Abstract
The regulation of hypoxia-inducible factor-1 (HIF-1) transcriptional activity in the nucleus is related to factor inhibiting HIF-1 (FIH-1). FIH-1 hydrolyzes asparagine at the C-terminal of HIF-1α, preventing the interaction between HIF-1α and its associated cofactors, and leading to suppressed activation of HIF-1. FIH-1 is a cytosolic protein and its entry to the nucleus has to be coordinated with HIF-1α. The present study was undertaken to examine the correlation between HIF-1α and FIH-1 in their nuclear entry. Human umbilical vein endothelial cells were treated with dimethyloxalylglycine at a final concentration of 100 µM for 4 h, resulting in an accumulation of HIF-1α and an increase of FIH-1 in the nucleus as determined by Western blot analysis. Pretreatment of the cells with copper (Cu) chelator tetraethylenepentamine at 50 µM in cultures for 24 h reduced both HIF-1α protein levels and the HIF-1α entry to the nucleus, along with decreased FIH-1 protein levels in the nucleus but no changes in the total FIH-1 protein levels in the cells. These effects were prevented by simultaneous addition of 50 µM CuSO4 with tetraethylenepentamine. Gene-silencing of HIF-1α significantly inhibited FIH-1 entry to the nucleus, but did not affect the total protein levels of FIH-1 in the cells. This work demonstrates that the nuclear entry of FIH-1 depends on HIF-1α. Cu deficiency caused a decrease of HIF-1α, leading to suppression of FIH-1 entry to the nucleus.
Keywords: Copper, tetraethylenepentamine, FIH-1, HIF-1, nuclear entry, gene silencing
Introduction
Factor inhibiting hypoxia-inducible factor-1 (FIH-1)1 is found in both vertebrates and invertebrates extending from human, mouse, and rat to worm and fly.2 FIH-1 has a 2-His-1-carboxylate facial triad of iron binding residues3–6 and is one of the 2-oxolutarate-dependent dioxygenases.7–9 The dioxygenases require Fe2+, 2-oxoglutarate (2-OG), O2, and ascorbate for enzymatic activity.1,3 However, different dioxygenases hydroxylate distinguished residues of hypoxia-inducible factor-1α (HIF-1α).10,11 In brief, FIH-1 hydroxylates an asparagine residue of HIF-1α,1,12 whereas the other dioxygenases, proline hydroxylases (PHDs), hydroxylate the proline residues of HIF-1α.13,14 In addition, FIH-1 functions in the nucleus, regulating the formation of HIF-1 transcription complex, whereas PHDs function in the cytoplasm,12 regulating the stability of HIF-1α.14
Previous studies demonstrated that the transcription activity of HIF-1 requires copper (Cu): one possible mechanism is that Cu inhibits the activity of FIH-1,15 protecting HIF-1α from the hydrolysis of asparagine at its C-terminal in the nucleus. The hydrolysis of asparagine prevents the interaction between HIF-1α and its associated cofactors, such as p300 and ARNT, leading to suppressed HIF-1α activation.16–19 Therefore, Cu inhibition of FIH-1 ensures the formation of HIF-1 transcription complex, leading to HIF-1 activation in the nucleus.15
FIH-1 is mainly localized in cytoplasm; however, a small but significant portion of FIH-1 was also found in nucleus.9 In particular, FIH-1 shuttles between cytoplasm and nucleus under hypoxia and reoxygenation condition.20 But, it remains unclear how FIH-1 enters the nucleus and how its entry coordinates with the nuclear entry of HIF-1α. Therefore, the present study was undertaken to address the correlation between HIF-1 and FIH-1 in their entry to the nucleus, and the effect of Cu on this process.
