Abstract
Thyroid cancer is the most common endocrine malignancy and accounts for the majority of endocrine cancer-related deaths each year. Our group and others have previously demonstrated dysfunctional microRNA (miRNA or miR) expression in the context of thyroid cancer. The objective of the present study was to investigate the impact of synthetic manipulation of expression of miR-25 and miR-222 in benign and malignant thyroid cells. miR-25 and miR-222 expression was upregulated in 8505C (an anaplastic thyroid cell line) and Nthy-ori (a SV40-immortalised thyroid cell line) cells, respectively. A transcriptomics-based approach was utilised to identify targets of the two miRNAs and real-time PCR and western blotting were used to validate a subset of the targets. Almost 100 mRNAs of diverse functions were found to be either directly or indirectly targeted by both miR-222 and miR-25 [fold change ≥2, false discovery rate (FDR) ≤0.05]. Gene ontology analysis showed the miR-25 gene target list to be significantly enriched for genes involved in cell adhesion. Fluidigm real-time PCR technologies were used to validate the downregulation of 23 and 22 genes in response to miR-25 and miR-222 overexpression, respectively. The reduction of the expression of two miR-25 protein targets, TNF-related apoptosis-inducing ligand (TRAIL) and mitogen-activated protein kinase kinase 4 (MEK4), was also validated. Manipulating the expression of both miR-222 and miR-25 influenced diverse gene expression changes in thyroid cells. Increased expression of miR-25 reduced MEK4 and TRAIL protein expression, and cell adhesion and apoptosis are important aspects of miR-25 functioning in thyroid cells.
Keywords: thyroid cancer, miR-222 and miR-25, microarray, target genes, mitogen-activated protein kinase kinase 4 and TNF-related apoptosis-inducing ligand
Introduction
Thyroid cancer is the most common endocrine malignancy, and incidences are on the rise worldwide. Thyroid tumours frequently possess genetic alterations which lead to the activation of the mitogen-activated protein kinase (MAPK) signalling pathway (1). The expression of microRNAs (miRNAs or miRs) has been studied in thyroid cancers, and as with other types of cancer, miRNA profiles have been found to be significantly different between tumours of the thyroid compared to normal thyroid tissue. For instance miR-146, miR-221, miR-222, miR-155, miR-181a and miR-181b have been shown to differentiate papillary thyroid cancer from normal thyroid tissue (2,3).
miRNAs have also shown differential expression between different types of thyroid cancer (4) and within a multifocal pluriform tumour from an individual thyroid gland (5), thus illustrating that miRNA profiles have demonstrated both intra- and inter-tumour variability. The functions of some of these differentially expressed miRNAs have also been elucidated: for instance, miR-221 has been shown to regulate HOXB5 (6) and the human telomerase reverse transcriptase (hTERT) gene is regulated by miR-138 (7). Furthermore, the potential utility of these small molecules to aid thyroid cancer diagnosis (8) and prognosis (9) has also been investigated.
The aim of the present study was to elucidate the target mRNAs of two miRNAs that we had previously found to be differentially expressed in thyroid cancer (5): miR-222 and miR-25. miR-222 is commonly found to be upregulated in thyroid cancer (2–4) and is part of an intergenic miRNA cluster (that also contains miR-221) on the p11.3 region of the X chromosome. It is one of the most well-known miRNAs linked to thyroid cancer, and some of its gene targets have been elucidated, including the KIT gene (2) and the p27Kip1 protein (10).
miR-25 is also located in a miRNA cluster, termed the mi-106b-25 cluster. miR-106b and miR-93 are the two other miRNAs in this highly conserved cluster which is located in a 515-bp region at chromosome 7q22, in intron 13 of the host gene MCM7. The miRNAs are co-transcribed in the context of the MCM7 primary transcript and have been found to accumulate in different types of cancer, including gastric, prostate, pancreatic neuroendocrine tumours, neuroblastoma and multiple myeloma (11). miR-25 has been shown to regulate p57 (12) and E2F1 as part of a negative feedback loop in gastric cancer (11). It has also been shown to promote cell invasion and migration in esophageal squamous cell carcinoma (13) and regulate apoptosis by targeting the Bim protein in ovarian (14) and eosophangeal cells (15). miR-25 has been found to be downregulated in anaplastic thyroid carcinoma (16) and, along with miR-30d, to target the polycomb protein enhancer of zeste 2 (EZH2) in this disease context (17).
In this study, we describe work in which we examined the impact of upregulating the thyroid cancer-associated miRNA miR-222 in benign Nthy-ori cells, and miR-25, a miRNA downregulated in anaplastic thyroid cancer, in the anaplastic cancer-derived 8505C cell line. Microarray technologies were utilised to monitor global gene expression changes in response to altered expression of miR-222 and miR-25. This unbiased genome-wide approach provided by the microarrays yielded the discovery of almost 100 mRNAs that are either directly or indirectly targeted by each miRNA in thyroid cells and have not been previously described to the best of our knowledge. These gene lists provide insights as to the functions of these miRNAs within thyroid cells; they contain both predicted and novel targets of the miRNAs, a subset of which were validated at the mRNA and protein level.
Materials and methods
Cell culture
The human thyroid follicular epithelial cell line Nthy-ori 3-1 (cat no. 90011609; ECACC, Salisbury, UK) was grown in RPMI media containing 10% foetal bovine serum (FBS), 2% penicillin/streptomycin (5,000 U/ml). An undifferentiated human thyroid carcinoma cell line, 8505C, (cat no. 94090184; ECACC) was grown in Eagle's minimum essential medium (EMEM) with Hank's buffered salt solution (HBSS) containing 2 mM glutamine and 1% non-essential amino acids (NEAA), 10% FBS, and 2% penicillin/streptomycin (5,000 U/ml). All cell culture reagents were purchased from Lonza (Basel, Switzerland) and cells were incubated at 37°C in a 5% CO2 humidified chamber (series II water jacketed CO2 incubator; Thermo Fisher Scientific, Waltham, MA, USA).
Transfections
For transfections, Nthy-ori 3-1 and 8505C cells were plated at a density of 1.5×105 cells/ml in 12-well plates (Nalge Nunc, Penfield, NY, USA) with three replicate wells for each condition. Cells were reverse transfected using Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA) according to the manufacturer's instructions with 50 nM pre-miR positive control (cat. # AM17150), pre-miR negative control #1 (cat. # AM17110), pre-miR-222 (cat. # PM11376) or pre-miR-25 (cat. # PM12401) (Ambion, Austin, TX, USA).
Transfection efficiency was evaluated using TaqMan real-time polymerase chain reaction (PCR) as follows. Pre-miR hsa-miR-1 miRNA precursor was used as a positive control in transfection experiments as, upon delivery into cells, it effectively downregulates the expression of PTK9 at the mRNA level. Effective delivery and activity of the pre-miR hsa-miR-1 miRNA precursor was detected by real-time PCR using a TaqMan Gene Expression assay to PTK9 (assay ID: Hs00702289_s1), and GAPDH mRNA was measured as an endogenous control (assay ID: Hs02758991_g1) (both from Applied Biosystems, Foster City, CA, USA). A high capacity cDNA reverse transcription kit (cat. # 4374966; Applied Biosystems) was used to convert total RNA to single-stranded cDNA for positive control analysis. Reactions contained 2 μl of RT buffer (10X), 0.8 μl of deoxynucleotide triphosphate (25X), 2 μl of random primers (10X), 1 μl of multiscribe RT enzyme (500 U/μl), 1 μl of RNase inhibitor, 3.2 μl of nuclease-free water and 10 μl of extracted total RNA. The reactions were incubated at 25°C for 10 min, 37°C for 2 h, and 85°C for 5 sec (Perkin Elmer 9600 GeneAmp PCR system; Applied Biosystems). Real-time PCR reactions were performed with TaqMan 2X Universal master mix (no AmpErase UNG) (cat. # 4304437; Applied Biosystems) and TaqMan gene expression assays (listed previously). Reactions contained 10 μl of TaqMan Universal PCR master mix (no AmpErase UNG), 1 μl of TaqMan gene expression assay (20X), 2 μl of cDNA template and 7 μl of nuclease-free water.
For miRNA analysis, total RNA from transfections was used to synthesise cDNA using the high-capacity cDNA reverse transcription kit and miRNA-specific RT primers for miR-222 (cat. # 4427975), miR-25 (cat. # 4427975), and endogenous control RNU6B (cat. # 4427975) (Applied Biosystems) in 15 μl reactions. Reactions contained 0.15 μl of 25X dNTP mixture (100 mM), 1 μl of multiScribe reverse transcriptase (50 U/μl), 1.5 μl of reverse transcription buffer (10X), 0.19 μl of RNase inhibitor, 4.16 μl of nuclease-free water, 3 μl of RT primer and 5 μl of extracted total RNA. The reactions were incubated at 16°C for 30 min, 42°C for 30 min, 85°C for 5 min and then an indefinite hold at 4°C (Perkin Elmer 9600 GeneAmp PCR system; Applied Biosystems). miRNA expression was then assessed using sequence specific primers from the TaqMan microRNA assays and TaqMan 2X Universal master mix (no AmpErase UNG) (cat. # 4304437; Applied Biosystems) in 20 μl reactions according to the manufacturer's instructions. Reactions contained 1 μl of TaqMan miRNA assay mix (20X), 1.33 μl of product from RT reaction (1:15 dilution), 10 μl of TaqMan 2X Universal master mix (no AmpErase UNG) and 7.67 μl of nuclease free water. Each assay was performed in triplicate for each sample.
