Abstract
Mesenchymal stem cells (MSCs) have the capacity to restore liver function by differentiating into hepatocyte like cells. However, the underlying mechanisms are not well understood. Here, we have investigated the signals involved in the hepatic differentiation of human umbilical cord-derived mesenchymal stem cells (hUCMSCs). hUCMSCs were treated with mouse fetal liver-conditioned medium (FLCM) to induce hepatic differentiation. Flow cytometry, reverse transcription PCR, real-time PCR, immunocytochemistry, and polymerase chain reaction (PCR) array were used to detect the expression of MSC- and hepotocyte-specific markers in FLCM-treated hUCMSCs. Urea production and cytochrome P450 3A4 (CYP3A4) activity were used as indicators to evaluate liver cell characteristics. Raf/mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) was analyzed in hUCMSCs by Western blotting. Following FLCM treatment, expression of MSC-specific markers decreased, while hepatocyte-specific gene expression was increased. Urea production, albumin secretion, glycogen storage, and CYP3A4 activity were significantly enhanced in FLCM-treated cells. In addition, ERK1/2 phosphorylation was increased in a time-dependent manner through Raf/MEK/ERK pathway, and phosphorylation was sustained at a high level during hepatic induction. Inhibition of ERK1/2 activation by U0126 (an ERK1/2 inhibitor) and pFLAG-CMV-ERK1(K71R) (negative mutant of ERK1) reversed the expression of liver-specific genes in hUCMSCs and affected hepatic function significantly. In summary, this work shows that ERK1/2 phosphorylation plays an important role in inducing hepatic differentiation of hUCMSCs in FLCM.
Keywords: ERK1/2, hepatocytes, umbilical cord-derived mesenchymal stem cell, fetal liver-conditioned medium
Introduction
Human umbilical cord-derived mesenchymal stem cells (hUCMSCs) are anticipated to be a potential source for cell-based therapies. Although bone marrow is an important source of mesenchymal stem cells (MSCs), the use of hUCMSCs is advantageous: no appreciable ethical problems,1 a lower risk of viral transmission,2 and is more primitive.3 In addition, hUCMSCs can be cryopreserved and thawed efficiently, which make them suitable for use in clinical cell banking. Most human umbilical cords are currently discarded as medical waste in delivery rooms. Recently, several studies have provided encouraging results. The use of hUCMSCs have been applicated in the treatment of various hepatic injuries.4–6
The clinical relevance of hUCMSCs is primarily dependent on their potential to differentiate. Like human bone marrow mesenchymal stem cells, hUCMSCs were demonstrated to be multipotent. hUCMSCs can differentiate into endodermal cell types as well as most mesodermal and neuroectodermal cell types.7 Under defined conditions, hUCMSCs can differentiate into functional hepatocytes and effectively rescue experimental liver failure.6,8,9 We had previously demonstrated that hUCMSCs can differentiate into hepatocyte-like cells in vivo.10 However, due to the low efficiency, this differentiation is not sufficient to produce enough functional hepatocytes for clinical purposes.
To gain a better understanding of the mechanisms controlling hepatic differentiation, different induction strategies have been used for hepatic differentiation of MSCs. Several studies11,12 have reported that the hepatic differentiation potential of MSCs is controlled by the combined action of several cytokines. Fetal liver-conditioned medium (FLCM) provides a more suitable environment for efficient bone marrow-derived MSC differentiation into functional hepatocytes.13 The fetal liver-specific cytokines (e.g. hepatocyte growth factor (HGF), basic Fibroblast Growth Factor (bFGF), and Oncostatin M (OSM)) involved in early maturation act as promoting factors for the differentiation and maturation of hepatocytes during embryogenesis. Whether FLCM can induce hepatic differentiation of hUCMSCs and the mechanism through which this differentiation may occur is not clear yet.
We believe that hepatic differentiation of MSCs is tightly regulated by many cytokines and growth factors through signaling events. In our previous study, we showed that hUCMSCs can differentiate into hepatocyte-like cells that express liver-specific genes and enhance recovery from hepatic injury. In this study, FLCM was used to induce the differentiation of hUCMSCs to hepatocyte-like cells. Our study indicates the signals that regulate hepatic differentiation of FLCM-stimulated hUCMSCs.
Materials and methods
hUCMSCs isolation and expansion
hUCMSCs were isolated as previously described.10 Briefly, human umbilical cords were obtained with donor consent and processed within 6 h after birth. For removal of the blood, cords were rinsed twice with phosphate-buffered saline (PBS). Cords were then cut into 1 mm3-sized pieces and cultured in low glucose Dulbecco’s modified Eagle’s medium (LG-DMEM) (containing 10% fetal bovine serum (FBS, Gibco, USA), penicillin, and streptomycin) at 37℃ with 5% CO2. Medium was changed in 3-day intervals and when cells reached 70–80% confluency, cultures were trypsinized with 0.25% trypsin–Ethylene Diamine Tetraacetic Acid (EDTA) (Invitrogen, USA) and passaged into new flasks for further expansion. Human hepatocte cell line HL7702 was bought from Chinese Academy of Sciences type culture collection cell library and cultured in LG-DMEM (containing 10% FBS (Gibco, USA), penicillin, and streptomycin) at 37℃ with 5% CO2.
Osteogenic and adipogenic differentiation of hUCMSCs
hUCMSCs in passage 3 were cultured in LG-DMEM that contained either osteogenic (0.1 mM dexamethasone, 10 mMb glycerophosphate, and 50 mM ascorbate-phosphate, Sigma-Aldrich) or adipogenic (0.5 mM isobutylmethylxanthine, 1 mM dexamethasone, 10 mM insulin, and 200 mM indomethacin, Sigma-Aldrich) reagents. hUCMSCs cultured in LG-DMEM were used as control. After 2 weeks induction, osteogenic differentiation was assessed by the examination of neutrophil alkaline phosphatase (NAP) with the NAP staining kit (Sun Bio) and adipogenic differentiation was examined via intracellular lipid accumulation, which was visualized using Oil-Red-O staining by an inverted microscope (Ti; NikonCorporation).
Hepatic differentiation
FLCM was prepared as described.13 Briefly, fetal livers of mice at 13.5 embryonic days were harvested and cut into approximately 1 mm3 pieces. Following adhesion to the dishes, 6 mL DMEM with 10% FBS was added and incubated at 37℃, 5% CO2 for 48 h. Supernatants were collected and filtered using a 0.25 µm filter. The filtrate, defined as FLCM, was stored in aliquots at –20℃ for further use. hUCMSCs of passage 3 were incubated in FLCM at 5 × 104 cells/cm2. Cultures were maintained by changing the medium every 2 days. For analysis of the mechanisms involved in the differentiation, U0126 (Promega, USA) was added simultaneously until cells were collected. Cell morphology was observed under a phase-contrast microscope (Nikon, Japan). Induced hUCMSCs were collected and stored at −70℃ for further investigation.
Flow cytometry
hUCMSCs (1 × 105) were washed with PBS to remove the medium and trypsin–EDTA, followed by incubation with phycoerythrin (PE)-conjugated mouse antihuman monoclonal antibodies (CD13, CD14, CD19, CD29, CD34, CD44, CD45, CD73, CD90, CD105, CD271 and HLA-I, HLA-DR) (Invitrogen, USA) for 30 min at 4℃ in PBS. PE-labeled mouse immunoglobulin Gs were used as negative controls. Labeled cells were washed and analyzed by flow cytometry (FACS Calibur, BD Biosiences, USA).
Microarray analysis
Total RNA was purified using Trizol and RNA samples were treated with DNase (Invitrogen, USA) treatment to remove any DNA contamination. After RNA yield and quality was assessed, cDNA was synthesized using SuperScript III Reverse Transcriptase (Invitrogen). cDNA was then loaded into the well of a PCR array to perform real-time PCR detection according to the manufacturer’s instructions (RT2 Profiler™ PCR Array Human Mesenchymal Stem Cell (PAHS-082 A), Qiagen, USA). Data analysis was performed using the ΔΔCt method. The expression of each gene was compared between hUCMSCs treated with and without FLCM and results were reported as ratios.