Materials and methods
Cell culture and treatment
Human umbilical vein endothelial cells (HUVECs, American Type Culture Collection, Virginia, USA) were used because of their role in angiogenesis and the demonstrated function of HIF-1 in these cells.21 Experimental procedures were described previously.22 For varying treatments: cells were seeded in 6-well plates or T25 cell culture flasks, and grown overnight. Thereafter, dimethyloxalylglycine (DMOG, Sigma, St. Louis, USA), tetraethylenepentamine (TEPA, Sigma, St. Louis, USA) or CuSO4 (Kelong, Chengdu, CN) were added to the cultures, followed by further incubation for a period of time as indicated in figure legends or the text of results section.
Nuclear and cytoplasmic extraction
The nucleus and cytoplasm were extracted using NE-PER® nuclear and cytoplasmic extraction reagents (Thermo Scientific, USA) according to the manufacturer’s instruction.
Western blotting analysis of proteins
Protein extracts from nuclei, cytoplasm, and whole cells were prepared as described previously,22 with some modifications. Briefly, 30 µg of protein from each sample was loaded onto a 10% SDS-polyacrylamide electrophoresis gel and transferred to a polyvinylidene difluoride membrane (Bio-RAD, USA). Blots were firstly probed with specific antibodies as follows: mouse anti-human HIF-1α monoclonal antibody (BD Biosciences, San Jose, USA), mouse anti-FIH-1 monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, USA), goat anti-H2AX polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, USA), mouse anti-GAPDH monoclonal antibody (ZSGB-BIO, Beijing, CN) and mouse anti-beta Actin monoclonal antibody (ZSGB-BIO, Beijing, CN). Then the membranes were incubated with a horseradish peroxidase-linked anti-mouse or anti-rabbit IgG antibody (ZSGB-BIO, Beijing, CN), and protein bands were visualized as described previously22 and analyzed by densitometry using Quantity-one 1-D analysis software.
Real-time quantitative RT-PCR
RNA extraction and quantification were performed according to the protocol reported previously.22 The primer sequences were designed as shown below:
| Gene name | Gene sequence | |
|---|---|---|
| HIF-1α | Forward | 5′-GCCGAGGAAGAACTATGAA-3′ |
| Reverse | 3′-CACTGAGGTTGGTTACTGT-5′ | |
| RPS18 | Forward | 5′-TTCGGAATGAGGCCATGAT-3′ |
| Reverse | 3′-TTTCGCTCTGGTCCGTCTTG-5′ | |
Gene silencing of HIF-1α
The siRNA targeting human HIF-1α and mismatched control were designed and synthesized from RiboBio (Guangzhou, China). The siRNA sequences for HIF-1α were as follows: sense, GGCCUCUGUGAUGAGGCUUtt; antisense, AAGCCUCACAGAGGCCtt. Mismatched control siRNA sequences were as follows: sense, UUCUCCGAACGUACGUtt; antisense, ACGUGACACGUUCGGAGAAtt. The optimal transfection efficiency was determined from our preliminary study testing 50 nM or 100 nM, and we selected the condition that the siRNA caused an optimal silencing effect with minimal cytotoxicity. After HUVECs were transfected with 50 nM annealed siRNA targeting human HIF-1α or negative mismatched control siRNA in serum- or antibiotics-free media, a Lipofectamine 2000 reagent (Invitrogen, California, USA) was used as the transfection reagent according to the manufacturer's instruction.
Statistical analysis
Data were obtained from three separate experiments and presented as mean values ± SEM. The significance of differences was determined by one-way ANOVA and further analyzed by Dunnett's T3 test for comparison among multiple groups. The level of significance was considered at P < 0.05.
