possibly in parts, cell mitosis.
Figure 8.
Effect of long chain fatty acid esters of Q3G on cell cycle distribution of HepG2 cells. After treatment with 100 μM of the test compounds for 24 h in complete medium, cells were fixed and stained with propidium iodide, and the cell cycle distribution was analyzed by flow cytometry. (a) Representative DNA histograms of the flow cytometric analysis are shown for control and each treatment. (b) The percentage of cells in G1, S, and G2/M phases was calculated and is summarized as a bar graph of the mean values (n = 3). (A color version of this figure is available in the online journal.)
SA-Q3G: Stearic acid ester of Q3G; OA-Q3G: Oleic acid ester of Q3G; LA-Q3G: Linoleic acid ester of Q3G; a-LA-Q3G: alpha-linolenic acid ester of Q3G; EPA-Q3G: EPA ester of Q3G; DHA-Q3G: DHA ester of Q3G
Long chain fatty acid esters of Q3G behave as potent topoisomerase II inhibitor
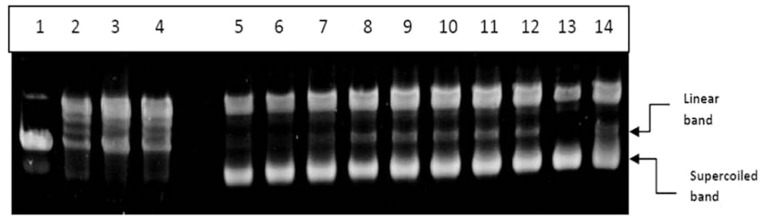
DNA topoisomearse II is essential for cell division and proliferation as it marks the completion of mitosis. To further examine whether the long chain fatty acid esters of Q3G were also able to catalytically inhibit DNA topoisomerase II activity which could in turn be a reason to activate caspases and drive cells to apoptosis, DNA topoisomerase II catalytic activity was monitored via gel electrophoresis. A representative gel image of the relaxation assay for determining the catalytic inhibition on human DNA topoisomerase II by the long chain fatty acid esters of Q3G (100 µM) and controls is presented in Figure 9. The figure presents the pHot1 DNA linear marker (lane 1) which is diagnostic for topoisomearse inhibitor (poison), relaxed pHot1 DNA (lane 2), positive control drug VP16 showing linear band (lane 3), negative control DMSO showing relaxed bands (lane 4), long chain fatty acid esters of Q3G with topoisomearse II (lane 5 to 10), quercetin and Q3G + topoisomearse II (lane 11 and 12 respectively), supercoiled substrate pHot1 DNA (lane 13) and Sorafenib + topoisomearse II (lane 14). The assay was performed to test whether long chain fatty acid esters of Q3G act as a poison and increase the DNA cleavage via topoisomearse II. As shown in Figure 9, the esters did not stabilize topoisomerase II cleavage complexes and failed to exhibit the formation of single linear DNA, and increased the supercoiled DNA intensity, whereas, positive control drug VP 16 increased the formation of linear DNA. This result suggests that long chain fatty acid esters of Q3G do not act as human topoisomearse II poison but as a catalytic inhibitor by inhibiting the DNA relaxation activity. The intensity of the supercoiled bands in comparison to the negative control (DMSO) appeared very high indicating the inability of DNA topoisomearse II to relax the supercoiled DNA (Figure 9).
Figure 9.