All real-time PCR reactions were incubated in a 96-well optical plate (cat. # N8010560; Applied Biosystems) at 95°C for 10 min, following by 40 cycles of 95°C for 15 sec and 60°C for 1 min on the Applied Biosystems 7900HT Real-Time PCR system (Applied Biosystems). Study files were generated using a fixed threshold of 0.1 on the SDS2.2.2 software (Applied Biosystems), and Microsoft Excel (Microsoft, Redmond, WA, USA) was used to perform ΔΔCt analysis on the real-time PCR output (18).
RNA isolation and mRNA microarray analysis
Total RNA was isolated from transfected cell lines using PARIS™ Protein and RNA isolation kit (cat. # AM1556; Ambion) and RNeasy mini kit (cat. # 74104; Qiagen, Venlo, The Netherlands). Briefly, the cells were lysed with PARIS Cell Disruption Buffer and 2X Lysis/Binding Solution. Cell lysates were then transferred to Qiagen QIAshredder columns followed by RNA extraction and on column DNase treatment (Cat. # 79254; Qiagen, Venlo, The Netherlands) according to the manufacturer's instructions. RNA nanogram concentration per microlitre was verified using a NanoDrop spectrophotometer (ND-1000; Labtech International, Lewes, UK) and RNA integrity was verified using a 2100 Bioanalyser (Agilent, Santa Clara, CA, USA).
Affymetrix GeneChip Human Gene 1.0 ST arrays were used for gene expression analysis according to the manufacturer's instructions (cat. # 902461; Affymetrix, Santa Clara, CA, USA). Three biological repeats were used for each treatment. Microarray statistical analysis was performed on. CEL files using the Partek Genomics Suite (Partek Inc., St. Louis, MO, USA; www.partek.com). Data were normalised and summarised using the robust multi-average method, as previously described (19). Paired t-tests were performed to compare the data from the pre-miR transfections to the negative control transfections. Comparisons were corrected for multiple testing using the false discovery rate (FDR). Genes were deemed to be differentially regulated in the pre-miR™ transfected cells if they possessed a FDR ≤0.05 and a fold change ≥2. Gene functional and pathway enrichment analysis was assessed by the PANTHER database (http://www.pantherdb.org/).
Fluidigm reverse transcription-PCR analysis of target genes downregulated by miR-222 and miR-25
Total RNA from transfections was converted to single-stranded cDNA using the High Capacity cDNA Reverse Transcription kit (cat. # 4374966; Applied Biosystems) in 20 μl reactions, as described in the transfection section of this manuscript.
Pre-amplification of cDNA was performed using the TaqMan PreAmp Master Mix kit (cat. # 4384267; Applied Biosystems). The assays used were as follows: MAL2; Hs00294541_m1, MAL; Hs00242748_m1, TLR3; Hs01551078_m1, ADM; Hs00181605_m1, RAB19; Hs01397748_m1, PDCD1LG2; Hs00228839_m1, RAD51; Hs00947968_m1, ADAM21; Hs01652548_s1, HYOU1; Hs01026180_m1, THBS1; Hs00962908_m1, ETS1; Hs00428287_m1, FGFR2; Hs03466165_gH, CYR61; Hs00609994_m1, PHLDB2; Hs01083801_m1, BIRC3; Hs00154109_m1, PRKAA2; Hs00178903_m1, CYP24A1; Hs00989011_g1, AOX1; Hs00154079_m1, MYO5C; Hs00218921_m1, GPR126; Hs01097890_m1, CDK6; Hs00608037_m1, PPME1; Hs00211693_m1, RBM24; Hs00290607_m1, TNFSF10/TRAIL; Hs00921974_m1, ITGA6; Hs01041012_m1, MPP1; Hs00609971_m1, TIMP2; Hs00234278_m1, ITGA3; Hs01076876_m1, ADAMTS1; Hs00199608_m1, ITGA5; Hs00233732_m1, TRIP13; Hs01020073_m1, MAP2K4; Hs00387426_m1, RAB23; Hs00212407_m1, ADAM10; Hs00153853_m1, TRHDE; Hs00183821_m1, RAB14; Hs00249440_m1, RARB; Hs00233405_m1, MAN2A1; Hs00159007_m1, RGS4; Hs00194501_m1, PTGS2; Hs00153133_m1, EPGN; Hs02385425_m1, SNAP23; Hs01047498_m1, LHFPL2; Hs00299613_m1, MYO1B; Hs01031676_m1, NFIA; Hs00906448_m1, RNF38; Hs01014398_m1 (all from Applied Biosystems). Before running the pre-amplification reaction, the intended assays were pooled together, as per the manufacturer's instructions, with 1X Tris-EDTA (TE) buffer in a 0.2X pooled assay mix. The pre-amplification reaction was performed in 5 μl reactions containing 2.5 μl of TaqMan PreAmp master mix (2X), 1.25 μl of the pooled assay mix (0.2X), and 1.25 μl of cDNA sample (100 ng). The pre-amplification reaction was performed for 10 min at 95°C, and 14 cycles of 15 sec at 95°C and 4 min at 60°C (PE 9600 GeneAmp PCR system; Applied Biosystems).
The Fluidigm Dynamic Arrays facilitate the testing of the expression of 48 genes in 48 samples by performing 2,304 PCR assays per chip. The TaqMan gene expression assays listed in the previous paragraph were diluted to a final concentration of 10X using Fluidigm DA Assay Loading Reagent (cat. # 100-7611; Fluidigm, South San Francisco, CA, USA), and the pre-amplified products, diluted to 1:5 using 1X TE buffer. The cDNA product was combined with real-time PCR reagents in 5 μl reactions as follows: 2.25 μl of diluted pre-amplified cDNA, 2.25 μl Universal PCR master mix (2X) (cat. # 4304437; Applied Biosystems), and 0.25 μl Fluidigm sample loading agent (cat. # 100-7610; Fluidigm). Samples, primers and probes were then loaded onto the Fluidigm 48.48 Dynamic Arrays and placed on the IFC controller (both from Fluidigm) which pressure-loads the assay components into the reaction chambers. Assay components are automatically combined on-chip. The Biomark Real-Time PCR system (Fluidigm) was then used for thermal cycling and fluorescence detection. cDNA from transfections was loaded in duplicate on three replicate dynamic arrays. Microsoft Excel (Microsoft) was used to perform ΔΔCt analysis on the real-time PCR output, as previously described (18).
Protein extraction and western blotting
Protein was isolated from transfections using ice-cold radioimmunoprecipitation assay (RIPA) buffer (cat. # R0278; Medical Supply Co., Dublin, Ireland) with added protease and phosphatase inhibitor cocktails (cat. # 04693116001 and 04693116001 respectively; Roche, Indianapolis, IN, USA). Samples were suspended in SDS-PAGE sample buffer (cat. # S3401; Sigma-Aldrich, St. Louis, MO, USA), boiled for 4 min, and resolved on 10% SDS-PAGE gels. The separated proteins were electrophoretically transferred to PVDF membranes by the semi-dry transfer technique for 1 h. PVDF membranes were subsequently blocked in 5% non-fat dried milk in TBS-0.1% Tween-2 (TBS-T) for 1 h at room temperature, washed three times in TBS-T, and incubated with primary antibodies (polyclonal rabbit anti-human mitogen-activated protein kinase kinase 4 (MEK4) (sc-964; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) diluted to 1:50, or monoclonal mouse anti-human TNF-related apoptosis-inducing ligand (TRAIL) (ab12124) at 2 μg/ml, or monoclonal mouse anti-human GAPDH (ab8245) (diluted to 1:25,000) (both from Abcam, Cambridge, UK) overnight at 4°C with gentle rocking. After three washes in TBS-T, the membranes were incubated with the appropriate HRP-labelled secondary antibodies [goat anti-mouse IgG-HRP, goat anti-rabbit IgG-HRP (cat. # 7076 and 7074 respectively; Cell Signaling Technology, Danvers, MA, USA)] for 1 h at room temperature. The membranes were washed three times in TBS-T and immunoreactive bands were visualised using a HRP development solution containing 100 mM Tris-HCl pH 8.8, 30% H2O2, 250 mM luminol, and 90 mM 4-iodophenylboronic acid (4-IPBA), and subsequent exposure to Kodak light-sensitive film (cat. # F5763-50EA; Sigma-Aldrich). Jurkat T cell lysates were a gift from Dr Michael Freeley and were used as postive control lysates in western blot experiments
Results
As has been previously noted, expression of miR-222 is upregulated in thyroid cancers compared to their normal counterparts (2–5), while expression of miR-25 is downregulated in anaplastic thyroid carcinoma (5,16). We therefore investigated the impact of overexpression of miR-222 in normal Nthy-ori cells, and of miR-25 in the 8505C cells. An average 2.9-fold upregulation of miR-222 and 10,939-fold upregulation of miR-25 was observed with the pre-miR transfections (n=3). The very large miR-25 fold change increase resulted from an average Ct change of 20.95 in the pre-miR-25 treated cells compared to 33.53 in the cells treated with the negative control pre-miR. Reduced transfection efficiency in the normal Nthy-ori cells may explain the more modest increase in miR-222 post-transfection; however, the positive control gene, PTK9, was downregulated to a similar extent in both cell lines; 82.9 and 76.9% in the 8505C and Nthy-ori cells respectively. Microarray analysis elucidated the genes significantly downregulated (FDR value ≤0.05 and fold-change ≥2) by pre-miR-222 in Nthy-ori cells and pre-miR-25 in 8505C cells. We found that overexpression of miR-222 in normal Nthy-ori cells resulted in the downregulation of 82 target genes, while overexpression of miR-25 resulted in the downregulation of 98 target genes in 8505C cells. Figs. 1A and 2A illustrate the genes that were significantly downregulated by pre-miR-222 and pre-miR-25, respectively (see Tables I and II for full gene lists). The PANTHER database was used to perform gene ontology analysis on these pre-miR target lists. This analysis illustrated that the two miRNAs target genes of diverse functions in the thyroid cells, as no pathway was significantly over- or under-represented in either list. This observation concurs with other studies on miRNA target prediction and elucidation (20–22). miR-222 and miR-25 target lists were also cross-referenced with in silico prediction results from three prediction algorithms: miRanda, PicTar and TargetScan.