Results
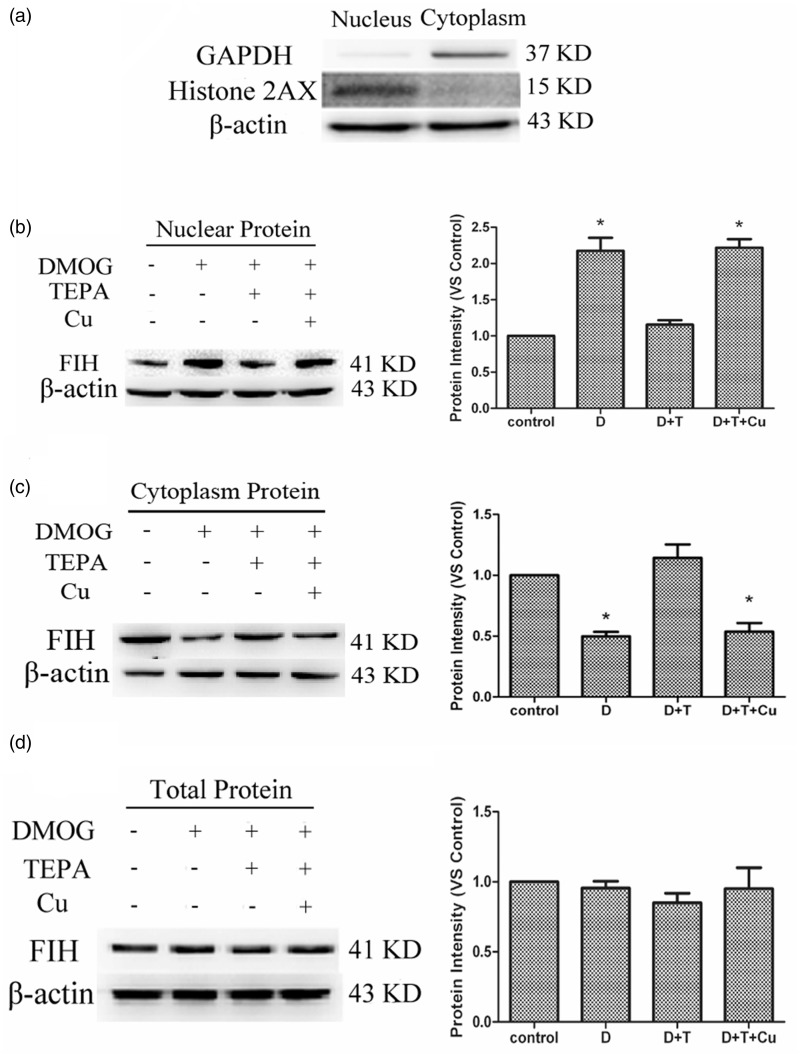
The nuclear purity was qualified using the nucleus specific marker histone 2AX and the cytoplasm specific marker GAPDH (Figure 1(a)). Histone 2AX was detected in the nuclear fraction, but not in the cytoplasm, and conversely, GAPDH was only found in the cytoplasm. Treatment of HUVECs with DMOG at a final concentration of 100 µM for 4 h significantly increased FIH-1 protein levels in the nucleus, as shown in Figure 1(b). When the cells were pretreated with 50 µM TEPA (final concentration in cultures) for 20 h, and then exposed to DMOG for 4 h, the nuclear FIH-1 protein levels were reduced. This reduction was prevented by a simultaneous addition of 50 µM CuSO4 with TEPA. In contrast, DMOG treatment decreased the protein levels of FIH-1 in the cytoplasm (Figure 1(c)); opposite to the increase in the nucleus (Figure 1(b)). TEPA pretreatment prevented the decrease in FIH-1 protein levels in the cytoplasm, and the TEPA effect was reversed by the addition of CuSO4 with TEPA simultaneously. Unlike the changes in the nuclear and cytoplasmic levels of FIH-1, the total protein levels of FIH-1 were not altered under different treatment conditions (Figure 1(d)).
Figure 1.
