SUMMARY
Centromeres are specialized chromatin domains specified by the centromere-specific CENP-A nucleosome. The stable inheritance of vertebrate centromeres is an epigenetic process requiring deposition of new CENP-A nucleosomes by HJURP. We show HJURP is recruited to centromeres through a direct interaction between the HJURP centromere targeting domain and the Mis18α-β C-terminal coiled-coil domains. We demonstrate Mis18α and Mis18β form a heterotetramer through their C-terminal coiled-coil domains. Mis18α-β heterotetramer formation is required for Mis18BP1 binding and centromere recognition. S. pombe contains a single Mis18 isoform that forms a homotetramer, showing tetrameric Mis18 is conserved from fission yeast to humans. HJURP binding disrupts the Mis18α-β heterotetramer and removes Mis18α from centromeres. We propose stable binding of Mis18 to centromeres in telophase licenses them for CENP-A deposition. Binding of HJURP deposits CENP-A at centromeres and facilitates the removal of Mis18, restricting CENP-A deposition to a single event per cell cycle.
Graphical Abstract
INTRODUCTION
The centromere is an epigenetically specified chromosomal locus in humans that orchestrates the segregation of chromosomes during cell division. The centromere is defined by the presence of the histone H3 variant, CENP-A (centromere protein A) (Cleveland et al., 2003). The presence of the CENP-A nucleosome is sufficient to recruit the constitutive centromere-associated network (CCAN) and mitotic kinetochore proteins that are required for proper chromosome segregation (Barnhart et al., 2011; Guse et al., 2011; Hori et al., 2013; Mendiburo et al., 2011).
New CENP-A nucleosomes must be deposited at the existing centromere every cell cycle to maintain centromere identity. CENP-A uses a conserved chromatin assembly factor HJURP (Holliday junction recognition protein), or Scm3 (suppressor of chromosome segregation) in yeast, to distinguish it from other histone H3 variants and facilitate its deposition into centromeric chromatin (Barnhart et al., 2011; Bernad et al., 2011; Camahort et al., 2007; Dechassa et al., 2011; Dunleavy et al., 2009; Foltz et al., 2009; Mizuguchi et al., 2007; Pidoux et al., 2009; Shuaib et al., 2010; Stoler et al., 2007; Williams et al., 2009). HJURP binds the CENP-A/H4 heterodimer through the N-terminal Scm3 homology domain (Bassett et al., 2012; Hu et al., 2011; Sanchez-Pulido et al., 2009; Shuaib et al., 2010) and contains a centromere-targeting domain (CenTD) within the first C-terminal repeat that is absent in yeast Scm3 (Wang et al., 2014; Zasadzińska et al., 2013).
The Mis18 proteins are required for HJURP/Scm3 recruitment, and therefore CENP-A/Cnp1 deposition in diverse eukaryotes with regional centromeres, but absent from yeast with point centromeres (Camahort et al., 2007; Fujita et al., 2007; Hayashi et al., 2004; Mizuguchi et al., 2007; Moree et al., 2011; Pidoux et al., 2009; Stoler et al., 1995; Williams et al., 2009). S. pombe contains a single Mis18 protein; however, vertebrates possess two Mis18 paralogs, Mis18α and Mis18β, that share ~27% amino acid identity and each display 30% identity with S. pombe Mis18. Humans possess a Mis18-associated protein Mis18BP1/Knl-2, which is required for Mis18 recruitment to centromeres (Fujita et al., 2007; Maddox et al., 2007). The human Mis18 proteins are recruited to centromeres during late telophase and remain associated with the centromere during early G1 phase when new CENP-A is deposited. Recruitment of the Mis18 complex is regulated by Cdk1 and Plk1 phosphorylation (Barnhart-Dailey and Foltz, 2014; McKinley and Cheeseman, 2014; Silva et al., 2012). Mis18 has been previously shown to affect histone modifications and the methylation status of underlying chromatin (Fujita et al., 2007; Kim et al., 2012). Eliminating histone methylation within the alpha satellite DNA repeats of a human artificial chromosome alters the recruitment of HJURP (Bergmann et al., 2011).
We demonstrate that human Mis18α and Mis18β associate through C-terminal coiled coils to form a heterotetramer (HT). The tetrameric stoichiometry of the Mis18 complex is conserved in fission yeast. We find that HT formation is essential for Mis18 centromere recruitment. Using FRAP, we observe that Mis18α is highly stable at centromeres during G1 phase. The Mis18α-β HT is disrupted upon binding of the HJURP CenTD to the Mis18α-β coiled coils, leading to the removal of Mis18α-β from centromeres. We propose these dynamics ensure each Mis18 complex bound per cell cycle directs the deposition of a single CENP-A nucleosome and thereby limits the amount of new CENP-A recruited.
RESULTS
Mis18 Proteins Form a Conserved Tetramer
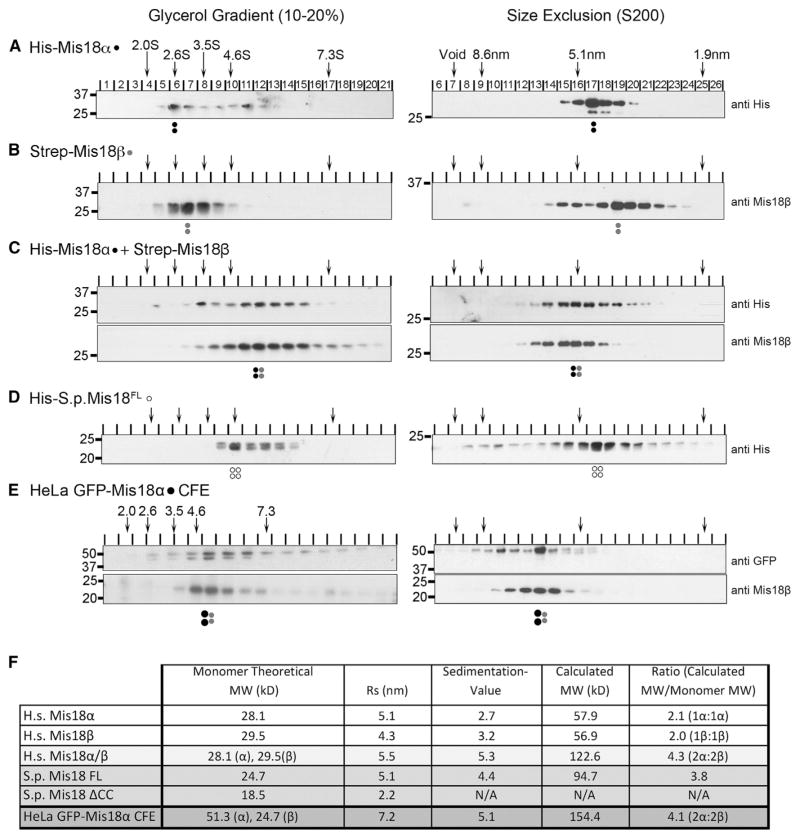
We performed size-exclusion chromatography (SEC) and glycerol gradient sedimentation using recombinant human Mis18α-β complex (Figures 1A–1D and S1A–S1D, available online). Mis18α or Mis18β alone migrated with a Stokes radius of 5.1 nm and 4.3 nm, respectively (Figures 1A, 1B, and 1F), and sedimented with S values of 2.7S and 3.2S (Figures 1A, 1B, and 1F). Based on this analysis, Mis18α and Mis18β alone exhibit native molecular weights twice the size of their predicted monomeric molecular weights, consistent with formation of a homodimer (Figure 1F). The calculated molecular weight of the combined Mis18α-β complex, formed by incubating equivalent amounts of Mis18α and Mis18β, shows that this complex exists as a HT (Figures 1C and 1F). S. pombe, as well as most nonvertebrate eukaryotes, contains a single Mis18 homolog. Recombinant S. pombe Mis18 formed a tetramer out of the single Mis18 protein (Figures 1D and S1A). Taken together, these data show Mis18 proteins form an evolutionally conserved tetramer.
Figure 1. Mis18α and Mis18β Form a Conserved Tetramer.
(A–D) Glycerol gradient sedimentation (left) and SEC (right) performed using recombinant (A) His-tagged Mis18α, (B) Strep-tagged Mis18β, (C) Mis18α and Mis18β combined, and (D) S. pombe Mis18. Arrows at the top indicate the sedimentation values (S) and Stokes radii (nm) of standards. Proteins were detected by immunoblot, and antibodies used are shown to the right. Dots signify the peak fractions and the proteins represented: black, Mis18α; gray, Mis18β; open circles, S. pombe Mis18.
