Abstract
Statins are potent cholesterol-lowering drugs and are generally well tolerated. Hepatotoxicity is a rare but serious adverse effect of statins; however, its mechanisms are not clear. Coenzyme Q10 deficiency has been suggested, and supplementation of reduced coenzyme Q10 (ubiquinol) has been shown to have hepatoprotective effects. MicroRNAs (miRNAs) are small nucleotides that have been shown to be up-regulated in drug-induced liver injury. We hypothesized that circulating miRNAs may be differentially regulated after simvastatin treatment and by comparing with that of simvastatin and ubiquinol supplementation could potentially uncover signatory miRNA profile for simvastatin-induced liver injury. In this double-blind, prospective, randomized-controlled trial, miRNA profiles and liver enzymes were compared between simvastatin-treated patients, with and without ubiquinol supplementation, over 12 weeks compared to baseline. miRNA expression was further validated in HepG2 liver cell lines by real-time PCR. Changes in miR-192, miR-146a, miR-148a, miR-15a, and miR-21 were positively correlated (p<0.05) with alanine aminotransferase in simvastatin-only treated patients. In ubiquinol supplementation group, alanine aminotransferase and alkaline phosphatase were significantly down-regulated after 12 weeks and changes in miR-15a, miR-21 and miR-33a were negatively correlated with alkaline phosphatase (p < 0.05). Bioinformatics analyses predicted that miRNA regulation in simvastatin group was related to reduce proliferation and adenosine triphosphate-binding cassette transporters. Ubiquinol supplementation additionally regulated miRNAs that inhibit apoptotic and inflammatory pathways, suggesting potential hepatoprotective effects. Our results suggest that 20 mg/day of simvastatin does not have significant risk of hepatotoxicity and ubiquinol supplementation may, at the miRNA level, provide potential beneficial changes to reduce the effects of coenzyme Q10 deficiency in the liver.
Keywords: Simvastatin, HMG-CoA reductase, cholesterol, hepatotoxicity, miR-192, miR-21
Introduction
Statins are 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase inhibitors which inhibit the synthesis of mevalonate, a precursor for cholesterol and coenzyme Q10 (CoQ10) biosynthesis.1,2 They are widely prescribed to patients with hypercholesterolemia to lower low-density lipoprotein cholesterol (LDL-C). Sustained statin therapy has been shown to lower LDL-C significantly with corresponding reduction of the risk of cardiovascular events by about 33%.3
Despite its well-known benefits, studies have reported that statins are also uncommonly associated with dose-dependent side effects. Compared to placebo, adverse effects such as lethargy and exertional fatigue have been reported by patients on simvastatin and pravastatin,4 with inhibition of mitochondrial respiration resulting in reducing oxygen consumption in C2C12 myotubes.5 Additionally, elevation of liver transaminases (e.g., alanine aminotransferase [ALT] and aspartate aminotransferase [AST]) has been documented in about 2% of the patients in clinical trials. Although several studies suggest that the elevation of transaminases is minor, case reports of patients with severe hepatotoxicity have been published.6 These included report of two patients treated with simvastatin (20 mg/day for 90 days), one of whom died while the other required liver transplant.7 Although the etiology of statin-induced toxicity has not been elucidated, it has been suggested that CoQ10 deficiency may play an important role. CoQ10 is involved in mitochondrial energy coupling, adenosine triphosphate (ATP) production, and anti-oxidative action.8 In humans, two major forms exist: the oxidized form, ubiquinone; and the reduced and active form ubiquinol. Using an in-vitro model, we previously showed that CoQ10 supplementation has a hepatoprotective role in simvastatin-treated HepG2 cells,1 resulting in increased ATP synthesis and reduced apoptosis and oxidative stress.
microRNAs (miRNAs) are small, non-coding RNA molecules that have been shown to regulate biological functions by repressing post-transcriptional processes through mRNA transcript destabilization or protein translational inhibition, often resulting in decreased protein expression.9 Some miRNAs (miR-192, miR-146a, miR-148a, miR-15a, and miR-21) have been reported to be up-regulated in drug-induced liver injury.10 Animal models11 and clinical studies12 suggest that serum miR-192 concentration may increase early in the disease process of acetaminophen-induced liver injury, even before rise in ALT, raising the potential for circulating miRNAs to be early biomarkers for drug-induced liver injury.
We hypothesize that circulating miRNAs profiles change in hypercholesterolemia patients, treated with simvastatin. By comparing their profiles to that in simvastatin-treated patients with ubiquinol supplementation (previously shown to be hepatoprotective), this may reveal a signatory miRNA profile to identify early biomarkers for rare cases with simvastatin-induced liver injury. Additionally, we explored changes in liver transaminases (ALT, AST), alkaline phosphatase (ALP), γ-glutamyltransferase (GGT), and additional hepatotoxic biomarkers (Arginase-1, Serum F, and glutathione S-transferase-α [GSTα]) in these two groups of patients.
Materials and methods
Patient recruitment and sample collection
This was a randomized, double-blinded, placebo-controlled study where 40 hypercholesterolemic patients with marginal elevations of liver enzymes at baseline were enrolled. After a two-week washout of lipid modifying medications, all patients received simvastatin (20 mg/day) and were randomized to receive supplementation with either ubiquinol 150 mg/day (Group 2, n = 20) or placebo of ubiquinol (Group 1, n = 20). Capsules of ubiquinol and placebo of ubiquinol were obtained from Kaneka Corporation (Japan). Randomization was first carried out by SPSS (version 16.0) and placed in sealed envelopes with a study administrator who had no direct patient contact.
Peripheral blood was collected before drug treatment (week 0) and at 12 weeks, while on treatment. Patients whose baseline AST and ALT were greater than three times upper limit of normal, who had active hepatitis or cirrhosis (shown by blood biomarkers, ultrasound or histology), and with history of abnormal liver function rising more than three times upper limit of normal while on statin therapy were excluded. This study was approved by our institutional ethics committee, and all patients signed written informed consent.
Clinical and biochemical measurements
Anthropometric measures and fasting blood were collected in serum-separating tubes after 10-h overnight fast. Serum triglyceride (TG), total cholesterol, high-density lipoprotein cholesterol (HDL-C), LDL-C levels, ALT, AST, ALP, and GGT were measured using automatized autoanalyzer (COBAS-Roche, Germany), while lactate was measured with Lactate Analyzer, specific for L-lactate (Roche, Switzerland). Pyruvate was measured using commercial kit (Boehringer, Germany), while Serum F-protein (USCN, China), Arginase-1 (Abnova, Taiwan), and GST-α (Argutus Medical, Ireland) were measured by ELISA kits. Intra- and inter-assay coefficients of variation were Serum-F (8.2% and 17.0%, respectively), Arginase-1 (5.4% and 3.0%, respectively), and GST-α (3.5% and 16.4%, respectively). RNAlater® solution (Ambion, USA) was added to ethylenediaminetetraacetic acid (EDTA)-collected whole blood and stored at −80℃ for RNA isolation.
Measurement of ubiquinone, ubiquinol, and total Q10 using high-performance liquid chromatography
Serum samples from all patients (baseline and week 12) collected were protected from light and kept frozen at −80℃ until analysis. High-performance liquid chromatography (HPLC) was used to simultaneously measure the serum concentrations of redox forms of CoQ10, as described by Lee et al.13
Total RNA isolation
Total RNA, including miRNA from whole blood, was extracted using RiboPure™-Blood kit (Ambion, USA) as previously described.14 For cell culture experiments, total RNA and miRNA were extracted from cell lysate using miRNeasy Mini Kit (Qiagen, Germany).
miRNA microarray and quantitative real-time PCR
RNA (including miRNA) was profiled from 32 patients (females and those with ethnicity indicated as “Others” were excluded; see details in Supplementary methods).
Validation of microarray results was done by quantitative real-time PCR (qPCR) on all 40 patients. Total RNA (20 ng) was reversed transcribed using cDNA synthesis kit (Exiqon, Denmark). miRNA expression was performed using Locked nucleic acid (LNA) qPCR with miRNA-specific primers of miR-192, miR-146a, miR-148a, miR-30b, miR-15a, miR-21, or miR-33a (Exiqon, Denmark). PCR protocol and dissociation stage were performed according to manufacturer’s instructions, using 7500 Fast PCR system (Applied Biosystems, USA). SNORD48 was used as an internal reference for relative expression, using the method 2−ΔΔCt.