Effects of TEPA on DMOG-induced FIH-1 protein levels. (a) Evaluation of nucleus and cytoplasm purity by Western blot. (b) Western blot analysis of changes of nuclear FIH-1 protein by different treatments. The HUVECs were untreated as negative controls (Control), treated for 4 h with 100 µM DMOG only (D), or pretreated with 50 µM TEPA (D + T) or 50 µM TEPA plus 50 µM CuSO4 (D + T + Cu) for 20 h before treated with DMOG for 4 h. (c) Western blot analysis of cytoplasm levels of FIH-1. (d) Western blot analysis of changes of total FIH-1 protein by the treatments above. Data were obtained from three independent experiments, and each experiment contains triplicate samples for each treatment. The comparison was done in relation to controls. Values are means ± S.E.M. *Significantly different from control group (p < 0.05)
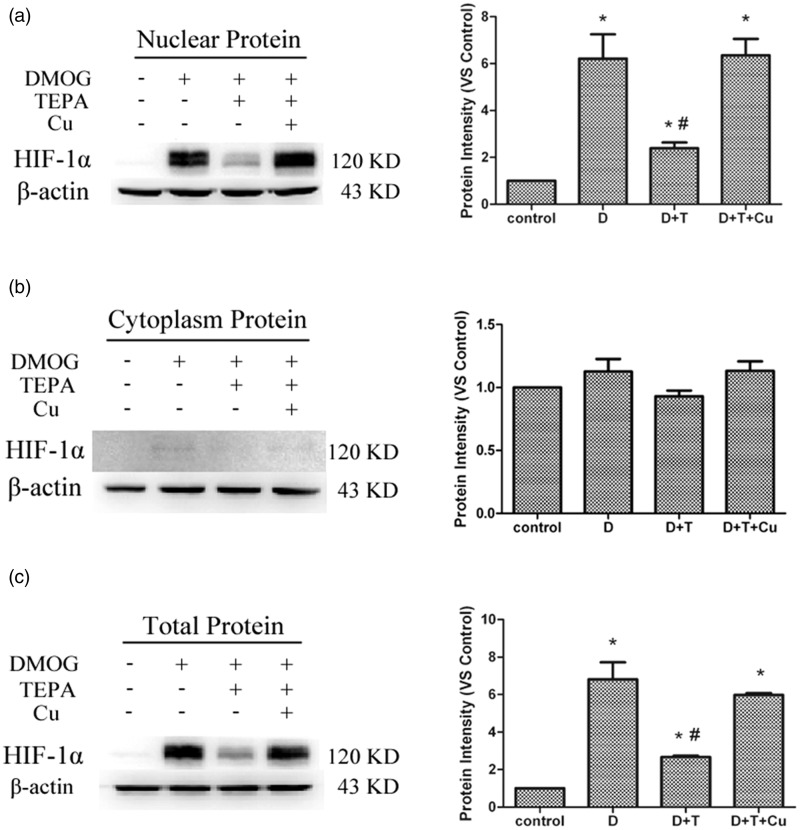
Under the same condition noted above, HIF-1α protein levels in the nucleus were increased after 4-h treatment with DMOG, and this increase was prevented by pretreatment with TEPA, which was preventable by a simultaneous addition of CuSO4 with TEPA (Figure 2(a)). The protein levels of HIF-1α in the cytoplasm were barely detectable under all the conditions (Figure 2(b)), but the changes in the total HIF-1α protein levels followed the same pattern as observed for the nuclear fraction (Figure 2(c)).
Figure 2.