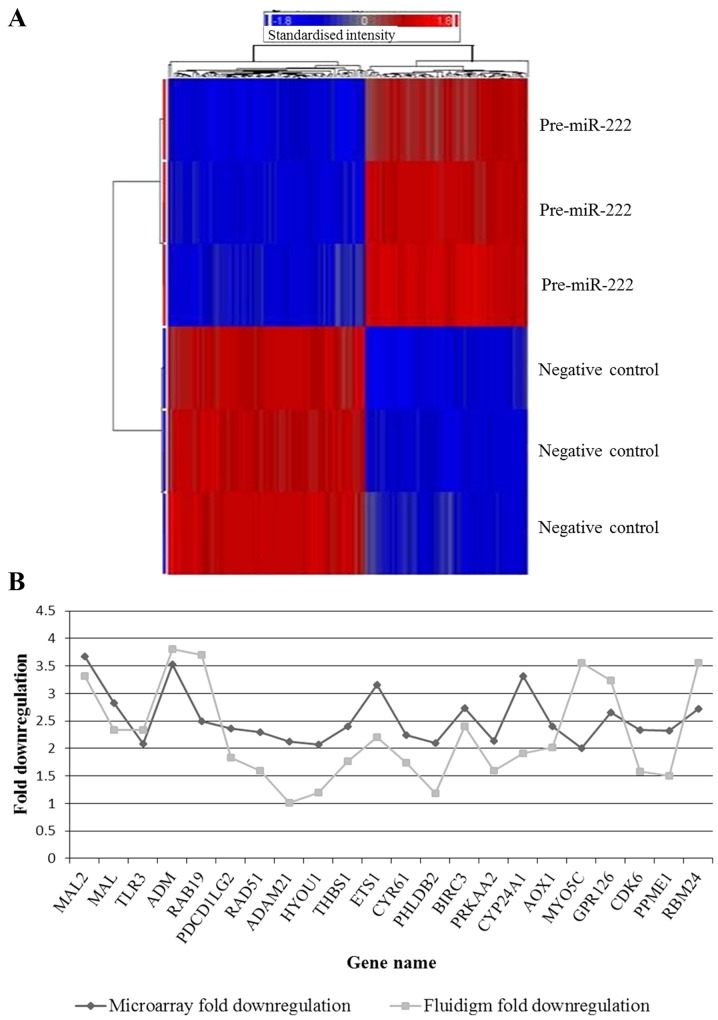
Figure 1.
miR-222 regulated genes. (A) Heat map of the significantly differentially regulated genes in pre-miR-222 treated Nthy-ori cells compared to negative control treated cells, false discovery rate (FDR) ≤0.05 and fold-change ≥2. A clear separation can be seen between the genes in pre-miR treated samples compared to negative control treated samples. (B) Line graph of the microarray and Fluidigm fold downregulation of the 22 genes validated by real-time PCR. All genes validated were found to be downregulated by both real-time PCR and microarray analyses (Pearson's correlation, 0.506).
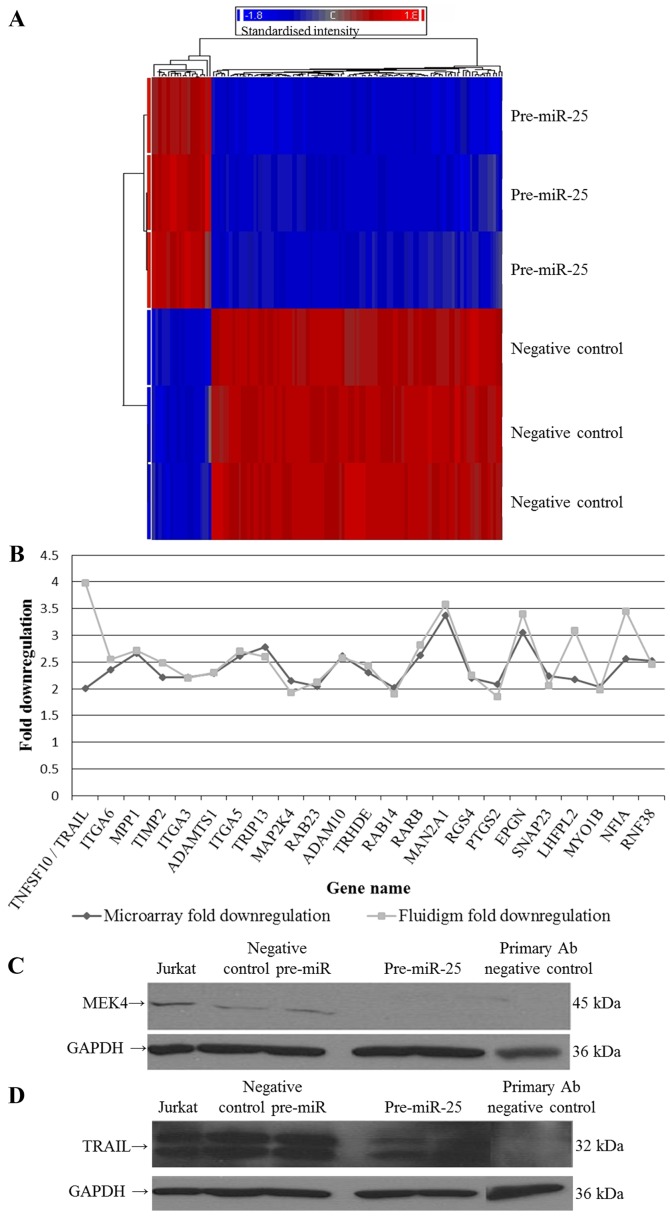
Figure 2.
miR-25 regulated genes. (A) Heat map of the significantly differentially regulated genes in pre-miR-25 treated 8505C cells compared to negative control treated cells; false discovery rate (FDR) ≤0.05 and fold-change ≥2. A clear separation can be seen between the genes in pre-miR treated samples compared to negative control treated samples. (B) Line graph of the microarray and Fluidigm fold downregulation of the 23 genes validated by real-time PCR. All genes validated were found to be downregulated by both real-time PCR and microarray analyses [Pearson's correlation, 0.538, TNF-related apoptosis-inducing ligand (TRAIL) removed, 0.810]. (C) Western blotting results for mitogen-activated protein kinase kinase 4 (MEK4) expression in 8505C cells. Jurkat T cell lysate acts as a positive control for expression of MEK4. There are two lanes where protein from negative control and pre-miR-25 transfected cells was loaded, and untreated 8505C protein was loaded and acted as a 'no primary antibody' negative control as it was only exposed to secondary antibody. GAPDH acts as a loading control. A band of the correct size for MEK4 (45 kDa) was detected on the blot. The reduction of MEK4 in the pre-miR-25 treated cells successfully validates the fact that pre-miR-25 decreases the expression of this protein. (D) Western blotting results for TRAIL expression in 8505C cells. Lane titles are the same as the previous blot. Bands of approximately the correct size for TRAIL (32 kDa) were detected in the cell lysates tested. The reduction of TRAIL in the pre-miR-25 treated cells successfully validates the fact that pre-miR-25 decreases the expression of this protein.
Table I.