DNA topoisomerase II activity. Supercoiled circular pHot1 DNA (0.25 µg) was taken in test and control tubes and incubated with 4 units of the DNA topoisomearse II enzyme. The reactions were kept at 37℃ for 30 min followed by 1% agarose gel run without DNA stain. The gel was stained with GelRed™ DNA staining solution for 2 h and then destained with TBE buffer for 15 to 20 min. The gel was imaged in BioRad Gel doc system. Lane 1, Linear DNA marker; Lane 2, DNA + topo II; Lane 3, VP-16 + DNA + topo II; Lane 4, DMSO control; Lane 5, stearic acid ester + DNA + topoII; Lane 6, oleic acid ester; Lane 7, linoleic acid ester + DNA + topo II; Lane 8, alpha-linolenic acid ester + DNA + topoII; Lane 9, EPA ester + DNA + topoII; Lane 10, DHA ester + DNA + topoII; Lane 11, Q3G + DNA + topo II; Lane 12, quercetin + DNA + topo II; Lane 13, Control (no topo); Lane 14- Sorafenib + DNA + topo II
Discussion
Flavonoids are polyphenolic plant secondary metabolites which have been shown by both epidemiological and in vitro studies to have strong antioxidant, antiproliferative and other biological activities beneficial to human health.26 Over the years, these properties of flavonoids have gained a lot of interest in scientific research to use them as drug leads. Antiproliferative property of flavonoids has prompted research throughout the world to develop alternative natural medicines to substitute synthetic chemotherapeutic agents which apparently are known to have many side effects. Quercetin and its glycosides have been studied extensively for their anti-cancerous and toxicological properties in various transformed cell lines making them promising candidates for cancer therapeutics.26 However, poor bioavailability limits their biological effects in vivo,13 due to low membrane permeability and hence, limits their applications as therapeutic agents.27 One of the effective approaches to enhance the membrane permeability is to increase the lipophilicity of these compounds by acylation with fatty acids.15 Q3G was acylated with long chain fatty acid esters as previously described in Ziaullah et al.18 In the current study, we investigated the antiproliferative and cytotoxic properties of six long chain fatty acid esters of Q3G to investigate whether the acylation enhances its biological action and finally to elucidate the mechanism of action of the novel compounds.
Over the years, natural product chemists have been focused essentially on the ability of flavonoids to influence cell cycle in cancer cells and driving cells to apoptosis. This makes cell cycle arrest and apoptosis induction a significant preventive approach. In this study, we showed that the novel synthesized long chain fatty acid esters of Q3G can inhibit liver cancer cell proliferation (HepG2) through induction of apoptosis by the activation of caspase-3 family followed by necrosis, through cell cycle changes, and possibly through inhibition of DNA topoisomerase II activity. Interestingly, as hypothesized, long chain fatty acid esters of Q3G exhibited much stronger anti-proliferative property than precursor compounds (quercetin, Q3G and free fatty acids) and two prescribed chemotherapy drugs, Sorafenib and Cisplatin. The long chain fatty acid esters of Q3G inhibited proliferation of HepG2 cells within 6 h of incubation in comparison to quercetin, Q3G, free fatty acids and chemo drugs at the similar concentration (100 µM). The cell proliferation was shown to further reduce by 24 h of incubation (Figure 1(a) and (b)). Also, at lower concentrations of 10, 30 and 50 µM the esters, 48 to 72 h incubations were necessary to obtain a significant reduction in cell viability (data not included). Based on the effects on cell viability and morphology, our data suggested that the test compounds caused cytotoxicty to the HepG2 cells resulting in the cell membrane shrinkage and eventually breakage (Figure 4). This result was further assessed by the membrane integrity test via LDH release assay which showed that there was clear membrane breakage when compared with untreated control cells (Figure 2(a) and (b)). Interestingly, the strong inhibition of cell proliferation by the fatty acid esters of Q3G when compared to the precursor compounds alone and the chemotherapy drugs is noteworthy (Figures 1 and 2).
Interestingly, oleic acid ester of Q3G appeared to show the strongest antiproliferative action whereas, stearic acid ester of Q3G showed the least growth inhibitory action among all the tested esters of Q3G. The experimental results support the assumption that there is a structure-activity relationship due to the fact that stearic acid is the only saturated fatty acid among the six fatty acids used for acylation of Q3G. Once the stearic acid is attached to the Q3G skeleton, the change in the orientation may not be favourable for membrane interaction thereby, getting less absorbed by cells and subsequently showing less activity. Overall, these data suggested the potential of long chain fatty acid esters of Q3G as strong antiproliferative agents. Interestingly, the precursor compounds (quercetin and Q3G) that have been shown to display strong antiproliferative action by previous studies are in fact concentration and time dependent. The peak growth inhibitory action displayed by the precursor compounds has usually been shown to range from 48–72 h.12,20 In this study we showed that the long chain fatty acid esters of Q3G display the growth inhibitory effect on HepG2 cells within 6 h of treatment (Figures 1 and 2). This data revealed that acylation of Q3G with unsaturated long chain fatty acid esters enhances its antiproliferative activity in vitro.