Effects of TEPA on DMOG-induced HIF-1α protein levels. (a) Western blot analysis of nuclear levels of HIF-1α under different conditions described for Figure 1. (b) Western blot analysis of cytoplasm levels of HIF-1α. (c) Western blot analysis of total protein levels of HIF-1α. Data were obtained from three independent experiments, and each experiment contains triplicate samples for each treatment. The comparison was done in relation to controls. Values are means ± S.E.M. *Significantly different from control group (p < 0.05); # significantly different from DMOG group (p < 0.05)
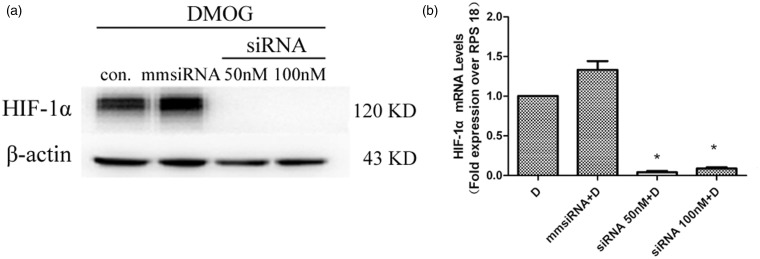
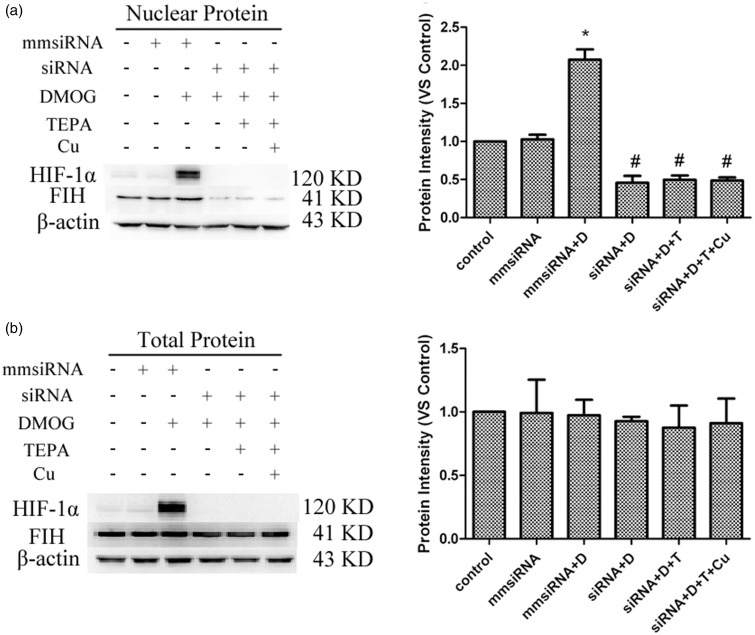
Gene silencing of HIF-1α resulted in a significant decrease in both protein and mRNA levels of HIF-1α (Figure 3), along with a significant decrease in the nuclear level of FIH-1 proteins (Figure 4(a)), but no significant changes in the total protein levels of FIH-1 in the cells (Figure 4(b)).
Figure 3.
Effects of HIF-1α gene silencing on HIF-1α protein and mRNA levels. The HUVECs treated with DMOG only for 4 h as control group (D), Control), or pretreated with 50 nM mismatched siRNA (mmsiRNA), 50 nM, or 100 nM HIF-1α siRNA for 72 h in serum-free media before exposed to DMOG for 4 h. a Changes in the total protein levels of HIF-1α. b HIF-1α mRNA levels measured by real-time RT-PCR. Data were obtained from three independent experiments, and each experiment contains triplicate samples for each treatment. Values are means ± SEM. *Significantly different from control group control group (p < 0.05)
Figure 4.
Effects of HIF-1α gene silencing on DMOG-induced FIH-1 nuclear entry. (a) Effects of HIF-1α gene silencing on FIH-1 protein levels in the nucleus detected by western blot. (b) Effects of HIF-1α gene silencing on total FIH-1 protein levels detected by Western blot. Data were obtained from three independent experiments, and each experiment contains triplicate samples for each treatment. The comparison was done in relation to controls. Values are means ± S.E.M. *Significantly different from mmsiRNA group (p < 0.05); # significantly different from mmsiRNA plus DMOG group (p < 0.05)
Discussion
FIH-1 is a protein encoded by the Hif1an gene and found in the cytoplasm.1 One of its functions is to hydrolyze asparagine at the C-terminal of HIF-1α in the nucleus.12,16 This raises a question: how is the nuclear entry of FIH-1 regulated? Since the function of FIH-1 is coupled to the nuclear location of HIF-1α, it is reasonable to speculate that the nuclear entry between HIF-1α and FIH-1 is coordinated. On the other hand, Cu also enters the nucleus and functions in the regulation of HIF-1 transcriptional activity through at least in part the inhibition of FIH-1 activity.15 Therefore, we made an attempt here to examine the correlation between HIf-1α and FIH-1 in their entry to the nucleus and the effect of Cu on this process.