All significantly downregulated genes in pre-miR-222 treated cells compared to cells treated with negative control pre-miRs.
| RefSeq | Gene symbol | Gene assignment | Fold change | P-value (FDR) |
|---|---|---|---|---|
| NM_015913 | TXNDC12 | Thioredoxin domain containing 12 (endoplasmic reticulum) | −4.807 | 1.02E-07 |
| NM_000700 | ANXA1 | Annexin A1 | −3.881 | 2.24E-08 |
| NM_052886 | MAL2 | Mal, T cell differentiation protein 2 | −3.681 | 1.16E-07 |
| NM_005139 | ANXA3 | Annexin A3 | −3.595 | 1.48E-09 |
| NM_004493 | HSD17B10 | Hydroxysteroid (17-beta) dehydrogenase 10 | −3.548 | 3.73E-10 |
| NM_001124 | ADMb | Adrenomedullin | −3.526 | 1.08E-09 |
| NM_020373 | ANO2 | Anoctamin 2 | −3.475 | 1.68E-07 |
| NM_000782 | CYP24A1 | Cytochrome P450, family 24, subfamily A, polypeptide 1 | −3.321 | 1.71E-06 |
| NM_005238 | ETS1b | V-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | −3.159 | 2.16E-08 |
| NM_003494 | DYSF | Dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) | −3.057 | 8.62E-09 |
| NM_019026 | TMCO1 | Transmembrane and coiled-coil domains 1 | −2.935 | 4.46E-07 |
| NM_007257 | PNMA2 | Paraneoplastic antigen MA2 | −2.916 | 4.12E-08 |
| NM_031302 | GLT8D2 | Glycosyltransferase 8 domain containing 2 | −2.853 | 1.19E-07 |
| NM_002371 | MAL | Mal, T cell differentiation protein | −2.830 | 4.19E-08 |
| NM_001165 | BIRC3 | Baculoviral IAP repeat-containing 3 | −2.734 | 6.86E-08 |
| NM_153020 | RBM24b | RNA binding motif protein 24 | −2.717 | 1.34E-05 |
| NM_020651 | PELI1 | Pellino homolog 1 (Drosophila) | −2.668 | 4.42E-06 |
| NM_020455 | GPR126 | G protein-coupled receptor 126 | −2.659 | 9.57E-08 |
| NM_004572 | PKP2 | Plakophilin 2 | −2.630 | 7.81E-10 |
| NM_012431 | SEMA3E | Sema domain, immunoglobulin domain (Ig), short basic doma in, secreted, (semaphorin) 3E | −2.625 | 1.83E-05 |
| NM_002780 | PSG4 | Pregnancy specific beta-1-glycoprotein 4 | −2.606 | 8.80E-08 |
| NM_002764 | PRPS1 | Phosphoribosyl pyrophosphate synthetase 1 | −2.599 | 1.99E-08 |
| NM_014322 | OPN3 | Opsin 3 | −2.581 | 9.04E-07 |
| NM_013322 | SNX10 | Sorting nexin 10 | −2.569 | 2.18E-07 |
| NM_182757 | RNF144B | Ring finger 144B | −2.519 | 1.61E-09 |
| NM_000716 | C4BPB | Complement component 4 binding protein beta | −2.519 | 6.66E-05 |
| NM_020127 | TUFT1 | Tuftelin 1 | −2.519 | 3.51E-08 |
| NM_001008749 | RAB19 | RAB19, member RAS oncogene family | −2.501 | 8.91E-07 |
| NM_017439 | PION | Pigeon homolog (Drosophila) | −2.499 | 3.05E-05 |
| NM_018639 | WSB2 | WD repeat and SOCS box-containing 2 | −2.482 | 4.86E-08 |
| NM_014143 | CD274 | CD274 molecule | −2.420 | 5.96E-07 |
| NM_003246 | THBS1 | Thrombospondin 1 | −2.403 | 1.78E-08 |
| NM_001159 | AOX1 | Aldehyde oxidase 1 | −2.403 | 2.50E-06 |
| NM_000141 | FGFR2 | Fibroblast growth factor receptor 2 | −2.394 | 2.56E-06 |
| NM_003901 | SGPL1 | Sphingosine-1-phosphate lyase 1 | −2.380 | 1.82E-06 |
| NM_002296 | LBR | Lamin B receptor | −2.368 | 2.36E-08 |
| NM_025239 | PDCD1LG2 | Programmed cell death 1 ligand 2 | −2.360 | 7.42E-06 |
| NM_004741 | NOLC1 | Nucleolar and coiled-body phosphoprotein 1 | −2.351 | 6.16E-07 |
| NM_002998 | SDC2a | Syndecan 2 | −2.333 | 2.69E-07 |
| NM_001259 | CDK6c | Cyclin-dependent kinase 6 | −2.332 | 2.46E-07 |
| NM_016147 | PPME1 | Protein phosphatase methylesterase 1 | −2.326 | 1.44E-08 |
| NM_004482 | GALNT3 | UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) | −2.314 | 4.48E-06 |
| NM_021069 | SORBS2 | Sorbin and SH3 domain containing 2 | −2.314 | 2.89E-07 |
| NM_005264 | GFRA1 | GDNF family receptor alpha 1 | −2.304 | 5.49E-07 |
| NM_022131 | CLSTN2 | Calsyntenin 2 | −2.298 | 7.38E-08 |
| NM_002875 | RAD51 | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | −2.295 | 2.71E-06 |
| BC066649 | C1orf198 | Chromosome 1 open reading frame 198 | −2.275 | 2.03E-07 |
| AK131040 | LOC388022 | Hypothetical gene supported by AK131040 | −2.259 | 8.18E-05 |
| BC016278 | LOH3CR2A | Loss of heterozygosity, 3, chromosomal region 2, gene A | −2.243 | 0.0004969 |
| NM_001554 | CYR61 | Cysteine-rich, angiogenic inducer, 61 | −2.241 | 6.15E-07 |
| NM_033393 | FHDC1 | FH2 domain containing 1 | −2.237 | 3.51E-07 |
| NM_002492 | NDUFB5 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5 | −2.225 | 1.36E-07 |
| NM_018327 | SPTLC3 | Serine palmitoyltransferase, long chain base subunit 3 | −2.225 | 6.97E-07 |
| NM_080670 | SLC35A4 | Solute carrier family 35, member A4 | −2.211 | 6.39E-07 |
| NM_022772 | EPS8L2 | EPS8-like 2 | −2.202 | 3.77E-08 |
| NM_014391 | ANKRD1 | Ankyrin repeat domain 1 (cardiac muscle) | −2.191 | 7.78E-08 |
| NM_016206 | VGLL3 | Vestigial like 3 (Drosophila) | −2.191 | 8.67E-07 |
| Ensembl no: ENST00000319426 | - | Partially transcribed sequence | −2.188 | 4.57E-06 |
| NM_004694 | SLC16A6 | Solute carrier family 16, member 6 (monocarboxylic acid transporter 7) | −2.184 | 0.0003082 |
| NM_021623 | PLEKHA2 | Pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 | −2.160 | 1.74E-06 |
| NR_002836 | PGM5P2 | Phosphoglucomutase 5 pseudogene 2 | −2.159 | 2.55E-07 |
| NM_001145204 | LOC729993 | Hypothetical protein LOC729993, transcript variant 1, mRNA | −2.148 | 2.73E-07 |
| NM_153345 | TMEM139 | Transmembrane protein 139 | −2.143 | 3.58E-06 |
| NR_002836 | PGM5P2 | Phosphoglucomutase 5 pseudogene 2 | −2.141 | 4.39E-08 |
| NM_006252 | PRKAA2 | Protein kinase, AMP-activated, alpha 2 catalytic subunit | −2.137 | 7.04E-07 |
| NM_002547 | OPHN1 | Oligophrenin 1 | −2.137 | 2.60E-07 |
| NM_016132 | MYEF2 | Myelin expression factor 2 | −2.136 | 4.39E-05 |
| NM_003813 | ADAM21 | ADAM metallopeptidase domain 21 | −2.121 | 0.0005355 |
| Ensembl no: ENST00000384701 | – | Expressed sequence tag (EST) | −2.116 | 0.0002848 |
| NM_145753 | PHLDB2 | Pleckstrin homology-like domain, family B, member 2 | −2.099 | 4.55E-05 |
| NM_001415 | EIF2S3 | Eukaryotic translation initiation factor 2, subunit 3 gamma, 52 kDa | −2.096 | 2.72E-07 |
| NM_000331 | SAA1 | Serum amyloid A1 | −2.086 | 0.0041182 |
| NM_003265 | TLR3 | Toll-like receptor 3 | −2.082 | 5.79E-05 |
| NM_006389 | HYOU1 | Hypoxia up-regulated 1 | −2.077 | 1.02E-07 |
| NM_015238 | WWC1 | WW and C2 domain containing 1 | −2.069 | 1.56E-07 |
| NM_001001522 | TAGLN | Transgelin | −2.056 | 9.60E-07 |
| Ensembl no: ENST00000322446 | – | Partially transcribed sequence | −2.042 | 0.0007311 |
| NM_006832 | FERMT2 | Fermitin family homolog 2 (Drosophila) | −2.005 | 3.07E-05 |
| NM_018728 | MYO5C | Myosin VC | −2.003 | 9.51E-07 |
| NM_002354 | EPCAM | Epithelial cell adhesion molecule | −2.018 | 0.0224988 |
Predicted target in miRanda, PicTar and TargetScan;
predicted target in PicTar and TargetScan;
predicted target in TargetScan.
Table II.