The long chain fatty acid esters of Q3G also showed significantly lower cytotoxic effect to normal hepatocytes as compared to the transformed HepG2 (Figure 3) suggesting their specific action on HepG2 cells. Additionally, fluorescence microscopy showed cell membrane breakage suggesting symptoms of late apoptosis and necrosis. However, distinction between the apoptosis and necrosis is very challenging and could be difficult to confirm as other mechanisms of various death routes and symptoms of cell death may involve common molecular target.28 To distinguish between apoptosis and necrosis, cells were analyzed after staining with Annexin V and PI through fluorescence microscopy. After 24 h of treatment of HepG2 cells with long chain fatty acid esters of Q3G showed that, some treated cells were positive for PI and some for both Annexin V and PI (Figure 7), suggesting that the cells at this time present necrotic and also apoptotic features presumably late apoptosis. This finding may be explained if treatment with the fatty acid esters of Q3G within 6 h triggers the activation of apoptotic proteins for phagocytosis and gradually with the overload of dying cells early apoptotic cells will progress to late apoptosis or secondary necrosis, where the membrane becomes more permeable weakening phagocytosis.29 Alternatively, this could also be explained as apoptosis, the programmed cell death taking over the necrosis caused by the initial response of HepG2 cells to incubation of long chain fatty acid esters of Q3G.
To further analyse and confirm the apoptotic effect of the Q3G esters, we examined the caspase-3 activity which is a key enzyme in apoptotic signalling.25 Consistent with the fluorescent data, a significant activation of caspase-3 was observed in long chain fatty acid esters of Q3G treated cells in comparison to the un-induced control and precursor compounds after 24 h of incubation (Figure 6). Additionally, these data were supported by the DNA fragmentation analysis. After 24 h and 48 h of treatment of HepG2 cells with long chain fatty acid esters of Q3G, the fragmented DNA like pattern was observed (Figure 5) which is a basic hallmark of apoptosis.30 These data suggested the apoptotic action of our novel long chain fatty acid esters of Q3G on HepG2 cells.
Cell cycle analysis showed that long chain fatty acid esters of Q3G blocked the HepG2 cells in the S phase (Figure 8), this effect being related with the possible inhibition of DNA synthesis. Previous studies have shown that quercetin can induce cell cycle arrest in the S phase31 as well as in the G1/S phase32 or G2/M phase33 depending on the cancer cell type. Apparently, a considerable increase in cell population in G2/M phase was also seen in the long chain fatty acid esters of Q3G treated HepG2 cells (Figure 8). This suggests that the cells that were in fact able to cross S phase got arrested in G2/M phase. These data are thus in accordance with the previous studies that the cell cycle alterations are cell type and treatment dependent.