Simvastatin and ubiquinol treatment in liver cell lines
HepG2 and THLE liver cell lines (ATCC, USA) were treated as described previously1 (Supplementary methods).
miR-192 and miR-21 transfection in HepG2 cells
HepG2 were seeded in a six-well plate at 0.35, 0.3, 0.2, or 0.175 million cells per well for 24 h, 48 h, 72 h, and 96 h, respectively, to achieve 90% confluence before harvesting. Twenty-four hours after seeding, 25 nM of miRIDIAN hsa-miR-192 mimic (CUGACCUAUGAAUUGACAGCC), hsa-miR-21 mimic (UAGCUUAUCAGACUGAUGUUGA), dual transfection of miR-192 and miR-21, or negative mimic (UCACAACCUCCUAGAAAGAGUAGA) (Dharmacon, USA) were transfected into HepG2 using transfection reagent (JetPrime, France). HepG2 were harvested after indicated times and stored at −80℃ before extraction. Experiments were conducted four to five times.
Cell viability assay
HepG2 were seeded in 96-well black opaque plates (Costar,USA) with proportional cell number to the 6-well plate mimic transfection for the same time points. Both CellTiter-Blue® Cell Viability and CellTiter-Glo® Luminescent Cell Viability Assays (Promega, USA) were carried out according to manufacturer’s protocol after transfection. Viability of the miRNA mimic-transfected cells was normalized against negative mimic-transfected HepG2.
Statistical analysis
Data were analyzed using SPSS version 21 and presented as mean ± standard deviation (SD) or median (interquartile range) or in percentages. Chi-square and independent t-test was employed to assess differences in distribution of categorical and continuous variables, respectively. Paired t-test (parametric) or Wilcoxon signed-rank test (non-parametric) was used to assess differences in log transformed signal intensities for microarray, Qpcr, and clinical variables at baseline and week 12. Associations of miRNAs with clinical parameters were analyzed using multiple linear regression analysis. Skewed data such as TG, pyruvate, lactate:pyruvate ratio, GGT, ALT, GSTα and miRNA expressions were analyzed after logarithmic transformation. ANOVA and Student’s t-test were used to assess differences in cell culture. A p-value < 0.05 was considered statistically significant. miRNA targets and pathway analyses were analyzed using miRsystem version20130328.15
Results
The 40 patients recruited had mean age of 46.15 ± 11.97 years and consisted of 35 males. Mean baseline total cholesterol was 6.34 ± 0.94 mmol/l, LDL-C 4.35 ± 0.92 mmol/l, TG 2.11 (1.92) mmol/l and HDL-C was 1.25 ± 0.65 mmol/l. Except for Group 2 having a higher proportion of patients with diabetes, baseline characteristics of patients in the placebo and ubiquinol groups were not significantly different (Table 1).
Table 1.
Baseline characteristics of study subjects in both simvastatin and ubiquinol; or simvastatin with placebo of ubiquinol groups
| Characteristics | Group 1 Simvastatin and Placebo (n = 20) | Group 2 Simvastatin and Ubiquinol (n = 20) | p-value |
|---|---|---|---|
| Age (years) | 49.2 ± 12.2 | 43.1 ± 11.3 | 0.108 |
| Gender (%male) | 85 | 90 | 0.5 |
| Ethnicity (%) | |||
| Chinese | 45.0 | 50 | |
| Malay | 20.0 | 30.0 | |
| Indian | 30.0 | 15.0 | |
| Others | 5.0 | 5.0 | 0.505 |
| Body mass index (kg/m2) | 31.2 ± 6.1 | 29.2 ± 3.8 | 0.229 |
| Fasting glucose (mmol/L) | 7.41 ± 3.41 | 8.39 ± 2.48 | 0.305 |
| Systolic blood pressure (mmHg) | 130.7 ± 16.2 | 128.1 ± 12.9 | 0.584 |
| Diastolic blood pressure (mmHg) | 78.2 ± 12.1 | 78.8 ± 10.2 | 0.866 |
| Total cholesterol (mmol/L) | 6.48 ± 1.05 | 6.19 ± 0.81 | 0.33 |
| Triglyceride (mmol/L)a | 2.27 (1.86) | 1.73 (2.37) | 0.966 |
| HDL-cholesterol (mmol/L) | 1.36 ± 0.89 | 1.13 ± 0.29 | 0.283 |
| LDL-cholesterol (mmol/L) | 4.51 ± 1.02 | 4.18 ± 0.82 | 0.266 |
| Lactate | 1.56 ± 0.51 | 1.78 ± 0.62 | 0.223 |
| Pyruvate | 48.75 ± 16.94 | 45.75 ± 18.21 | 0.474 |
| Lactate:Pyruvatea | 0.027 (0.03) | 0.34 (0.04) | 0.249 |
| Ubiquinol | 0.61 ± 0.20 | 0.64 ± 0.21 | 0.652 |
| Ubiquinone | 0.21 ± 0.07 | 0.22 ± 0.07 | 0.719 |
| Total Q10 | 0.81 ± 0.26 | 0.85 ± 0.26 | 0.656 |
| Ubiquinol/Total Cholesterol (%) | 9.39 ± 2.79 | 10.35 ± 3.37 | 0.335 |
| GGTa (U/L) | 54.5 (36.25) | 71.0 (80.3) | 0.327 |
| ALP (U/L) | 79.7 ± 26.7 | 78.7 ± 18.8 | 0.892 |
| ALTa (U/L) | 63.5 (22.5) | 64.5 (53.0) | 0.485 |
| AST (U/L) | 36.7 ± 9.5 | 36.8 ± 14.2 | 0.981 |
| Creatine Kinase (U/L) | 174.6 ± 108.6 | 186.2 ± 98.7 | 0.821 |
| SerumF1 (ng/ml) | 332.4 ± 62.3 | 342.0 ± 67.0 | 0.643 |
| GSTαa (ng/ml) | 10.3 (16.1) | 22.0 (19.2) | 0.088 |
| Arginasea (ng/ml) | 17.39 (57.07) | 25.25 (57.07) | 0.643 |
| % on medication | |||
| Fibrates | 10 | 15 | 0.5 |
| Aspirin | 20 | 25 | 0.5 |
| Blood pressure lowering | 55 | 55 | 0.5 |
| Insulin | 10 | 30 | 0.118 |
| % Disease | |||
| Diabetes | 40 | 80 | 0.011 |
| Hypertension | 55 | 65 | 0.374 |
| Smoking | 10 | 0 | 0.198 |
| Exercise | 75 | 65 | 0.442 |
Data are expressed as mean ± SD or median (interquartile range). HDL: high-density lipoprotein; LDL: low-density lipoprotein; GGT: γ-glutamyltransferase; ALP: alkaline phosphatase; ALT: alanine aminotransferase; AST: aspartate aminotransferase; CK: ▪; GSTα: glutathione S-transferase-α. Bold text indicates a statistically significant difference with a p-value < 0.05
Log transformed before analysis.
Effects on lipids, lactate, pyruvate, and redox forms of Q10 after treatment
Total cholesterol, LDL-C, and TG were significantly reduced after 12 weeks of treatment in both groups (Table 2). Lactate/Pyruvate ratios were significantly higher (p = 0.035) after simvastatin with placebo treatment (Group 1), but not with ubiquinol supplementation (Group 2). Ubiquinol was significantly reduced by 48% in Group 1 patients (p < 0.0001), but with ubiquinol supplementation (Group 2), both redox forms of Q10 were significantly increased in the serum after 12 weeks.
Table 2.