Plasmids and extracellular signal-regulated kinase1-K71R (ERK1-K71R) transfection
hUCMSCs were plated in 9.5 cm2 wells of a six-well plate. After an overnight incubation, the cells were washed once with PBS and then transfected using Lipofectomine2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The cells were transfected with 0.5 µg of pFLAG-CMV7.1 vector or pFLAG-CMV-Erk1(K71R) (kinase-deficient form of ERK-1). The pFLAG-CMV-Erk1(K71R) plasmids were bought from Addgene (plasmid 49329) and deposited by Dr. Melanie Cobb, University of Texas Southwestern Medical Center.14 After 5 h the cells were replaced with complete DMEM and incubated for 60 h. The cells were then washed once with PBS and cultured in DMEM or FLCM. Expression of P-ERK and T-ERK was confirmed by Western Blotting analysis.
RT-PCR and real-time PCR
Total RNA extracted from hUCMSCs was used in RT-PCR reactions to determine the expression of liver- and MSC-specific genes by a PCR thermal cycler (PCR Express, ThermoHybaid, USA). Expression of tryptophan 2,3-dioxygenase (TDO) and cytochrome P450 1A1 (CYP1A1) genes was detected by real-time PCR. Reaction mixtures contained 10 µl 2 × SYBG mix (ToYoBo, Japan), 0.4 µl 10 µM of each primer, 8.2 µl ddH2O, and 1 µl of first-strand cDNA. Samples were run in triplicate, and each reaction was independently repeated three times to ensure reproducibility using Rotor-Gene Real-time analyzer (Rotor-Gene 6000, CR, Australia). All the primers were gene specific (Table 1) and were produced by Shanghai Bio-Engineering Company (Shanghai, China).
Table 1.
Primers for RT-PCR and real-time PCR
| Genes | Primer Sequence (5′–3′) | Annealing Temperature(℃) | Product size (bp) |
|---|---|---|---|
| AFP | For: TGCAGCCAAAGTGAAGAGGGAAGA | 63 | 216 |
| Rev: CATAGCGAGCAGCCCAAAGAAGAA | |||
| TDO | For: ACCTCCGTGCTTCTCAGACA | 55 | 493 |
| Rev: GGAAGCCTGATGCTGGAGAT | |||
| CK18 | For: GGAGGCATCCAGAACGAGAA | 58 | 376 |
| Rev: CCAGCTGCAGTCGTGTGATA | |||
| Nanog | For: ATGCCTCACACGGAGACTG | 60 | 369 |
| Rev: CTGCGTCACACCATTGCTA | |||
| CYP1A1 | For: ACCATGACCAGAAGCTATGGGT | 60 | 215 |
| Rev: TTAACACCTTGTCGATAGCACCA | |||
| ALB | For: TTGCCAAGC TGCTGATAAGG | 58.5 | 238 |
| Rev: ATGGCAGCA TTCCGTGTG | |||
| ABCG2 | For: CGGCTTGCAACAACTATGAC | 60 | 537 |
| Rev: ATCCTGCTTGGAAGGCTCTA | |||
| OCT4 | For: TATACACAGGCCGATGTGG | 60 | 397 |
| Rev: GTGCATAGTCGCTGCTTGA | |||
| CD44 | For: TCACAGGTGGAAGAAGAGAC | 57 | 447 |
| Rev: CATTGCCACTGTTGATCACT | |||
| CYP3A4 | For:CCAAGCTATGCTCTTTCACCG | 60 | 324 |
| Rev:TCAGGCTCCACTTACGGTGC | |||
| GAPDH | For: GGATTTGGTCGTATTGGG | 55 | 205 |
| Rev: GGAAGATGGTGATGGGATT |
Immunocytochemistry
hUCMSCs collected by enzymatic methods were plated onto coverslips, fixed in methanol for 10 min, and permeabilized with PBS for another 10 min. After blocking with 5% bovine serum albumin for 1 h, samples were incubated with anti-neuron-specific enolase (NSE) (1:50), antiglial fibrillary acidic protein (GFAP) (1:50), antidesmin (1:100) and cardiac troponin T (1:100), antihuman albumin (ALB) (1:50), antihuman Cytokeratin 18 (CK-18) (1:100), and anti-a-fetoprotein (AFP) (1:100) antibodies (Boster Bioengineering Company Limited, Wuhan, China) for 1.5 h at 37℃, followed by incubation with a secondary antibody for 20 min. Slides were visualized using diaminobenzidine (Boster Bioengineering Company Limited, Wuhan, China). Hematoxylin was utilized for nuclear counterstaining. Removal of the primary antibody from the procedure provided a negative control.
Western blotting
Western blotting was performed to analyze expression of P-ERK/T-ERK (1:1000; Kang Cheng, Shanghai, China), P-Raf/T-Raf, P-MEK/T-MEK, P-P38/P38, and phosphorylated c-Jun N-terminal kinas (p-JNK)/c-Jun N-terminal kinas (JNK) (1:500, SAB, China) along with GAPDH (1:3000; Kang Cheng, Shanghai, China) in different groups. Cells were harvested and lysed in RIPA buffer (10 mM Tris, pH 7.4, 150 mM NaCl, and 1 µg/mL leupeptin). Aliquots containing equal amounts of protein were fractionated by sodium dodecyl sulfate-Polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to methanol preactivated Polyvinylidene fluoride (PVDF) membranes. After blocking with 5% (w/v) milk, membranes were incubated with primary and secondary antibodies. Blots were developed and detected by enhanced chemiluminescence (GE Healthcare, Little Chalfont, UK).
CYP3A4 activity assay
Cell-based assays were used to determine the cellular activities of CYP3A4 according to manufacturer’s instructions (Galen Biopharm International Co., Ltd). Briefly, the cultured cells were washed twice with PBS and incubated with cell lysis buffer for 10 min at room temperature. After incubation, an aliquot of 20 µL cell lysate was transferred to a 1.5 mL eppendorf tube at room temperature. Then, Luciferase assay reagent (100 µL) was added and mixed with cell lysate. The resulting luminescence was read using a luminometer (GloMax® 20/20, Promega). Following luminescence determinations, the luminescence data Relative Luminescence Unit (RLU) were normalized to the respective DMEM-cultured hUCMSCs to represent various CYP3A4 activities in FLCM or FLCM/U0126-treated hUCMSCs.
Enzyme-linked immunosorbent assay
The concentration of HGF in the DMEM and FLCM was quantified by a human HGF enzyme-linked immunosorbent assay kit (Senxiong Technology Limit Company, China). The level of HGF in the medium was determined according to the manufacturer’s protocol using recombinant HGF as a control standard. The range of detection is from 62.5 to 8000 pg/mL, so DMEM and FLCM were optimally diluted for detection.
Periodic acid-Schiff staining
Glycogen storage in hUCMSCs was detected by a periodic acid-Schiff staining kit according to the manufacturer’s protocol (SUNBIO Company, China). Briefly, hUCMSC were fixed with methanol for 5 min and rinsed in distilled water. Then the cells were oxidized in periodic acid solution for 5 min and placed in Schiff reagent for 30 min, respectively. After counterstain in hematoxylin for 1 min, stained cells were visualized by an inverted microscope (Ti; Nikon Corporation).
Albumin secretion
Albumin levels in the culture medium of DMEM or FLCM-treated hUCMSCs were measured with an automated biochemical analyzer.
Urea assay
Untreated, FLCM-treated, and FLCM/U0126-treated hUCMSCs were plated at 3 × 104 cells/well. After incubation with 5 mM ammonium chloride, the amount of urea secreted into the medium was measured every 24 h for up to 96 h. Urea concentrations were determined with an automated biochemical analyzer.