(E) Chromatin-free extracts (CFE) from mitotically arrested cells stably expressing GFP-tagged Mis18α were separated on glycerol gradient (left) and SEC S200 column (right) and immunoblotted using anti-GFP and anti-Mis18β.
(F) Table summarizing the hydrodynamic analyses in (A)–(D). Calculated molecular weights were calculated from Stokes radii (Rs) and sedimentation coefficients (Siegel and Monty, 1966). Stokes radii and sedimentation values were calculated from linear standard curves (Figures S1A–S1D). All values are the average of two replicates. Stoichiometry of the individual complexes was determined by dividing the calculated molecular weight of the complex by the theoretical molecular weights of the monomeric proteins based on amino acid content.
To assess whether the human Mis18α-β forms a tetramer in vivo, we determined the native molecular weight of the Mis18 complex from mitotic HeLa cells stably expressing GFP-tagged Mis18α. Chromatin-free extracts (CFEs) from nocodazole-blocked cells were analyzed by SEC and glycerol gradient sedimentation (Figures 1E, 1F, S1B, and S1C). The native molecular weight of the cell-derived Mis18 complex was 154 kD, close to the 152 kD predicted molecular weight of a Mis18 tetramer containing two GFP-Mis18α (51 kD) and two Mis18β molecules.
Mis18 Proteins Interact through Conserved Coiled Coils
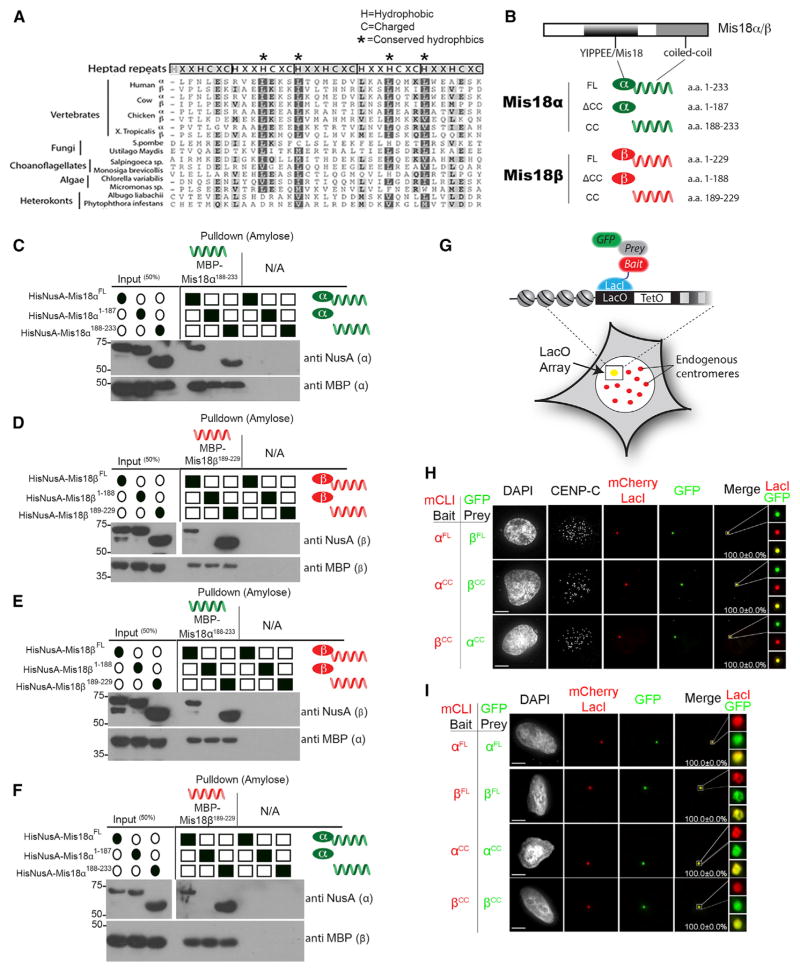
Sequence alignment of Mis18 proteins from a wide variety of eukaryotes identified the previously recognized YIPPEE domain and a second conserved region within the Mis18 C termini (Figures 2A and S2A). The newly identified C-terminal conserved domains span amino acids 198–227 of human Mis18α and 189–219 of Mis18β and are predicted to form coiled coils based on the MARCOIL program (Figures S2B and S2C) (Delorenzi and Speed, 2002). To determine the contribution of the coiled coils to Mis18 homodimer and HT formation, we purified recombinant full-length (FL), coiled-coil deletion (ΔCC), and coiled-coil alone (CC) fragments of Mis18α and Mis18β (Figures 2B and S1A). MBP-Mis18αCC interacted with HisNusA-Mis18αFL and HisNusA-Mis18αCC fragments, but not with the YIPPEE/Mis18 domain alone (HisNusA-Mis18αΔCC) (Figure 2C). Likewise, MBP-Mis18βCC interacted with the Mis18βFL and the Mis18βCC, but not the YIPPEE/Mis18 domain alone (Figure 2D). Therefore, Mis18α and Mis18β form homodimers through the coiled-coil domain.
Figure 2. Mis18 Proteins Multimerize through the Conserved Coiled-Coil Domains.
(A) Diagram showing the conservation of the predicted C-terminal coiled coils. The positions of the characteristic hydrophobic (H) and charged (C) residues are shown. Shading indicates the degree of amino acid similarity (see also Figure S2A).
(B) Schematic of the Mis18 fragments used (CC, coiled coil).
(C) In vitro pull-downs using differentially tagged recombinant Mis18α fragments (Figure S1A) showing that Mis18α can homomultimerize through its predicted C-terminal coiled coil. Filled ovals or squares indicate proteins present. Proteins or affinity tags were detected by immunoblot.
(D) In vitro pull-downs of differentially tagged recombinant Mis18β fragments (Figure S1A) showing that Mis18β can homomultimerize through its predicted C-terminal coiled coil.
(E and F) In vitro pull-down using the (E) Mis18α and (F) Mis18β coiled-coil domains alone showing this domain can mediate Mis18α and Mis18β heterotypic interactions. Proteins or affinity tags were detected by immunoblot.
(G) Schematic of the LacO array system in U2OS cells. GFP-tagged prey proteins were assayed for recruitment to the noncentromeric LacO array by the mCherry LacI-fused (mCLI) bait protein.
(H) Heterotypic interactions were assayed in U2OS-LacO cells cotransfected with the indicated mCLI-Mis18α/β and GFP-Mis18α/βCC/FL constructs. Cells were stained with an anti-CENP-C antibody to mark centromeres.
(I) Homodimeric interactions were assayed in U2OS-LacO cells cotransfected with the indicated mCLI-Mis18α/β and GFP-Mis18α/βCC/FL constructs. Percentages of cells with recruitment of the prey proteins to the array are shown. Scale bars, 5 μm (see also Figures S3A–S3D and S4E).
To assess the role of the coiled coil in mediating HT formation, we determined if the C-terminal domains of Mis18α could bind Mis18β. We found that MBP-Mis18αCC associated with HisNusA-Mis18βFL and HisNusA-Mis18βCC, but not HisNusA-Mis18βΔCC (Figure 2E). The same held true for the reciprocal experiment when we used the coiled-coil domain of Mis18β (MBP-Mis18βCC) to pull down HisNusA-Mis18α (Figure 2F). No interaction was detected between the coiled-coil and YIPPEE/Mis18 domains. Together these results show the Mis18 C-terminal coiled coils contribute to both the homodimerization and the formation of the HT, potentially forming a four-helix coiled coil.
To assess whether the coiled coils mediate homodimer and HT formation of Mis18α and Mis18β in vivo, we targeted Mis18α to a noncentromeric site in U2OS cells containing a LacO array by fusing Mis18α to mCherry and the LacI repressor (mCLI) (Figure 2G) (Janicki et al., 2004). mCLI-Mis18α was coexpressed with GFP-Mis18α or GFP-Mis18β, and the recruitment of the GFP-tagged protein to the LacO array was analyzed. Mis18α recruited Mis18β to the array in 100% of transfected cells (Figure 2H). Consistent with our in vitro experiments, the minimal coiled-coil domains were equally efficient at mediating the Mis18α-β interaction at the array (Figure 2H). Likewise, we observed efficient formation of the Mis18α or Mis18β homodimer at the array by targeting either paralog and expressing the GFP version of the same protein (Figures 2I and S4E). Mutation of the conserved cysteine residues or other highly conserved amino acids within the YIPPEE/Mis18 domain (Mis18αY176A, Mis18βY172A) did not alter the ability of Mis18α and Mis18β to interact in vitro or in vivo (Figures S3A–S3D). Therefore, the coiled-coil domains were sufficient to mediate the homo- as well as the heteromultimerization of Mis18α and Mis18β.
