Abstract
To determine the metabolic response to food deprivation, cuttlefish (Sepia officinalis) juveniles were either fed, fasted (3 to 5 days food deprivation), or starved (12 days food deprivation). Fasting resulted in a decrease in triglyceride levels in the digestive gland, and after 12 days, these lipid reserves were essentially depleted. Oxygen consumption was decreased to 53% and NH4 excretion to 36% of the fed group following 3–5 days of food deprivation. Oxygen consumption remained low in the starved group, but NH4 excretion returned to the level recorded for fed animals during starvation. The fractional rate of protein synthesis of fasting animals decreased to 25% in both mantle and gill compared with fed animals and remained low in the mantle with the onset of starvation. In gill, however, protein synthesis rate increased to a level that was 45% of the fed group during starvation. In mantle, starvation led to an increase in cathepsin A-, B-, H-, and L-like enzyme activity and a 2.3-fold increase in polyubiquitin mRNA that suggested an increase in ubiquitin-proteasome activity. In gill, there was a transient increase in the polyubiquitin transcript levels in the transition from fed through fasted to the starved state and cathepsin A-, B-, H-, and L-like activity was lower in starved compared with fed animals. The response in gill appears more complex, as they better maintain rates of protein synthesis and show no evidence of enhanced protein breakdown through recognized catabolic processes.
Keywords: NH4 production, ammonia quotient, cathepsin, triglyceride, digestive gland, proteasome, polyubiquitin
in the marine environment, cephalopods are positioned at the top of the invertebrates in terms of size, intelligence, and trophic rank. These molluscs present some functional convergence with fishes, while diverging in important ways, such as whole animal locomotion and metabolic fuel preferences (28). Cephalopods have a “live fast and die young” life strategy. They are mostly carnivorous animals that have a protein-based metabolism (21, 27, 28, 33). Contrary to many other animals, cephalopods do not store substantial amounts of lipids and carbohydrates as energy reserves, although triglycerides in the digestive gland may serve as a short-term fuel to support aerobic metabolism during food limitation. In these animals, surplus energy is instead used for somatic growth, and the protein in the mantle may, thus, be viewed as a form of stored energy (6, 21, 27). A consequence of this metabolic strategy is that, during starvation, cephalopods need to rely rapidly on this protein reserve to sustain metabolism. The physiological response to food deprivation is relatively similar in most vertebrates and invertebrates and is typically described as a three-phase process (4). Phase I is a transient phase occurring during the first few days of food deprivation, where diet-derived carbohydrates, lipids, and proteins are used to maintain basal metabolism. During phase II, the animals mainly mobilize their lipid reserves, with the duration mainly dependent on initial lipid mass. Once lipids are almost depleted, the animals are forced into phase III and oxidize their proteins as a fuel of last resort, and only then, are they considered to be in a true phase of starvation (23). Cuttlefish (Sepia officinalis) reach phase III following 7 days of food deprivation (19).
We have previously shown that in the mantle of food-deprived cuttlefish, at a time when digestive gland triglycerides were being mobilized, total RNA levels were decreased and cellular signaling pathways stimulating protein synthesis were disengaged (21). More specifically, the phosphorylation of AKT and 4EBP1 decreased. Phosphorylation of AKT can result in phosphorylation of 4EBP1, leading to an increase of protein synthesis, and dephosphorylation of these proteins should act in the opposite direction (10, 17). Under conditions where triglyceride levels in the digestive gland were almost totally exhausted, some protein degradation pathways in the mantle were activated. Cathepsin A-, B-, H-, and L-like proteases were elevated, as were indices of the ubiquitin-proteasome pathway. Most notably, there were increases in proteasome (β-subunit) and polyubiquitin transcript levels, and polyubiquinated protein (21). These biochemical indices imply that protein synthesis is curtailed during the early stages of food deprivation in the mantle of S. officinalis, and as starvation ensues, mantle protein is catabolized. Protein turnover in gill appears to be more complex. The phosphorylation of AKT and 4EBP1 increased under some conditions, suggesting an increase in the rate of protein synthesis. As well, for any given triglyceride level in digestive gland, the total RNA content was higher in food-deprived than fed animals, again consistent with increases in rates of protein synthesis. At the same time, it appears that protein catabolism is activated, as evidenced by increases in proteasome enzyme activity and polyubiquitin transcript levels. Although provocative, the interpretive power of the aforementioned experiment (21) is limited because the initial nutritional state of the animals was not known with certainty, as these were wild-caught specimens sampled over two different years, and there was no direct measure of rates of protein synthesis.
The current experiment aims to further characterize the effects of starvation on protein metabolism in S. officinalis by combining whole animal respirometry and rates of ammonia excretion, measurements of the fractional rate of protein synthesis, activities of enzymes, and expression of genes involved in protein degradation. This work was conducted with animals grown since hatching under aquaculture conditions, so the initial nutritional status was well regulated. Measurements of oxygen consumption rate (ṀO2) allow an assessment of the whole animal metabolic rate. Ammonia production rates (ṀNH4) reveal rates of amino acid catabolism based on the premise that the NH4 is the primary nitrogenous end product (2, 3, 32), further supported by the finding that urea cannot be detected as an excretion product (Sykes AV, personal observation). The ammonia quotient (A.Q.: ṀNH4/ṀO2) can then be used to calculate what percentage of aerobic metabolism is supported by protein (18). We determined the fractional rate of protein synthesis, as well as indicators of protein degradation pathways in the gills and mantle to gather information on protein metabolism. The mantle is viewed as the main protein reserve, so protein metabolism in this tissue during starvation is of interest. The gills are a metabolically important and active tissue and were previously shown to respond differently than the mantle to starvation.
The response of the fractional rate of protein synthesis in the mantle follows the same pattern as a sustained decrease in oxygen consumption; however, in gill increases in 12-day relative to 3-day food-deprived animals suggests maintenance of gill function and/or tissue remodeling.
MATERIALS AND METHODS
Animals.
The experiments were conducted during May 2014, at Centro de Ciências do Mar do Algarve's Ramalhete Aquaculture Station (Ria Formosa, South of Portugal, 37°00′22.39′′N and 7°58′02.69′′W). The cuttlefish (Sepia officinalis) were obtained from eggs laid by an F3 captive stock that hatched and were reared at Ramalhete facilities, according to the latest culture technology described by Sykes et al. (40). Temperature (°C), salinity, and dissolved oxygen (%) were measured every day, at 0930, in all experimental tanks. Both temperature and dissolved oxygen were measured with an OxiGuard Handy Gamma probe, while salinity was measured with a VWR EC300 salinity meter. Water temperature was 21.0 ± 2.1°C, salinity was 36.3 ± 0.9 g/l, and dissolved oxygen level was 94.8 ± 4.6% air saturation. Animals were individually housed in plastic baskets (5.5 liter water volume; 31 cm × 22 cm × 8 cm) with 1-mm mesh size all around, which were placed inside rectangular 500-liter tanks of a flow-through system. Fed cuttlefish were given frozen grass shrimp (Palaemonetes varians) ad libitum on a daily basis. The average mass of the animals was 48.99 ± 1.60 g. This project was approved by the Institutional Animal Care Committee, Memorial University of Newfoundland, St. John's, Newfoundland, Canada. This experiment was performed under project SEPIATECH (PROMAR 31.03.05.FEP.002), which was approved before the entry into force of Directive 2010/63/EU as national legislation in Portugal.
Experimental protocol.