Statistical analysis
Data are expressed as mean ± (standard deviation) SD. Statistical significance was determined by using Student’s t-test or Analysis of variance (ANOVA) using Prism software (Graph Pad, San Diego, USA). P-values less than 0.05 were considered significant.
Results
Characteristics of hUCMSCs
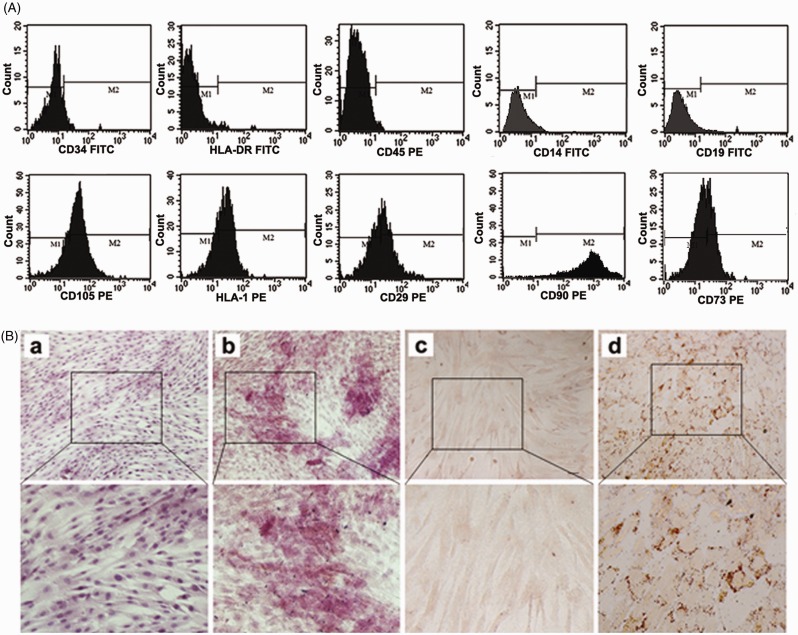
hUCMSCs presented a homogeneous population of fibroblast-like cells and positively expressed CD105, HLA-I, CD29, CD73, CD90, and negative for CD14, CD19, CD34, HLA-DR, CD45 (Figure 1(a)). hUCMSCs possess the ability for multidifferentiation. Non-treated control culture did not show any adipocyte or osteoblast formation (Figure 1(B)a and (c)). When addition of osteogenic and adipogenic supplementation, hUCMSCs showed positively staining for alkaline phosphatase (Figure 1Bb) and Oil-Red-O (Figure 1Bd) when cultured in osteogenic and adipogenic supplemented DMEM.
Figure 1.
Characterization of hUCMSCs. A: Flow cytometry analysis of hUCMSCs. hUCMSCs were negative for CD34, HLA-DR, CD45, CD14, CD19 and positive for CD105, HLA-I, CD29, CD73, CD90. The results confirmed that the cells were a kind of MSCs but non-hematopoietic cells; B: Immunocytochemistry with osteoblast-specific and oil red staining marker. Control culture of hUCMSCs did not exhibit such osteocytic features (a). Osteogenic potential of hUCMSCs was shown by alkaline phosphatase (b). hUCMSCs cultured in control culture did not exhibit such adipocytic features (c). Adipogenic potential of hUCMSCs was positively stained with Oil Red O (d). (a, b, c, d: 100×). hUCMSCs, human umbilical cord-derived mesenchymal stem cells. (A color version of this figure is available in the online journal.)
Hepatic differentiation of FLCM-induced hUCMSCs
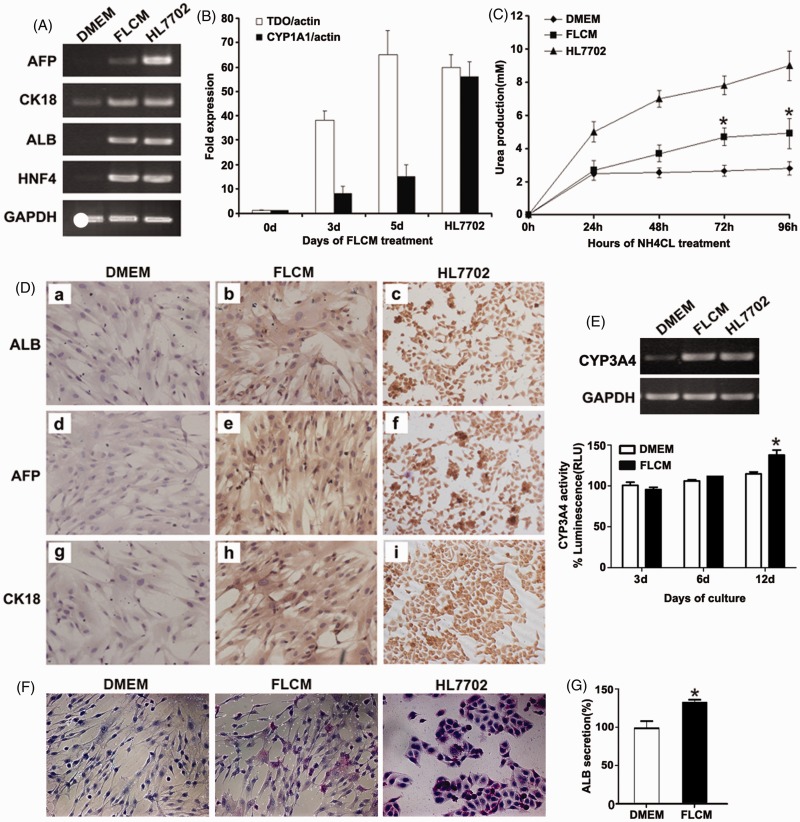
hUCMSCs were cultured in DMEM and FLCM separately. Human hepatocyte cell line HL7702 was used as a positive control. Three days after FLCM treatment, expression of liver-specific genes such as ALB, AFP, and human nuclear factor 4 (HNF4) was strongly induced in FLCM group (Figure 2(a)). No obvious expression was found in DMEM-cultured hUCMSCs except CK-18 expression, which was also enhanced after FLCM treatment. Real-time PCR of FLCM-treated hUCMSCs samples revealed increased expression of TDO and CYP1A1 in a time-dependent manner (Figure 2(b)).
Figure 2.
FLCM-induced hepatic differentiation of hUCMSCs. A: Expression of hepatic-specific genes evaluated by RT-PCR at day 3 post-FLCM induction; B: Expression of hepatic-specific genes evaluated by real-time PCR. The level of TDO and CYP1A1 expression is shown as a fold-change compared with 0 day. Human hepatocyte cell line HL7702 was used as a positive control. Data are expressed as mean + SD of three experiments; C: Urea production in the medium of induced hUCMSCs at different time. Data are expressed as mean + SD of six experiments. *P < 0.05, compared with DMEM-treated hUCMSCs. D: Immunocytochemistry analysis of hepatic-specific genes ALB, AFP, CK-18 expression in DMEM (a, d, g) and FLCM (b, e, h) treated hUCMSCs on day 3. Human hepatocyte cell line HL7702 was used as a positive control (c, f, i). (magnification, ×200). E: Expression of CYP3A4 gene at 12 days and CYP3A4 activity at 3, 6, and 12 days during FLCM culture. F: Periodic Acid-Schiff staining of FLCM-treated hUCMSCs on day 12. DMEM-cultured hUCMSCs and HL7702 were used as control. G: ALB secretion in the medium of induced hUCMSCs at day 12. Data are expressed as mean + SD of three experiments. *P < 0.05, compared with DMEM-treated hUCMSCs. FLCM, fetal liver conditioned medium. TDO, tryptophan 2,3-dioxygenase. AFP, a-fetoprotein; ALB, albumin; CK-18, Cytokeratin 18; HNF4, human nuclear factor 4; CYP1A1, cytochrome P450 1A1; CYP3A4, cytochrome P450 3A4. (A color version of this figure is available in the online journal.)