Lipid, liver markers, lactate, pyruvate, and redox forms of Q10 before and 12 weeks after treatment
| Characteristics | Group 1 |
Group 2 |
p-value between Group 1 and 2 after treatment | |||||
|---|---|---|---|---|---|---|---|---|
| Placebo (n = 20) |
Ubiquinol (n = 20) |
|||||||
| Baseline | 12 weeks | p-value | Baseline | 12 weeks | p-value | |||
| Total cholesterol (mmol/L) | 6.48 ± 1.05 | 4.78 ± 1.02 | <0.0001 | 6.19 ± 0.81 | 4.52 ± 0.82 | <0.0001 | 0.648 | |
| Triglyceride (mmol/L)a | 2.27 (1.86) | 1.52 (1.1) | 0.008 | 1.71 (2.61) | 1.68 (2.02) | 0.027 | 0.595 | |
| HDL-cholesterol (mmol/L) | 1.36 ± 0.89 | 1.21 ± 0.31 | 0.453 | 1.13 ± 0.29 | 1.18 ± 0.25 | 0.3 | 0.622 | |
| LDL-cholesterol (mmol/L) | 4.51 ± 1.02 | 3.08 ± 0.75 | <0.0001 | 4.18 ± 0.82 | 2.57 ± 0.58 | <0.0001 | 0.125 | |
| Lactate:Pyruvatea | 0.027 (0.03) | 0.046 (0.04) | 0.035 | 0.034 (0.04) | 0.036 (0.020) | 0.52 | 0.124 | |
| Ubiquinol | 0.61 ± 0.20 | 0.32 ± 0.10 | <0.0001 | 0.64 ± 0.21 | 1.74 ± 0.95 | <0.0001 | <0.0001 | |
| Ubiquinone | 0.21 ± 0.07 | 0.18 ± 0.05 | 0.215 | 0.22 ± 0.07 | 0.65 ± 0.26 | <0.0001 | <0.0001 | |
| Total Q10 | 0.81 ± 0.26 | 0.50 ± 0.12 | <0.0001 | 0.85 ± 0.26 | 2.39 ± 1.18 | <0.0001 | <0.0001 | |
| Ubiquinol/Total Cholesterol (%) | 9.39 ± 2.79 | 6.72 ± 2.01 | <0.0001 | 10.35 ± 3.37 | 40.20 ± 23.11 | <0.0001 | <0.0001 | |
| GGTa (U/L) | 54.5 (36.3) | 54.5 (36.3) | 0.759 | 69.0 (77.0) | 64.0 (50.0) | 0.35 | 0.629 | |
| ALP (U/L) | 79.7 ± 26.7 | 77.6 ± 23.3 | 0.499 | 78.7 ± 18.8 | 73.3 ± 15.8 | 0.011 | 0.085 | |
| ALTa (U/L) | 63.5 (22.5) | 52.0 (29.5) | 0.262 | 65.6 (55.0) | 56.0 (42.0) | 0.017 | 0.45 | |
| AST (U/L) | 36.7 ± 9.5 | 35.9 ± 11.6 | 0.704 | 36.8 ± 14.2 | 38.7 ± 14.4 | 0.291 | 0.158 | |
| SerumF (ng/ml) | 332.4 ± 62.3 | 329.14 ± 82.0 | 0.766 | 342.0 ± 67.0 | 324.3 ± 80.5 | 0.339 | 0.707 | |
| GSTαa (ng/ml) | 10.29 (16.12) | 9.44 (15.19) | 0.435 | 20.1 (20.3) | 18.5 (30.1) | 0.642 | 0.692 | |
| Arginase1a (ng/ml) | 17.39 (57.07) | 24.26 (52.05) | 0.792 | 25.25 (50.0) | 21.25 (29.63) | 0.169 | 0.137 | |
| Creatine Kinase (U/L) | 174.6 ± 108.6 | 184.8 ± 100.4 | 0.628 | 186.2 ± 98.7 | 267.3 ± 282.8 | 0.142 | 0.265 | |
Data are expressed as mean ± SD or median (interquartile range). HDL: high-density lipoprotein; LDL: low-density lipoprotein; GGT: γ-glutamyltransferase; ALP: alkaline phosphatase; ALT: alanine aminotransferase; AST: aspartate aminotransferase; CK: ▪; GSTα: glutathione S-transferase-α.
Log transformed before analysis. Bold text indicates a statistically significant difference with a p-value < 0.05
Effects on liver enzymes after treatment
Baseline ALT was marginally elevated and was not significantly different between Groups 1 and 2 (Table 1). All measured liver enzymes were not significantly different after 12 weeks in Group 1 (Table 2). Instead, ALP and ALT were significantly reduced after 12 weeks of ubiquinol supplementation (p = 0.011 and p = 0.017, respectively). The percentage change of ALT (p = 0.45) and ALP (p = 0.085), with respect to baseline, was however not significantly different between the two groups.
Effects on miRNAs after treatment
In sum, 207 miRNAs were detected above threshold. There were no significant differences in miRNA microarray expression (not shown) and qPCR (Supplementary Table 1(c)), between both groups at baseline. Microarray data are summarized in Supplementary Figure 1. In Group 1, 13 miRNAs were significantly down-regulated and 28 miRNAs were significantly up-regulated after 12 weeks of treatment (Supplementary Figure 1(a)). In Group 2, 8 miRNAs were significantly down-regulated and 18 miRNAs were significantly up-regulated after 12 weeks of treatment. Of the down-regulated miRNAs, 6 miRNAs overlap between the two groups and of up-regulated miRNAs, 16 miRNAs overlap between the two groups. Supplementary Figure 1(b) shows the heatmap of the microarray, and Supplementary Table 1 shows the list of miRNAs differentially regulated in week 12. We subjected circulating miRNA expression from both groups to global miRNA prediction and pathway analyses. miRNAs affecting fatty acid, amino acid, and tricarboxylic acid cycle (TCA) cycle metabolism pathways had high enrichment scores irrespective of ubiquinol use (Supplementary Table 2(a)), while pathways related to oxidative phosphorylation and cholesterol transport were highly enriched in group with ubiquinol (Supplementary Table 2(b)).
miR-33a was shown to be implicated in cholesterol16 and hence included in analyses even though it was below detection limits in the microarray. Relative expression of six selected miRNAs by qPCR was highly correlated to microarray data (p < 0.0001) (Supplementary Figure 2).
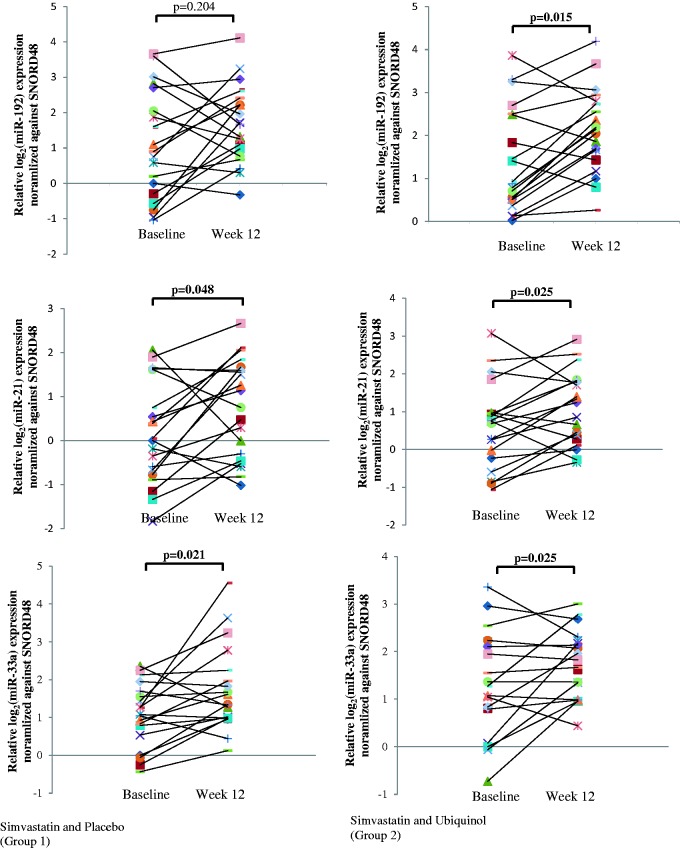
miR-21, miR-33a (Figure 1), and miR-15a (not shown) were significantly up-regulated in both groups after treatment, as determined by qPCR. After 12 weeks of treatment, there were no significant changes in miR-192 (Figure 1), miR-146a, miR-148a, and miR-30b (not shown) expression in Group 1 (simvastatin with placebo) (miR-192, p = 0.204; miR-146a, p = 0.279; miR-148a, p = 0.391; and miR-30b, p = 0.179), but these miRNAs were up-regulated in Group 2, where patients received simvastatin, with ubiquinol supplementation (miR-192, p = 0.015; miR-146a, p = 0.021; miR-148a, p = 0.025; and miR-30b, p = 0.044).