Experiments were conducted with animals that had been fed, fasted (3 days without food, 3–5 days in the case of ṀO2 and ṀNH4; n = 6), or starved (12 days without food; n = 6) following a week of adaptation to solitary conditions inside the baskets. In most cases, all of the measurements were taken on the same day in animals that were previously fed or food-deprived for the stated number of days. The rates of oxygen consumption and ammonia excretion were measured in different animals but from the same experimental treatments (fed, fasted, and starved; n = 6 per treatment).
Triglyceride content.
Triglycerides from the digestive gland were extracted using a chloroform:methanol procedure, as previously described (7, 21) and the level of glycerol determined using Sigma kit TR0100.
ṀO2. Resting ṀO2 was assessed using an automated intermittent flow respirometry system (Q-Box AQUA, Qubit Systems, Kingston, ON, Canada). Animals were removed from their holding tank, weighed, and quickly transferred to a 1.5-liter cylindrical respirometry chamber (8.9 cm diameter). The chamber was housed in a darkened 50-liter reservoir of continuously aerated seawater at 23.5 ± 0.5°C. Animals that had been fed, fasted for 3–5 days, or starved for 12 days were transferred into the respirometry chamber at around 1600, and oxygen consumption was monitored overnight. Oxygen levels were recorded at 15-min intervals throughout the experiment: 5 min with the system in closed loop followed by a 10-min flush cycle between each reading. Animals were housed in the respirometer for 8 to 16 h, and ṀO2 returned to baseline levels within the first 2–3 h after transfer into the system.
Ammonia excretion.
NH4 excretion (ṀNH4) rates for individual animals were determined by quantifying changes in water NH4 levels over time in a static tank system. Animals were removed from their holding tank, weighed, and transferred into darkened 3.0-liter plastic chambers with continuously aerated seawater at 23.5 ± 0.5°C. A 1.5-ml water sample was collected immediately after transfer into the chamber and after 60 min. Results from preliminary studies employing more frequent sampling indicated that ammonia excretion was pulsatile over short periods, but rates were consistent when averaged over the full 60-min exposure period. Ammonia levels were determined by the phenolhypochlorite method, according to Solórzano (38).
Protein synthesis.
The fractional rate of protein synthesis was measured using the flooding dose method (9) modified to use a stable isotope tracer (20). Preliminary experiments are required to validate the methodology, with the key criteria being that 1) the presence of a high concentration of the amino acid does not affect the rate of protein synthesis, 2) the labeled amino acid equilibrates rapidly with the precursor pool, 3) the enrichment of the labeled amino acid remains elevated and constant during the incorporation period (9, 20), and 4) the incorporation of the labeled amino acid in the protein pool is linear for the duration of the experiment. The method was validated using fed animals (the animals used for the validation were 33.38 ± 7.81 g, n = 10). Each cuttlefish received an injection of a 150 mM solution of phenylalanine (PHE) containing 50% ring-D5 L-phenylalanine (D5-PHE, Cambridge Isotope Laboratories, Tewksbury, MA) in 0.2 μm-filtered seawater at a dosage of 1 ml/100 g body mass. The labeled amino acid was injected at the base of the arm on the dorsal side, as previously described for another cephalopod species (5). Immediately after the injection, the animals were returned to their respective container. Two animals were sampled after 60, 120, 240, 360, and 480 min following the injection of the tracer. Animals were killed by performing anesthesia in 5% ethanol in seawater and then bisecting the brain downward and forward, followed by two lateral cuts to sever the brain from the optical lobes (22). A sample of mantle (on the ventral side), digestive gland, and gill was collected and quickly frozen on dry ice before further processing. Following the validation experiment, we used the same technique to measure the fractional rate of protein synthesis in the fed, fasted, and starved animals (n = 6 per group) using a tracer incorporation time of ∼120 min (see validation results below). All of the tissue-based measurements [i.e., triglycerides (TG), enzyme assays, and quantitative PCR] were conducted using the same animals. Approximately 75 mg of tissue was homogenized in 1 ml ice-cold perchloric acid (PCA). For the mantle, we used a mini-BeadBeater (BioSpec 3110BX with glass beads of 1 mm) at a maximum speed for two bursts of 1 min separated by 30 s on ice, while the gills and digestive gland samples were homogenized using a Polytron homogenizer. The treatment of the homogenized samples to measure protein-bound and free-pool enrichment of phenylalanine was as described before (20). The analyses were performed on an Agilent 5973 mass spectrometer equipped with a 6890 gas chromatograph. The capillary column was a 30-m Agilent DB-5MS (0.25 mm ID, 0.25-μm film thickness). The carrier gas was helium at 1 ml/min. The initial oven temperature was 70°C; 1 min following the injection, the temperature was increased to 280°C at a rate of 25°C/min, and the final temperature was maintained for 5 min (total run time 14.4 min). The mass spectrometer was operated in SIM mode with m/z 300 and 305 ions selected for PHE and D5-PHE, respectively. Fractional rates of protein synthesis (ks, %/day) were calculated from the phenylalanine enrichment of the protein pool [Sb = [D5-PHE]/([D5-PHE] + [PHE])] and the enrichment of the free-pool [Sa = [D5-PHE]/([D5-PHE] + [PHE])], according to ks = 100·[(Sb/Sa)·(1440/t)], where t is the incorporation time (min) and 1440 is used to convert from min to day (9, 20). In the gills, the free-pool phenylalanine enrichment declined significantly over time, and in this case, we used an alternate formula to account for this and calculate ks; ks = 100·[(Sbt2 − Sbt1)/Sa (t2 − t1)]·(1440/t2 − t1) where Sbt2 is the final protein bound D5-PHE enrichment and Sbt2 is the average incorporation at an earlier time (9, 13, 20).
In vitro protein degradation pathways.
Tissue samples were homogenized in 5 volumes of ice-cold 50 mM Tris buffer containing 0.1 mM EDTA and 0.007% β-mercaptoethanol (pH 8.0) using a Polytron homogenizer. The samples were centrifuged 13,000 g at 4°C for 60 min, and then the protein concentration was measured using a Bradford protein assay kit from Bio-Rad. The chymotrypsin-like activity of the 20S proteasome was measured using a microplate fluorescence assay (21). Briefly, each well contained 100 μl of 100 mM Tris buffer with 0.0285% SDS, 40 μM LLVY-AMC, and 50 μg of supernatant protein. Blanks were prepared by adding MG-132 at a final concentration of 50 μM. The MG-132-sensitive activity is reported in relative fluorescence units·min−1·50 μg protein−1. For the various protease assays, the homogenates were diluted to 1% using ice-cold homogenization buffer. Cathepsin-like proteases activities were measured using the McIlvaine's buffer system (24), and the activity of calpain-like proteases was determined in medium containing 20 mM Tris, 1 mM EDTA, 10 mM CaCl2, 100 mM KCl, and 0.1% mercaptoethanol, at pH 7.5. These assays were based on the degradation of a BODIPY-labeled casein (Molecular Probes, kit E6638), as previously described (16, 21, 41). All assays were performed at 25°C on a BioTek Synergy HT microplate reader.
RNA preparation and cDNA synthesis.