Mis18 Localization Requires Formation of the HT via the Coiled-Coil Domain
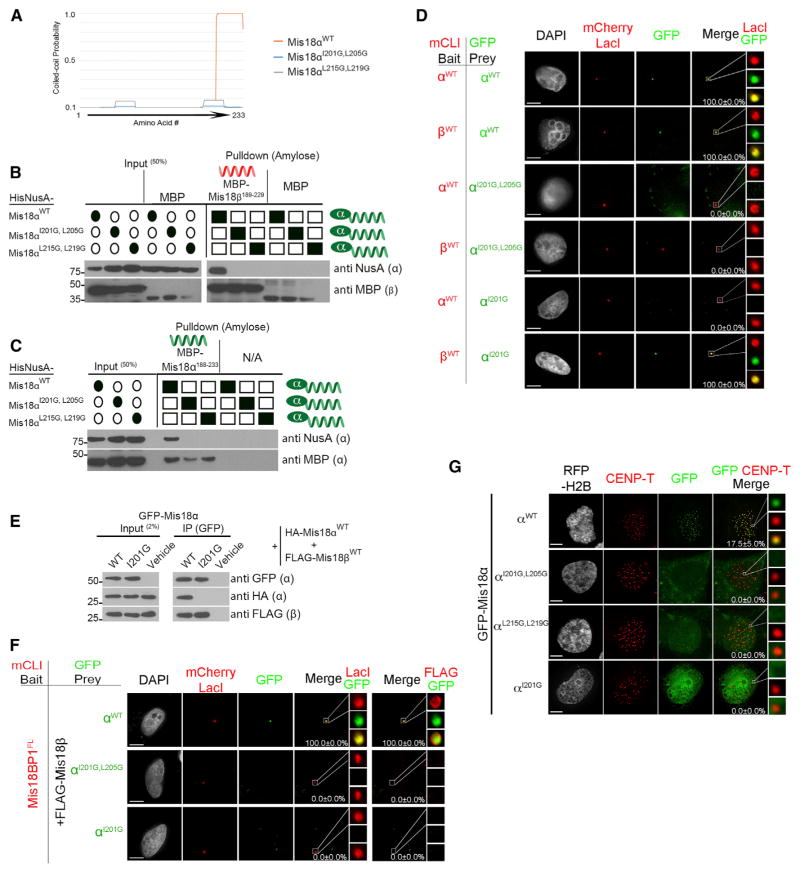
Hydrophobic amino acids contribute to formation of the coiled coils (Figures 2A and S2A–S2C) (Grigoryan and Keating, 2008). We made two double mutants of highly conserved hydrophobic residues at the “a” and “d” positions within the Mis18α coiled coil (I201G/L205G and L215G/L219G) (Figure S2C) and tested their effect on Mis18 complex formation. Both mutants abolished the predicted coiled-coil domain in Mis18α but are not expected to affect the alpha helix (Figure 3A). The effect of these mutations on Mis18 homodimer and tetramer formation was tested using the in vitro pull-down assay. HisNusA-tagged mutant Mis18 proteins, along with their wild-type (WT) counterpart, were pre-assembled with either MBP-Mis18αCC or MBP-Mis18βCC (Figures 3B, 3C, and S1A). The Mis18α coiled-coil mutants were not able to pull down either Mis18α or Mis18β WT or the coiled-coil domains. The coiled-coil domain mutants severely reduced the ability of Mis18α and Mis18β to homo- or heteromultimerize in vivo when assayed at the LacO array (Figure 3D). These data demonstrate that the coiled-coil motif is responsible for the interaction between C-terminal alpha helixes of Mis18α and Mis18β.
Figure 3. Mis18 Coiled Coils Are Required for Proper Centromere Localization.
(A) MARCOIL coiled-coil prediction for Mis18α with indicated mutations in the conserved hydrophobic amino acids.
(B and C) In vitro pull-down assays were conducted between recombinant (B) Mis18β or (C) Mis18α and differentially tagged Mis18α WT and coiled-coil mutants (Figure S1A). Filled ovals and squares indicate proteins present in the input and pull-downs. Proteins or affinity tags were detected by immunoblot.
(D) U2OS-LacO cells were cotransfected with mCLI-Mis18α/β and GFP-Mis18αWT/I201G,L205G/I201G or Mis18αI201G. Percentages of cells with recruitment of the prey proteins to the array are shown. Scale bars, 5 μm.
(E) Anti-GFP immunoprecipitation conducted from U2OS cells transfected with GFP-Mis18α (WT or Mis18αI205G mutant), HA-Mis18α (WT), and FLAG-Mis18β (WT).
(F) U2OS-LacO cells were cotransfected with mCLI-Mis18BP1, GFP-Mis18αWT/I201G,L205G/I201G or Mis18αI201G, and FLAG-Mis18βWT. Percentages of cells with recruitment of the prey proteins to the array are shown. Scale bars, 5 μm.
(G) U2OS cells were cotransfected with GFP-Mis18α WT or mutant constructs. RFP-histone H2B was used as a transfection marker. Centromeres were identified using anti-CENP-T antibody. The percentage of cells with GFP-Mis18 recruited to centromeres is shown ± SD. Scale bars, 5 μm (see also Figures S4A–S4D and S3E).
In contrast, we found that the single amino acid mutation I201G selectively eliminated the ability of Mis18α to form the homodimer but did not alter its ability to bind Mis18β. At the LacO array, mCLI-Mis18αI201G was able to recruit GFP-Mis18β but failed to recruit GFP-Mis18α (Figure 3D). Likewise, Mis18αI201G coimmunoprecipitated Mis18β, but not Mis18α (Figure 3E). Therefore, the Mis18αI201G mutant would be expected to form a Mis18α-β heterodimer.
The interaction of Mis18α and Mis18β with Mis18BP1 is critical for recruitment of the complex to centromeres (Fujita et al., 2007). We tested whether the association of the Mis18α-β complex with Mis18BP1 was affected by HT formation using the LacO array (Figure 3F). mCLI-tagged Mis18BP1 was tethered to the LacO array, and the recruitment of FLAG-Mis18β and GFP-tagged Mis18αWT or coiled-coil mutants was examined. As expected, Mis18BP1 recruited Mis18αWT and Mis18β. In contrast, mutations that completely disrupted the Mis18 HT (Mis18αI201G/L205G) eliminated the ability of Mis18BP1 to recruit Mis18α and Mis18β. Likewise, the Mis18αI201G mutant that selectively disrupts homodimer formation was unable to interact with mCLI-Mis18BP1 at the array (Figure 3F).
GFP-tagged Mis18αI201G/L205G, Mis18αL215G/L219G, or Mis18I201G was transfected into U2OS cells to examine the role of HT formation in centromere localization. Localization of the coiled-coil mutants to centromeres was completely abolished (Figures 3G and S4F). GFP-tagged deletions containing either the YIPPEE/Mis18 or coiled-coil domains of Mis18α or Mis18β did not localize to centromeres; therefore, the coiled-coil domains are not sufficient to determine centromeric localization (Figures S4A–S4D). Mutation of the conserved cysteine residues (Fujita et al., 2007) or other highly conserved amino acids within the YIPPEE domain also results in a loss of centromere localization (Figure S3E). Together these data show the functional YIPPEE/Mis18 domain and HT formation through the coiled coil are both required for the complex to associate with centromeres. The Mis18α-β heterodimer is not sufficient to recruit Mis18 to centromeres or to interact with Mis18BP1. This suggests that multiple YIPPEE/Mis18α motifs are required for the Mis18 complex to stably bind centromeres.