All significantly downregulated genes in pre-miR-25 treated cells compared to cells treated with negative control pre-miRs.
| RefSeq | Gene symbol | Gene assignment | Fold change | P-value (FDR) |
|---|---|---|---|---|
| NM_000210 | ITGA6 | Integrin alpha 6 | −2.352 | 6.17E-09 |
| NM_002372 | MAN2A1b | Mannosidase, alpha, class 2A, member 1 | −3.370 | 4.98E-10 |
| NM_002436 | MPP1 | Membrane protein, palmitoylated 1 | −2.669 | 1.08E-09 |
| NM_003255 | TIMP2 | TIMP metallopeptidase inhibitor 2 | −2.218 | 1.75E-09 |
| NM_021913 | AXLd | AXL receptor tyrosine kinase | −2.594 | 2.49E-09 |
| NM_015881 | DKK3 | Dickkopf homolog 3 (Xenopus laevis) | −3.428 | 2.89E-09 |
| NM_018442 | IQWD1c | IQ motif and WD repeats 1 | −2.134 | 2.97E-09 |
| NM_002204 | ITGA3 | Integrin alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) | −2.209 | 7.00E-09 |
| NM_004035 | ACOX1 | Acyl-Coenzyme A oxidase 1, palmitoyl | −2.206 | 1.42E-08 |
| NM_005562 | LAMC2 | Laminin gamma 2 | −2.803 | 1.12E-08 |
| NM_006520 | DYNLT3d | Dynein, light chain, Tctex-type 3 | −2.694 | 1.07E-08 |
| NM_006988 | ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif 1 | −2.285 | 1.34E-08 |
| NM_006080 | SEMA3Ae | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A | −2.450 | 2.61E-08 |
| NM_000351 | STS | Steroid sulfatase (microsomal), isozyme S | −2.410 | 2.36E-08 |
| NM_019004 | ANKIB1 | Ankyrin repeat and IBR domain containing 1 | −2.304 | 1.15E-08 |
| NM_181659 | NCOA3 | Nuclear receptor coactivator 3 | −2.006 | 1.41E-08 |
| NM_002844 | PTPRK | Protein tyrosine phosphatase receptor type K | −2.152 | 1.97E-08 |
| NM_002205 | ITGA5a | Integrin alpha 5 (fibronectin receptor, alpha polypeptide) | −2.614 | 9.07E-09 |
| NM_000916 | OXTR | Oxytocin receptor | −2.473 | 4.16E-08 |
| NM_005349 | RBPJ | Recombination signal binding protein for immunoglobulin kappa J region | −2.033 | 4.83E-08 |
| NM_005471 | GNPDA1 | Glucosamine-6-phosphate deaminase 1 | −2.236 | 1.42E-08 |
| NM_004237 | TRIP13 | Thyroid hormone receptor interactor 13 | −2.781 | 1.02E-08 |
| NM_001418 | EIF4G2e | Eukaryotic translation initiation factor 4 gamma, 2 | −2.494 | 1.95E-08 |
| NM_021999 | ITM2B | Integral membrane protein 2B | −2.070 | 3.95E-08 |
| NM_000138 | FBN1b | Fibrillin 1 | −2.355 | 2.18E-07 |
| NM_003010 | MAP2K4b | Mitogen-activated protein kinase kinase 4 | −2.147 | 1.34E-08 |
| NM_016275 | SELT | Selenoprotein T | −3.175 | 2.35E-08 |
| NM_001102445 | RGS4 | Regulator of G-protein signaling 4 | −2.194 | 3.62E-08 |
| NM_001001521 | UGP2b | UDP-glucose pyrophosphorylase 2 | −3.032 | 6.61E-08 |
| NM_002948 | RPL15 | Ribosomal protein L15 | −2.659 | 4.97E-08 |
| NM_003139 | SRPRb | Signal recognition particle receptor ('docking protein') | −2.195 | 1.33E-07 |
| NM_001190 | BCAT2b | Branched chain aminotransferase 2, mitochondrial | −3.040 | 7.52E-08 |
| NM_000693 | ALDH1A3 | Aldehyde dehydrogenase 1 family, member A3 | −2.168 | 2.58E-06 |
| NM_012242 | DKK1 | Dickkopf homolog 1 (Xenopus laevis) | −2.060 | 3.20E-07 |
| NM_006178 | NSF | N-ethylmaleimide-sensitive factor | −2.202 | 4.09E-07 |
| NM_199511 | CCDC80 | Coiled-coil domain containing 80 | −2.016 | 1.25E-07 |
| NM_014674 | EDEM1b | ER degradation enhancer, mannosidase alpha-like 1 | −2.391 | 8.84E-08 |
| NM_213611 | SLC25A3 | Solute carrier family 25 (mitochondrial carrier; phosphate carrier) | −2.086 | 1.17E-07 |
| NM_133494 | NEK7 | NIMA (never in mitosis gene a)-related kinase 7 | −2.424 | 3.00E-07 |
| NM_017958 | PLEKHB2 | Pleckstrin homology domain containing, family B2 (evectins) member | −3.420 | 8.84E-08 |
| NM_000259 | MYO5A | Myosin VA (heavy chain 12, myoxin) | −2.426 | 2.11E-07 |
| NM_016277 | RAB23b | RAB23, member RAS oncogene family | −2.051 | 1.26E-07 |
| NM_001110 | ADAM10b | ADAM metallopeptidase domain 10 | −2.616 | 3.21E-07 |
| NM_001693 | ATP6V1B2 | ATPase, H+ transporting, lysosomal 56/58 kDa, V1 subunit B2 | −2.064 | 1.30E-07 |
| NM_005779 | LHFPL2a | Lipoma HMGIC fusion partner-like 2 | −2.173 | 1.72E-07 |
| NM_016422 | RNF141e | Ring finger protein 141 | −2.260 | 8.57E-08 |
| NM_002095 | GTF2E2 | General transcription factor IIE, polypeptide 2, beta 34k | −2.570 | 7.22E-07 |
| NM_003483 | HMGA2e | High mobility group AT-hook 2 | −2.654 | 1.24E-07 |
| NM_014223 | NFYCc | Nuclear transcription factor Y, gamma | −2.112 | 1.57E-07 |
| NM_013381 | TRHDE | Thyrotropin-releasing hormone degrading enzyme | −2.306 | 2.96E-07 |
| NM_006287 | TFPI | Tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) | −2.145 | 5.36E-07 |
| NM_033540 | MFN1 | Mitofusin 1 | −2.014 | 3.46E-07 |
| NM_024664 | PPCSb | Phosphopantothenoylcysteine synthetase | −2.386 | 3.87E-07 |
| NM_023080 | C8orf33 | Chromosome 8 open reading frame 33 | −2.006 | 1.98E-07 |
| NM_016352 | CPA4 | Carboxypeptidase A4 | −2.060 | 1.26E-06 |
| NM_012223 | MYO1Bb | Myosin IB | −2.030 | 1.35E-06 |
| NM_020223 | FAM20C | Family with sequence similarity 20 member C | −2.387 | 1.16E-06 |
| NM_004670 | PAPSS2 | 3′-phosphoadenosine 5′-phosphosulfate synthase 2 | −2.347 | 9.66E-07 |
| NM_005896 | IDH1 | Isocitrate dehydrogenase 1 (NADP+), soluble | −2.599 | 3.89E-06 |
| NM_016322 | RAB14b | RAB14, member RAS oncogene family | −2.021 | 4.30E-07 |
| NM_002971 | SATB1 | SATB homeobox 1 | −2.080 | 1.43E-06 |
| NM_000405 | GM2A | GM2 ganglioside activator | −2.249 | 4.07E-07 |
| NM_001547 | IFIT2 | Interferon-induced protein with tetratricopeptide repeats | −4.287 | 8.87E-07 |
| NM_016570 | ERGIC2 | ERGIC and golgi 2 | −2.858 | 9.73E-07 |
| NM_005595 | NFIAb | Nuclear factor I/A | −2.567 | 6.45E-07 |
| NM_005981 | TSPAN31 | Tetraspanin 31 | −2.127 | 6.55E-07 |
| NM_001085471 | FOXN3 | Forkhead box N3 | −2.314 | 1.11E-06 |
| BC016048 | LRRC38 | Leucine rich repeat containing 38 | −2.394 | 5.78E-07 |
| NM_005314 | GRPR | Gastrin-releasing peptide receptor | −2.545 | 6.82E-06 |
| NM_002783 | PSG7 | Pregnancy specific beta-1-glycoprotein 7 | −2.015 | 3.89E-06 |
| NM_002781 | PSG5 | Pregnancy specific beta-1-glycoprotein 5 | −2.222 | 4.73E-06 |
| NM_003884 | KAT2B | K(lysine) acetyltransferase 2B | −2.108 | 4.43E-06 |
| NM_001040409 | MTHFD2 | Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase | −2.312 | 1.92E-05 |
| NM_003580 | NSMAFb | Neutral sphingomyelinase (N-SMase) activation associated factor | −2.072 | 2.33E-06 |
| NM_001013442 | EPGN | Epithelial mitogen homolog (mouse) | −3.057 | 1.89E-06 |
| NM_012319 | SLC39A6 | Solute carrier family 39 (zinc transporter), member 6 | −2.026 | 2.95E-06 |
| NM_000965 | RARB | Retinoic acid receptor, beta | −2.624 | 5.42E-06 |
| NM_152772 | TCP11L2 | T-complex 11 (mouse)-like 2 | −2.105 | 4.63E-05 |
| BC017297 | FAM49B | Family with sequence similarity 49 member B | −2.051 | 1.87E-06 |
| NM_001002265 | MARCH8 | Membrane-associated ring finger (C3HC4) 8 | −2.232 | 7.22E-06 |
| NM_015509 | NECAP1b | NECAP endocytosis associated 1 | −2.013 | 1.85E-06 |
| NM_001080512 | BICC1 | Bicaudal C homolog 1 (Drosophila) | −2.193 | 9.59E-06 |
| NM_001031850 | PSG6 | Pregnancy specific beta-1-glycoprotein 6 | −2.806 | 7.76E-06 |
| NM_020841 | OSBPL8 | Oxysterol binding protein-like 8 | −2.135 | 8.37E-06 |
| NM_007036 | ESM1 | Endothelial cell-specific molecule 1 | −2.059 | 5.81E-06 |
| NM_016075 | VPS36 | Vacuolar protein sorting 36 homolog (S. cerevisiae) | −2.021 | 3.55E-06 |
| NM_000963 | PTGS2 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | −2.084 | 9.13E-06 |
| NM_017946 | FKBP14 | FK506 binding protein 14, 22 kDa | −2.240 | 1.13E-05 |
| NM_003825 | SNAP23 | Synaptosomal-associated protein, 23 kDa | −2.233 | 6.08E-06 |
| NM_194328 | RNF38a | Ring finger protein 38 | −2.526 | 7.91E-06 |
| NM_006636 | MTHFD2 | Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase | −2.362 | 0.0001745 |
| NM_006905 | PSG1 | Pregnancy specific β-1-glycoprotein 1 | −2.016 | 0.0001079 |
| Ensembl no: ENST00000390952 | – | Expressed Sequence Tag (EST) | −2.078 | 0.0001661 |
| NM_030920 | ANP32Ee | Acidic (leucine-rich) nuclear phosphoprotein 32 family, member E | −2.663 | 6.01E-05 |
| NM_000262 | NAGA | N-acetylgalactosaminidase, alpha | −2.055 | 4.77E-05 |
| NM_001548 | IFIT1 | Interferon-induced protein with tetratricopeptide repeats | −2.439 | 3.00E-05 |
| NM_001039569 | AP1S3 | Adaptor-related protein complex 1, sigma 3 subunit | −2.193 | 0.0003395 |
| NM_003810 | TNFSF10 | Tumour necrosis factor (ligand) superfamily, member 10 | −2.003 | 0.0011012 |
Predicted target in miRanda, PicTar and TargetScan;
predicted target in PicTar and TargetScan;
predicted target in miRanda and TargetScan;
predicted target in miRanda;
predicted target in TargetScan.