Total RNA was extracted using TRIzol Reagent (Invitrogen/Life Technologies, Burlington, ON, Canada). Tissues were homogenized in TRIzol reagent using a motorized Kontes RNase-Free Pellet Pestle Grinder (Kimble Chase, Vineland, NJ). The remainder of the protocol was carried out following the manufacturer's instructions. Total RNA was treated with TURBO DNA-free (Ambion/Life Technologies) following the manufacturer's instructions. RNA integrity was verified by 1% agarose gel electrophoresis, and purity was assessed by A260/280 and A260/230 UV NanoDrop spectrophotometry.
First-strand cDNA was synthesized from 1 μg of DNaseI-treated total RNA in a 20 μl reaction using random primers [250 ng (Invitrogen/Life Technologies)] and M-MLV reverse transcriptase [200 U (Invitrogen/Life Technologies)] with the manufacturer's first-strand buffer (1× final concentration) and DTT (10 mM final concentration) at 37°C for 50 min.
Quantitative PCR analysis of ubiquitin-proteasome pathway related transcript levels.
Quantitative PCR (qPCR) analyses were performed using the ViiA 7 real-time PCR system (Applied Biosystems/Life Technologies). The 96-well platform was used for primer quality and normalizer testing, and the 384-well platform was used for the experimental plates. In all cases, reaction volume for the PCR amplifications was 13 μl and contained 1× Power SYBR Green PCR Master Mix (Applied Biosystems/Life Technologies), 50 nM of both the forward and reverse primers, and the indicated cDNA quantity (see below). The real-time analysis program consisted of one cycle of 50°C for 2 min, 1 cycle of 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 1 min, with fluorescence detection at the end of each 60°C step.
The sequences of all primer pairs used in qPCR analyses are presented in Table 1. Primer sequences for polyubiquitin and proteasome (β-subunit) were reported previously (21). The additional primer sequences were based on the following sequences from GenBank; ubiquitin-activating enzyme (FO182945), ubiquitin-conjugating enzyme (HM157280), elongation factor 1-α (HM157271), and cleavage and polyadenylation specificity factor (CPSF) (HM157279). Each primer pair was quality tested to ensure that a single product was amplified (dissociation curve analysis) and that there was no primer-dimer present in the no-template control. Amplicons were electrophoretically separated on 2% agarose gels and compared with a 1 kb plus ladder (Invitrogen/Life Technologies) to verify that the correct size fragment was being amplified. Amplification efficiencies (30) were calculated using cDNA synthesized from gill and mantle from animal no. 8. The reported efficiencies (Table 1) are an average of the two values. Standard curves were generated using a five-point 1:3 dilution series starting with cDNA representing 10 ng of input total RNA.
Table 1.
Primers used in qPCR studies
| Gene Name | Direction | Nucleotide Sequence (5′-3′) | Efficiency, %a | Amplicon Size, bp |
|---|---|---|---|---|
| Ubiquitin-activating enzyme (E1) | Forward | ccttgatgggacttgcttgt | 97 | 136 |
| Reverse | gcacctgcacactgtgactt | |||
| Ubiquitin-conjugating enzyme (E2A) | Forward | atggcagtatttgcctggac | 97 | 122 |
| Reverse | attattggctgggctgtttg | |||
| Polyubiquitin | Forward | caactctcctactggacgaaagc | 93 | 70 |
| Reverse | tgtccagagcgctaaacaact | |||
| Proteasome β | Forward | ataatgctggctgccgacact | 96 | 102 |
| Reverse | tgcggcaatcacagtagcatc | |||
| Elongation factor 1-α | Forward | gtctcccattgcaggatgtt | 95 | 104 |
| Reverse | ggcaaaggtcaccaccatac | |||
| Cleavage and polyadenylation specificity factor | Forward | caggtcgagtggatgagtga | 100 | 100 |
| Reverse | cggcaagattatggcagagt |
Amplification efficiencies were calculated using a five-point 1:3 dilution series starting with cDNA representing 10 ng of input RNA.
Transcript levels of the genes of interest (GOIs) were normalized to two endogenous control genes. To select these endogenous controls, transcript levels were measured for four candidate normalizers [CPSF, EF1-α, eukaryotic translation initiation factor (ETIF), and 16S ribosomal RNA] using cDNA representing 5 ng of input total RNA synthesized from gill and mantle from two fed, two short-fasted, and two starved S. officinalis. Ct values were analyzed using geNorm to select the most stably expressed transcripts. Using this software, EF1-α (geNorm M = 0.34) and CPSF (geNorm M = 0.36) were determined to be the most stable.
Transcript (mRNA) expression levels of the GOIs were then assessed by qPCR. In all cases, cDNA representing 5 ng of input RNA was used as a template in the PCR reactions. On each plate, for every sample, the target gene and endogenous controls were tested in triplicate, and a plate linker sample (i.e., a sample that was run on all plates in a given study) and a no-template control were included. The relative quantity (RQ) of each transcript was determined using the ViiA 7 Software Relative Quantification Study Application (version 1.2.3) (Applied Biosystems/Life Technologies), with normalization to CPSF and EF1-α transcript levels, and with amplification efficiencies incorporated. For each GOI, the sample with the lowest normalized expression (mRNA) level was set as the calibrator sample (i.e., assigned an RQ value = 1).
Statistical analysis.
All values are expressed as means ± SE and were compared using one-way ANOVA and Tukey post hoc test using GraphPad Prism 6. The data were log10 transformed when necessary. All differences were considered significant when P < 0.05.
RESULTS
Nutritional status.
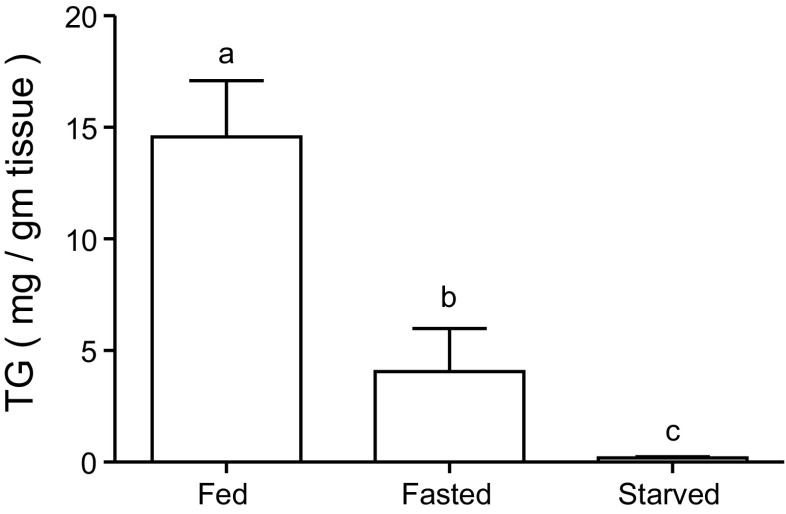
As food deprivation persisted, the digestive gland TG content decreased significantly among the fed, short-term fasted (3–5 days of food deprivation 28% of the fed group), and starved (12 days of food deprivation, 1% of the fed group) S. officinalis (Fig. 1) (ANOVA, F2,19 = 15.64; P < 0.001).
Fig. 1.
Triglycerides (TG) levels in the digestive gland of fed, fasted (3 days of food deprivation), and starved (12 days of food deprivation) Sepia officinalis. Values are given as means ± SE; n = 8 for fed and n = 7 for fasted and starved cuttlefish. Different letters indicate significant difference (P < 0.05).
Oxygen uptake and ammonia excretion.