Figure 1.
Real-time PCR validation of microRNA expression of individual patients at baseline and after 12 weeks of treatment. microRNAs are as indicated. Each experiment was repeated at least twice and data were normalized to SNORD48 expression. Paired t-test was used to compute statistical significance of log microRNA expression (A color version of this figure is available in the online journal)
As TG and LDL-C were significantly reduced in both groups (Table 2), we analyzed the association of changes in TGs and LDL-C on miRNAs expression. In bivariate analyses, changes in miR-192 (ΔmiR-192) (Rho = −0.317, p = 0.046), ΔmiR-146a (Rho = −0.384, p = 0.014), ΔmiR-148a (Rho = −0.317, p = 0.046), ΔmiR-15a (Rho = −0.389, p = 0.013), and ΔmiR-21 (Rho = −0.406, p = 0.009) were negatively and significantly associated with changes in TGs (Supplementary Figure 3), while changes in LDL-C were not significantly associated with all seven candidate miRNAs (not shown).
After adjustment for age, gender, and ethnic group, changes in miR-192 and miR-148a (ΔmiR-192: B = 0.076, p = 0.015 and ΔmiR-148a: B = 0.075, p = 0.043) remained significantly associated with changes in ALT, while ΔmiR-33a (B = 0.049, p = 0.002) remained significantly associated with changes in ALP (Table 3). When segregated into respective treatment groups, changes in ALT were significantly associated with ΔmiR-192 (B = 0.117, p = 0.001), ΔmiR-146a (B = 0.151, p = 0.012), ΔmiR-148a (B = 0.153, p < 0.0001), ΔmiR-15a (B = 0.178, p = 0.006), and ΔmiR-21 (B = 0.130, p = 0.022), after adjustment for age, gender, and ethnicity (Table 4) in Group 1 (simvastatin + placebo) but not Group 2. There were no significant correlation of changes in seven candidate miRNAs with changes in ALP in Group 1 while ΔmiR-15a (B = −0.069, p = 0.014), ΔmiR-21 (B = −0.059, p = 0.005), and ΔmiR-33a (B = −0.043, p = 0.018) correlated negatively with changes in ALP, after adjustment for age, gender, and ethnicity (Table 4) in Group 2. All seven candidate miRNAs correlated significantly with one another (Supplementary Table 3).
Table 3.
Correlations of changes in ALT and ALP with changes in microRNAs and other liver biomarkers, adjusted for age, gender, and ethnicity (n = 40)
| Change in ALT (%) |
Change in ALP (%) |
||||
|---|---|---|---|---|---|
| B | p-value | B | p-value | ||
| Changes in miR-192 (%) | 0.076 | 0.015 | Changes in miR-192 (%) | −0.016 | 0.397 |
| Changes in miR-146a (%) | 0.057 | 0.119 | Changes in miR-146a (%) | −0.012 | 0.555 |
| Changes in miR-148a (%) | 0.075 | 0.043 | Changes in miR-148a (%) | 0.033 | 0.121 |
| Changes in miR-30b (%) | 0.027 | 0.421 | Changes in miR-30b (%) | −0.019 | 0.310 |
| Changes in miR-15a (%) | 0.088 | 0.092 | Changes in miR-15a (%) | 0.013 | 0.676 |
| Changes in miR-21 (%) | 0.074 | 0.097 | Changes in miR-21 (%) | 0.011 | 0.668 |
| Changes in miR-33a (%) | −0.002 | 0.948 | Changes in miR-33a (%) | 0.049 | 0.002 |
| Changes in ALP (%) | 0.111 | 0.671 | Changes in ALT (%) | 0.085 | 0.334 |
| Changes in AST (%) | 0.539 | <0.0001 | Changes in AST (%) | −0.040 | 0.635 |
| Changes in GST-α (%) | 0.152 | 0.125 | Changes in GST-α (%) | −0.081 | 0.147 |
| Changes in GGT (%) | 0.389 | 0.001 | Changes in GGT (%) | 0.207 | 0.003 |
| Changes in Arginase (%) | 0.011 | 0.875 | Changes in Arginase (%) | −0.003 | 0.943 |
| Changes in Serum F (%) | −0.049 | 0.852 | Changes in Serum F (%) | 0.098 | 0.495 |
Generalized linear model was used to assess changes with ALT and ALP with adjustments for age, gender, and ethnicity. GGT: γ-glutamyltransferase; ALP: alkaline phosphatase; ALT: alanine aminotransferase; AST: aspartate aminotransferase. Bold text indicates a statistically significant difference with a p-value < 0.05
Table 4.
Correlations of changes in ALT and ALP with changes in microRNAs and other liver biomarkers, adjusted for age, gender, and ethnicity, segregated by treatment groups (n = 20/group)
| Group 1 (Simvastatin and Placebo) |
Group 2 (Simvastatin and Ubiquinol) |
||||
|---|---|---|---|---|---|
| B | p-value | B | p-value | ||
| Change in ALT (%) | |||||
| Changes in miR-192 (%) | 0.117 | 0.001 | 0.02 | 0.697 | |
| Changes in miR-146a (%) | 0.151 | 0.012 | 0.017 | 0.568 | |
| Changes in miR-148a (%) | 0.153 | <0.0001 | −0.031 | 0.577 | |
| Changes in miR-30b (%) | 0.094 | 0.054 | −0.010 | 0.763 | |
| Changes in miR-15a (%) | 0.178 | 0.006 | 0.013 | 0.860 | |
| Changes in miR-21 (%) | 0.130 | 0.022 | 0.02 | 0.730 | |
| Changes in miR-33a (%) | <0.001 | 0.994 | 0.073 | 0.106 | |
| Change in ALP (%) | |||||
| Changes in miR-192 (%) | −0.032 | 0.253 | −0.034 | 0.102 | |
| Changes in miR-146a (%) | −0.020 | 0.654 | −0.18 | 0.131 | |
| Changes in miR-148a (%) | 0.024 | 0.500 | −0.041 | 0.063 | |
| Changes in miR-30b (%) | −0.056 | 0.089 | −0.023 | 0.075 | |
| Changes in miR-15a (%) | −0.021 | 0.671 | −0.069 | 0.014 | |
| Changes in miR-21 (%) | −0.002 | 0.957 | −0.059 | 0.005 | |
| Changes in miR-33a (%) | 0.048 | 0.058 | −0.043 | 0.018 | |
Generalized linear model was used to assess changes with ALT and ALP with adjustments for age, gender, and ethnicity. ALP: alkaline phosphatase; ALT: alanine aminotransferase. Bold text indicates a statistically significant difference with a p-value < 0.05
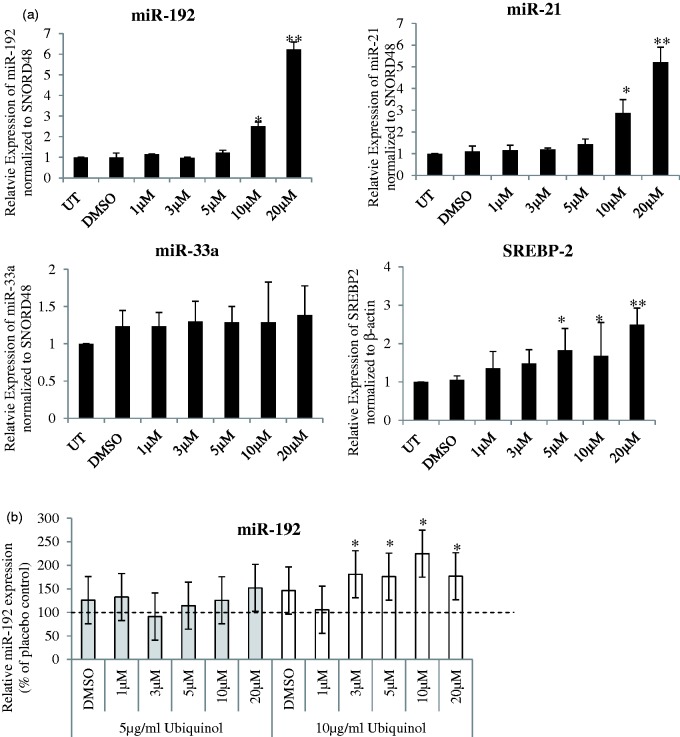
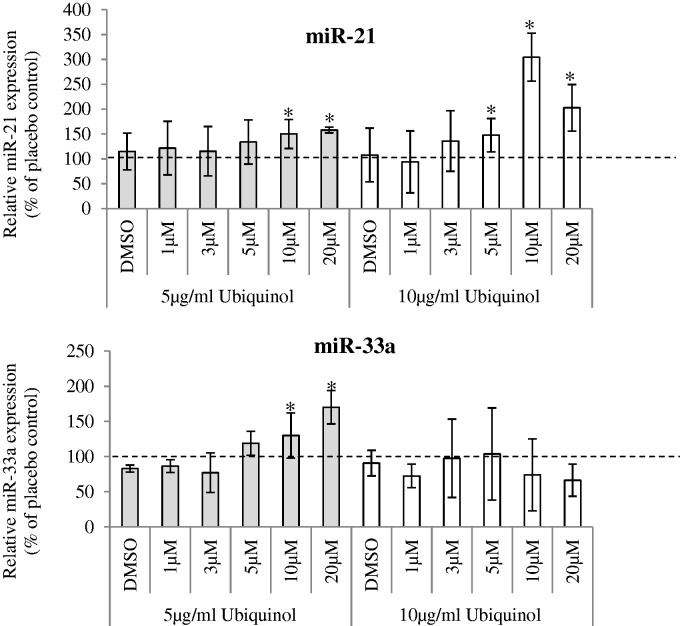
In HepG2 cells treated with different doses of simvastatin, with or without ubiquinol supplementation (Figure 2), we observed significant dose-dependent increase in miR-192 and miR-21 after 16 hours of simvastatin treatment (Figure 2(a)). miR-146a expression was low in HepG2 cells (Ct ≈ 40) and hence, not further evaluated. Supplementation of 5 µg/ml or 10 µg/ml ubiquinol increased the expression of all six candidate miRNAs (other than miR-146a) (data for miR-192, miR-21, and miR33a shown in Figure 2(b)). Similarly, reduced cell viability and increased miR-192 and miR-21 expressions were observed in THLE-2 cells (derived from normal liver) at both 20 µM and IC50 dose (60 µM) of simvastatin (Supplementary Figure 4).