Mis18 Recruits HJURP through a Direct Interaction with the Coiled-Coil Domain
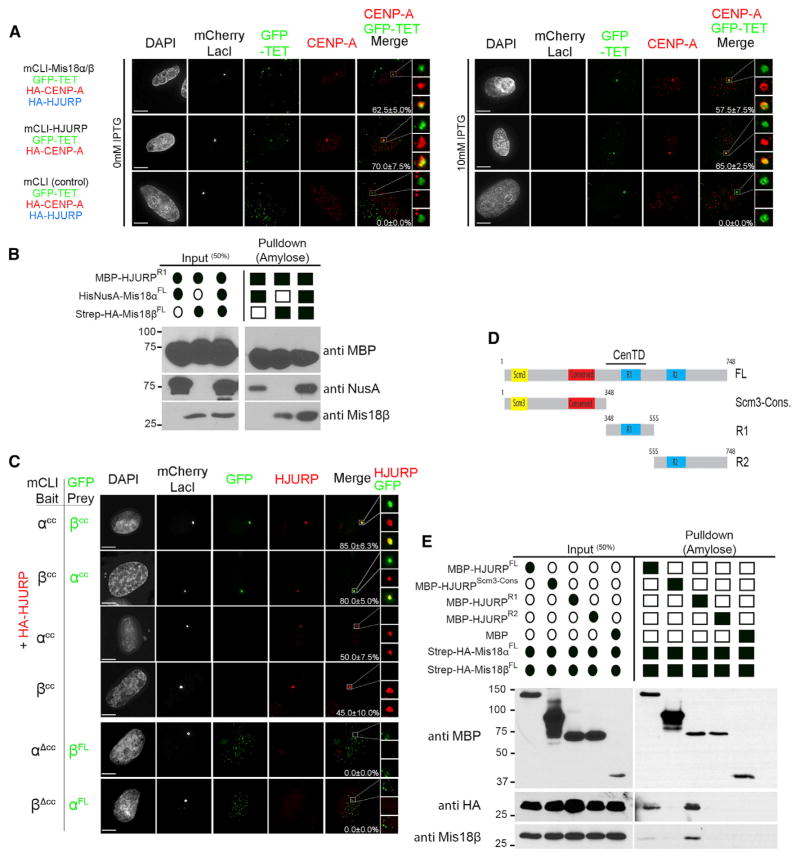
Mis18 precedes HJURP to centromeres and is required for HJURP centromere recruitment (Barnhart et al., 2011; Bernad et al., 2011; Fujita et al., 2007). To determine if Mis18α-β are sufficient to recruit HJURP and deposit CENP-A independently of other centromere proteins, we relocalized Mis18α-β to the noncentromeric LacO array (Figure 4A). Cells were cotransfected with mCLI-Mis18α, mCLI-Mis18β, HA-HJURP, and HA-CENP-A. Under these conditions, CENP-A was robustly recruited to the mCLI-Mis18-bound arrays (Figure 4A, left panel). CENP-A was retained at the LacO array following disruption of LacI binding with IPTG treatment, consistent with stable CENP-A nucleosome assembly at the array (Figure 4A, right panel). We showed previously that mCLI-HJURP was able to assemble CENP-A nucleosome, and these domains facilitated the formation of active centromeres and kinetochores during mitosis (Barnhart et al., 2011; Bassett et al., 2012). CENP-A was retained in a percentage of IPTG-treated cells expressing mCLI-Mis18α-β similar to that in cells expressing mCLI-HJURP.
Figure 4. The Mis18α/β Complex Promotes CENP-A Deposition through a Direct Physical Interaction with HJURP.
(A) U2OS-LacO cells were cotransfected with the indicated mCLI, GFP-TET, HA-CENP-A, and HA-HJURP constructs with/without 10 mM IPTG treatment to disrupt the LacI interaction with the LacO array. Cells were stained with anti-CENP-A antibody. The LacO array was marked by GFP-TET. Recruitment of CENP-A to the LacO array can be seen in the magnified boxed regions to the right. Percentages of cells with CENP-A recruitment to the array are shown. Scale bars, 5 μm.
(B) In vitro pull-down of recombinant HJURP and differentially tagged Mis18α and Mis18β. Filled ovals and squares indicate proteins present in the input and pull-downs. Proteins or affinity tags were detected by immunoblot.
(C) U2OS-LacO cells were cotransfected with mCLI-Mis18α/βCC, GFP-Mis18α/βCC, and HA-HJURP. Cells were stained with an anti-HA antibody. Percentages of cells with recruitment of the prey proteins to the array are shown. Scale bars, 5 μm (see also Figure S5A).
(D) Schematic of the domain structure of HJURP and the recombinant fragments used in this experiment. CenTD, centromere-targeting domain.
(E) In vitro pull-down between recombinant HJURP fragments, Mis18α, and Mis18β (Figure S1A). Proteins or affinity tags were detected by immunoblot.
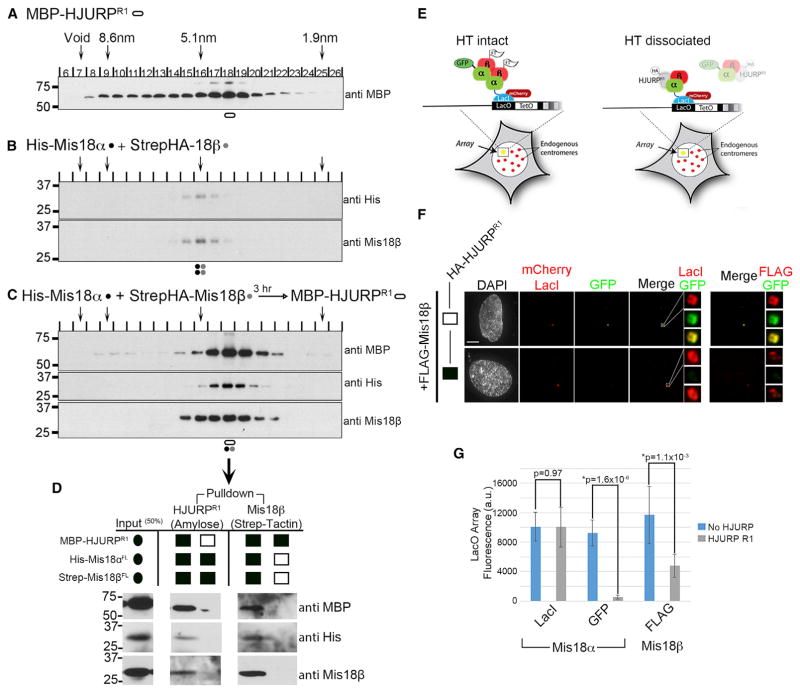
To determine if the recruitment of HJURP to centromeres by Mis18α–β is due to a direct interaction, we expressed and purified recombinant MBP-HJURPR1 (Figures 4B and S1A). MBP-HJURPR1 was incubated with differentially tagged Mis18α and Mis18β separately or with the preformed Mis18α-β HT. In vitro MPB pull-downs showed that both Mis18 proteins alone were able to bind HJURP (Figure 4B). However, binding of HJURP was more robust (~2-fold) when Mis18α and Mis18β were present. This is in contrast to previous reports suggesting that HJURP binds only Mis18β (Wang et al., 2014).
The Mis18 Coiled Coils Recruit HJURP
In order to determine the domain of Mis18α-β required to recruit HJURP, mCLI- and GFP-tagged Mis18 truncation mutants were coexpressed with HA-HJURP in the LacO-U2OS cells. Targeting of the Mis18α-β coiled coils together to the array by coexpressing mCLI-Mis18αCC and GFP-Mis18βCC or vice versa was sufficient to recruit HJURP to the array and as efficient as full-length Mis18 (Figure 4C). Targeting the YIPPEE domain alone (Mis18α/βΔCC) was not sufficient to recruit HJURP. Mis18α/β mutants that disrupt the YIPPEE domains (but not the coiled coil) and abolish Mis18 centromere recruitment (Figures S3A and S3E) were tethered to the array and recruited HJURP similarly to their WT counterparts (Figure S5A). This further supports the conclusion that the coiled coils are the minimal domain sufficient to recruit HJURP to chromatin. Each coiled coil alone also recruited HJURP to the array, but to a lesser degree than when Mis18α and Mis18β were present (Figure 4C). This recapitulates our in vitro results showing Mis18α or Mis18β alone both bind HJURP, albeit not as efficiently when compared to the complete Mis18α-β complex (Figure 4B).
HJURP Binding Disrupts the Mis18 HT
Previously we identified a CenTD of HJURP (aa 348–555) (Zasadzińska et al., 2013). In vitro MBP pull-downs revealed that the Mis18 complex bound exclusively to the R1 fragment containing the HJURP CenTD (Figures 4D, 4E, and S1A). This demonstrates that the HJURP CenTD functions as the only direct binding site for the Mis18 complex, consistent with a report from Wang et al. (2014).