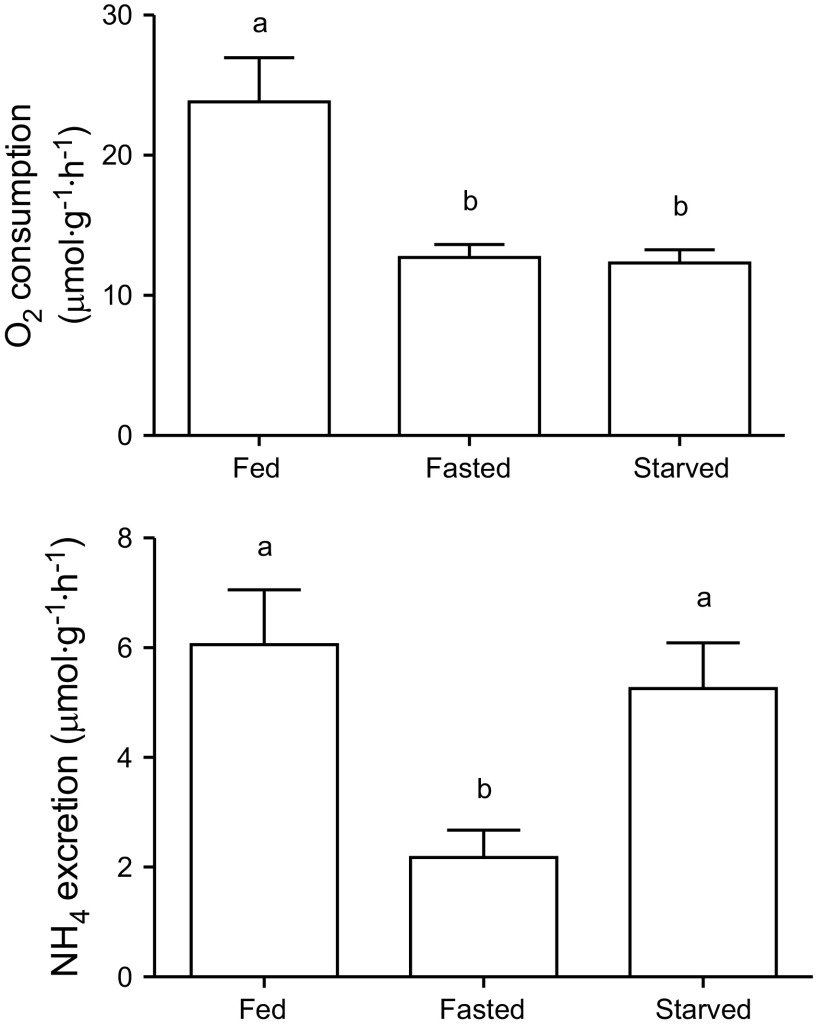
Food deprivation was associated with a decrease in ṀO2 to 53% of the fed group (Fig. 2, top) (n = 6 per group, F2,14 = 12.56, P < 0.001). The duration of the food restriction period (3, 5, or 12 days) did not have a significant effect on ṀO2. Short-term (3–5 days) food deprivation was associated with a decrease in ṀNH4 to 36% of the fed group. As the food restriction (12 days) persisted, ṀNH4 returned to control levels (Fig. 2, bottom). Because ṀNH4 and ṀO2 were not measured in the same animals, the A.Q. was determined by calculating ṀNH4 using group averages of each measurements. A.Q. in the control group was 0.285, and following three to five days of fasting decreased to 0.171. After 12 days of food deprivation, the A.Q. increased to 0.426.
Fig. 2.
Oxygen consumption and ammonia excretion of fed, fasted (3–5 days of food deprivation) and starved (12 days of food deprivation) S. officinalis. Values are given as means ± SE; n = 6. Different letters indicate significant difference (P < 0.05).
Validation of protein synthesis.
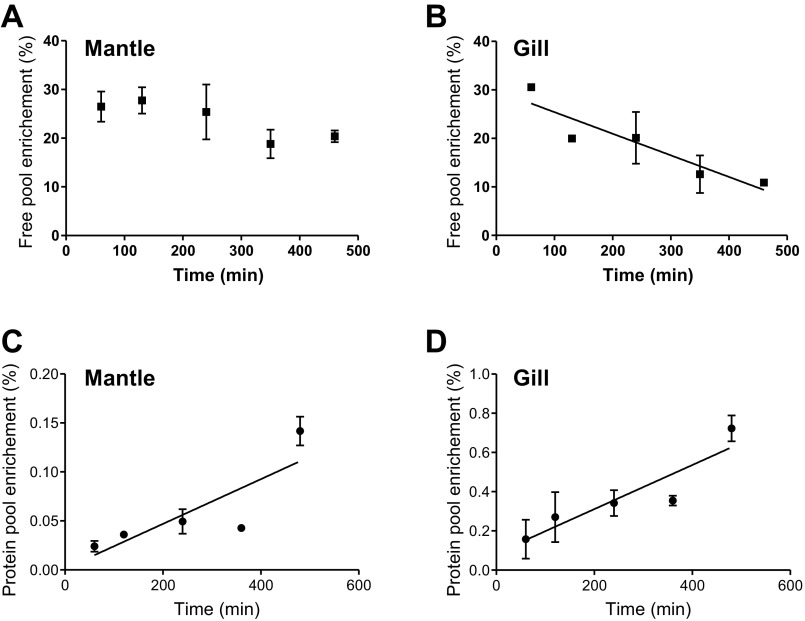
The flooding dose technique can only be used when the four criteria underlying the technique are met. The first validation criterion, that a high concentration of phenylalanine does not affect the rate of protein synthesis, could not be tested in this experiment, but we assumed that it was met as usually observed in vertebrates and invertebrates (8). Injected D5-PHE rapidly flooded the tissues to reach ∼30% enrichment of the free phenylalanine pool in both the mantle and the gills (Fig. 3, A and B), thus fulfilling the second criterion that the tracer must equilibrate rapidly with the precursor pool.
Fig. 3.
Phenylalanine protein-free pool enrichment in mantle and gill (A and B, respectively) and enrichment of the protein-bound phenylalanine in mantle and gill (C and D, respectively) following an injection of 150 mM phenylalanine containing 50% ring-D5-phenylalanine. Values are expressed as means ± SD (n = 2 for each point; n = 10 for each panel). Slopes of the regression lines are significantly different from 0.
The third criterion of sustained enrichment of the tracer was met since Sa remained elevated for a period of over 480 min in the mantle (regression line did not deviate from 0, F1,8 = 5.35, P > 0.05), while a slight decrease was observed in the gills (slope −0.04 ± 0.01%/min, F1,8 = 12.66, P = 0.007). The fourth criterion for validation of the technique was also met, as Sb increased in a linear fashion in the two tissues (Fig. 3, C and D). We deemed that the three last criteria required for the validation of the flooding dose technique were fulfilled. From these data, in further experiments, we elected to use an incorporation period of 120 min, which provided sufficient time for the tracer to accumulate in the protein pool to a detectable level while minimizing the decrease of Sa and recycling of the tracer due to protein degradation.
Protein synthesis in fed, fasted, and starved cuttlefish.