Figure 2.
Simvastatin treatment in HepG2 cells. (a) Respective microRNA expression are shown after varying doses of (i) simvastatin treatment only and (ii) co-treatment of varying doses of simvastatin and 5 µg/ml ubiquinol or 10 µg/ml ubiquinol for 16 h. Relative expression were normalized to the housekeeping small RNA, SNORD48, and expressed with respect to untreated control. UT: Untreated and DMSO refers to cells treated with simvastatin solvent, DMSO, for the same amount of time. In (a), SREBP2 gene expression was normalized to β-actin. In (b), relative expression of microRNA was normalized with respect to placebo control, which is the solvent for ubiquinol. microRNA are as indicated. Data are mean values ± SD of three to four independent experiments, each repeated at least twice. *p < 0.05, **p < 0.01 compared with untreated
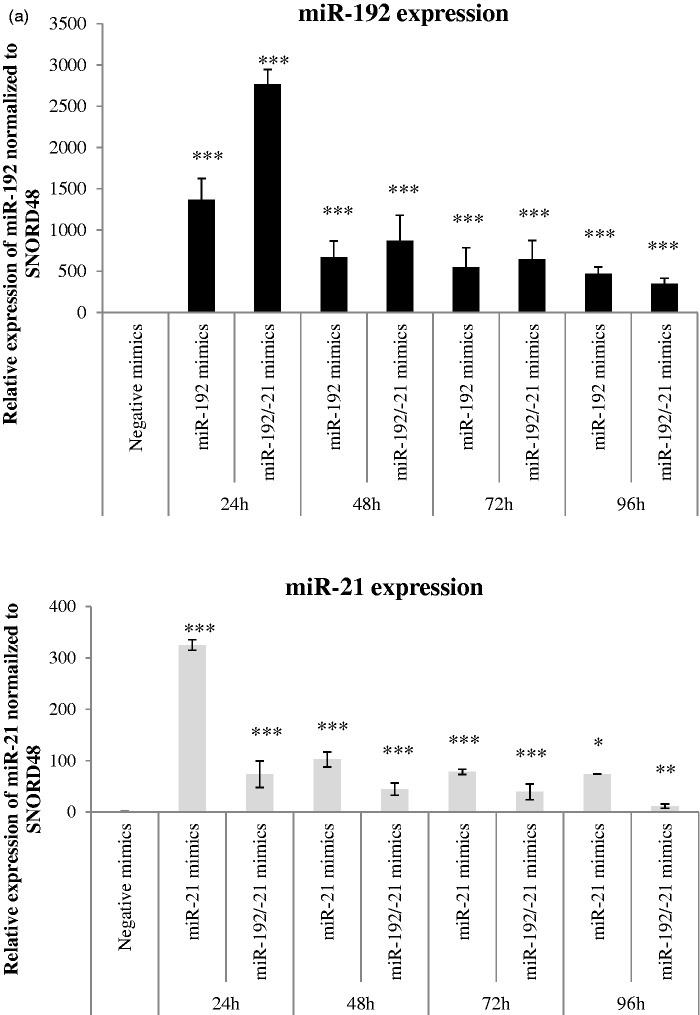
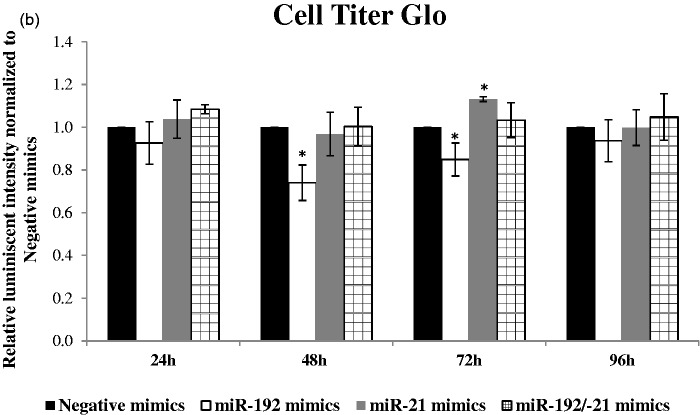
In HepG2 transfected with miR-192 or miR-21 or dual transfection of both mimics (Figure 3(a)), over-expression of miR-192 or miR-21 over various time-points indicated no significant difference in cell viability using CellTiter-Blue assay (not shown). Using an alternative method, CellTiter-Glo, which measures ATP production as a surrogate for metabolically active cells, there was a reduction at 48 h and 72 h, after miR-192 transfection (Figure 3(b)). Conversely, there was an increase in cell viability, 72 h after miR-21 transfection, while dual transfection showed no significant changes in viability.
Figure 3.
Transfection of HepG2 liver cells with microRNA mimics. (a) Real-time PCR validation of miR-192 and miR-21 or dual transfection of miR-192 and miR-21 over time points 24 h, 48 h, 72 h, and 96 h are shown. (Top) miR-192 expression normalized to housekeeping control SNORD48 and (bottom) miR-21 expression normalized to housekeeping control SNORD48. miRNA expression were relative to negative mimics at indicated time points. *p < 0.05, **p < 0.01, ***p < 0.0001 compared with negative mimics transfected HepG2. (b) Cell Titer Blue (not shown) and Cell Titer Glo were used to assess cell viability after transfection
Discussion
We report, for the first time, changes in a panel of liver function enzymes with miRNA changes to assess the safety of simvastatin treatment in patients randomized to ubiquinol or placebo of ubiquinol. Expectedly, simvastatin reduced LDL-C significantly, while ubiquinol was significantly reduced in Group 1, with supplementation of ubiquinol (Group 2) significantly increasing ubiquinol and ubiquinone. Simvastatin dose at 20 mg/day is relatively safe in our patients as it did not raise liver enzymes, similar to recent clinical trials.17 Ubiquinol supplementation did not affect LDL-lowering effect of simvastatin and attenuated lactate/pyruvate ratio elevation, possibly from an improvement in mitochondrial function. This finding is similar to a recent study18 and is in agreement with our previous in-vitro data showing Q10 supplementation in hepatocytes to reduce cell death and DNA oxidative stress and improve ATP synthesis.1
Circulating ALT and ALP were reduced significantly in Group 2, suggesting potential benefits of ubiquinol at molecular level although its clinical significance is uncertain. In a previous clinical trial,17 plasma ALT and AST were not significantly different in CoQ10 supplemented group from patients treated with atorvastatin. Animal studies,19 however, showed significant down-regulation of ALT and AST in atorvastatin-treated rats supplemented with CoQ10 compared to atorvastatin alone, with authors hypothesizing that CoQ10’s role in mitochondrial bioenergy transfer of ATP and its antioxidative effects to be potentially hepatoprotective.