Overexpression of miR-222 in Nthy-ori cells
Five of the 82 genes (6.1%) significantly downregulated by miR-222 overexpression in the Nthy-ori 3-1 cells were predicted gene targets of the miRNA in miRanda, PicTar or TargetScan, or a combination of the databases. Gene ontology analysis revealed that no molecular function or biological process was significantly over- or underrepresented in the miR-222 target list. Notwithstanding this observation, several genes that were found to be targets of this miRNA have interesting molecular functions. For instance, seven receptors are in the lists (MAL, OPN3, PSG4, FGFR2, GPR126, TLR3 and LBR), four transcription factors (PSG4, ETS1, MAL and RBM24), five signalling molecules (THBS1, ETS1, CYR61, SEMA3E and ADM), and five genes involved in defence and immunity (PNMA2, C4BPB, SDC2, MAL, TLR3 and SAA1).
The downregulation of 22 genes in response to miR-222 overexpression identified by the microarray analysis (both predicted and novel targets) was successfully validated using Fluidigm real-time PCR technologies (Table III and Fig. 1B) (Pearson's correlation, 0.506).
Table III.
Downregulated genes following expression of pre-miR-222 in Nthy-ori 3-1.
| Gene | Microarray fold downregulation | Microarray P-value (FDR) | Fluidigm qPCR fold downregulation | Fluidigm P-value (t-test) |
|---|---|---|---|---|
| MAL2 | 3.681 | 1.16E-07 | 3.314 | 3.55E-05 |
| MAL | 2.83 | 4.19E-08 | 2.332 | 1.54E-05 |
| TLR3 | 2.082 | 5.79E-05 | 2.341 | 0.0001 |
| ADMa | 3.526 | 1.08E-09 | 3.81 | 2.93E-05 |
| RAB19 | 2.501 | 8.91E-07 | 3.7 | 4.48E-06 |
| PDCD1LG2 | 2.36 | 7.42E-06 | 1.827 | 0.004 |
| RAD51 | 2.295 | 2.71E-06 | 1.597 | 9.59E-05 |
| ADAM21 | 2.121 | 0.0005 | 1.008 | 0.9517 |
| HYOU1 | 2.077 | 1.02E-07 | 1.198 | 0.0001 |
| THBS1 | 2.403 | 1.78E-08 | 1.769 | 0.0002 |
| ETS1a | 3.159 | 2.16E-08 | 2.208 | 0.0003 |
| CYR61 | 2.241 | 6.15E-07 | 1.745 | 0.0006 |
| PHLDB2 | 2.099 | 4.55E-05 | 1.178 | 0.003 |
| BIRC3 | 2.734 | 6.86E-08 | 2.406 | 0.0005 |
| PRKAA2 | 2.137 | 7.04E-07 | 1.595 | 0.001 |
| CYP24A1 | 3.321 | 1.71E-06 | 1.911 | 0.0006 |
| AOX1 | 2.403 | 2.50E-06 | 2.024 | 0.0006 |
| MYO5C | 2.003 | 9.51E-07 | 3.549 | 0.0002 |
| GPR126 | 2.659 | 9.57E-08 | 3.24 | 6.8E-05 |
| CDK6b | 2.332 | 2.46E-07 | 1.588 | 0.0004 |
| PPME1 | 2.326 | 1.44E-08 | 1.5 | 0.0006 |
| RBM24a | 2.717 | 1.34E-05 | 3.55 | 3.11E-06 |
Predicted target in PicTar and TargetScan;
predicted target in TargetScan.
Overexpression of miR-25 in 8505C cells
The list of 98 miR-25 target genes was also cross-referenced with in silico prediction results. This comparison revealed that the list of genes that were significantly downregulated by pre-miR-25 in the 8505C cells is enriched with 27/98 or 27.55% predicted gene targets for miR-25. Gene ontology analysis of the miR-25 gene target list suggests that cell adhesion is an important aspect of miR-25 functioning in the thyroid cells as biological processes including cell adhesion, cell communication, signal transduction, and cell adhesion-mediated signalling were significantly enriched for in this list (P-value 1.47E-02, 1.18E-03, 3.13E-02 and 1.59E-02, respectively). The list is also significantly enriched for cell adhesion molecules: ITGA3, ITGA5 (which is also a predicted target of the miRNA in all three databases used); and ITGA6 (P-value 3.10E-03) and CAM family adhesion molecules: PSG1, PSG5, PSG6 and PSG7 (P-value 2.47E-02). Seven receptors are also in the miR-25 list: TCP11L2, RARB, GRPR, PTPRK, AXL, LRRC38 and OXTR; six transcription factors: RARB, FOXN3, SATB1, NCOA3, GTF2E2 and NFIA; and five kinases: MPP1, AXL, MAP2K4, PAPSS2 and NEK7, and a member of the tumour necrosis family, TNFSF10/TRAIL.
Fluidigm real-time PCR technologies were used to confirm the downregulation of 23 predicted and novel gene targets in response to miR-25 expression in the 8505C cells (Table IV and Fig. 2B). All genes tested were downregulated in both the microarray and Fluidigm analyses (Pearson's correlation, 0.538; TRAIL removed, 0.810). In addition, two novel targets of miR-25 were successfully validated at the protein level by western blotting; predicted target MEK4 and novel target TRAIL. Fig. 2C and D illustrate the reduction in MEK4 and TRAIL protein expression following treatment of anaplastic thyroid cells with pre-miR-25, demonstrating that they are direct or indirect targets of this miRNA.
Table IV.
Downregulated genes following expression of pre-miR-25 in 8505C cells.
| Gene | Microarray fold downregulation | Microarray P-value (FDR) | Fluidigm qPCR fold downregulation | Fluidigm P-value (t-test) |
|---|---|---|---|---|
| TNFSF10/TRAIL | 2.003 | 0.001 | 3.982 | 0.003 |
| ITGA6 | 2.352 | 6.17E-09 | 2.552 | 0.0002 |
| MPP1 | 2.669 | 1.08E-09 | 2.718 | 7.3E-06 |
| TIMP2 | 2.218 | 1.75E-09 | 2.483 | 2.19E-05 |
| ITGA3 | 2.209 | 7.00E-09 | 2.194 | 2.40E-07 |
| ADAMTS1 | 2.285 | 1.34E-08 | 2.304 | 0.002 |
| ITGA5b | 2.614 | 9.07E-09 | 2.699 | 4.68E-07 |
| TRIP13 | 2.781 | 1.02E-08 | 2.606 | 4.92E-06 |
| MAP2K4a | 2.147 | 1.34E-08 | 1.925 | 7.70E-05 |
| RAB23a | 2.051 | 1.26E-07 | 2.122 | 8.45E-05 |
| ADAM10a | 2.616 | 3.21E-07 | 2.571 | 1.57E-05 |
| TRHDE | 2.306 | 2.96E-07 | 2.43 | 2.09E-06 |
| RAB14a | 2.021 | 4.30E-07 | 1.907 | 2.91E-05 |
| RARB | 2.624 | 5.42E-06 | 2.816 | 7.68E-06 |
| MAN2A1a | 3.37 | 4.98E-10 | 3.576 | 2.19E-07 |
| RGS4 | 2.194 | 3.62E-08 | 2.255 | 0.0009 |
| PTGS2 | 2.084 | 9.13E-06 | 1.853 | 0.0098 |
| EPGN | 3.057 | 1.89E-06 | 3.394 | 0.0003 |
| SNAP23 | 2.233 | 6.08E-06 | 2.057 | 8.58E-05 |
| LHFPL2b | 2.173 | 1.72E-07 | 3.092 | 0.0002 |
| MYO1Ba | 2.03 | 1.35E-06 | 1.986 | 1.997E-06 |
| NFIAa | 2.567 | 6.45E-07 | 3.445 | 0.015 |
Predicted target in PicTar and TargetScan;
predicted target in miRanda, PicTar and TargetScan.