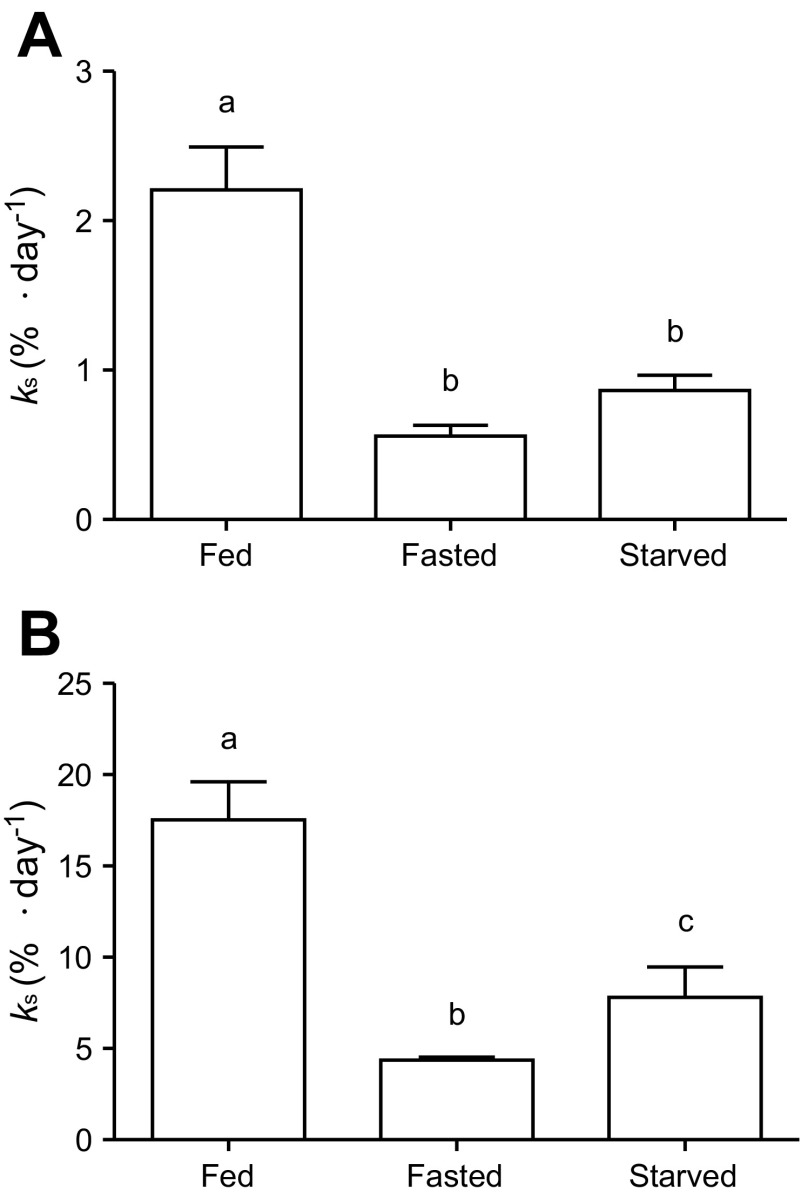
The fractional rate of protein synthesis in the mantle and gills of fed, fasted, and starved cuttlefish was measured (Fig. 4). In all groups, the rate of protein synthesis was ∼8–9 times higher in the gills compared with the mantle. The effect of fasting was similar in the two tissues; food deprivation decreased the rate of protein synthesis to 25% of that observed in the fed animals. As fasting progressed into starvation, there was no further significant change in the rate of protein synthesis in mantle; however, in gill, the rate of protein synthesis significantly increased to a level that was 45% of the fed group. We also attempted to measure the rate of protein synthesis in the digestive gland; however, Sa decreased considerably during the 120-min incorporation period (data not shown). After the incorporation period, Sa was only 3.24 ± 1.1% in the digestive gland of the fed animals, while it was still 26.0 ± 5.30% in the starved animals. As such, it was impossible to accurately measure the rate of protein synthesis in digestive glands of the fed animals.
Fig. 4.
Fractional rate of protein synthesis (ks) in mantle and gill of S. officinalis that were either fed, fasted (3 days of food deprivation), or starved (12 days of food deprivation). Values are given as means ± SE; n = 6. Different letters indicate significant difference (P < 0.05).
In vitro protein degradation enzyme activities.
The enzyme activity of two classes of cathepsins (pH 2.5 and pH 5.5), the calpain-like proteases, and the 20S proteasome was measured in the fed, fasted, and starved animals (Table 2). For the cathepsins at pH 2.5, the only observable difference was in the digestive gland where the starved S. officinalis had lower enzyme activity. For the cathepsins at pH 5.5, we observed differences in the three studied tissues; however, the direction of these differences varied. Enzyme activity was significantly lower in gills and digestive gland of the starved animals with respect to the fed group but was higher in the mantle of the starved than the fed S. officinalis. There was no effect of starvation on the calpain-like proteases. The 20S proteasome activity was unaffected by food deprivation in the gills and mantle but was significantly lower in the digestive gland of the starved than fed animals.
Table 2.
Proteases enzyme activity in gill, mantle, and digestive gland of Sepia officinalis that were either fed or starved for 7 days
| Fed | Starved | |
|---|---|---|
| Cathepsin 2.5 | ||
| Gill | 3555 ± 278.0 | 3887 ± 225.6 |
| Mantle | 7175 ± 290.0 | 6941 ± 159.5 |
| Digestive gland | 11767 ± 152.8 | 8644 ± 184.8* |
| Cathepsin 5.5 | ||
| Gill | 16012 ± 257.4 | 13291 ± 595.6* |
| Mantle | 11369 ± 326.4 | 14232 ± 1256* |
| Digestive gland | 216622 ± 11959 | 116893 ± 9299* |
| Calpain | ||
| Gill | 21213 ± 1167 | 20545 ± 1126 |
| Mantle | 35159 ± 6049 | 36797 ± 4227 |
| Digestive gland | 247236 ± 23462 | 259753 ± 11602 |
| 20S Proteasome | ||
| Gill | 615847 ± 66536 | 676633 ± 29758 |
| Mantle | 101810 ± 7318 | 87803 ± 10310 |
| Digestive gland | 962202 ± 149639 | 552593 ± 25005* |
Values are expressed as means ± SE (n = 6). Data are expressed as fluorescence units·min−1·mg tissue−1 for cathepsin pH 2.5, cathepsin pH 5.5, and calpain: fluorescence units·min−1·50 μg protein−1 for 20 S proteasome.
Significant difference between fed and starved cuttlefish (P < 0.05).
Transcript levels of genes related to the ubiquitin-proteasome pathway.
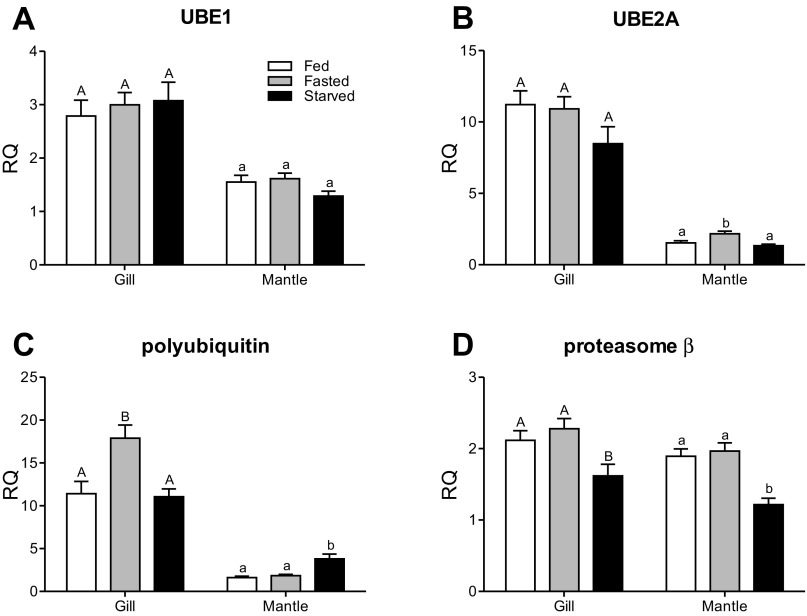
UBE1 transcript levels were not affected by food deprivation (Fig. 5A). UBE2A transcript levels were significantly higher in mantle of the 3 days fasted than either fed or starved animals (Fig. 5B); however, there was no change in gill. Polyubiquitin transcript levels were transiently elevated in the gills of the fasted S. officinalis, while this increase was observed in the mantle of the starving animals (Fig. 5C). Finally, proteasome (β-subunit) transcript levels decreased in both the gills and mantle of the starving animals with respect to the fed or fasted groups (Fig. 5D).