Although serum ALT is a frequently used clinical indicator of hepatotoxicity, it does not always correlate well with preclinical liver histology data. Therefore, additional markers may be useful as early biomarkers to detect and distinguish liver injuries of different etiologies.10 When segregated by treatment groups, five miRNAs (miR-192, miR-146a, miR-148a, miR-15a, and miR-21) were positively correlated to ALT in the simvastatin (with placebo) but not in the ubiquinol supplementation group. None of the candidate miRNAs correlated with changes in ALP in Group 1, while miR-15a, miR-21, and miR-33a were negatively correlated with ALP in Group 2. ALP is known to be a marker of hepatobiliary effects and cholestasis; negative correlation of miR-15a, miR-21, and miR-33a in the ubiquinol group may suggest protective effect of these three miRNAs. It was previously hypothesized that miRNAs, including the seven miRNAs evaluated, were potentially more sensitive than ALT20 as biomarkers. Although no clinical and biochemical evidence of toxicity was detected based on liver function tests at 20 mg/day of simvastatin, the positive correlation of the five miRNAs with ALT in Group 1 and negative correlation of three miRNAs with ALP in Group 2 may again suggest potential molecular mechanism of the hepatoprotective effect of ubiquinol.
While we aimed to investigate circulating miRNAs as early biomarkers for simvastatin-induced injury, changes of miRNAs could be secondary to lipid modifying effects of simvastatin. As miR-192 antagomirs do not lower plasma cholesterol,21 changes in circulating miR-192 are unlikely due to lipid lowering effect of simvastatin. miR-192 was found to be increased in patients and animal models before significant rises in ALT activity in acetaminophen-induced, alcohol-, and chemical-related hepatic diseases.12 Although our data did not show any increase in all liver enzymes, miR-192 was positively correlated with ALT only in Group 1 (simvastatin + placebo) but not in Group 2 (simvastatin + ubiquinol). One possible consequence of increased miR-192 was a reduction in cell viability as seen in our in-vitro assay, after miR-192 transfection. miR-192 was increased in a dose-dependent manner with a concomitant reduction in cell viability as observed in THLE-2 liver cells (Supplementary Figure 4). This observation is similar to studies in other cell types where miR-192 was shown to enhance apoptosis due to reduced Bcl-2 expression.22 Whether increased miR-192 may be useful as a marker for liver injury in statin requires more investigation.
miR-21 was positively correlated with ALT in Group 1 and negatively correlated with ALP in Group 2. Patients with non-alcoholic fatty liver disease showed increased circulating miR-21,23 suggesting an association of miR-21 with liver injury. A recent publication24 postulated that increased miR-21 during liver regeneration can accelerate cell cycle through facilitation of cyclin D1 translation in mice models. Corroborating this finding, we observed increased cell viability in miR-21 mimics-transfected HepG2 at 72 h. In addition, miR-21 has been implicated in anti-inflammatory responses,25 by targeting programmed cell death 4 protein (PDCD4), positively influencing IL-10 (anti-inflammatory cytokine) and negatively influencing NFKB activity (Figure 4). The apparent opposing effects in cell viability for miR-192 and miR-21 may suggest miR-21 to be up-regulated as a response to simvastatin treatment.
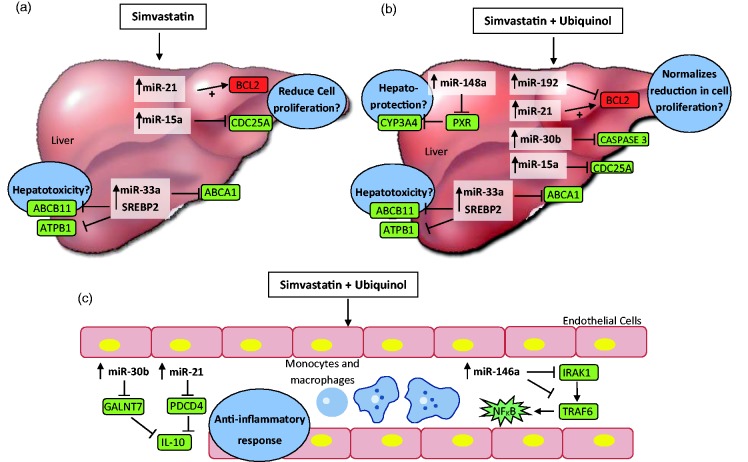
Figure 4.
Proposed model of miRNA regulation by simvastation and possible protective role of ubiquinol. (a, b) Possible targets of miRNAs regulated in the liver. (c) miRNAs regulated in macrophages and monocytes. Targets of miRNA were hypothesized by published literature as described in text. denotes negative regulation resulting in target molecules which may be down-regulated (in green) and denotes a positive regulation resulting in target molecules which may be up-regulated (red). (a) Patients treated with 20 mg simvastatin/day had increased circulating miR-15a, miR-21, and miR-33a. miR-15a was shown to target and inhibit CDC25A, a cell-cycle regulator known for G1 to S phase progression. miR-21 has been shown to potentiate expression of Bcl-2, an anti-apoptotic protein. The balance of miR-15a and miR-21 expression in the liver may determine deterimental effects on its proliferation. (b) In addition to miR-15a and miR-21, mir-192 and miR-30b were up-regulated in patients receiving ubiquinol supplementation and in HepG2 cell lines. Their roles in proliferation and anti-apoptosis might be one possible mechanism of hepatoprotection and observed by reduced ALP and ALT. (c) In the circulation, miR-146a, miR-15a, and miR-21 were up-regulated in patients treated with simvastatin supplementated with ubiquinol. Consistent with its anti-inflammatory function, miRNAs regulated with ubiquinol supplementation reduce pro-inflammatory cytokines (NFKB) and increase anti-inflammatory cytokines (e.g., IL-10). miR-146a target molecules in the innate immune response (TLR4 signaling) pathway, directly targeting IRAK1 and TRAF6.
IRAK1: interleukin-1 receptor-associated kinase; TRAF6: tumor necrosis factor receptor-assicated family; GALNT7: GalNAc transferase 7; PDCD4: programmed cell death 4 protein
miR-146a, a potent inflammation mediator, acts as regulator of Toll-like receptor 4 (TLR4) signaling,26,27 was unchanged after simvastatin (with placebo) treatment28 but combinatorial treatment with ubiquinol increased miR-146a significantly. One possible mechanism for the beneficial action of ubiquinol was a reduction in pro-inflammatory cytokines, by increase in miR-146a.29 By inhibiting TLR4 pathways, miR-146a reduces cytokine secretion in stimulated macrophages. Up-regulation of miR-146a, though usually related to inflammation, may be increased in non-inflammatory processes. As reported,10 increase in miR-146a was unexpected as acetaminophen injury is not associated with significant inflammation. Others have reported a down-regulation of miR-146a in liver cells, after pre-treatment with ubiquinol, before lipo-polysaccahride (LPS) stimulation in endothelial, monocytes, and liver in mice model.30 We were not able to elucidate the regulation of miR-146a in HepG2 cells after simvastatin and ubiquinol treatment as the Ct-values were ≥ 40. The up-regulation of circulating miR-146a in simvastatin-treated patients with ubiquinol supplementation requires further investigation in other cell models.