DNA topoisomearse II is essential for the basic cellular function and proliferation including mitosis and has been shown to be expressed in much higher levels in proliferating tumor cells34 which make it a promising target for chemotherapy drugs. Additionally, apoptosis has been shown to be the most efficient death-pathway in tumor cells after topoisomerase II inhibition.35 This prompted us to think that whether long chain fatty acid esters of Q3G were causing topoisomerase II inhibition which possibly was driving cells to apoptosis. As expected, our data showed that long chain fatty acid esters of Q3G are behaving as strong topoisomearse II inhibitors. It was also interesting to note that stearic acid ester of Q3G which failed to show any significant response in any of the assays showed a significant reduction in topoisomerase II activity. This shows that the compound is an active and potent topoisomerase II inhibitor but since the drug screening topoisomerase II assay was a cell free assay, stearic acid ester of Q3G showed its direct inhibitory action without the interaction with the cells or cellular membranes. Nonetheless, a reduction in DNA topoisomerase II activity observed with a reduced ability to relax the supercoiled DNA, indicates that the reduced cell proliferation may be, in parts, a result of DNA topoisomearse II inhibition which may have generated double strand breaks in DNA. It is also possible that DNA topoisomearse II-mediated DNA damage activated the cell cycle checkpoint causing growth arrest which in turn triggered and activated apoptotic signalling through caspase-3 and hence caused cell death which agrees with the literature.36 Apoptosis is regulated via the action of several oncogenes and subsequently oncoproteins that display inhibiting or promoting action. The bax protein is a member of the bcl-2 family that promotes apoptosis. The ratio of bax to bcl-2 determines the susceptibility of a cell to apoptosis. Therefore, future investigations should be continued to identify the mechanism of activation of caspase-3 and whether the long chain fatty acid esters of Q3G induced apoptosis is involved in intrinsic- or extrinsic-pathway. Antiproliferative effect of a similar molecule, decosahexaenoic acid ester of phloridzin, has been shown to down-regulate anti-apoptotic gene (BCL2), growth factor receptors (EBFR family, IGF1R/IGF2, PDGFR) and its downstream signalling partners (PI3k/AKT/mTOR, Ras/Raf/MAPK), cell cycle machinery (CDKs, TERT, TOP2A, TOP2B) as well as epigenetics regulators (HDACs).37
Conclusions
In conclusion, the novel unsaturated long chain fatty acid esters of Q3G exhibit antiproliferative action on liver cancer cells (HepG2) and very low cytotoxic action on normal cells as compared to the precursor compounds and currently used chemotherapy drugs at the same concentration. Additional studies using other established transformed cell lines should be performed to confirm the anti-proliferative effects on other cancer types.
Acknowledgement
This research was supported by Canada Research Chair program and the Discovery Grant program of the Natural Sciences and Engineering Research Council (NSERC) of Canada.
Author contributions
SS conducted the experiments, analyzed, and interpreted data and drafted the manuscript. HPVR is the principal investigator who generated the funds, designed the overall experimental approach, and reviewed the article for content.
References
- 1.Rupasinghe HPV, Nair S, Robinson R. Chemopreventive properties of fruit phenolics and their possible mode of actions. In: Atta-Ur-Rahman (ed.). Studies in natural products chemistry 2014; Vol 42Amsterdam: Elsevier Science Publishers, pp. 229–66. [Google Scholar]
- 2.Arts IC, Hollman PC, De Mesquita HB, Feskens EJ, Kromhout D. Dietary catechins and epithelial cancer incidence: the Zutphen elderly study. Int J Cancer 2001; 92: 298–302. [DOI] [PubMed] [Google Scholar]
- 3.Knekt P, Kumpulainen J, Järvinen R, Rissanen H, Heliövaara M, Reunanen A, Hakulinen T, Aromaa A. Flavonoid intake and risk of chronic diseases. Am J Clin Nutr 2002; 76: 560–8. [DOI] [PubMed] [Google Scholar]
- 4.Le Marchand L, Murphy SP, Hankin JH, Wilkens LR, Kolonel LN. Intake of flavonoids and lung cancer. J Natl Cancer Inst 2000; 92: 154–60. [DOI] [PubMed] [Google Scholar]