The measurement of cytoplasmic versus nuclear levels of FIH-1 protein through isolation of nuclear fraction was effective as demonstrated by the purity test of the nuclear marker histone-2AX and the cytoplasm marker GAPDH in the isolated fractions. DMOG inhibits the PHDs leading to HIF-1α accumulation in the cells.10,23 Under this DMOG treatment condition, the level of nucleus FIH-1 was significantly increased and the pretreatment with Cu chelator TEPA prevented this increase. The TEPA effect was due to Cu deficiency, as evidenced by the antagonistic effect of simultaneous addition of equal molar Cu with TEPA. Under these treatment conditions, the total protein levels of FIH-1 were not changed, as further evidenced by the increased nuclear levels along with decreased cytoplasm levels of FIH-1, and vice versa. These together suggest that Cu is related to the nuclear entry of FIH-1 but does not affect the turnover of the FIH-1 proteins.
How does Cu affect the nuclear entry of FIH-1? We observed here that under Cu deficiency the total protein levels and the nuclear levels of HIF-1α induced by DMOG were both depressed, but the cytoplasm levels remained undetectable. The nuclear depression of HIF-1α protein paralleled the decrease of FIH-1 nuclear entry. Both effects were produced by TEPA and prevented by a simultaneous Cu addition with TEPA.
Gene silencing using siRNA targeting HIF-1α effectively depressed total and nuclear levels of HIF-1α proteins. Under this condition, Cu levels in the cells would not be affected. But, we observed that with the depletion of HIF-1α, DMOG-induced nuclear entry of FIH-1 was completely depressed. Therefore, it suggests that the accumulation of HIF-1α and its nuclear translocation most likely trigger the nuclear entry of FIH-1.
One of the limitations of this study is that we do not understand how Cu deficiency reduces the protein levels of HIF-1α induced by DMOG. We previously observed that TEPA treatment after HIF-1α accumulation induced by hypoxia15 or cobalt chloride22 does not affect the total protein levels of HIF-1α. But this study here demonstrates that Cu deficiency prior to DMOG treatment prevents the increase of HIF-1α in the cells, suggesting an inhibition of HIF-1α synthesis by Cu deficiency may take place, which needs to be investigated in the future.
In summary, the present study demonstrates that FIH-1 nuclear entry is dependent on HIF-1α accumulation and nuclear entry, and Cu deficiency prevents FIH-1 nuclear entry through suppression of total protein levels of HIF-1α.
ACKNOWLEDGEMENTS
This study was supported by National Science Foundation of China (grant number: 81230004 to YJK) and Sichuan University West China Hospital.
Author contributions
All authors participated in the design, interpretation and analysis of the data, and review of the manuscript; KL, XD, and CL carried out the experiments; YJK wrote the manuscript. KL and XD made equal contributions to this study.