miR-148a was shown to correlate positively to ALT in simvastatin (placebo)-treated patients but not in patients with ubiquinol supplementation. Some studies have identified miRNAs that affect Cytochrome P450 family (CYP) genes by targeting their upstream transcription factors. The metabolism of more than 50% of all clinically relevant drugs, including simvastatin is catalyzed by CYP3A4 in human.31 Expression of CYP3A4 is predominantly regulated by pregnane X receptor (PXR), which can be negatively regulated by miR-148a. The association of miR-148a and ALT with its direct role in P450 cytochromes may substantiate future studies in larger cohorts of patients treated with simvastatin.
miR-30b showed an up-regulation only in Group 2 (ubiquinol supplementation) but did not correlate with ALT or ALP, suggesting this up-regulation maybe specific for ubiquinol supplementation. miR-30b was shown to up-regulate anti-inflammatory IL-10, via suppression of GalNAc transferase 7 (GALNT7) in melanoma cell lines. miR-30b was also found to target caspase 3 directly in glioma cell lines, supporting a possible anti-apoptotic function.32 Its potential anti-inflammatory and anti-apoptotic function suggests that miR-30b might be up-regulated as a protective mechanism in simvastatin-treated patients supplemented with ubiquinol.
miR-15a was shown to be up-regulated in the plasma of acetaminophen overdose-induced mouse model.11 Increased miR-15a expression was found to directly bind and reduce Cdc25A (a cell-cycle regulator), thereby inhibiting cell proliferation in hepatic cystogenesis.33 Despite the observation that miR-15a was positively correlated with ALT in simvastatin (with placebo)-treated patients and negatively correlated with ALP in simvastatin (with ubiquinol)-treated patients, miR-15a was not modulated by simvastatin treatment in HepG2 cells, suggesting the source of circulating miR-15a in patients may be from other organs.
Up-regulation of miR-33a after simvastatin treatment in our study is consistent with animal models. miR-33 was shown to be involved in the hepatotoxic effect of simvastatin, as miR-33 silencing before simvastatin treatment, rescued liver damage in mice, on cholate-rich diet.34 In contrast to animal studies, simvastatin dosage in our clinical trial was relatively low and did not elevate serum liver enzymes. However, we observed that miR-33a changes was only positively significantly associated with changes in ALP and not other liver enzymes (Table 3). Increased levels of ALP were found to be associated with primary biliary cirrhosis, without known increases in ALT levels.35 Animal models have shown that miR-33 targets ABCB11 (ATP-binding cassette, sub-family B, member 11) and ATPB1 (ATPase, aminophospholipid transporter, class I, type 8B, member 1), both of which are major biliary lipid transporters in hepatocytes. Combination of lithogenic diet and statin treatment induced cholestasis and liver steatosis in mice, which could be rescued upon anti-miR-33 oligonucleotide treatment.34 Thus, up-regulation of miR-33a in both groups and its potential hepatotoxic effect in biliary damage demand further investigation in simvastatin treatment. Similarly, the negative association of miR-33a and ALP in Group 2 and the potential hepatoprotective effect of ubiquinol supplementation could be elucidated in future studies.
Mechanistic effects of simvastatin-induced hepatotoxicity have been previously analyzed by transcriptomic and proteomic profiling.36 Pathways related to NRF2-mediated oxidative stress, metabolism of cytochrome P450, fatty acid, bile, urea, and inflammation metabolism were modulated by simvastatin in rat hepatocytes. Independently, gene array analyses were conducted in the livers of ubiquinol-treated mice.37 Ubiquinol-regulated genes were functionally categorized to play a role in anti-oxidative processes, cholesterol, lipid metabolism, and PPARα pathways in mice livers. Our global analyses on miRNAs after simvastatin treatment corroborates with published data where miRNAs related to fatty acid and amino acid metabolism were highly regulated and that ubiquinol did not disturb these pathways. miRNAs that differed between the two groups, presumably regulated by ubiquinol, had additionally affected oxidative phosphorylation, cytokine signaling and ABC transporters, further suggesting its hepatoprotective role.
Although molecular events occurring in patient liver are unknown, we propose a model for simvastatin treatment and possible protective effects with ubiquinol (Figure 4). Treatment of simvastatin at 20 mg/day may possibly affect molecular changes involving reduced proliferation and ABC transporters by miR-15a, miR-21, and miR-33a (Figure 4(a)). Ubiquinol supplementation additionally regulated miRNAs (e.g., miR-30b) which have been reported to reduce apoptotic proteins (caspase-3) (Figure 4(b)). Known to exert anti-inflammatory effects, ubiquinol supplementation also up-regulated miR-30b, miR-21, and miR-146a, which may target inflammatory pathways (Figure 4(c)).
The strengths of our study include our ability to evaluate the association between miRNAs and liver enzymes in a prospective trial where inter-individual variability can be minimized and effects of intervention directly assessed. Our in-vitro findings support clinical observations, raising the hypotheses that simvastatin, with or without ubiquinol treatment affects liver cells and miRNA changes in the liver could possibly explain, in part, differences in circulating miRNAs. Some differences were observed in our microarray and qPCR data. Performing miR-33a qPCR on the samples, we detected significant up-regulation of this transcript after 12 weeks of treatment in both groups, although miR-33a signals were not detected in our arrays. This is likely due to the difference in the dynamic range of detection in both techniques, with qPCR being a more sensitive method.38 One limitation in the study is that our sample size was small. Another limitation was that circulating miRNAs have unknown origins. A recent review concluded that less than a third of reported miRNA biomarkers are expressed with some exclusivity in diseased cell type, suggesting that many circulating miRNAs are non-specific markers of organ injury.20 Therefore, while many miRNAs have been identified as potential markers, current utility of miRNAs has not been able to differentiate liver damage of different etiologies. Our current study therefore adds to existing knowledge of hepatoprotection.
In conclusion, we show that simvastatin at 20 mg/day does not have significant risk of hepatotoxicity for patients with hypercholesterolemia and that ubiquinol supplementation may, at the molecular level, provide potential beneficial changes to reduce the effects of CoQ10 deficiency in the liver, as shown by both diagnostic liver function tests and potential miRNA markers.
Acknowledgements
The authors would like to thank Dr Eric Wee Wei Long (Consultant Gastroenterologist, Head, Division of Gastroenterology, Khoo Teck Puat Hospital) for his critical comments on the manuscript; Liew Yiting (Research Nurse, Clinical Research Unit, Khoo Teck Puat Hospital) for her assistance in recruiting patients for this study; Ms Ong Bee Lan (National University of Singapore) for her technical assistance in HPLC analyses; and Kaneka Corporation (Japan) for their sponsorship of Ubiquinol and placebo of ubiquinol. This study is supported by Small Innovative Grant 2 (SIGII/09005) and Enabling Grant (KPREF12EG020) from Alexandra Health Pte Ltd (AHPL), Singapore. This trial is registered with Singapore Health Science Authority (HSA), Registration number: CTC0900356.