Differentiation was also evaluated at the functional level. The levels of cell secretion were measured every 24 h following the addition of ammonium chloride. Urea in the medium increased in a time-dependent manner both in FLCM and DMEM group. Seventy-two h and 96 h after NH4Cl addition, production of urea was significantly higher in the FLCM-treated cells compared with DMEM-cultured cells (Figure 2(c)). Furthermore, immunocytochemistry analysis showed that the number of cells expressing AFP, ALB, and CK-18 was increased in FLCM-treated hUCMSCs (Figure 2(d)). Expression of CYP3A4 and CYP3A4 activity was not increased until 12 days during FLCM treatment (Figure 2(e)). Glycogen storage and albumin secretion were also enhanced in FLCM-treated hUCMSCs (Figure 2(f) and (g)).
Specificity of FLCM-induced hepatic differentiation
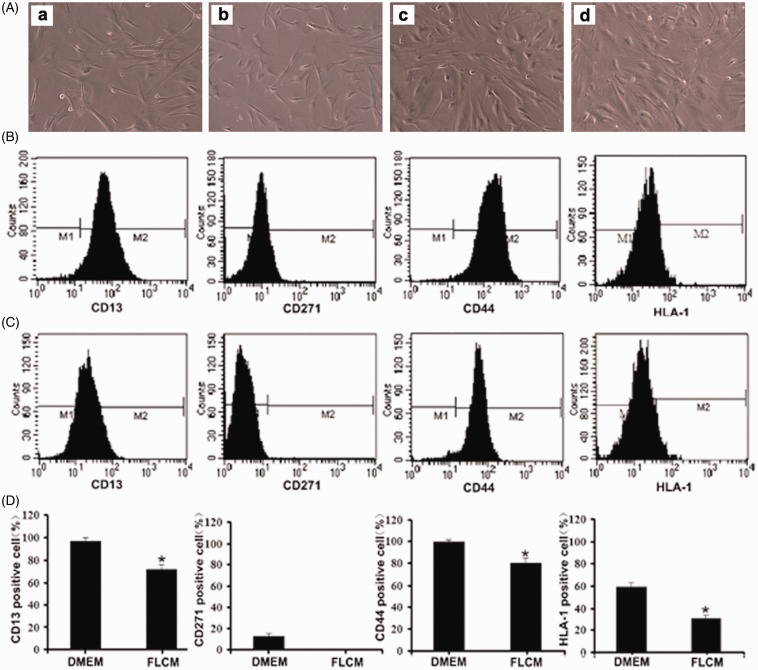
Untreated hUCMSCs displayed a fibroblast-like appearance at 0 and 3 days during FLCM treatment. After being induced with FLCM for 14 days, hUCMSCs lost their spindle-shaped appearance and adopted a polygonal shape with a granular cytoplasm (Figure 3(a)). Flow cytometry analysis of MSC-specific surface markers showed that the percentage of hUCMSCs positive for CD13, CD271, CD44, and HLA-1 were 96.32, 13.8, 99.05, and 60.58%, respectively. On day 3, these MSC-specific markers decreased to 71.43, 0.03, 81.2, and 31.13% (Figure 3(b) to (d)).
Figure 3.
MSC-specific markers in FLCM-treated hUCMSCs. A: The morphology of hUCMSCs at 0 (a), 3 (b), 7 (c), 14 (d) days during FLCM treatment; B and C: Expression of MSC-specific surface markers in hUCMSCs before (B) and after (C) FLCM treatment; D: The percentage of hUCMSCs positive for CD13, CD271, CD44, and HLA-I were 96.32, 13.8, 99.05, and 60.58%, respectively. On day 3 after FLCM treatment, cells expressing MSC-specific markers decreased to 71.43, 0.03, 81.2, and 31.13%, respectively. FLCM, fetal liver conditioned medium.*P < 0.05, compared with DMEM group. (A color version of this figure is available in the online journal.)
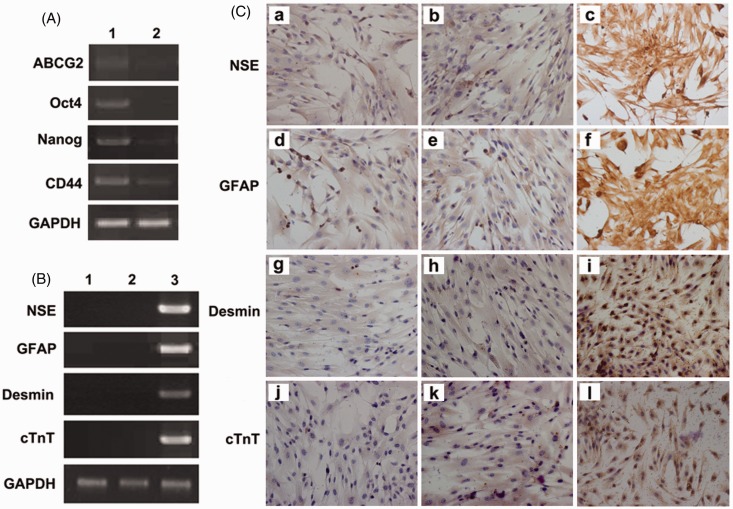
RT-PCR showed a decrease in expression of MSC specific and stemness markers such as ATP-binding cassette sub-family G member 2 (ABCG2), CD44, OCT4, and Nanog (Figure 4(a)). PCR array analysis also confirmed that MSC specific and stemness markers were down-regulated on day 3 in FLCM-treated hUCMSCs (Table 2). These findings indicated a downregulation of MSC-specific markers. At the same time, both DMEM- and FLCM-treated hUCMSCs were negative for neuron-specific genes NSE, GFAP and cardiomyocyte related genes cTNT, desmin. These results were confirmed by RT-PCR and immunocytochemical staining (Figure 4(b) and (c)).
Figure 4.
Analysis of FLCM-induced hepatic specificity. A: Expression of MSC-specific genes ABCG2, OCT4, Nanog, CD44 by RT-PCR in DMEM and FLCM-induced hUCMSCs. (Lane 1, DMEM; Lane 2, FLCM); B: Expression of nerve cell-specific genes NSE, GFAP and cardiocyte-specific genes desmin, cTnT by RT-PCR in DMEM and FLCM-induced hUCMSCs. (Lane 1, DMEM; Lane 2, FLCM; 3, Positive control of primary nerve cell for NSE, GFAP and human cardiomyocyte for desmin, cTnT). C: Immunocytochemistry analysis of cardiocyte- and nerve cell-specific proteins in DMEM and FLCM-cultured hUCMSCs. hUCMSCs cultured in DMEM (a, d, g, j) and FLCM (b, e, h, k) were negatively stained for NSE, GFAP, desmin, and cTnT. Primary nerve cell and human cardiomyocyte served as a positive control (c, f, i, l). (magnification × 200). cTnT, cardiac troponin T; NSE, neuron-specific enolase; ABCG2, ATP-binding cassette sub-family G member 2; GFAP, glial fibrillary acidic protein. (A color version of this figure is available in the online journal.)
Table 2.