- 5.Shen SC, Chen YC, Hsu FL, Lee WR. Differential apoptosis-inducing effect of quercetin and its glycosides in human promyeloleukemic HL-60 cells by alternative activation of the caspase 3 cascade. J Cell Biochem 2003; 89: 1044–55. [DOI] [PubMed] [Google Scholar]
- 6.Singhal R, Yeh YA, Praja N, Olah E, Slege GW, Weber G. Quercetin down-regulates signal transduction in human breast carcinoma cells. Biochem Biophys Res Commun 1995; 208: 425–31. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen T, Tran E, Nguyen T, Do PT, Huynh TH, Huynh H. The role of activated MEK-ERK pathway in quercetin-induced growth inhibition and apoptosis in A549 lung cancer cells. Carcinogenesis 2004; 25: 647–59. [DOI] [PubMed] [Google Scholar]
- 8.Agullo G, Gamet-Payrastre L, Manenti S, Viala C, Remesy C, Chap H, Payrastre B. Relationship between flavonoid structure and inhibition of phosphatidylinositol 3-kinase: a comparison with tyrosine kinase and protein kinase C inhibition. Biochem Pharmacol 1997; 53: 1649–57. [DOI] [PubMed] [Google Scholar]
- 9.Granado-Serrano AB, Martín MA, Bravo L, Goya L, Ramos S. Quercetin induces apoptosis via caspase activation, regulation of Bcl-2, and inhibition of PI-3-kinase/Akt and ERK pathways in a human hepatoma cell line (HepG2). J Nutr 2006; 136: 2715–21. [DOI] [PubMed] [Google Scholar]
- 10.Lanoue L, Green K, Kwik-Uribe C, Keen CL. Dietary factors and the risk for acute infant leukemia: evaluating the effects of cocoa-derived flavanols on DNA topoisomerase activity. Exp Biol Med 2010; 235: 77–89. [DOI] [PubMed] [Google Scholar]
- 11.Razavi SM, Zahri S, Zarrini G, Nazemiyeh H, Mohammadi S. Biological activity of quercetin-3-O-glucoside, a known plant flavonoid. Bioorg Khim 2009; 35: 414–6. [DOI] [PubMed] [Google Scholar]
- 12.Sudan S, Rupasinghe HPV. Quercetin-3-O-glucoside induces human DNA topoisomerase II inhibition, cell cycle arrest and apoptosis in hepatocellular carcinoma cells. Anti-cancer Res 2014; 34: 1691–9. [PubMed] [Google Scholar]
- 13.Passamonti S, Terdoslavich M, Franca R, Vanzo A, Tramer F, Braidot E, Petrussa E, Vianello A. Bioavailability of flavonoids: a review of their membrane transport and the function of bilitranslocase in animal and plant organisms. Curr Drug Metabol 2009; 10: 369–94. [DOI] [PubMed] [Google Scholar]
- 14.Lin YT, Hsiu SL, Hou YC, Chen HY, Lee CP. Degradation of flavonoid aglycones by rabbit, rat and human fecal flora. Biol Pharm Bull 2003; 26: 747–51. [DOI] [PubMed] [Google Scholar]
- 15.Salem J, Chevalot I, Schiavo CH, Paris C, Fick M, Humeau C. Biological activities of flavonoids from Nitraria retusa (Forssk.) Asch. and their acylated derivatives. Food Chem 2011; 124: 486–94. [Google Scholar]
- 16.Mainou-Fowler T, Proctor SJ, Dickinson AM. Gamma-linolenic acid induces apoptosis in B-chronic lymphocytic leukaemia cells in vitro. Leuk Lymph 2001; 40: 393–403. [DOI] [PubMed] [Google Scholar]
- 17.Hardman WE. Omega-3 fatty acids to augment cancer therapy. J Nutr 2002; 132: 3508–12. [DOI] [PubMed] [Google Scholar]
- 18.Ziaullah, Bhullar KS, Warnakulasuriya SN, Rupasinghe HPV. Biocatalytic synthesis structural elucidation, antioxidant capacity and tyrosinase inhibition activity of long chain fatty acid acylated derivatives of phloridzin and isoquercitrin. Bioorg Med Chem 2013; 21: 684–92. [DOI] [PubMed] [Google Scholar]
- 19.Talib W, Mahasneh A. Antiproliferative activity of plant extracts used against cancer in traditional medicine. Sci Pharm 2010; 78: 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shan B, Wang M, Li R. Quercetin inhibit human SW480 colon cancer growth in association with inhibition of cyclin D1 and survivin expression through Wnt/β-catenin signaling pathway. Cancer Invest 2009; 27: 604–12. [DOI] [PubMed] [Google Scholar]
- 21.Patra N, De U, Kang JA, Kim JM, Ahn MY, Lee J. A novel epoxypropoxy flavonoid derivative and topoisomerase II inhibitor, MHY336, induces apoptosis in prostate cancer cells. Eur J Pharmacol 2011; 658: 98–107. [DOI] [PubMed] [Google Scholar]
- 22.Van de Loosdrecht AA, Beelen RHJ, Ossenkoppele GJ, Broekhoven MG, Langenhuijsen MM. A tetrazolium-based colorimetric MTT assay to quantitate human monocyte mediated cytotoxicity against leukemic cells from cell lines and patients with acute myeloid leukemia. J Immunol Methods 1994; 174: 311–20. [DOI] [PubMed] [Google Scholar]
- 23.Lisa KM. Handbook of assay development in drug discovery. 2004: Series 5. 385–406.