To assess the stoichiometry of the Mis18 complex bound to the HJURP CenTD, we performed SEC and glycerol gradient sedimentation on the purified Mis18α-β complex bound to the MBP-HJURPR1 fragment (Figures 5A–5C and S1A). The MBP-HJURPR1 fragment alone migrated as a monomer (Figures 5A, S1D, S1F, and S1G), consistent with previously reported data (Zasadzińska et al., 2013). To recapitulate the endogenous assembly of the Mis18-HJURP complex, we combined Mis18α and Mis18β prior to the addition of MBP-HJURPR1. We observed the Mis18α-β complex shifted its migration to the same fraction as MBP-HJURPR1 following fractionation by SEC, consistent with a direct interaction between these proteins. However, the HJURP-bound Mis18 complex displayed a smaller Stokes radius compared to the Mis18α-β HT (4.7 nm versus 5.5 nm) (Figures 5B and 5C). Based on the SEC and glycerol gradient sedimentation, we calculated the size of the complex formed by Mis18α, Mis18β, and MBP-HJURPR1 to be 114 kD, which is consistent with a three-protein complex containing a single copy of Mis18α, Mis18β, and MBP-HJURPR1 (Figures 5C and S1E–S1I). An in vitro pull-down that was performed from the peak size-exclusion fraction containing the Mis18-HJURPR1 complex showed that both Mis18α and Mis18β are bound to HJURP and bound to one another (Figure 5D). Taken together, these results suggest the Mis18 HT is reduced to a Mis18 dimer upon binding to HJURP, although we cannot completely eliminate the possibility that subcomplexes containing only one Mis18 paralog are also present.
Figure 5. HJURP Disrupts the Mis18α/β HT.
(A–C) SEC was performed on recombinant (A) HJURP fragment (aa 348–555) MBP-HJURPR1, (B) Mis18α + Mis18β, or (C) MBP-HJURPR1 + Mis18α/β (see also Figures S1A, S1D, S1F, and S1G). Stokes radii of standards are indicated by arrows. Proteins were detected by immunoblot. Shapes under the blots identify the peak fractions and the proteins represented: black, Mis18α; gray, Mis18β; open oval, HJURPR1.
(D) In vitro pull-downs of the peak SEC fraction on either amylose or strep-tactin beads showing that the peak complex contains Mis18α, Mis18β, and HJURPR1.
(E) Predicted model of Mis18α/β recruitment and HT disruption in the presence of HJURPR1 at the LacO array.
(F) U2OS-LacO cells were cotransfected with mCLI-Mis18α, GFP-Mis18α, FLAG-Mis18β, and HA-HJURPR1. Cells were stained with an anti-FLAG antibody to determine Mis18β recruitment. The LacO arrays are magnified in the boxed regions to right with either GFP-Mis18α (left) or FLAG-Mis18β (right). Scale bars, 5 μm.
(G) Fluorescence quantitation of the mCherry (mCLI-Mis18α), GFP (GFP-Mis18α), and FLAG (FLAG-Mis18β) channels without HJURPR1 transfected into the cells. A Student’s two-tailed t test was performed comparing the two conditions in each channel ± SD. p values are noted (see also Figure S5B).
To confirm that our in vitro observations held true in vivo, HEK293T cells were transfected with HA-Mis18α/β with or without GFP-HJURPR1. Chromatin-free extracts were analyzed by SEC. Similar to the in vitro results, when HJURPR1 was present there was a characteristic decrease in Stokes radius of the Mis18 complex, suggesting the Mis18 complex was being disrupted by HJURPR1 binding (Figure S5B).
To determine if HJURP interaction in vivo disrupts the hetero- or homodimerization of Mis18, we used the LacO array. mCLI-Mis18α was tethered to the array and cotransfected with GFP-Mis18α and FLAG-Mis18β with or without HA-HJURPR1. Cells transfected with mCLI-Mis18α at the array recruited both GFP-Mis18α and FLAG-Mis18β, consistent with the formation of the heteromeric complex (Figure 5E, left panel and Figure 5F). If the HT is disrupted into heterodimers by HJURPR1, we expect a loss of GFP-Mis18α fluorescence at the array and reduction of FLAG-Mis18β when HJURPR1 is transfected (Figure 5E, right panel). As predicted, transfection of HJURPR1 almost completely abolished the GFP-Mis18α signal and reduced the FLAG-Mis18β fluorescence by 50% when compared to cells with no HJURPR1 transfected (Figures 5F and 5G). These results are consistent with disruption of the Mis18 complex into heterodimers and recapitulate our in vitro pull-down results (Figures 5C and 5D). These data show that the Mis18α-β complex recruits HJURP to the centromere through direct binding of the CenTD and that HJURP disrupts the Mis18α-β HT into heterodimers upon binding.
Mis18α Is Stable and Does Not Turn Over at the Centromere during Early G1
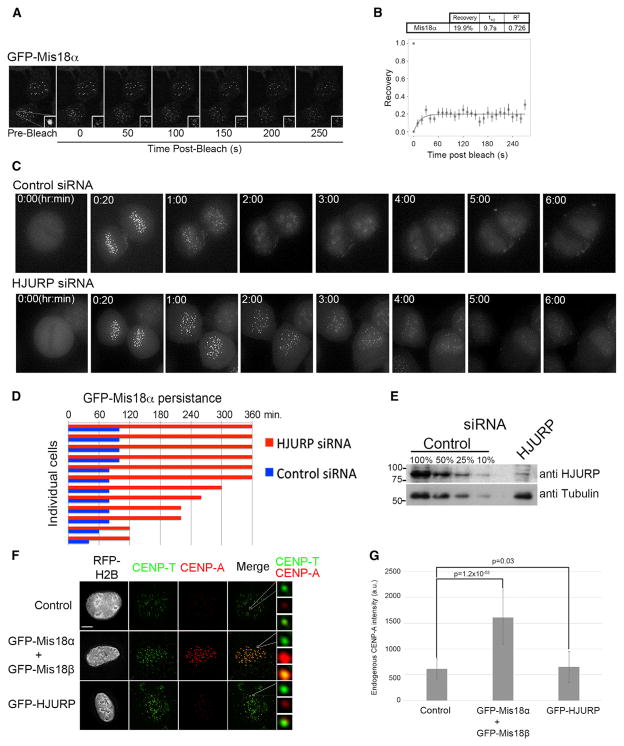
We hypothesized that for the Mis18α-β complex to license centromeres, new Mis18 proteins should not be recruited to centromeres following its recruitment in late telophase. To determine if the Mis18α-β HT is stably bound to centromeres or if it cycles on and off during early G1, we performed a fluorescence recovery assay using a HeLa cell line stably expressing GFP-Mis18α. We selectively photobleached Mis18α-positive centromeres in G1 and allowed them to recover for 5 min (Figure 6A). GFP Mis18α was only able to recover 20.8% of the prebleach level (Figure 6B), showing that once the Mis18α-β complex is bound to the centromere in late telophase it does not actively turn over. This shows the Mis18α-β complex is not replaced at centromeres after binding of the complex in late mitosis.
Figure 6. The Stable Pool Mis18α Is Removed from Centromeres Following HJURP Binding.
(A) Representative images of fluorescence recovery after photobleaching (FRAP) of GFP-Mis18α.
(B) Averaged fluorescence recovery values were fitted to an exponential recovery curve, ± SEM.
(C) Live-cell imaging of GFP Mis18α HeLa cells treated with either control or HJURP siRNA. Cells were imaged starting in metaphase when GFP-Mis18α is absent.
(D) Quantitation of the persistence of GFP-Mis18α in centromeric foci in control and HJURP siRNA-treated cells. Cells were examined for Mis18α foci at 20 min intervals following metaphase. Bars indicate the time points during which Mis18 was visible (see also Figure S5C).
(E) Western blot of cell populations from control siRNA and HJURP siRNA-treated cells to assess the efficiency of HJURP suppression.
(F) Parental U2OS cells were transfected with the indicated GFP constructs along with RFP-H2B as a transfection marker. Cells were stained with anti-CENP-T and CENP-A antibodies. Scale bars, 5 μm.
(G) Bar graph of total CENP-A fluorescence between the different conditions, ± SD. p values obtained from a two-tailed t test with a significance level >0.05 (see also Figures S6A–S6G).
HJURP Facilitates Removal of Mis18 from Centromeres
We showed that HJURP binding disrupted the Mis18 HT into heterodimers and that Mis18 mutants that favor heterodimer formation cannot bind to centromeres. We hypothesize HJURP binding and disruption of the Mis18α-β complex is the final step in centromere licensing that promotes removal of the Mis18 complex from the centromere. Therefore, we predicted that in the absence of HJURP, the Mis18 complex should persist at centromeres. To assess this hypothesis, we performed HJURP or control siRNA depletion in an asynchronously dividing GFP-Mis18α HeLa cell line (Figures 6C–6E). Twenty-four hours postdepletion, cells progressing from metaphase to G1 were imaged for 6 hr (Figures 6C and 6D). Consistent with our hypothesis, GFP-Mis18α persisted for longer time periods into G1 when HJURP was suppressed (Figure 6C). At 3 hr postmetaphase, 80% of cells treated with HJURP siRNA showed GFP-Mis18α puncta compared to control cells in which GFP-Mis18α persisted at centromeres for less than 2 hr following metaphase (Figures 6C, 6D, and S5C). The rate of GFP-Mis18α loss in the HJURP siRNA condition was five times slower than in controls. In the siRNA-treated cells, small amounts of HJURP may still be present despite significant suppression of HJURP, and this may contribute to removal of Mis18α over time. Alternatively, a secondary pathway may exist for the slow removal of Mis18α from centromeres that is independent of HJURP. Taken together, these data show that HJURP is required for rapid dissociation of the Mis18 complex from the centromere in G1 phase.