PCR array analysis of human mesenchymal stem cell-related genes
| Genes | Description | Accession Number | Ratio |
|---|---|---|---|
| MSC-specific markers | |||
| ALCAMc | Activated leukocyte cell adhesion molecule | NM_001627 | 0.29 |
| ANPEP | Alanyl (membrane) aminopeptidase | NM_001150 | 0.39 |
| BMP2 | Bone morphogenetic protein 2 | NM_001200 | 0.65 |
| CASP3 | Caspase 3, apoptosis-related cysteine peptidase | NM_004346 | 0.76 |
| CD44 | CD44 molecule (Indian blood group) | NM_000610 | 0.51 |
| ENG | Endoglin | NM_000118 | 0.42 |
| ERBB2 | V-erb-b2 erythroblastic leukemia viral oncogene homolog 2 | NM_004448 | 0.58 |
| FUT4 | Fucosyltransferase 4 | NM_002033 | 0.34 |
| FZD9 | Frizzled homolog 9 (Drosophila) | NM_003508 | 3.60 |
| ITGA6 | Integrin, alpha 6 | NM_000210 | 3.48 |
| ITGAV | Integrin, alpha V | NM_002210 | 0.47 |
| KDR | Kinase insert domain receptor | NM_002253 | 0.43 |
| MCAM | Melanoma cell adhesion molecule | NM_006500 | 0.37 |
| NGFR | Nerve growth factor receptor | NM_002507 | 0.77 |
| NT5E | 5'-nucleotidase, ecto (CD73) | NM_002526 | 0.40 |
| PROM1 | Prominin 1 | NM_006017 | 0.04 |
| THY1 | Thy-1 cell surface antigen | NM_006288 | 0.25 |
| VCAM1 | Vascular cell adhesion molecule 1 | NM_001078 | 0.45 |
| Stemness markers | |||
| INS | Insulin | NM_000207 | 0.34 |
| POU5F1 | POU class 5 homeobox 1 | NM_002701 | 0.33 |
| SMURF1 | SMAD specific E3 ubiquitin protein ligase 1 | NM_020429 | 0.31 |
| SOX2 | SRY (sex determining region Y)-box 2 | NM_003106 | 0.34 |
| SOX9 | SRY (sex determining region Y)-box 9 | NM_000346 | 0.36 |
| TERT | Telomerase reverse transcriptase | NM_198253 | 0.51 |
| WNT3A | Wingless-type MMTV integration site family, member 3 A | NM_033131 | 0.34 |
| ZFP42 | Zinc finger protein 42 homolog (mouse) | NM_174900 | 0.34 |
FLCM provokes ERK1/2 phosphorylation in hUCMSCs
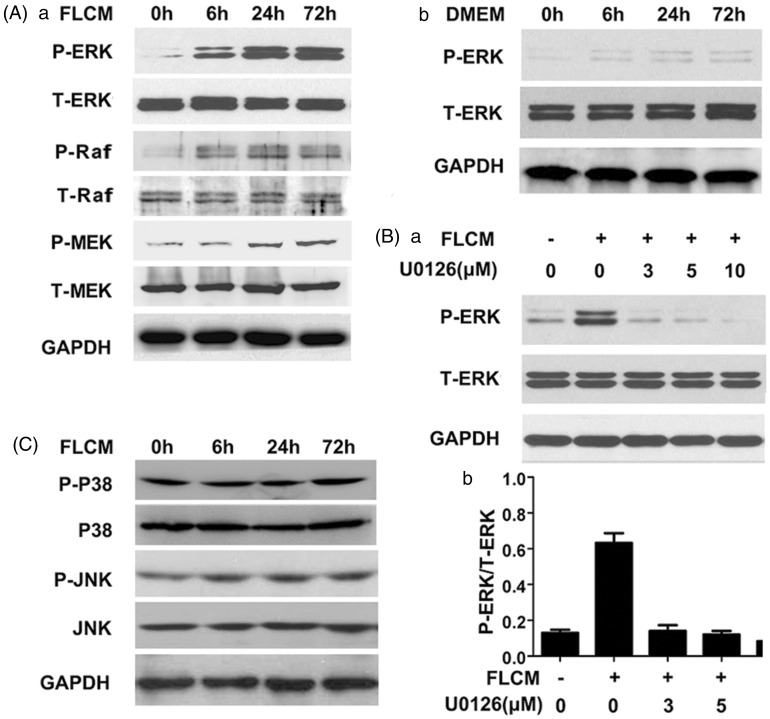
To investigate the pathways involved in hepatic differentiation of hUCMSCs, Raf/MEK/ERK pathway was investigated during FLCM treatment. We found expression of P-ERK, P-Raf, and P-MEK increased significantly in the FLCM-treated group (Figure 5(A)a) and P-ERK was not evident in DMEM group (Figure 5(A)b). We further blocked the ERK1/2 phosphorylation with U0126, an ERK1/2 inhibitor. Our results indicated a dose-dependent inhibition of U0126 on P-ERK expression during FLCM treatment (Figure 5(B)a and (b)). However, expression of p-P38, P38, p-JNK, and JNK was not changed in FLCM-treated hUCMSCs (Figure 3(c)). Our results indicated that FLCM can activate ERK1/2 phosphorylation through Raf/MEK/ERK pathway.
Figure 5.
FLCM provoked ERK1/2 phosphorylation in hUCMSCs. A: Western blotting analysis of P-ERK, P-Raf, P-MEK, and T-ERK, T-Raf, T-MEK in FLCM-treated (a) and DMEM-treated (b) hUCMSCs at 0, 6, 24, and 72 h; B: Effect of U0126(ERK1/2 inhibitor) on the ERK1/2 phosphorylation at 3, 5, and 10 µM after FLCM treatment for 24 h (a). Density analysis of Western blotting bands (b). Data are expressed as mean + SD of three experiments. C: Western blotting analysis of P-JNK, JNK, P-P38, and P38 in FLCM-treated hUCMSCs at 0, 6, 24, and 72 h. P-ERK, phosphorylated ERK1/2; T-ERK, total ERK 1/2; P-Raf, phosphorylated Raf; T-Raf, total Raf; P-MEK, phosphorylated MEK; T-MEK, total MEK
Blockade of ERK1/2 phosphorylation inhibits the hepatic differentiation of hUCMSCs
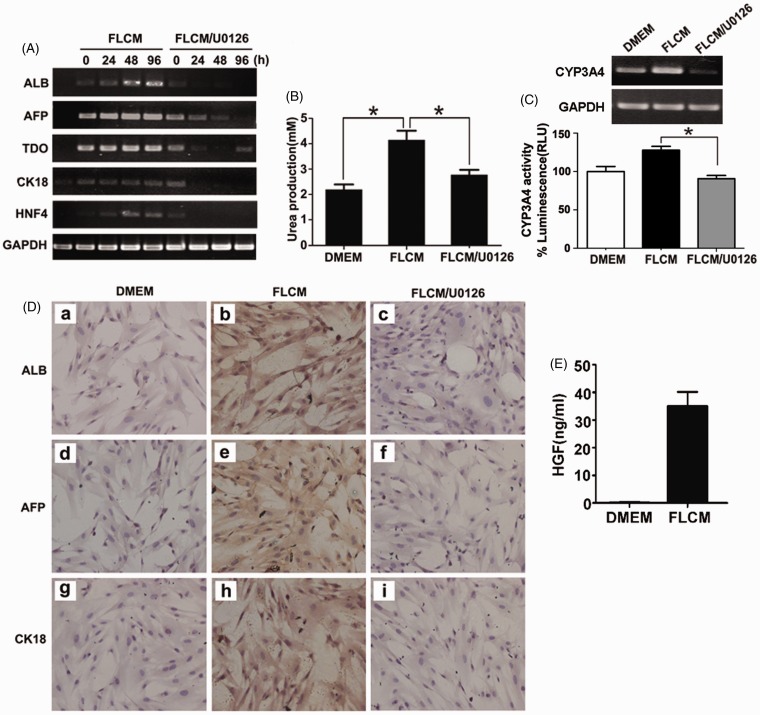
To study the role of ERK1/2 phosphorylation in hepatic differentiation in FLCM-treated hUCMSCs, we investigated the expression of liver-specific genes in FLCM-treated hUCMSCs after ERK1/2 was blocked. U0126 and ERK1-K71R transfection were used to block ERK1/2 phosphorylation. Expression of hepatic specific genes and proteins was detected by RT-PCR and immunocytochemistry at 24, 48, 72, and 96 h followed by U0126 (5 µM) treatment. RT-PCR results revealed that expression of hepatic-specific genes including ALB, AFP, TDO, CK-18, and HNF4 was reduced in the FLCM/U0126 group (Figure 6(a)). Urea secretion in the medium was decreased at 72 h in the FLCM/U0126-treated group, while increased in the FLCM-treated group (Figure 6(b)). Furthermore, CYP3A4 activity was suppressed in FLCM/U0126 group compared with FLCM alone (Figure 6(c)). Immunocytochemistry also confirmed that upon U0126 treatment, FLCM-treated cells negatively expressed those markers mentioned earlier (Figure 6(d)). To investigate the component that activates Raf/MEK/ERK pathway, we tested the concentration of HGF in FLCM. Result showed that HGF was highly increased in FLCM (Figure 6(e)).