Fig. 5.
Quantitative PCR analysis of transcript levels of ubiquitin-proteasome pathway-related genes in gill and mantle of S. officinalis that were either fed, fasted (3 days of food deprivation), or starved (12 days of food deprivation). Transcript expression data are presented as means ± SE (n = 6 or 7) relative quantity (RQ) values for the transcript of interest normalized to cleavage and polyadenylation specificity factor and elongation factor 1-α transcript levels, and they were calibrated to the individual with the lowest normalized expression of that given gene (i.e., assigned an RQ value = 1). Different letters indicate significant difference (P < 0.05).
DISCUSSION
On-board protein ultimately fuels aerobic metabolism.
Consistent with previous studies on food restriction in cephalopods, the triglyceride stores of the digestive gland were decreased to one-third that of the fed S. officinalis after 3 days of fasting and were virtually depleted following 12 days of fasting (6, 21). Following 3–5 days of fasting, at a temperature of 21°C, ṀO2 was about one half that of the fed animals and remained the same during starvation. Consistent with this, in a similar experiment, Grigoriou and Richardson (11) found that, at a lower temperature of 15°C, metabolic rate began to differ from that of fed animals after ∼10 days of food deprivation. The ṀNH4 reported here are in the same range as that of the Chinese cuttlefish (Sepiella maindroni) (42); however, lower rates of ṀNH4 for S. officinalis were reported earlier (2). We have no explanation for the discrepancy, but we believe that the values reported here for this population of S. officinalis are accurate as ṀNH4 are compatible with ṀO2. The rate of ammonia excretion returned to the fed level after the TG reserves were exhausted. This is in agreement with the view that, when fasting, cephalopods switch from an amino acid-dominated metabolism to a lipid-dominated metabolism by mobilizing the triglycerides stored in the digestive gland (6, 21, 27). This assertion is further supported by the A.Q. of the fed animals that is close to the theoretical maximum calculated to be 0.27 to 0.33 (18) for aerobic oxidation of amino acids. This suggests that virtually 100% of their energy needs are met using amino acids during resting metabolism. In the fasted S. officinalis, the A.Q. reveals that ∼50% of their aerobically based metabolism is met using amino acids as a fuel source. The source of the amino acids is probably body protein. Cephalopods have very little carbohydrate stores (28, 39), and on the basis of the triglyceride decrease in the digestive gland in food-deprived cephalopods (this study; Refs. 6, 21, 33), the latter likely contributes to the remaining ∼50% of aerobic energy metabolism. Following the depletion of triglycerides in the digestive gland after 12 days of starvation, the A.Q. goes above 0.27, which suggests that aerobic metabolism is met almost exclusively using amino acids, and further that anaerobic degradation of amino acids is ongoing and/or that the carbon backbone of deaminated amino acids is used in anabolic reactions (18). We are not aware of any other study in which A.Q. went above 0.27 in aerobic conditions. This novel finding though must be considered with caution as ṀO2 and ṀNH4 were determined on different animals, but, nevertheless, it is a provocative discovery that requires confirmation. Regardless, the changes in triglyceride content in the digestive gland along with the decrease and subsequent increase in ṀNH4 leads us to conclude that the current experimental groups were suitable to test the effects of the duration of the restriction period on protein metabolism. Furthermore, in the starved group, there must be catabolism of body protein, presumably from mantle, given it is by far the largest tissue by mass.
Control of protein synthesis.
Prior to our study, the fractional rate of protein synthesis had only been measured in two species of cephalopods; the common octopus (Octopus vulgaris) (14) and the Southern dumpling squid (Euprymna tasmanica) (5, 25, 26), but never in a cuttlefish. Our work is the first to measure the fractional rate of protein synthesis in S. officinalis. The flooding dose technique was validated by using a time course and proved to respect all the assumptions of the technique: injected isotope rapidly flooded the tissues, the free pool of phenylalanine remained elevated for the course of the experiment, and phenylalanine enrichment of the protein pool increased in a linear fashion. The fractional rates of protein synthesis in mantle and gill of fed and food-deprived S. officinalis are almost identical to what Houlihan et al. (14) calculated for an octopus growing at 6%/day or having a growth rate of zero, respectively. The rates of protein synthesis in S. officinalis are lower than those in the southern dumpling squid (5), the only other decapod cephalopod for which comparable data are available. This finding is likely to be a result of the fact that the squid were much smaller than the animals used here (2.8 g and 9.01 g vs. 48.99 g in this experiment) and that these two groups have very different life histories and growth profiles. It was impossible to measure the rate of protein synthesis in the digestive gland because the specific enrichment of free amino acid pool decreased too quickly to allow calculation of the rate of protein synthesis in the fed animals (data not shown). This suggests that the fed animals were still in a postprandial state 24 h following their last meal. Information on the time needed to digest a meal in cephalopods is scarce; however, it is generally accepted that these animals are geared for rapid digestion (36). Furthermore, the digestive gland may be involved in providing precursors for melanin synthesis for the production of ink by the ink gland. In this pathway, phenylalanine is hydroxylated to tyrosine that is then used for the biosynthesis of melanin by the ink gland (29). Thus, it is likely that a great proportion of the tracer phenylalanine was converted to labeled tyrosine, which was not detected by our mass spectrometry analysis.
The measured decrease in rates of protein synthesis in mantle are consistent with previously measured decreases in starved cuttlefish of the phosphorylation of AKT and 4EBP1, which are components of cellular signaling pathways controlling protein synthesis (21). Similarly, the increase in the rate of protein synthesis in the gill as the triglyceride stores in the digestive gland are depleted matches the observed increase in the phosphorylation of 4EBP1. These observations are important beyond S. officinalis per se because they help confirm that deductions made from the biochemical analysis of cellular signaling pathways concerning the rate of protein synthesis are valid (15, 21, 34, 35).
Indices of protein catabolism.