CENP-A Levels at Centromeres Are Controlled by Mis18
A licensing model predicts that CENP-A levels remain relatively constant across generations. To test this we performed live-cell fluorescence imaging of YFP-CENP-A across three cellular generations. Cells were imaged at the G1/S boundary in the first (one cell), second (two cell), and third (four cell) generations (Figures S6A–S6C). We chose to measure YFP-CENP-A intensity at the G1/S boundary to ensure that centromere assembly was completed, and all cells contain a 2N DNA content. The mean CENP-A intensity between cells varied less than the distribution of centromeres within the same cell. There was no significant difference in the YFP-CENP-A intensities between generations I, II, and III within the same lineage (Figure S6C). This is consistent with a mechanism that pegs the amount of new CENP-A deposited in each cell cycle to the amount of existing CENP-A.
To test if the availability of Mis18 determines how much CENP-A is loaded, we overexpressed GFP-Mis18α/β in U2OS cells and assessed the amount of CENP-A present at centromeres after 48 hr (Figures 6F, 6G, and S6D). There was a clear and significant increase in total CENP-A fluorescence at centromeres in cells that had the Mis18 complex overexpressed. In contrast, when HJURP was overexpressed the CENP-A levels were not significantly different from that of the control. This indicates that although HJURP is the CENP-A chaperone responsible for shuttling it to the centromere, it is the amount of Mis18 present that controls how much CENP-A is deposited every cell cycle.
DISCUSSION
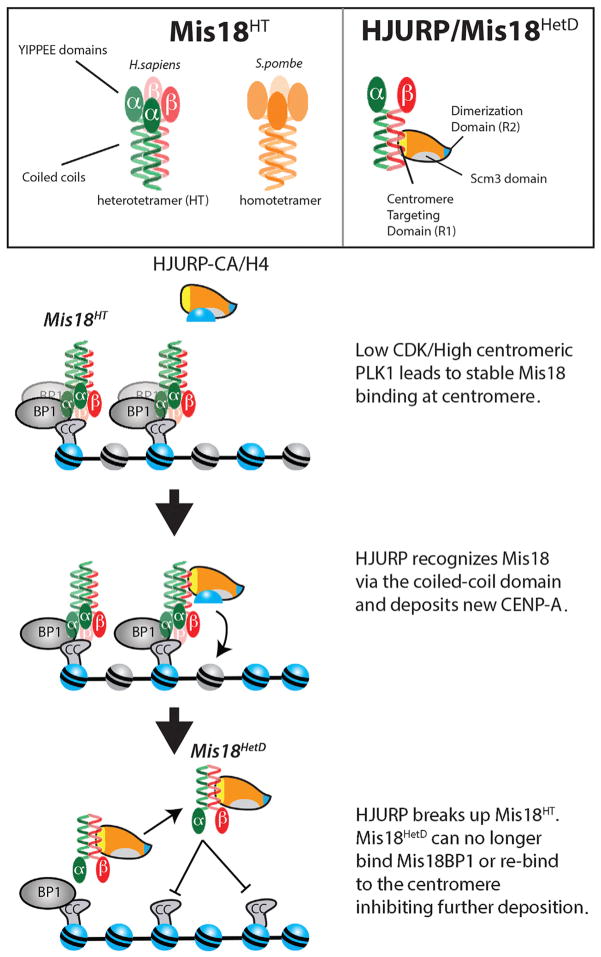
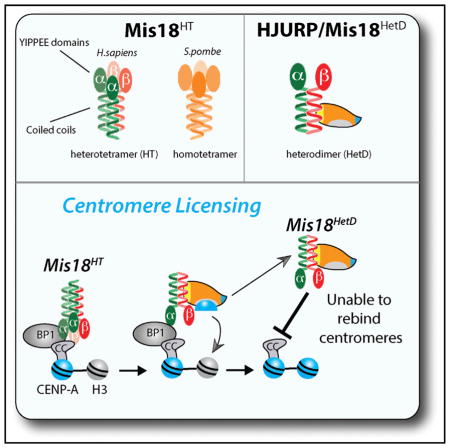
Here we demonstrate that human Mis18α and Mis18β form a Mis18α-β HT through a conserved coiled-coil domain within their C termini (Figure 7). Using S. pombe Mis18 we show that tetramer formation is an evolutionarily conserved feature of the Mis18 complex across divergent eukaryotes. Consistent with these observations, recent work using SEC-MALS also demonstrated that the S. pombe and human Mis18 complexes form a tetramer (Subramanian et al., 2016). We find once Mis18α is bound to centromeres in early G1, it is highly stable. Mutations that completely eliminate coiled-coil formation, or favor Mis18 heterodimer formation (Mis18HetD), show that Mis18HT formation is required for the complex to recognize Mis18BP1 and stably bind to centromeres. We observe that HJURP binding dissociated the Mis18HT into heterodimers. We propose a model where HJURP binding to Mis18α-β disrupts the HT into the Mis18HetD and eliminates the ability of the Mis18 complex to be retained at the centromere. The stable association of Mis18 at centromeres and its removal by HJURP binding limits CENP-A deposition to a single round and couples the amount of new CENP-A to the pre-existing size of the centromere.
Figure 7. CENP-A Levels at Centromeres Are Controlled by Mis18.
A model of centromeric licensing by the Mis18α-β HT. Mis18α-β is recruited to centromeres during late telophase, at which time it becomes stably bound to the centromere. The prenucleosomal complex of CENP-A and HJURP recognizes the centromere through a direct interaction between the Mis18α-β coiled-coil domains and the CenTD domain of HJURP. Binding of HJURP disrupts the Mis18α-β HT and reduces Mis18 binding to Mis18BP1 and the centromere. Degradation of Mis18α and Mis18β may further limit re-recruitment of the complex to centromeres.
We and others have shown that mutations within the Mis18α or Mis18β YIPPEE/Mis18 domain, including the CXXC motifs, eliminate their ability to bind to centromeres (Fujita et al., 2007; Subramanian et al., 2016). The YIPPEE/Mis18 domains must be required for recognition of centromere proteins and are likely to include Mis18BP1 and CENP-C, which are known to be involved in Mis18 centromere localization in humans, and the Eic1 (a.k.a. Mis19) protein in S. pombe, which bridges recognition of spMis18 to the CCAN (Dambacher et al., 2012; Fujita et al., 2007; Hayashi et al., 2004; Maddox et al., 2007; Moree et al., 2011; Subramanian et al., 2014). In addition, we demonstrate that the YIPPEE/Mis18 domain must be organized into the tetramer by the C-terminal coiled coil for centromere localization to occur, as point mutations of the residues that contribute to coiled-coil formation eliminate Mis18BP1 recognition and recruitment of Mis18α to centromeres.
The formation of the Mis18HT and the requirement for the coiled-coil domain for hetero- and homotypic interactions suggest that the C termini of Mis18α and Mis18β form a four-helix coiled coil. Based on the observation that both Mis18 paralogs can form homodimers in isolation, we predict that the Mis18α:Mis18β stoichiometry of the complex is 2:2. However, we cannot eliminate the possibility that the endogenous complex contains a 3:1 ratio. The exact arrangement of the subunits within the Mis18HT is not known. Coiled coils can form parallel or antiparallel (Grigoryan and Keating, 2008), so it is conceivable that YIPPEE/Mis18 domains of the alpha subunits may be closely juxtaposed or at opposite ends of the coiled-coil structure. Given that Mis18α and Mis18β form both homo- and heterodimers, we favor a pattern in which each coiled coil makes homotypic and heterotypic interactions within the structure (Figure S2). In favor of this model, we see that mutations in Mis18α at the “a” and “d” position affect both homodimer and heterodimer formation; however, a single mutation at position d (Mis18αI201G) disrupts only homodimer formation, suggesting homodimer and heterodimer interactions may take place on different “faces” of the coiled coil. In-depth structural analysis of the coiled coil of Mis18α and Mis18β will be required to fully address these questions. HJURP and Scm3 directly interact with Mis18 (Pidoux et al., 2009; Wang et al., 2014) (Figure 4). However, Scm3 lacks the CenTD required for HJURP localization. This suggests that HJURP and Scm3 use different domains to recognize a conserved feature of the Mis18, and we propose that the conserved feature of Mis18 is the C-terminal coiled coil that we described here.