Figure 6.
Blockade of ERK1/2 phosphorylation with U0126 reversed FLCM-induced hepatic differentiation of hUCMSCs. hUCMSCs were treated with FLCM in the presence or absence of U0126 (5 μM) for different times. A: Expression of hepatic-specific genes in hUCMSCs detected by RT-PCR. B: Urea production in the medium of hUCMSCs at 72 h. Data are expressed as mean + SD of three experiments. C: CYP3A4 gene expression and CYP3A4 activity at 12 days during FLCM or FLCM/U0126 treatment; D: Expression of hepatic-specific proteins AFP, ALB, CK-18 in hUCMSCs by immunocytochemistry. hUCMSCs cultured in DMEM were used as a control (a, d, g). hUCMSCs were treated with FLCM (b, e, h) and FLCM/U0126 (c, f, i) for 72 h (magnification × 200). E: ELISA assay of HGF concentration in DMEM and FLCM. HGF, hepatocyte growth factor. *P < 0.05. (A color version of this figure is available in the online journal.)
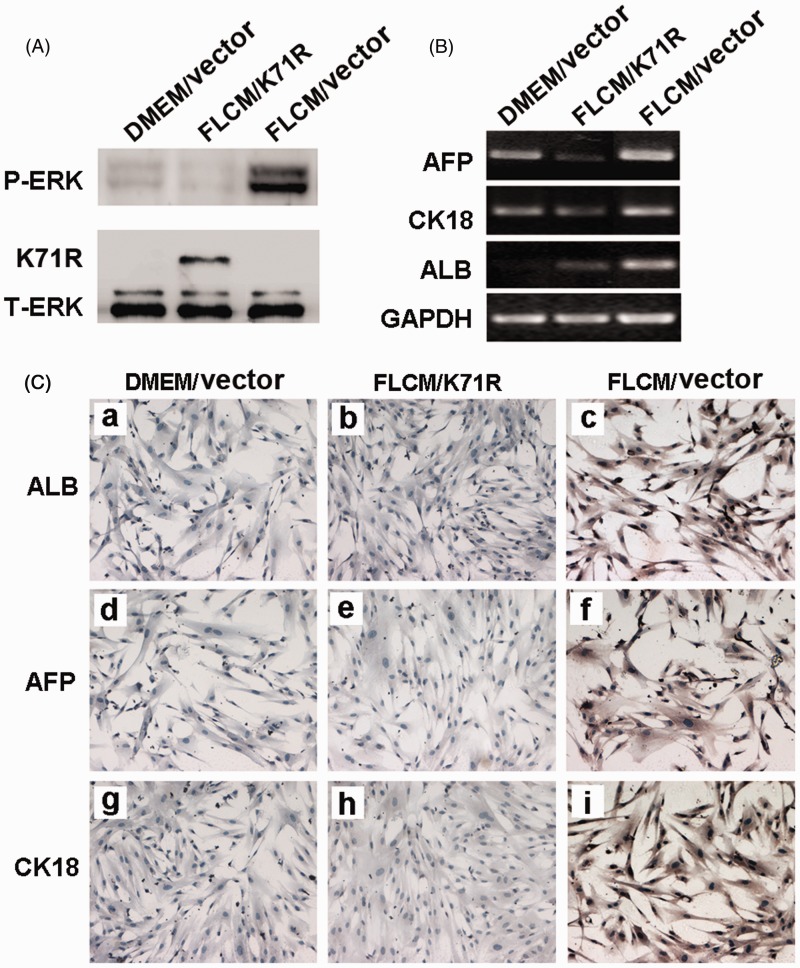
To further identify the role of ERK1/2 phosphorylation in hepatic differentiation induced by FLCM treatment, hUCMSCs were transfected with control vector or pFLAG-CMV-Erk1(K71R) plasmids. Result showed that P-ERK expression was highly suppressed in hUCMSCs transfected with ERK1(K71R) during FLCM treatment (Figure 7(a)). Moreover, expression of hepatic-specific genes was also reduced in ERK1(K71R) transfected hUCMSCs (Figure 7(b) and (c)).
Figure 7.
Blockade of ERK1/2 phosphorylation with kinase deficient ERK1(K71R) reversed FLCM-induced hepatic differentiation of hUCMSCs. hUCMSCs were transfected with control vector or pFLAG-CMV-ERK1(K71R) and cultured in DMEM or FLCM for different times. A: Expression of P-ERK and T-ERK in hUCMSCs transfected with control vector or pFLAG-CMV-ERK1(K71R) by Western blotting analysis at day 2 during FLCM treatment; B and C: Expression of hepatocyte-specific genes evaluated by RT-PCR(B) and immunocytochemistry analysis(C). (A color version of this figure is available in the online journal.)
Discussion
Several reports have demonstrated that the efficiency of hepatic differentiation of MSCs can be improved by modifying the culture condition or by adding various growth factors and cytokines. However, the efficiency of hepatic differentiation of MSCs is still insufficient for clinical application. It is necessary to understand the mechanism of hepatic differentiation of MSCs to achieve transdifferentiation with high efficiency. Therefore, we tried to identify the signals important for hepatic specification by using FLCM to induce hepatic differentiation of hUCMSCs in vitro. Fetal livers have cytokines for early maturation, which act as promoting factors in the differentiation and maturation of hepatocytes during embryogenesis. For instance, the initiation of murine liver ontogeny requires secretion of FGF from the precardiac mesoderm.15 This is then followed by secretion of HGF and OSM by the surrounding mesenchymal stromal cells and hematopoietic stem cells, respectively,16–18 which promote hepatic differentiation and maturation. Indeed, several studies have demonstrated that the combination of HGF, OSM, and bFGF can induce hepatic differentiation in MSCs from different sources.11,19,20 However, differentiation was accelerated in FLCM-treated cells compared with HGF, OSM, and bFGF induction.13 Therefore, the FLCM may be a more suitable inducer for hepatic differentiation.
Previously we established a method to isolate hUCMSCs.21 The characteristics of hUCMSCs isolated in this manner corroborate results from previous studies and the documented expression of a consensus marker set of MSCs. hUCMSCs displayed a fibroblast-like appearance in culture and were positive for MSCs-specific markers such as CD13, CD44, CD271, and HLA-I. Following FLCM treatment, hUCMSCs lost their spindle-shaped appearance, and the expression of MSC-specific markers changed significantly on day 3. Moreover, the expression of stem cell-associated markers ABCG2, CD44, OCT4, and Nanog decreased in FLCM-induced cells. Expression of early stage and mature liver-specific markers such as AFP, ALB, CK-18, HNF4, TDO, and CYP1A1 were increased in FLCM-treated hUCMSCs. In addition, FLCM-induced hUCMSCs displayed functional characteristics of liver cells. Urea production was significantly higher in differentiated hUCMSCs than in undifferentiated cells. These results showed that FLCM can induce hepatic differentiation in hUCMSCs. Furthermore, FLCM treatment did not induce expression of neuron- and cardiomyocyte-specific genes, which means the specificity of FLCM-induced hepatic differentiation. Thus, the use of FLCM may serve as a useful inducing reagent, allowing detailed investigation of the signals involved in hepatic differentiation of hUCMSCs.