The findings that digestive gland triglyceride reserves are used to the point of depletion, followed by an increase in NH4 excretion with an exceptionally high A.Q. value provides compelling evidence that during food deprivation, S. officinalis mainly relies on amino acids from onboard protein to fuel their metabolism. Therefore, it is of interest to learn how protein catabolism is regulated. It is generally accepted that protein stored in the mantle is the primary source of amino acids during starvation (6, 21, 27, 28). The cathepsin A-, B-, H-, and L-like enzyme activity is stimulated in the mantle during starvation while the activity of the other measured proteases remains unchanged. This pattern was previously noted in starving S. officinalis (21) and strengthens the putative role of the mantle as a source of amino acids through the degradation of protein involving cathepsin-like proteases. The ubiquitin-proteasome system is a highly regulated protein degradation pathway that requires the coordinated work of many proteins (12) and might also be involved in the food deprivation response. The enzyme activity of the 20S proteasome was not affected by starvation in mantle, again consistent with previous findings (21), but this information alone is not sufficient to declare that this pathway does not play a role in protein catabolism. Accordingly, we measured the transcript levels of UBE1, UBE2A, polyubiquitin and the proteasome (β-subunit). UBE1 transcript levels were not influenced by food restriction; however, UBE2A transcript levels showed a transient elevation in mantle of fasting animals. This protein is responsible for the ubiquitination of the proteins recognized by E3 proteins for degradation (1, 31). After 12 days of food restriction, polyubiquitin transcript levels increased 2.3-fold, once more consistent with earlier findings that reported a massive increase in polyubiquinated protein (21). Proteasome (β-subunit) transcript levels, however, decreased, which is in contrast to our earlier work (21). Overall, the current data set does not convincingly reveal an enhanced ubiquitin-proteasome system in mantle during food deprivation, although the significant increase noted here and the five-fold increase in polyubiquitin transcript levels previously reported (21) warrant further investigation.
Gill appears to present a more complex situation with respect to protein breakdown just as it does for protein synthesis. Cathepsin A-, B-, H-, and L-like activity and the proteasome (β-subunit) transcript levels were lower in starved animals than the other two groups. This suggests decreased rates of protein breakdown at this time point. The transient increase in polyubiquitin transcript levels in the fasted animal suggests similar or even decreased rates of protein breakdown in gill between starved and fed animals. The very clear and significant increase in polyubiquitin transcript levels upon the transition from feeding to fasting confirms earlier findings (21) and implies a transient elevation in the ubiquitin-proteasome system.
In the digestive gland, the activities of the cathepsins and the 20S proteasome were all lower in starved than in fed animals. The simplest explanation for this is that by the time the animals entered the starvation state, any protein that could be mobilized in the tissue had already been catabolized.
Perspectives and Significance
Consistent with numerous studies on cephalopods, aerobic metabolism in fed animals is fueled primarily by dietary protein. Upon food deprivation, aerobic metabolism is decreased. NH4 production is decreased to a relatively greater extent than ṀO2, indicating that lipids are being called upon, thus initially sparing body protein. As food deprivation persists, lipids of the digestive gland are depleted, and body protein serves as the aerobic metabolic fuel. This is evidenced by an increase in ṀNH4 resulting in an A.Q. that fully supports ṀO2 and tentatively even leads to an excess production of NH4. The mantle seems to be the major source of amino acids; this is supported by an increase in protein breakdown by the cathepsin A-, B-, H-, and L-like proteases. It is likely that the ubiquitin-proteasome system is also activated, but this contention requires additional support. Although it is clear that mantle protein could support aerobic metabolism in mantle through the provision of amino acids, it is not known whether amino acids are released to fuel other tissues as well. We previously proposed that food deprivation in S. officinalis results in gill remodeling (21), as observed in many fish species under adverse conditions (37). The new information on the rate of protein synthesis continues to support the proposition. There is no evidence on the basis of either enzyme activities or transcript levels that protein catabolism in gill of starved S. officinalis is higher than that of fed animals. It may be that gill restructuring during food deprivation is associated with the maintenance of ionic balance, protection of water soluble plasma metabolites, or excretion ammonia.
GRANTS
This work was supported, in part, by Natural Sciences and Engineering Research Council (NSERC) of Canada Discovery Grants (to W. R. Driedzic, T. J. MacCormack, and S. G. Lamarre), and the Fundação para a Ciência e a Tecnologia through Programa Investigador FCT 2014 (IF/00576/2014) and project SEPIATECH (31-03-05-FEP-2 Program PROMAR) funded by the Portuguese Government (to A. V. Sykes). W. R. Driedzic holds the Canada Research Chair in Marine Bioscience. B. Speers-Roesch was supported by a NSERC Postdoctoral Fellowship. N. I. Callaghan was supported by an NSERC CGS-M award and a New Brunswick Innovation Foundation Graduate Fellowship.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
S.G.L., T.J.M., A.V.S., B.S.-R., N.I.C., and W.R.D. conception and design of research; S.G.L., T.J.M., A.V.S., J.R.H., B.S.-R., N.I.C., and W.R.D. performed experiments; S.G.L., T.J.M., J.R.H., and W.R.D. analyzed data; S.G.L., T.J.M., J.R.H., and W.R.D. interpreted results of experiments; S.G.L. and W.R.D. prepared figures; S.G.L. and W.R.D. drafted manuscript; S.G.L., T.J.M., A.V.S., J.R.H., B.S.-R., N.I.C., and W.R.D. edited and revised manuscript; S.G.L., T.J.M., A.V.S., J.R.H., B.S.-R., N.I.C., and W.R.D. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors are thankful to João Reis, Juan C. Capaz, Ana T. Couto, and staff at Ramalhete Station for valuable input and logistical assistance with the study. The authors also thank Dr. Matthew Rise, Department of Ocean Sciences, Memorial University of Newfoundland, for use of the ViiA 7 Real-Time PCR System.
REFERENCES
- 1.Bodine SC, Baehr LM. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am J Physiol Endocrinol Metab 307: E469–E484, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boucher-Rodoni R, Mangold K. Comparative aspects of ammonia excretion in cephalopods. Malacologia 29: 145–151, 1988. [Google Scholar]
- 3.Boucher-Rodoni R, Mangold K. Ammonia production in cephalopods, physiological and evolutionary aspects. Mar Freshwater Behav Physiol 25: 53–60, 1995. [Google Scholar]
- 4.Caloin M. Modeling of lipid and protein depletion during total starvation. Am J Physiol Endocrinol Metab 287: E790–E798, 2004. [DOI] [PubMed] [Google Scholar]