References
- 1.Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev 2002; 16: 1466–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Targeting of HIF-alfa to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolylhydroxylation. Science 2001; 292: 468–72. [DOI] [PubMed] [Google Scholar]
- 3.Hewitson KS, McNeill LA, Riordan MV, Tian YM, Bullock AN, Welford RW, Elkins JM, Oldham NJ, Bhattacharya S, Gleadle JM, Ratcliffe PJ, Pugh CW, Schofield CJ. Hypoxia-induciblefactor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J BiolChem 2002; 277: 26351–55. [DOI] [PubMed] [Google Scholar]
- 4.Lee C, Kim SJ, Jeong DG, Lee SM, Ryu SE. Structure of human FIH-1 reveals a unique active site pocket and interaction sites for HIF-1 and von Hippel-Lindau. J Biol Chem 2003; 278: 7558–63. [DOI] [PubMed] [Google Scholar]
- 5.Que L. One motif—many different reactions. Nat Struct Biol 2000; 7: 182–84. [DOI] [PubMed] [Google Scholar]
- 6.Ryle MJ, Hausinger RP. Non-heme iron oxygenases. Curr Opin Chem Biol 2002; 6: 193–201. [DOI] [PubMed] [Google Scholar]
- 7.Dann CE, Bruick RK, Deisenhofer J. Structure of factor-inhibiting hypoxia-inducible-factor1: an asparaginyl hydroxylase involved in the hypoxic response pathway. Proc Natl Acad Sci USA 2002; 99: 15351–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elkins JM, Hewitson KS, McNeill LA, Seibel JF, Schlemminger I, Pugh CW, Ratcliffe PJ, Schofield CJ. Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1 alpha. J Biol Chem 2003; 278: 1802–06. [DOI] [PubMed] [Google Scholar]
- 9.Metzen E, Berchner-Pfannschmidt U, Stengel P, Marxsen JH, Stolze I, Klinger M, Huang WQ, Wotzlaw C, Hellwig-Bürgel T, Jelkmann W, Acker H, Fandrey J. Intracellular localization of human HIF-1α hydroxylases: implication for oxygen sensing. J Cell Sci 2002; 116: 1319–26. [DOI] [PubMed] [Google Scholar]
- 10.Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 2001; 107: 43–54. [DOI] [PubMed] [Google Scholar]
- 11.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 2001; 294: 1337–40. [DOI] [PubMed] [Google Scholar]
- 12.Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science 2002; 295: 858–61. [DOI] [PubMed] [Google Scholar]
- 13.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxiainducible-factor-1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA 1998; 95: 7987–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIF-1alpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 2001; 292: 464–68. [DOI] [PubMed] [Google Scholar]
- 15.Feng W, Ye F, Xue W, Zhou Z, Kang YJ. Copper regulation of hypoxia-inducible factor-1 activity. Mol Pharm 2009; 75: 174–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev 2009; 15: 2675–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNeill LA, Hewitson KS, Claridge TD, Seibel JF, Horsfall LE, Schofield CJ. Hypoxia-inducible factor asparaginyl hydroxylase (FIH-1) catalyses hydroxylation at the beta-carbon of asparagine-803. Biochem J 2002; 367: 571–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freedman SJ, Sun ZY, Poy F, Kung AL, Livingston DM, Wagner G, Eck MJ. Structural basis for recruitment of CBP/p300 by hypoxiainducible factor-1 alpha. Proc Natl Acad Sci USA 2002; 99: 5367–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dames SA, Martinez-Yamout M, De Guzman RN, Dyson HJ, Wright PE. Structural basis for Hif-1 alpha/CBP recognition in the cellular hypoxic response. Proc Natl Acad Sci USA 2002; 99: 5271–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan Z, Michalopoulos GK, Stolz DB. Peroxisomal localization of hypoxia-inducible factors and hypoxia inducible factor regulatory hydroxylases in primary rat hepatocytes exposed to hypoxia reoxygenation. Am J Pathol 2006; 169: 1251–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature 2011; 473: 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu L, Ding X, Zhang Z, Kang YJ. Copper is required for cobalt-induced transcriptional activity of hypoxia-inducible factor-1. J Pharmacol Exp Ther 2012; 342: 561–67. [DOI] [PubMed] [Google Scholar]
- 23.Elvidge GP, Glenny L, Appelhoff RJ, Ratcliffe PJ, Ragoussis J, Gleadle JM. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: the role of HIF-1a, HIF-2a, and other pathways. J Biol Chem 2006; 281: 15215–26. [DOI] [PubMed] [Google Scholar]