ERK1/2 is preferentially activated by mitogens such as serum or growth factors and is an important regulator of cellular growth, differentiation, and survival.22 Recent studies have demonstrated that ERK1/2 plays a crucial role in the regulation of stem cell function. ERK1/2 signaling maintains the self-renewing properties of MSCs.23–25 Ablation of ERK2 gene expression by RNA interference significantly reduces proliferation of hMSCs.26 In addition to promoting proliferation of stem cells, ERK1/2 also influences the lineage adopted by stem cells. A requirement for ERK1/2 signaling was observed for neuronal and mesendodermal differentiation in human ES cells.27 ERK1/2 phosphorylation is essential to adipogenic differentiation of bone marrow-derived MSC and endothelial cell differentiation of multipotent adult progenitor cells.28–30 Inhibition of the ERK1/2 pathway can increase ALP activity, and that mineralization in skeletal muscle-derived stem cells leads to osteogenesis.31
Hepatic differentiation is tightly regulated by cytokines and growth factors through signaling events. It has been reported that the fetal liver might provide factors such as HGF and bFGF for development and maturation of liver embryogenesis.13 HGF treatment can induce ERK1/2 phosphorylation in vitro.32,33 HGF can induce mitogenesis by ERK1/2 phosphorylation function in an autocrine manner.34 In fact, we found that HGF was enhanced in the FLCM group. Furthermore, ERKs are activated by many other growth factors and cytokines, including EGFR ligands and transforming growth factor-β (TGF-β) family members, some of which have been also reported to be expressed in the developing liver and to play a role in hepatocyte expansion, differentiation, and/or survival during liver development. To elucidate this, we devised a protocol using a combination of HGF and bFGF, and demonstrated its facilitation on ERK1/2 phosphorylation. However, ERK1/2 phosphorylation was not shown to increase until 28 days after HGF and bFGF treatment (data not shown). It is therefore implicated that some unknown factors are also involved in ERK1/2 phosphorylation during FLCM treatment on hUCMSCs.
In this study, we identified for the first time that ERK1/2 was an important signal in FLCM-induced hepatic differentiation of hUCMSCs. During FLCM induction, ERK1/2 phosphorylation was increased at 6 h and sustained at a high level even after 3 days. These results indicate that ERK1/2 phosphorylation was not transient and could be involved in hepatic differentiation of hUCMSCs in vitro. Treatment with U0126 (ERK1/2 inhibitor) strongly reversed the FLCM-induced expression of hepatic-specific genes including AFP, ALB, CK-18, HNF4, and TDO. Furthermore, the differentiation efficiency of FLCM-induced hUCMSCs decreased after ERK1/2 phosphorylation was blocked. HL 7702 is one kind of hepatocyte cell line and it has been used to study the function of human hepatocytes.35,36 Human hepatocyte cell HL7702 was used as a positive control. Taken together, ERK1/2 phosphorylation is associated with hepatic differentiation of hUCMSCs.
In summary, we demonstrated that FLCM could induce hUCMSCs to differentiate into hepatocyte-like cells and ERK1/2 signals play an important role in this differentiation. During hepatic differentiation, ERK1/2 phosphorylation was increased, and inhibition of ERK1/2 reduced the hepatic differentiation of hUCMSCs. These findings may provide useful information on stem cell biology, which should contribute to the development of regenerative medicine for liver diseases.
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (Grant no. 81200312, 81272481, 81270214), the Natural Science Foundation for Young Scholars of Jiangsu Province (Grant no. BK2012288), Jiangsu Province for Outstanding Sci-tech Innovation Team in Colleges and Universities (Grant no. SJK2013-10), Jiangsu Province’s Outstanding Medical Academic Leader and Sci-tech Innovation Team Program (Grant no. LJ201117), the Scientific Research Foundation of Jiangsu University (grant no. 11JDG062, 07JDG056).
Authors’ contributions
All authors participated in the design, interpretation of the studies, and analysis of the data and review of the manuscript; YY, YZ, FS, and LL conducted the experiments, BZ and WZ supplied critical reagents, ZS and WL wrote the manuscript, HQ and WX designed and supervised the experiments and revised the manuscript. YY, YZ, and FS contributed equally to this paper.
REFERENCES
- 1.Forraz N, McGuckin CP. The umbilical cord: A rich and ethical stem cell source to advance regenerative medicine. Cell Prolif Suppl 2011; 1: 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu KH, Liu YL, Zhou B, Han ZC. Cellular therapy and myocardial tissue engineering: The role of adult stem and progenitor cells. Eur J Cardiothorac Surg 2006; 30: 770–770. [DOI] [PubMed] [Google Scholar]
- 3.Jo CH, Kim OS, Park EY, Kim BJ, Lee JH, Kang SB, Lee JH, Han HS, Rhee SH, Yoon KS. Fetal mesenchymal stem cells derived from human umbilical cord sustain primitive characteristics during extensive expansion. Cell Tissue Res 2008; 334: 423–33. [DOI] [PubMed] [Google Scholar]
- 4.Lin SZ, Chang YJ, Liu JW, Chang LF, Sun LY, Li YS, Luo GH, Liao CH, Chen PH, Chen TM, Lee RP, Yang KL, Harn HJ, Chiou TW. Transplantation of human Wharton’s Jelly-derived stem cells alleviates chemically induced liver fibrosis in rats. Cell Transplant 2010; 19: 1451–63. [DOI] [PubMed] [Google Scholar]
- 5.Tsai PC, Fu TW, Chen YM, Ko TL, Chen TH, Shih YH, Hung SC, Fu YS. The therapeutic potential of human umbilical mesenchymal stem cells from Wharton’s jelly in the treatment of rat liver fibrosis. Liver Transpl 2009; 15: 484–95. [DOI] [PubMed] [Google Scholar]
- 6.Campard D, Lysy PA, Najimi M, Sokal EM. Native umbilical cord matrix stem cells express hepatic markers and differentiate into hepatocyte-like cells. Gastroenterology 2008; 134: 833–48. [DOI] [PubMed] [Google Scholar]
- 7.Bieback K, Brinkmann I. Mesenchymal stromal cells from human perinatal tissues: From biology to cell therapy. World J Stem Cells 2010; 2: 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ren H, Zhao Q, Cheng T, Lu S, Chen Z, Meng L, Zhu X, Yang S, Xing W, Xiao Y, Ren Q, Chi Y, Gu D, Yang R, Han ZC. No contribution of umbilical cord mesenchymal stromal cells to capillarization and venularization of hepatic sinusoids accompanied by hepatic differentiation in carbon tetrachloride-induced mouse liver fibrosis. Cytotherapy 2010; 12: 371–83. [DOI] [PubMed] [Google Scholar]
- 9.Zhang YN, Lie PC, Wei X. Differentiation of mesenchymal stromal cells derived from umbilical cord Wharton’s jelly into hepatocyte-like cells. Cytotherapy 2009; 11: 548–58. [DOI] [PubMed] [Google Scholar]
- 10.Yan Y, Xu W, Qian H, Si Y, Zhu W, Cao H, Zhou H, Mao F. Mesenchymal stem cells from human umbilical cords ameliorate mouse hepatic injury in vivo. Liver Int 2009; 29: 356–65. [DOI] [PubMed] [Google Scholar]