We have shown that total CENP-A levels at centromeres do not vary across multiple generations (Figures S6A–S6C). This suggests the process of new CENP-A deposition is restricted in order to ensure the amount of new CENP-A deposited in G1 is equal to the amount of CENP-A already present in the centromere. This can be achieved through a mechanism whereby a single de novo nucleosome determines the de novo deposition of a new CENP-A nucleosome once and only once per cell cycle. This process is reminiscent of replication licensing, which allows each replication origin to be activated only once per cell cycle (Machida et al., 2005).
Here we provide direct evidence that Mis18 acts as a licensing factor for centromere deposition, similar to regulation of DNA replication by the pre-replication complex (pre-RC). Replication origins are licensed by the formation of a stable pre-RC, which binds throughout G1 phase until replication is initiated in S phase. Similar to the pre-RC, we show that the Mis18HT complex is stably recruited to the centromeres prior to new CENP-A deposition. Temporal control of Mis18 recruitment and the assembly of the pre-RC are inhibited by Cdk activity (Dahmann et al., 1995; Hua et al., 1997; Noton and Diffley, 2000; Silva et al., 2012). The drop in Cdk activity upon exit from mitosis allows both the pre-RC and Mis18 complexes to bind their respective sites within chromatin.
In vitro and in cells, the binding of HJURP disrupts the Mis18 HT, forming a Mis18HetD, and eliminates the ability of Mis18 to continue to recognize Mis18BP1 and localize to the centromere. Likewise, following activation of the MCM2–MCM7 complex to initiate origin firing and DNA replication, several mechanisms inhibit reassociation of the MCM loaders Cdt1 and Cdc6, including binding to Geminin and degradation by ubiquitylation, to inhibit relicensing until the following M/G1 phase (Aladjem, 2007; Diffley and Labib, 2002; Truong and Wu, 2011; Wohlschlegel et al., 2000). Following the disruption of the Mis18HT, additional interactions with the centromere may contribute to association of HJURP and the final deposition of CENP-A. Müller and colleagues (Müller et al., 2014) identified DNA-binding activity of the “conserved” domain of HJURP that may contribute to the association of HJURP with the centromere once Mis18 has recruited it.
Consistent with our observation in human cells, artificially decreasing the amount of CENP-A (a.k.a. CID or CenH3) levels in Drosophila sperm causes a heritable reduction in the amount of CENP-A at centromeres (Raychaudhuri et al., 2012). This demonstrates that the mechanism that pegs the amount of new CENP-A to the amount pre-existing at centromeres is conserved in drosophila despite the divergent centromere deposition pathway involving the Cal1 chromatin assembly factor (Chen et al., 2014; Erhardt et al., 2008; Mellone et al., 2011). Chronic overexpression of CENP-A can lead to increased CENP-A at centromeres, suggesting that over time high levels of CENP-A can push this system toward overdeposition (Bodor et al., 2014). However, the same experiment showed no increase in CENP-C with increased CENP-A, leaving open the possibility that a core set of CENP-A nucleosomes is differentially recognized and subject to licensing as we describe here.
The highest degree of Mis18α recruitment occurs in late telophase, and the amount of Mis18 present steadily decreases through early G1 (Figures 6A and 6C), suggesting Mis18 is loaded onto chromatin in a single event and is gradually removed from the centromere as HJURP binds Mis18 and new CENP-A is deposited. In the HJURP siRNA-treated condition, Mis18α was still lost from centromeres but at a much lower rate. This may be due to incomplete removal of HJURP or, through additional mechanisms, to inhibit Mis18 rebinding to centromeres, such as ubiquitylation-mediated degradation. Ubiquitylation by βTrCP has been shown previously to regulate Mis18β protein levels (Kim et al., 2014). HJURP binding may eliminate the Mis18 HT and directly affect the ability of Mis18 to recognize the centromere, but it may also facilitate the proteasome-mediated degradation of Mis18. In addition, βTrCP-mediated ubiquitylation may also degrade Mis18 that is not bound to chromatin following the initial binding phase in late telophase. We observed that overexpression of Mis18 resulted in increased CENP-A deposition within one or two cell cycles. Increasing Mis18 levels (~9-fold; Figure S6D) may swamp the ubiquitylation system and result in more CENP-A assembly because Mis18 that would usually be degraded is available to rebind an already assembled site.
Centromeric licensing through the Mis18α-β HT may cooperate with other mechanisms to restrict CENP-A to pre-existing centromeres. The ubiquitylation and degradation of CENP-A by Psh1 in yeast and Ppa in Drosophila at noncentromeric sites avoids the accumulation of CENP-A outside of the pre-existing centromere and ensures the propagation of CENP-A chromatin exclusively at existing centromeres (Hewawasam et al., 2010; Moreno-Moreno et al., 2011; Ranjitkar et al., 2010). In addition, Histone H3K9 dimethylation makes heterochromatin regions refractory to centromere formation and helps to spatially restrict CENP-A nucleosome deposition to the centromere (Lam et al., 2006). Together these mechanisms ensure the stable inheritance of CENP-A-containing chromatin.
EXPERIMENTAL PROCEDURES
Cell Transfections and Immunocytochemistry
Parental U2OS and LacO-TRE (Janicki et al., 2004) cells were transfected with Lipofectamine-2000 (Invitrogen) according to the manufacturer’s protocol. Cells were processed for immunocytochemistry 48 hr after transfection. Immunocytochemistry and immunoprecipitation experiments were conducted as described previously (Zasadzińska et al., 2013). Images of fixed cells were collected using a 100× oil immersion objective lens on a DeltaVision deconvolution microscope (Applied Precision) using SoftWoRX acquisition software. Images were deconvolved and presented as stacked images. Images within experiments were collected with identical exposure times and scaled equally.
In Vitro Pull-Downs
Recombinant proteins were purified as described in the Supplemental Experimental Procedures. Proteins were combined for 3 hr at room temperature at 1:1 molar ratio in binding buffer: 50 mM Tris-HCl (pH 7.5), 250 mM NaCl, 20 mM MgCl2, 0.5% NP-40, 10% glycerol, and 5 mM BME. Pull-downs between Mis18 proteins were conducted using 160 nM recombinant protein in each condition. For pull-downs involving HJURP, 125 nM Mis18α dimer, 125 nM Mis18β dimer, or 125 nM Mis18 α-β tetramer were preincubated for 3 hr to form the complex prior to adding 62.5 nM HJURP. Affinity matrices were preincubated in the binding buffer with 0.2 mg/ml BSA for 1 hr and added to preformed complexes for 40 min. Complexes were washed six times in 1 ml of binding buffer, and 40 mM imidazole was added in the case of Ni-NTA purifications. Bound complexes were eluted in SDS sample buffer and boiled. Western blots were performed using antibodies against the 6× His (Santa Cruz), MBP (NEB), Mis18β (Bethyl), and the HA epitope (Covance).
Hydrodynamics
Stokes radii were determined by SEC on Superdex 200 10/300 column (GE Healthcare) in buffer containing 50 mM Tris-HCl (pH 7.5), 350 mM NaCl, 2.5% glycerol, 0.05% NP-40, and 5 mM BME. Proteins were premixed with one another at a 1:1 molar ratio at a final concentration of 7.4 μM for all conditions. Fractions (1 ml) were analyzed by western blot. Gradient sedimentation was performed on a 12 ml 10%–20% glycerol gradient made using a BioCOMP gradient station in buffer containing 50 mM Tris-HCl (pH 7.5), 350 mM NaCl, and 0.05% NP-40. Gradients were centrifuged at 40,000 rpm for 36 hr at 4°C in a SW41Ti rotor, and 500 μL fractions collected. Native molecular weights of Mis18 proteins were calculated based on the measured S values and Stokes radii (Siegel and Monty, 1966).
Live-Cell Imaging and siRNA Depletion
GFP-Mis18α-expressing HeLa cells were plated into eight-well coverglass (Thermo) (1.0 × 104 cells/well) and transfected with 20 nM HJURP (Ambion, #S30814) or control siRNA (Ambion, #4390846) using RNAiMax (Thermo). Cells were imaged 24 hr later at 37°C in 5% CO2 using a 60× oil immersion objective lens (numerical aperture = 1.40; Olympus) on a deconvolution microscope (DeltaVision) using a camera (CoolSNAP HQ2; Photometrics). Images were collected beginning in metaphase at 20 min intervals for 6 hr.