- 24.Riss TL, Moravec RA. Use of multiple assay endpoints to investigate the effects of incubation time, dose of toxin, and plating density in cell-based cytotoxicity assays. Assay Drug Dev Technol 2004; 2: 51–62. [DOI] [PubMed] [Google Scholar]
- 25.Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol 2004; 5: 897–907. [DOI] [PubMed] [Google Scholar]
- 26.Gibellini L, Pinti M, Nasi M. Quercetin and cancer chemoprevention. Evid Based Complement Alternat Med 2011, pp. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitagawa S, Tanaka Y, Tanaka M, Endo K, Yoshii A. Enhanced skin delivery of quercetin by microemulsion. J Pharm 2009; 61: 855–60. [DOI] [PubMed] [Google Scholar]
- 28.Bröker LE, Kruyt FAE, Giaccone G. Cell death independent of caspases: a review. Clin Cancer Res 2005; 11: 3155–62. [DOI] [PubMed] [Google Scholar]
- 29.Poon IKH, Hulett MD, Parish CR. Molecular mechanisms of late apoptotic/necrotic cell clearance. Cell Death Differ 2010; 17: 381–97. [DOI] [PubMed] [Google Scholar]
- 30.Saraste A, Pulkki K. Morphologic and biochemical hallmarks of apoptosis. Cardiovasc Res 2000; 45: 528–37. [DOI] [PubMed] [Google Scholar]
- 31.Cheong E, Ivory K, Doleman J, Parker ML, Rhodes A. Synthetic and naturally occurring COX-2 inhibitors suppress proliferation in a human oesophageal adenocarcinoma cell line (OE33) by inducing apoptosis and cell cycle arrest. Carcinogenesis 2004; 25: 1945–52. [DOI] [PubMed] [Google Scholar]
- 32.Hosokawa N, Hosokawa Y, Sakai T, Yoshida M, Marui N. Inhibitory effect of quercetin on the synthesis of a possibly cell-cycle-related 17-kDa protein, in human colon cancer cells. Int J Cancer 1990; 45: 1119–24. [DOI] [PubMed] [Google Scholar]
- 33.Choi J-A, Kim JY, Lee JY, Kang CM, Kwon HJ. Induction of cell cycle arrest and apoptosis in human breast cancer cells by quercetin. Int J Oncol 2001; 19: 837–44. [DOI] [PubMed] [Google Scholar]
- 34.Coss A, Tosetto M, Fox EJ, Sapetto-Rebow B, Gorman S, Kennedy BN, Lloyd AT, Hyland JM, O'Donoghue DP, Sheahan K, Leahy DT, Mulcahy HE, O'Sullivan JN. Increased topoisomerase IIα expression in colorectal cancer is associated with advanced disease and chemotherapeutic resistance via inhibition of apoptosis. Cancer Lett 2009; 276: 228–38. [DOI] [PubMed] [Google Scholar]
- 35.El-Awady A, Ali MM, Saleh EM, Ghaleb FM. Apoptosis is the most efficient death-pathway in tumor cells after topoisomerase II inhibition. Saudi Med J 2008; 29: 558–64. [PubMed] [Google Scholar]
- 36.Niida H, Nakanishi M. DNA damage checkpoints in mammals. Mutagenesis 2006; 21: 3–9. [DOI] [PubMed] [Google Scholar]
- 37.Nair SVG, Ziaullah, Rupasinghe HPV. Fatty acid esters of phloridzin induce apoptosis of human liver cancer cells through altered gene expression. PLoS ONE 2014; 9: e107149. [DOI] [PMC free article] [PubMed] [Google Scholar]