Discussion
The aim of this study was to elucidate the mRNA targets of two miRNAs that we had previously found to be differentially expressed in thyroid cancer (5): miR-222 and miR-25. This was achieved by overexpressing the thyroid cancer-associated miRNA miR-222 in the transformed normal Nthy-ori cells, and miR-25 in the anaplastic cancer-derived 8505C cells. The use of microarrays to analyse the RNA from these cells exploited an unbiased genome-wide approach and yielded the discovery of a set of mRNAs that are either directly or indirectly targeted by each miRNA in thyroid cells, and have not been previously described.
The gene target lists produced by this genome-wide investigation compare favourably with previously published studies on miRNA target elucidation and prediction. Similar to prior studies (21,23), the two pre-miRs used in this body of study each downregulated almost 100 target transcripts: 98 by pre-miR-25 in the 8505C cells and 82 by pre-miR-222 in the Nthy-ori 3-1 cells. The subtle change in gene expression observed in the genes targeted by pre-miR-222 and pre-miR-25 (between 2- and 4-fold) is also similar to other studies on the subject (6,21). Finally, the relatively low number of predicted miRNA targets significantly differentially expressed has been noted previously (6), and the divergent assortment of molecular functions and biological processes encompassed within the gene target lists is also reflective of published accounts of target prediction and elucidation (20,21). These diverse lists allow us insight into the functions of these two miRNAs within the respective thyroid cell lines. For instance, upregulation of miR-222 in Nthy-ori cells impacts on several transcription factors, cell signalling molecules, and genes involved in defence and immunity (PNMA2, C4BPB, SDC2, MAL, TLR3 and SAA1).
Expression of miR-25 is downregulated in anaplastic thyroid carcinoma, so we upregulated the expression of miR-25 in the anaplastic thyroid carcinoma 8505C cell line. Although miR-25 was found to target genes with a wide variety of functions, the gene ontology analysis of this list highlights the predilection of this miRNA for regulating genes involved in cell adhesion, with both this cellular process and molecules of this molecular function being significantly over-represented in this list. Two main groups of cell adhesion molecules are over-represented in the miR-25 target list; inte-grin α genes ITGA3, ITGA5 (which is a predicted target of the miR-25 in miRanda, PicTar and TargetScan) and ITGA6, and the pregnancy-specific glycoproteins (PSGs); PSG1, PSG5, PSG6 and PSG7. Loss of miR-25 expression in anaplastic thyroid carcinoma is therefore associated with upregulation of genes encoding integrins and glycoproteins. The ITGA genes appear to be markers of aggression in the cancers in which they have been explored. For instance, ITGA3 and ITGA5 have been shown to be markers of invasiveness in head and neck squamous cell carcinoma (24), ITGA3, along with ITGB4 and 5 were found to be candidate biomarkers for cervical lymph node metastasis or death in tongue squamous cell carcinoma (25) and ITGA6 was found to be necessary for the tumourigenicity of a stem cell-like subpopulation within the MCF7 breast cancer cell line (26).
This is not the first occasion miR-25 has been linked to a role in cell adhesion, as Xu et al have previously highlighted the role of miR-25 in oesophageal squamous cell carcinoma (ESCC). They found upregulation of this miRNA in ESCC cells promoted migration and invasion, and also found that miR-25 directly targets E-Cadherin, an important cellular adhesion protein in the ESCC cells (13). Gerson et al have recently identified miR-25, and indeed miR-222, to be responsive to β4 integrin expression in breast carcinoma cell lines (27). Our observation, along with these previous studies in breast cancer and ESCC, indicates a role for miR-25 in tumour progression through the disruption of cell adhesion.
Additional functions of miR-25 are highlighted with the two proteins (MEK4 and TRAIL) that were shown to be down-regulated in response to the expression of miR-25. MEK4 is a member of the MAP kinase kinase family that directly phosphorylates and activates c-Jun NH2-terminal kinase (JNK) in response to cellular stresses and pro-inflammatory cytokines, and can also activate p38 (28). MAP kinase signalling is often disrupted in thyroid cancer through RAS, BRAF or RET/PTC mutations (29). Both tumour suppressor and oncogenic functions have been attributed to MEK4 in cancer (30,31). Little is known of the role of MEK4 in thyroid cancer; however, Chiariello et al demonstrated that Ret signalling involves MEK4 (32). It remains unclear as to whether MEK4 expression influences anaplastic thyroid carcinoma in a tumour suppressive or oncogenic manner. However, as pre-miR-25 was shown to downregulate MEK4 mRNA and protein in ATC 8505C cells in this study, it is interesting to speculate that the endogenous downregulation of miR-25 may lead to the upregulation of MEK4 expression and its pro-oncogenic characteristics in ATC cells.
TRAIL expression was also decreased in response to miR-25 expression in 8505C cells. TRAIL has five cellular receptors and can activate the extrinsic and intrinsic pathways to regulate intercellular apoptotic responses in the immune system (33). Approximately a decade and a half ago it was noted that TRAIL could induce apoptosis in transformed and malignant cells but not in normal cells (34). TRAIL was subsequently found to be capable of triggering apoptosis in eight PTC and two ATC derived thyroid carcinoma cell lines but not in normal thyrocytes (35). As a result of this tumour-specific effect, a great deal of work was done to develop anticancer therapies to mimic the effect of TRAIL. The TRAIL and MAP kinase pathways appear to overlap in TRAIL therapy signalling. Ohtsuka et al reported that the combination of anti-death receptor antibodies and chemotherapy agents led to a synergistic activation of the JNK/p38 MAP kinase which was mediated by MEK4 in breast, prostate and colon cancer cells (36). Moreover, Söderström et al showed that MAP/extracellular regulated kinase (ERK) signalling (which is frequently over-activated in thyroid cancer) protected activated T cells from TRAIL-induced apoptosis (37).
miR-25 involvement in the TRAIL-mediated apoptosis pathway was confirmed further with the observation by Razumilava et al that this miRNA targets TRAIL death receptor-4 (DR4) and promotes apoptosis resistance in cholangiocarcinoma (38). If one considers the pro-apoptotic role of TRAIL, it is difficult to elucidate how an upregulation of this gene through the endogenous downregulation of miR-25 would be beneficial to the progress of anaplastic thyroid carcinoma. However, a review by Newsom-Davis et al outlines in vitro and in vivo experiments in which TRAIL expression induced proliferation, migration and invasion of tumour cells which were resistant to TRAIL-mediated apoptosis. They describe how secondary intracellular signalling complexes, following TRAIL DISC formation, can activate NF-κB via the inhibitor of κB kinase complex (IKK complex) which signals through MAPK, JNK and p38 (39). Other groups have shown that TRAIL-induced survival and proliferation does not involve the p38 kinase pathway but is dependent on the MAP kinase ERKs. Therefore, in the anaplastic thyroid cancer cells, the endogenous downregulation of miR-25 may enable the upregulation of TRAIL to activate its pro-survival MAP kinase responses (perhaps involving MEK4). Future studies may also benefit from investigating whether miR-25 is involved in regulating MEK4 and TRAIL in response to TRAIL cancer therapies.
In conclusion, this study used an unbiased approach to elucidate almost 100 genes that are either directly or indirectly targeted by miR-25 and miR-222 in thyroid cells. The number of genes in these target lists, the extent to which the genes are regulated and the diversity of their functions is reflective of other published accounts of miRNA target prediction and elucidation. The gene targets of these two miRNAs confirm the diverse nature of miRNA-target interactions within cells. A considerable proportion of these targets have been validated using RT-PCR with a further two, MEK4 and TRAIL, being confirmed at the protein level. We provide an interesting insight into the functions of these two miRNAs in thyroid cells, in particular that cell adhesion and apoptosis are important aspects of miR-25 functioning in thyroid cells. In addition to this, there is broad scope for further investigation of the many results produced by this study.
Acknowledgments
The authors would like to acknowledge grants from the Health Research Board Molecular Medicine PhD training programme and the Science Foundation Ireland funded Molecular Therapeutics for Cancer Ireland.