Fluorescence Recovery after Photobleaching
Stable HeLa GFP-Mis18α-expressing cells were grown on glass-bottomed culture dishes (MatTek Corporation). Prior to imaging, growth media were replaced with Leibovitz’s L-15 medium without phenol red (GIBCO) with 10% FBS (Optima, Atlanta Biologicals). Photobleaching was conducted using a Zeiss LSM 510 UV confocal microscope. Two prebleach images were collected. Individual centromeres were bleached with 70 iterations of the 488 nm laser, and the fluorescence recovery at the centromere was assessed at 10 s intervals. Fluorescence recovery at photobleached centromeres was analyzed using ImageJ and normalized to account for sample bleaching (Phair et al., 2004), and average fluorescence recovery data (GFP-Mis18α, n = 18) was fit to a single exponent curve A × (1 − e−kt).
Supplementary Material
Highlights.
Human Mis18α and Mis18β form an evolutionarily conserved heterotetramer
The Mis18 heterotetramer formation is required to bind Mis18BP1 and centromeres
Mis18 binds the HJURP CenTD through the Mis18 C-terminal coiled-coil domain
HJURP binding disrupts the Mis18 heterotetramer, mislocalizing it from centromeres
Acknowledgments
We thank I. Cheeseman and D. Cleveland for reagents and D. Burke, P.T. Stukenberg, and members of the D.R.F. lab for helpful comments. D.R.F. was supported by an American Cancer Society Research Scholar Award and NIH R01GM111907. M.E.S. and I.K.N. were supported by NIH T32CA00910937. C.M.K. was supported by a Harrison Award from the University of Virginia.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2016.02.014.
AUTHOR CONTRIBUTIONS
Conceptualization, I.K.N., E.Z., and D.R.F.; Methodology, I.K.N. and D.R.F.; Investigation, I.K.N., C.M.K., and M.E.S.; Writing – Original Draft, I.K.N. and D.R.F.; Writing – Reviewing & Editing, I.K.N., M.E.S., E.Z., and D.R.F.; Funding Acquisition, D.R.F.
References
- Aladjem MI. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet. 2007;8:588–600. doi: 10.1038/nrg2143. [DOI] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart-Dailey MC, Foltz DR. Centromere licensing: Mis18 is required to Polo-ver. Curr Biol. 2014;24:R808–R810. doi: 10.1016/j.cub.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, DeNizio J, Barnhart-Dailey MC, Panchenko T, Sekulic N, Rogers DJ, Foltz DR, Black BE. HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev Cell. 2012;22:749–762. doi: 10.1016/j.devcel.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann JH, Rodríguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad R, Sánchez P, Rivera T, Rodríguez-Corsino M, Boyarchuk E, Vassias I, Ray-Gallet D, Arnaoutov A, Dasso M, Almouzni G, Losada A. Xenopus HJURP and condensin II are required for CENP-A assembly. J Cell Biol. 2011;192:569–582. doi: 10.1083/jcb.201005136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, Jansen LE. The quantitative architecture of centromeric chromatin. eLife. 2014;3:e02137. doi: 10.7554/eLife.02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Chen CC, Dechassa ML, Bettini E, Ledoux MB, Belisario C, Heun P, Luger K, Mellone BG. CAL1 is the Drosophila CENP-A assembly factor. J Cell Biol. 2014;204:313–329. doi: 10.1083/jcb.201305036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Dahmann C, Diffley JF, Nasmyth KA. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- Dambacher S, Deng W, Hahn M, Sadic D, Fröhlich J, Nuber A, Hoischen C, Diekmann S, Leonhardt H, Schotta G. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus. 2012;3:101–110. doi: 10.4161/nucl.18955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechassa ML, Wyns K, Li M, Hall MA, Wang MD, Luger K. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun. 2011;2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorenzi M, Speed T. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics. 2002;18:617–625. doi: 10.1093/bioinformatics/18.4.617. [DOI] [PubMed] [Google Scholar]
- Diffley JF, Labib K. The chromosome replication cycle. J Cell Sci. 2002;115:869–872. doi: 10.1242/jcs.115.5.869. [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, Straight AF. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Grigoryan G, Keating AE. Structural specificity in coiled-coil interactions. Curr Opin Struct Biol. 2008;18:477–483. doi: 10.1016/j.sbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell. 2010;40:444–454. doi: 10.1016/j.molcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Shang WH, Takeuchi K, Fukagawa T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J Cell Biol. 2013;200:45–60. doi: 10.1083/jcb.201210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua XH, Yan H, Newport J. A role for Cdk2 kinase in negatively regulating DNA replication during S phase of the cell cycle. J Cell Biol. 1997;137:183–192. doi: 10.1083/jcb.137.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, Spector DL. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IS, Lee M, Park KC, Jeon Y, Park JH, Hwang EJ, Jeon TI, Ko S, Lee H, Baek SH, Kim KI. Roles of Mis18α in epigenetic regulation of centromeric chromatin and CENP-A loading. Mol Cell. 2012;46:260–273. doi: 10.1016/j.molcel.2012.03.021. [DOI] [PubMed] [Google Scholar]
- Kim IS, Lee M, Park JH, Jeon R, Baek SH, Kim KI. βTrCP-mediated ubiquitylation regulates protein stability of Mis18β in a cell cycle-dependent manner. Biochem Biophys Res Commun. 2014;443:62–67. doi: 10.1016/j.bbrc.2013.11.058. [DOI] [PubMed] [Google Scholar]
- Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA. Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc Natl Acad Sci USA. 2006;103:4186–4191. doi: 10.1073/pnas.0507947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida YJ, Hamlin JL, Dutta A. Right place, right time, and only once: replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Cheeseman IM. Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell. 2014;158:397–411. doi: 10.1016/j.cell.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, Karpen GH. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 2011;7:e1002068. doi: 10.1371/journal.pgen.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fülöp S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Moreno O, Medina-Giró S, Torras-Llort M, Azorín F. The F box protein partner of paired regulates stability of Drosophila centromeric histone H3, CenH3(CID) Curr Biol. 2011;21:1488–1493. doi: 10.1016/j.cub.2011.07.041. [DOI] [PubMed] [Google Scholar]
- Müller S, Montes de Oca R, Lacoste N, Dingli F, Loew D, Almouzni G. Phosphorylation and DNA binding of HJURP determine its centromeric recruitment and function in CenH3(CENP-A) loading. Cell Rep. 2014;8:190–203. doi: 10.1016/j.celrep.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Noton E, Diffley JF. CDK inactivation is the only essential function of the APC/C and the mitotic exit network proteins for origin resetting during mitosis. Mol Cell. 2000;5:85–95. doi: 10.1016/s1097-2765(00)80405-0. [DOI] [PubMed] [Google Scholar]
- Phair RD, Gorski SA, Misteli T. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 2004;375:393–414. doi: 10.1016/s0076-6879(03)75025-3. [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, Allshire RC. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell. 2010;40:455–464. doi: 10.1016/j.molcel.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri N, Dubruille R, Orsi GA, Bagheri HC, Loppin B, Lehner CF. Transgenerational propagation and quantitative maintenance of paternal centromeres depends on Cid/Cenp-A presence in Drosophila sperm. PLoS Biol. 2012;10:e1001434. doi: 10.1371/journal.pbio.1001434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci USA. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel LM, Monty KJ. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell. 2012;22:52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci USA. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L, Toda NR, Rappsilber J, Allshire RC. Eic1 links Mis18 with the CCAN/Mis6/Ctf19 complex to promote CENP-A assembly. Open Biol. 2014;4:140043. doi: 10.1098/rsob.140043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L, Medina-Pritchard B, Barton R, Spiller F, Kulasegaran-Shylini R, Radaviciute G, Allshire RC, Jeyaprakash AA. Centromere localisation and function of Mis18 requires Yippee-like domain-mediated oligomerisation. EMBO Rep. 2016 doi: 10.15252/embr.201541520. http://dx.doi.org/10.15252/embr.201541520. [DOI] [PMC free article] [PubMed]
- Truong LN, Wu X. Prevention of DNA re-replication in eukaryotic cells. J Mol Cell Biol. 2011;3:13–22. doi: 10.1093/jmcb/mjq052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Liu X, Dou Z, Chen L, Jiang H, Fu C, Fu G, Liu D, Zhang J, Zhu T, et al. Mitotic regulator Mis18β interacts with and specifies the centromeric assembly of molecular chaperone holliday junction recognition protein (HJURP) J Biol Chem. 2014;289:8326–8336. doi: 10.1074/jbc.M113.529958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- Zasadzińska E, Barnhart-Dailey MC, Kuich PH, Foltz DR. Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition. EMBO J. 2013;32:2113–2124. doi: 10.1038/emboj.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.