Author contributions
All authors contributed to the intellectual development of this paper. SLTP conducted data analyses and wrote the manuscript. KW, SCL, and CFS participated in the planning of the study and key discussions and edited the manuscript. LL performed miRNA assays and cell culture experiments and participated in key discussions of the manuscript. CNO performed HPLC assays and participated in key discussions of the manuscript. ST conceived and coordinated the whole work and prepared the manuscript; The guarantor, ST accepts full responsibility for the work and/or the conduct of the study, had access to the data, and controlled the decision to publish.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Tavintharan S, Ong CN, Jeyaseelan K, Sivakumar M, Lim SC, Sum CF. Reduced mitochondrial coenzyme Q10 levels in HepG2 cells treated with high-dose simvastatin: a possible role in statin-induced hepatotoxicity? Toxicol Appl Pharmacol 2007; 223: 173–9. [DOI] [PubMed] [Google Scholar]
- 2.Schachter M. Chemical, pharmacokinetic and pharmacodynamic properties of statins: an update. Fundam Clin Pharmacol 2005; 19: 117–25. [DOI] [PubMed] [Google Scholar]
- 3.Baigent C, Keech A, Kearney P, Blackwell L, Buck G, Pollicino C, Kirby A, Sourjina T, Peto R, Collins R, Simes J. Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. The Lancet 2005; 366: 1267–78. [DOI] [PubMed] [Google Scholar]
- 4.Golomb BA, Evans MA, Dimsdale JE, White HL. Effects of statins on energy and fatigue with exertion: results from a randomized controlled trial. Arch Intern Med 2012; 172: 1180–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mullen PJ, Zahno A, Lindinger P, Maseneni S, Felser A, Krahenbuhl S, Brecht K. Susceptibility to simvastatin-induced toxicity is partly determined by mitochondrial respiration and phosphorylation state of Akt. Biochim Biophys Acta 2011; 1813: 2079–87. [DOI] [PubMed] [Google Scholar]
- 6.Russo MW, Scobey M, Bonkovsky HL. Drug-induced liver injury associated with statins. Semin Liver Dis 2009; 29: 412–22. [DOI] [PubMed] [Google Scholar]
- 7.Lammert C, Einarsson S, Saha C, Niklasson A, Bjornsson E, Chalasani N. Relationship between daily dose of oral medications and idiosyncratic drug-induced liver injury: search for signals. Hepatology 2008; 47: 2003–9. [DOI] [PubMed] [Google Scholar]
- 8.Crane FL. Biochemical functions of coenzyme Q10. J Am Coll Nutr 2001; 20: 591–8. [DOI] [PubMed] [Google Scholar]
- 9.Moore KJ, Rayner KJ, Suárez Y, Fernández-Hernando C. microRNAs and cholesterol metabolism. Trends Endocrinol Metabol 2010; 21: 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, Alao H, Kodys K, Szabo G. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology 2012; 56: 1946–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A 2009; 106: 4402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Starkey Lewis PJ, Dear J, Platt V, Simpson KJ, Craig DG, Antoine DJ, French NS, Dhaun N, Webb DJ, Costello EM, Neoptolemos JP, Moggs J, Goldring CE, Park BK. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology 2011; 54: 1767–76. [DOI] [PubMed] [Google Scholar]
- 13.Lee BL, New AL, Ong CN. Simultaneous determination of tocotrienols, tocopherols, retinol, and major carotenoids in human plasma. Clin Chem 2003; 49: 2056–66. [DOI] [PubMed] [Google Scholar]
- 14.Karolina DS, Armugam A, Tavintharan S, Wong MT, Lim SC, Sum CF, Jeyaseelan K. MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitus. PLoS One 2011; 6: e22839–e22839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu TP, Lee CY, Tsai MH, Chiu YC, Hsiao CK, Lai LC, Chuang EY. miRSystem: an integrated system for characterizing enriched functions and pathways of microRNA targets. PLoS One 2012; 7: e42390–e42390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 2010; 328: 1566–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mabuchi H, Nohara A, Kobayashi J, Kawashiri MA, Katsuda S, Inazu A, Koizumi J. Effects of CoQ10 supplementation on plasma lipoprotein lipid, CoQ10 and liver and muscle enzyme levels in hypercholesterolemic patients treated with atorvastatin: a randomized double-blind study. Atherosclerosis 2007; 195: e182–9. [DOI] [PubMed] [Google Scholar]
- 18.Dai YL, Luk TH, Yiu KH, Wang M, Yip PM, Lee SW, Li SW, Tam S, Fong B, Lau CP, Siu CW, Tse HF. Reversal of mitochondrial dysfunction by coenzyme Q10 supplement improves endothelial function in patients with ischaemic left ventricular systolic dysfunction: a randomized controlled trial. Atherosclerosis 2011; 216: 395–401. [DOI] [PubMed] [Google Scholar]
- 19.Schaefer WH, Lawrence JW, Loughlin AF, Stoffregen DA, Mixson LA, Dean DC, Raab CE, Yu NX, Lankas GR, Frederick CB. Evaluation of ubiquinone concentration and mitochondrial function relative to cerivastatin-induced skeletal myopathy in rats. Toxicol Appl Pharmacol 2004; 194: 10–23. [DOI] [PubMed] [Google Scholar]
- 20.Haider BA, Baras AS, McCall MN, Hertel JA, Cornish TC, Halushka MK. A critical evaluation of microRNA biomarkers in non-neoplastic disease. PLoS One 2014; 9: e89565–e89565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005; 438: 685–9. [DOI] [PubMed] [Google Scholar]
- 22.Geng L, Chaudhuri A, Talmon G, Wisecarver JL, Are C, Brattain M, Wang J. MicroRNA-192 suppresses liver metastasis of colon cancer. Oncogene 2014; 33: 5332–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamada H, Suzuki K, Ichino N, Ando Y, Sawada A, Osakabe K, Sugimoto K, Ohashi K, Teradaira R, Inoue T, Hamajima N, Hashimoto S. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin Chim Acta 2013; 424: 99–103. [DOI] [PubMed] [Google Scholar]
- 24.Ng R, Song G, Roll GR, Frandsen NM, Willenbring H. A microRNA-21 surge facilitates rapid cyclin D1 translation and cell cycle progression in mouse liver regeneration. J Clin Invest 2012; 122: 1097–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary JJ, Ruan Q, Johnson DS, Chen Y, O'Neill LA. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol 2010; 11: 141–7. [DOI] [PubMed] [Google Scholar]
- 26.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A 2006; 103: 12481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi Y, Satoh M, Minami Y, Tabuchi T, Itoh T, Nakamura M. Expression of miR-146a/b is associated with the Toll-like receptor 4 signal in coronary artery disease: effect of renin-angiotensin system blockade and statins on miRNA-146a/b and Toll-like receptor 4 levels. Clin Sci (Lond) 2010; 119: 395–405. [DOI] [PubMed] [Google Scholar]
- 28.Dinarello CA. Anti-inflammatory agents: present and future. Cell 2010; 140: 935–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang K, He YS, Wang XQ, Lu L, Chen QJ, Liu J, Sun Z, Shen WF. MiR-146a inhibits oxidized low-density lipoprotein-induced lipid accumulation and inflammatory response via targeting toll-like receptor 4. FEBS Lett 2011; 585: 854–60. [DOI] [PubMed] [Google Scholar]
- 30.Schmelzer C, Kitano M, Rimbach G, Niklowitz P, Menke T, Hosoe K, Doring F. Effects of ubiquinol-10 on microRNA-146a expression in vitro and in vivo. Mediators Inflamm 2009; 2009: 415437–415437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rieger JK, Klein K, Winter S, Zanger UM. Expression variability of absorption, distribution, metabolism, excretion-related microRNAs in human liver: influence of nongenetic factors and association with gene expression. Drug Metab Dispos 2013; 41: 1752–62. [DOI] [PubMed] [Google Scholar]
- 32.Quintavalle C, Donnarumma E, Iaboni M, Roscigno G, Garofalo M, Romano G, Fiore D, De Marinis P, Croce CM, Condorelli G. Effect of miR-21 and miR-30b/c on TRAIL-induced apoptosis in glioma cells. Oncogene 2013; 32: 4001–8. [DOI] [PubMed] [Google Scholar]
- 33.Lee SO, Masyuk T, Splinter P, Banales JM, Masyuk A, Stroope A, Larusso N. MicroRNA15a modulates expression of the cell-cycle regulator Cdc25A and affects hepatic cystogenesis in a rat model of polycystic kidney disease. J Clin Invest 2008; 118: 3714–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allen RM, Marquart TJ, Albert CJ, Suchy FJ, Wang DQ, Ananthanarayanan M, Ford DA, Baldan A. miR-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO Mol Med 2012; 4: 882–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishio A, Keeffe EB, Ishibashi H, Gershwin EM. Diagnosis and treatment of primary biliary cirrhosis. Med Sci Monit 2000; 6: 181–93. [PubMed] [Google Scholar]
- 36.Cho YE, Moon PG, Lee JE, Singh TS, Kang W, Lee HC, Lee MH, Kim SH, Baek MC. Integrative analysis of proteomic and transcriptomic data for identification of pathways related to simvastatin-induced hepatotoxicity. Proteomics 2013; 13: 1257–75. [DOI] [PubMed] [Google Scholar]
- 37.Schmelzer C, Kitano M, Hosoe K, Doring F. Ubiquinol affects the expression of genes involved in PPARalpha signalling and lipid metabolism without changes in methylation of CpG promoter islands in the liver of mice. J Clin Biochem Nutr 2012; 50: 119–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 2005; 33: e179–e179. [DOI] [PMC free article] [PubMed] [Google Scholar]