- 5.Carter CG, Lynch KA, Moltschaniwskyj NA. Protein synthesis in a solitary benthic cephalopod, the Southern dumpling squid (Euprymna tasmanica). Comp Biochem Physiol Part A Mol Integr Physiol 153: 185–190, 2009. [DOI] [PubMed] [Google Scholar]
- 6.Castro BG, Garrido JL, Sotelo CG. Changes in composition of digestive gland and mantle muscle of the cuttlefish Sepia officinalis during starvation. Mar Biol 114: 11–20, 1992. [Google Scholar]
- 7.Collison KS, Maqbool Z, Saleh SM, Inglis A, Makhoul NJ, Bakheet R, Al-Johi M, Al-Rabiah R, Zaidi MZ, Al-Mohanna FA. Effect of dietary monosodium glutamate on trans fat-induced nonalcoholic fatty liver disease. J Lipid Res 50: 1521–1537, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraser KPP, Rogers AD. Protein metabolism in marine animals: The underlying mechanism of growth. Adv Mar Biol 52: 267–362, 2007. [DOI] [PubMed] [Google Scholar]
- 9.Garlick PJ, McNurlan MA, Preedy VR. A rapid and convenient technique for measuring the rate of protein synthesis in tissues by injection of [3H]phenylalanine. Biochem J 192: 719–723, 1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodman CA, Mayhew DL, Hornberger TA. Recent progress toward understanding the molecular mechanisms that regulate skeletal muscle mass. Cell Signal 23: 1896–1906, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grigoriou P, Richardson CA. Effect of body mass, temperature and food deprivation on oxygen consumption rate of common cuttlefish Sepia officinalis. Mar Biol 156: 2473–2481, 2009. [Google Scholar]
- 12.Hershko A, Ciechanover A, Varshavsky A. The ubiquitin system. Nat Med 6: 1073–1081, 2000. [DOI] [PubMed] [Google Scholar]
- 13.Houlihan DF, Carter CG, McCarthy ID. Protein turnover in animals. In: Nitrogen Metabolism and Excretion, edited by Walsh PJ and Wright P. New York: CRC Press, 1995. [Google Scholar]
- 14.Houlihan DF, McMillan DN, Agnisola C, Genoino IT, Foti L. Protein synthesis and growth in Octopus vulgaris. Mar Biol 106: 251–259, 1990. [Google Scholar]
- 15.Johnston IA, Bower NI, Macqueen DJ. Growth and the regulation of myotomal muscle mass in teleost fish. J Exp Biol 214: 1617–1628, 2011. [DOI] [PubMed] [Google Scholar]
- 16.Jones LJ, Upson RH, Haugland RP, Panchuk-Voloshina N, Zhou M, Haugland RP. Quenched BODIPY dye-labeled casein substrates for the assay of protease activity by direct fluorescence measurement. Anal Biochem 251: 144–152, 1997. [DOI] [PubMed] [Google Scholar]
- 17.Kasinath BS, Mariappan MM, Sataranatarajan K, Lee MJ, Feliers D. mRNA translation: unexplored territory in renal science. J Am Soc Nephrol 17: 3281–3292, 2006. [DOI] [PubMed] [Google Scholar]
- 18.Kutty MN. Ammonia quotient in sockeye salmon (Oncorhynchus nerka). J Fish Res Board Can 35: 1003–1005, 1978. [Google Scholar]
- 19.Lamarre S, Ditlecadet D, McKenzie DJ, Bonnaud L, Driedzic WR. Mechanisms of protein degradation in mantle muscle and proposed gill remodeling in starved Sepia officinalis. Am J Physiol Regul Integr Comp Physiol 303: R427–R437, 2012. [DOI] [PubMed] [Google Scholar]
- 20.Lamarre S, Saulnier RJ, Blier PU, Driedzic WR. A rapid and convenient method for measuring the fractional rate of protein synthesis in ectothermic animal tissues using a stable isotope tracer. Comp Biochem Physiol Part B Biochem Mol Biol 182: 1–5, 2015. [DOI] [PubMed] [Google Scholar]
- 21.Lamarre SG, Ditlecadet D, McKenzie DJ, Bonnaud L, Driedzic WR. Mechanisms of protein degradation in mantle muscle and proposed gill remodeling in starved Sepia officinalis. Am J Physiol Regul Integr Comp Physiol 303: R427–R437, 2012. [DOI] [PubMed] [Google Scholar]
- 22.Lewbart GA, Mosley C. Clinical anesthesia and analgesia in invertebrates. J Exotic Pet Med 21: 59–70, 2012. [Google Scholar]
- 23.McCue MD. Starvation physiology: reviewing the different strategies animals use to survive a common challenge. Comp Biochem Physiol Part A Mol Integr Physiol 156: 1–18, 2010. [DOI] [PubMed] [Google Scholar]
- 24.McIlvaine T. A buffer solution for colorimetric comparison. J Biol Chem 49: 183–186, 1921. [Google Scholar]
- 25.Moltschaniwskyj NA, Carter CG. The adaptive response of protein turnover to the energetic demands of reproduction in a cephalopod. Physiol Biochem Zool 86: 119–126, 2013. [DOI] [PubMed] [Google Scholar]
- 26.Moltschaniwskyj NA, Carter CG. Protein synthesis, degradation, and retention: mechanisms of indeterminate growth in cephalopods. Physiol Biochem Zool 83: 997–1008, 2010. [DOI] [PubMed] [Google Scholar]
- 27.O'Dor RK, Mangold K, Boucher-Rodoni R, Wells MJ, Wells J. Nutrient absorption, storage and remobilization in Octopus vulgaris. Mar Behav Physiol 11: 239–258, 1984. [Google Scholar]
- 28.O'Dor RK, Webber DM. The constraints on cephalopods—why squid aren't fish. Can J Zool 64: 1591–1605, 1986. [Google Scholar]
- 29.Palumbo A. Melanogenesis in the ink gland of Sepia officinalis. Pigment Cell Res 16: 517–522, 2003. [DOI] [PubMed] [Google Scholar]
- 30.Pfaffl M. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polge C, Attaix D, Taillandier D. Role of E2-Ub-conjugating enzymes during skeletal muscle atrophy. Front Physiol 6: 59, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Potts WTW. Excretion in the molluscs. Biol Rev 42: 1–41, 1967. [Google Scholar]
- 33.Segawa S, Hanlon R. Oxygen consumption and ammonia excretion rates in Octopus maya, Loligo forbesi, and Lolliguncula brevis (Mollusca: Cephalopoda). Mar Behav Physiol 13: 389–400, 1988. [Google Scholar]
- 34.Seiliez I, Gutierrez J, Salmerón C, Skiba-Cassy S, Chauvin C, Dias K, Kaushik S, Tesseraud S, Panserat S. An in vivo and in vitro assessment of autophagy-related gene expression in muscle of rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol Part B Biochem Mol Biol 157: 258–266, 2010. [DOI] [PubMed] [Google Scholar]
- 35.Seiliez I, Panserat S, Skiba-Cassy S, Fricot A, Vachot C, Kaushik S, Tesseraud S. Feeding status regulates the polyubiquitination step of the ubiquitin-proteasome-dependent proteolysis in rainbow trout (Oncorhynchus mykiss) muscle. J Nutr 138: 487–491, 2008. [DOI] [PubMed] [Google Scholar]
- 36.Semmens JM. Changes in the digestive gland of the loliginid squid Sepioteuthis lessoniana (Lesson 1830) associated with feeding. J Exp Mar Biol Ecol 274: 19–39, 2002. [Google Scholar]
- 37.Sollid J, Nilsson GE. Plasticity of respiratory structures—adaptive remodeling of fish gills induced by ambient oxygen and temperature. Respir Physiol Neurobiol 154: 241–251, 2006. [DOI] [PubMed] [Google Scholar]
- 38.Solórzano L. Determination of ammonia in natural waters by the phenolhypochlorite method. Limnol Oceanogr 14: 799–801, 1969. [Google Scholar]
- 39.Suryanarayanan H, Alexander KM. Fuel reserves of molluscan muscle. Comp Biochem Physiol Part A Physiol 40: 55–60, 1971. [Google Scholar]
- 40.Sykes AV, Domingues P, Andrade JP. Sepia officinalis In: Cephalopod Culture, edited by Iglesias J, Fuentes L and Villanueva R. Amsterdam, Netherlands: Springer, 2014, p. 175–204. [Google Scholar]
- 41.Thompson VF, Saldana S, Cong JY, Goll DE. A BODIPY fluorescent microplate assay for measuring activity of calpains and other proteases. Anal Biochem 279: 170–178, 2000. [DOI] [PubMed] [Google Scholar]
- 42.Yin F, Sun P, Peng S, Tang B, Zhang D, Wang C, Mu C, Shi Z. The respiration, excretion and biochemical response of the juvenile common Chinese cuttlefish, Sepiella maindroni at different temperatures. Aquaculture 402–403: 127–132, 2013. [Google Scholar]