Abbreviations
- MAPK
mitogen-activated protein kinase
- hTERT
human telomerase reverse transcriptase
- FDR
false discovery rate
- RIPA
radioimmunoprecipitation assay
- DR4
TRAIL death receptor-4
- PSG
pregnancy-specific glycoprotein
- TRAIL
TNF-related apoptosis-inducing ligand
- IKK complex
inhibitor of κB kinase complex
- ERK
extracellular regulated kinases
- JNK
c-Jun NH2-terminal kinase
References
- 1.Smallridge RC, Marlow LA, Copland JA. Anaplastic thyroid cancer: molecular pathogenesis and emerging therapies. Endocr Relat Cancer. 2009;16:17–44. doi: 10.1677/ERC-08-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, Troncone G, Chiappetta G, Liu CG, Santoro M, Negrini M, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 4.Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93:1600–1608. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aherne ST, Smyth PC, Flavin RJ, Russell SM, Denning KM, Li JH, Guenther SM, O'Leary JJ, Sheils OM. Geographical mapping of a multifocal thyroid tumour using genetic alteration analysis and miRNA profiling. Mol Cancer. 2008;7:89. doi: 10.1186/1476-4598-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HJ, Kim YH, Lee DS, Chung J-K, Kim S. In vivo imaging of functional targeting of miR-221 in papillary thyroid carcinoma. J Nucl Med. 2008;49:1686–1693. doi: 10.2967/jnumed.108.052894. [DOI] [PubMed] [Google Scholar]
- 7.Mitomo S, Maesawa C, Ogasawara S, Iwaya T, Shibazaki M, Yashima-Abo A, Kotani K, Oikawa H, Sakurai E, Izutsu N, et al. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci. 2008;99:280–286. doi: 10.1111/j.1349-7006.2007.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keutgen XM, Filicori F, Crowley MJ, Wang Y, Scognamiglio T, Hoda R, Buitrago D, Cooper D, Zeiger MA, Zarnegar R, et al. A panel of four miRNAs accurately differentiates malignant from benign indeterminate thyroid lesions on fine needle aspiration. Clin Cancer Res. 2012;18:2032–2038. doi: 10.1158/1078-0432.CCR-11-2487. [DOI] [PubMed] [Google Scholar]
- 9.Chou CK, Yang KD, Chou FF, Huang CC, Lan YW, Lee YF, Kang HY, Liu RT. Prognostic implications of miR-146b expression and its functional role in papillary thyroid carcinoma. J Clin Endocrinol Metab. 2013;98:E196–E205. doi: 10.1210/jc.2012-2666. [DOI] [PubMed] [Google Scholar]
- 10.Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, Borbone E, Petrocca F, Alder H, Croce CM, Fusco A. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 11.Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68:8191–8194. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- 12.Kim Y-K, Yu J, Han TS, Park SY, Namkoong B, Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK, Kim VN. Functional links between clustered microRNAs: Suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37:1672–1681. doi: 10.1093/nar/gkp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu X, Chen Z, Zhao X, Wang J, Ding D, Wang Z, Tan F, Tan X, Zhou F, Sun J, et al. MicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2012;421:640–645. doi: 10.1016/j.bbrc.2012.03.048. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, Zuo Z, Lu X, Wang L, Wang H, Zhu Z. MiR-25 regulates apoptosis by targeting Bim in human ovarian cancer. Oncol Rep. 2012;27:594–598. doi: 10.3892/or.2011.1530. [DOI] [PubMed] [Google Scholar]
- 15.Kan T, Sato F, Ito T, Matsumura N, David S, Cheng Y, Agarwal R, Paun BC, Jin Z, Olaru AV, et al. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136:1689–1700. doi: 10.1053/j.gastro.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Visone R, Pallante P, Vecchione A, Cirombella R, Ferracin M, Ferraro A, Volinia S, Coluzzi S, Leone V, Borbone E, et al. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2007;26:7590–7595. doi: 10.1038/sj.onc.1210564. [DOI] [PubMed] [Google Scholar]
- 17.Esposito F, Tornincasa M, Pallante P, Federico A, Borbone E, Pierantoni GM, Fusco A. Down-regulation of the miR-25 and miR-30d contributes to the development of anaplastic thyroid carcinoma targeting the polycomb protein EZH2. J Clin Endocrinol Metab. 2012;97:E710–E718. doi: 10.1210/jc.2011-3068. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 20.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 21.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 22.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu YH, Kuo HK, Chang KW. The evolving transcriptome of head and neck squamous cell carcinoma: a systematic review. PLoS One. 2008;3:e3215. doi: 10.1371/journal.pone.0003215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurokawa A, Nagata M, Kitamura N, Noman AA, Ohnishi M, Ohyama T, Kobayashi T, Shingaki S, Takagi R, Oral, Maxillofacial Pathology, and Surgery Group Diagnostic value of integrin alpha3, beta4, and beta5 gene expression levels for the clinical outcome of tongue squamous cell carcinoma. Cancer. 2008;112:1272–1281. doi: 10.1002/cncr.23295. [DOI] [PubMed] [Google Scholar]
- 26.Cariati M, Naderi A, Brown JP, Smalley MJ, Pinder SE, Caldas C, Purushotham AD. Alpha-6 integrin is necessary for the tumourigenicity of a stem cell-like subpopulation within the MCF7 breast cancer cell line. Int J Cancer. 2008;122:298–304. doi: 10.1002/ijc.23103. [DOI] [PubMed] [Google Scholar]
- 27.Gerson KD, Maddula VSRK, Seligmann BE, Shearstone JR, Khan A, Mercurio AM. Effects of β4 integrin expression on microRNA patterns in breast cancer. Biol Open. 2012;1:658–666. doi: 10.1242/bio.20121628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuenda A. Mitogen-activated protein kinase kinase 4 (MKK4) Int J Biochem Cell Biol. 2000;32:581–587. doi: 10.1016/S1357-2725(00)00003-0. [DOI] [PubMed] [Google Scholar]
- 29.Fagin JA, Mitsiades N. Molecular pathology of thyroid cancer: diagnostic and clinical implications. Best Pract Res Clin Endocrinol Metab. 2008;22:955–969. doi: 10.1016/j.beem.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Pan Y, Dai JL. Evidence of MKK4 pro-oncogenic activity in breast and pancreatic tumors. Oncogene. 2004;23:5978–5985. doi: 10.1038/sj.onc.1207802. [DOI] [PubMed] [Google Scholar]
- 31.Xin W, Yun KJ, Ricci F, Zahurak M, Qiu W, Su GH, Yeo CJ, Hruban RH, Kern SE, Iacobuzio-Donahue CA. MAP2K4/MKK4 expression in pancreatic cancer: genetic validation of immunohistochemistry and relationship to disease course. Clin Cancer Res. 2004;10:8516–8520. doi: 10.1158/1078-0432.CCR-04-0885. [DOI] [PubMed] [Google Scholar]
- 32.Chiariello M, Visconti R, Carlomagno F, Melillo RM, Bucci C, de Franciscis V, Fox GM, Jing S, Coso OA, Gutkind JS, et al. Signalling of the Ret receptor tyrosine kinase through the c-Jun NH2-terminal protein kinases (JNKS): evidence for a divergence of the ERKs and JNKs pathways induced by Ret. Oncogene. 1998;16:2435–2445. doi: 10.1038/sj.onc.1201778. [DOI] [PubMed] [Google Scholar]
- 33.Mahalingam D, Szegezdi E, Keane M, de Jong S, Samali A. TRAIL receptor signalling and modulation: are we on the right TRAIL? Cancer Treat Rev. 2009;35:280–288. doi: 10.1016/j.ctrv.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 34.Ashkenazi A, Dixit VM. Apoptosis control by death and decoy receptors. Curr Opin Cell Biol. 1999;11:255–260. doi: 10.1016/S0955-0674(99)80034-9. [DOI] [PubMed] [Google Scholar]
- 35.Mitsiades N, Poulaki V, Tseleni-Balafouta S, Koutras DA, Stamenkovic I. Thyroid carcinoma cells are resistant to FAS-mediated apoptosis but sensitive to tumor necrosis factor-related apoptosis-inducing ligand. Cancer Res. 2000;60:4122–4129. [PubMed] [Google Scholar]
- 36.Ohtsuka T, Buchsbaum D, Oliver P, Makhija S, Kimberly R, Zhou T. Synergistic induction of tumor cell apoptosis by death receptor antibody and chemotherapy agent through JNK/p38 and mitochondrial death pathway. Oncogene. 2003;22:2034–2044. doi: 10.1038/sj.onc.1206290. [DOI] [PubMed] [Google Scholar]
- 37.Söderström TS, Poukkula M, Holmström TH, Heiskanen KM, Eriksson JE. Mitogen-activated protein kinase/extracellular signal-regulated kinase signaling in activated T cells abrogates TRAIL-induced apoptosis upstream of the mitochondrial amplification loop and caspase-8. J Immunol. 2002;169:2851–2860. doi: 10.4049/jimmunol.169.6.2851. [DOI] [PubMed] [Google Scholar]
- 38.Razumilava N, Bronk SF, Smoot RL, Fingas CD, Werneburg NW, Roberts LR, Mott JL. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55:465–475. doi: 10.1002/hep.24698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newsom-Davis T, Prieske S, Walczak H. Is TRAIL the holy grail of cancer therapy? Apoptosis. 2009;14:607–623. doi: 10.1007/s10495-009-0321-2. [DOI] [PubMed] [Google Scholar]