- 11.Snykers S, De Kock J, Tamara V, Rogiers V. Hepatic differentiation of mesenchymal stem cells: In vitro strategies. Methods Mol Biol 2011; 698: 305–14. [DOI] [PubMed] [Google Scholar]
- 12.Waclawczyk S, Buchheiser A, Flögel U, Radke TF, Kögler G. In vitro differentiation of unrestricted somatic stem cells into functional hepatic-like cells displaying a hepatocyte-like glucose metabolism. J Cell Physiol 2010; 225: 545–54. [DOI] [PubMed] [Google Scholar]
- 13.Pan RL, Chen Y, Xiang LX, Shao JZ, Dong XJ, Zhang GR. Fetal liver-conditioned medium induces hepatic specification from mouse bone marrow mesenchymal stromal cells: A novel strategy for hepatic transdifferentiation. Cytotherapy 2008; 10: 668–75. [DOI] [PubMed] [Google Scholar]
- 14.Frost JA, Geppert TD, Cobb MH, Feramisco JR. A requirement for extracellular signal-regulated kinase (ERK) function in the activation of AP-1 by Ha-Ras, phorbol 12-myristate 13-acetate, and serum. Proc Natl Acad Sci USA 1994; 91: 3844–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tremblay KD. Inducing the liver: Understanding the signals that promote murine liver budding. J Cell Physiol 2011; 226: 1727–31. [DOI] [PubMed] [Google Scholar]
- 16.Sonnenberg E, Meyer D, Weidner KM, Birchmeier C. Scatter factor/hepatocyte growth factor and its receptor, the c-met tyrosine kinase, can mediate a signal exchange between mesenchyme and epithelia during mouse development. J Cell Biol 1993; 123: 223–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sonnenberg E, Weidner KM, Birchmeier C. Expression of the met-receptor and its ligand, HGF-SF during mouse embryogenesis. Exs 1993; 65: 381–94. [PubMed] [Google Scholar]
- 18.Koyama T, Ehashi T, Ohshima N, Miyoshi H. Efficient proliferation and maturation of fetal liver cells in three-dimensional culture by stimulation of oncostatin M, epidermal growth factor, and dimethyl sulfoxide. Tissue Eng Part A 2009; 15: 1099–107. [DOI] [PubMed] [Google Scholar]
- 19.Bonora-Centelles A, Jover R, Mirabet V, Lahoz A, Carbonell F, Castell JV, Gómez-Lechón MJ. Sequential hepatogenic transdifferentiation of adipose tissue-derived stem cells: Relevance of different extracellular signaling molecules, transcription factors involved, and expression of new key marker genes. Cell Transplant 2009; 18: 1319–40. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida Y, Shimomura T, Sakabe T, Ishii K, Gonda K, Matsuoka S, Watanabe Y, Takubo K, Tsuchiya H, Hoshikawa Y, Kurimasa A, Hisatome I, Uyama T, Terai M, Umezawa A, Shiota G. A role of Wnt/beta-catenin signals in hepatic fate specification of human umbilical cord blood-derived mesenchymal stem cells. Am J Physiol Gastrointest Liver Physiol 2007; 293: G1089–98. [DOI] [PubMed] [Google Scholar]
- 21.Qiao C, Xu W, Zhu W, Hu J, Qian H, Yin Q, Jiang R, Yan Y, Mao F, Yang H, Wang X, Chen Y. Human mesenchymal stem cells isolated from the umbilical cord. Cell Biol Int 2008; 32: 8–15. [DOI] [PubMed] [Google Scholar]
- 22.Keshet Y, Seger R. The MAP kinase signaling cascades: A system of hundreds of components regulates a diverse array of physiological functions. Methods Mol Biol 2010; 661: 3–38. [DOI] [PubMed] [Google Scholar]
- 23.Wang B, Gao Y, Xiao Z, Chen B, Han J, Zhang J, Wang X, Dai J. ERK1/2 promotes proliferation and inhibits neuronal differentiation of neural stem cells. Neurosci Lett 2009; 461: 252–7. [DOI] [PubMed] [Google Scholar]
- 24.Lee JA, Jang DJ, Kang BK. Two major gate-keepers in the self-renewal of neural stem cells: ERK1/2 and PLCgamma1 in FGFR signaling. Mol Brain 2009; 2: 15–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi SC, Kim SJ, Choi JH, Park CY, Shim WJ, Lim DS. Fibroblast growth factor-2 and -4 promote the proliferation of bone marrow mesenchymal stem cells by the activation of the PI3K-Akt and ERK1/2 signaling pathways. Stem Cells Dev 2008; 17: 725–36. [DOI] [PubMed] [Google Scholar]
- 26.Cárcamo-Orive I, Tejados N, Delgado J, Gaztelumendi A, Otaegui D, Lang V, Trigueros C. ERK2 protein regulates the proliferation of human mesenchymal stem cells without affecting their mobilization and differentiation potential. Exp Cell Res 2008; 314: 1777–88. [DOI] [PubMed] [Google Scholar]
- 27.Na J, Furue MK, Andrews PW. Inhibition of ERK1/2 prevents neural and mesendodermal differentiation and promotes human embryonic stem cell self-renewal. Stem Cell Res 2010; 5: 157–69. [DOI] [PubMed] [Google Scholar]
- 28.Liao QC, Li YL, Qin YF, Quarles LD, Xu KK, Li R, Zhou HH, Xiao ZS. Inhibition of adipocyte differentiation by phytoestrogen genistein through a potential downregulation of extracellular signal-regulated kinases 1/2 activity. J Cell Biochem 2008; 104: 1853–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donzelli E, Lucchini C, Ballarini E, Scuteri A, Carini F, Tredici G, Miloso M. ERK1 and ERK2 are involved in recruitment and maturation of human mesenchymal stem cells induced to adipogenic differentiation. J Mol Cell Biol 2011; 3: 123–31. [DOI] [PubMed] [Google Scholar]
- 30.Xu J, Liu X, Jiang Y, Chu L, Hao H, Liua Z, Verfaillie C, Zweier J, Gupta K, Liu Z. MAPK/ERK signalling mediates VEGF-induced bone marrow stem cell differentiation into endothelial cell. J Cell Mol Med 2008; 12: 2395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Payne KA, Meszaros LB, Phillippi JA, Huard J. Effect of phosphatidyl inositol 3-kinase, extracellular signal-regulated kinases 1/2, and p38 mitogen-activated protein kinase inhibition on osteogenic differentiation of muscle-derived stem cells. Tissue Eng Part A 2010; 16: 3647–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taniguchi F, Harada T, Deura I, Iwabe T, Tsukihara S, Terakawa N. Hepatocyte growth factor promotes cell proliferation and inhibits progesterone secretion via PKA and MAPK pathways in a human granulosa cell line. Mol Reprod Dev 2004; 68: 335–44. [DOI] [PubMed] [Google Scholar]
- 33.He F, Wu LX, Shu KX, Liu FY, Yang LJ, Zhou X, Zhang Y, Huang BS, Huang D, Deng XL. HGF protects cultured cortical neurons against hypoxia/reoxygenation induced cell injury via ERK1/2 and PI3K/Akt pathways. Colloids Surf B Biointerfaces 2008; 61: 290–7. [DOI] [PubMed] [Google Scholar]
- 34.Chattopadhyay N, Tfelt-Hansen J, Brown EM. PKC, p42/44 MAPK and p38 MAPK regulate hepatocyte growth factor secretion from human astrocytoma cells. Brain Res Mol Brain Res 2002; 102: 73–82. [DOI] [PubMed] [Google Scholar]
- 35.Xie S, Zhu M, Lv G, Geng Y, Chen G, Ma J, Wang G. Overexpression of Ras homologous C (RhoC) induces malignant transformation of hepatocytes in vitro and in nude mouse xenografts. PLoS One 2013; 8: e54493–e54493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao J, Chen J, Lu B, Dong L, Wang H, Bi C, Wu G, Guo H, Wu M, Guo Y. TIP30 induces apoptosis under oxidative stress through stabilization of p53 messenger RNA in human hepatocellular carcinoma. Cancer Res 2008; 68: 4133–41. [DOI] [PubMed] [Google Scholar]