Abstract
OBJECTIVES
With ENCODE epigenomic data and results from published genome-wide association studies (GWASs), we aimed to find regulatory signatures of obesity genes and discover novel susceptibility genes.
METHODS
Obesity genes were obtained from public GWASs databases and their promoters were annotated based on the regulatory elements information. Significantly enriched or depleted epigenomic elements in the promoters of obesity genes were evaluated and all human genes were then prioritized according to the existence of the selected elements to predict new candidate genes. Top ranked genes were subsequently applied to validate their associations with obesity-related traits in three independent in-house GWASs samples.
RESULTS
We identified RAD21 and EZH2 as over-represented, STAT2 and IRF3 as depleted transcription factors. Histone modification of H3K9me3 and chromatin state segmentation of “poised promoter” and “repressed” were overrepresented. All genes were prioritized and we selected the top five genes for validation at population level. Combined results from the three GWASs samples, rs7522101 in ESRRG remained significantly associated with BMI after multiple testing corrections (P = 7.25 × 10−5). It was also associated with β-cell function (P = 1.99 × 10−3) and fasting glucose level (P < 0.05) in the meta-analyses of glucose and insulin-related traits consortium (MAGIC) dataset.
CONCLUSIONS
In summary, we identified epigenomic characteristics for obesity genes and suggested ESRRG as a novel obesity susceptibility gene.
Keywords: Obesity, Epigenomic elements, ENCODE, ESRRG, Susceptibility
INTRODUCTION
Obesity is a global health problem and it increases the likelihood of various diseases, particularly cardiovascular disease and type 2 diabetes. Moreover, worldwide obesity has more than doubled since 1980 according to the WHO report (http://www.who.int/mediacentre/factsheets/fs311/en/) and it is becoming more widespread with a global projection of more than 1.12 billion obese individuals by 20301. Finding genetic components of obesity are becoming more critical in determining the risk of obesity.
Twin and family studies have established that obesity is highly heritable. The heritability of body mass index (BMI) was estimated to be 40–70%,2, 3 and other anthropometric measures of obesity show similar heritability.2-6 With the help of genome-wide association studies (GWASs), many obesity-susceptibility genes have been defined.7 Up to now, more than 100 loci have been identified to be associated with obesity.8, 9 However, all these variants only explain a small proportion of the heritability for obesity.8 Association signals may be missed at genome-wide significance level due to the modest genetic effect size and inadequate statistical power.10, 11 Thus, novel methods are needed to detect such associations.
Recently, functional and regulatory data for the entire human genome have been generated rapidly.12-14 In particular, the Encyclopedia of DNA Elements (ENCODE) project has provided a large amount of regulatory data (epigenomic elements).15, 16 Considering the indisputable association of epigenomics with obesity,17-19 investigating the regulatory features of obesity-susceptibility genes may deepen our understanding of the causes on obesity. However, the understanding of common factors that regulate the expression of obesity genes is still lacking. A previous study suggested that autoimmune-related gene sets shared similar epigenomic features and other gene promoters (besides the known autoimmunity susceptibility genes) containing the same features also tended to be associated with the immune response.20 Therefore in this study, we hypothesized that obesity gene promoters may share similar regulatory features and prioritizing genes with these regulatory features may identify novel candidates that may play a role in obesity.
Investigations in this study were carried out with the following steps: 1) we annotated the promoter regions of known obesity genes with three groups of epigenomic elements: transcription factor binding sites (TFBSs), chromatin segmentation states, and histone modification marks; 2) we obtained the epigenomic features of obesity genes using enrichment analysis; 3) we prioritized all genes with the selected epigenomic features in their promoters to identify novel candidates that may be associated with obesity; 4) For predicted novel obesity genes with top scores, association studies were carried out to investigate the potential effects of their genetic variants on BMI or fat mass using three GWAS samples from in-house studies. We also assessed the associations with glycaemic traits using data from the meta-analyses of glucose and insulin-related traits consortium (MAGIC). Our results would reveal the epigenomic character of obesity-related genes and identify novel genes that may contribute to the development of obesity.
MATERIALS AND METHODS
Acquisition of obesity associated genes
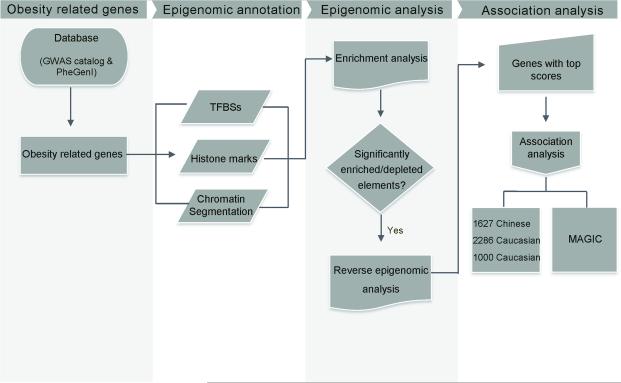
The analysis strategy is shown in Figure 1. Genes associated with obesity were obtained from the National Human Genome Research Institute (NHGRI) GWAS Catalog (www.genome.gov/gwastudies) database21 and phenotype-genotype integrator (PheGenI) database (http://www.ncbi.nlm.nih.gov/gap/phegeni), which are curated resources of SNP-trait associations. With P value < 5 ×10−8, we searched the databases to obtain genes that associated with obesity related phenotypes (including obesity, adiposity, overweight, waist circumference, waist-hip ratio, body mass index, body fat mass, and anthropometric traits). Recently identified novel loci for body mass index (BMI)8, 9 were also included in subsequent analysis. Promoters were defined as 2,000 bases upstream of a gene's transcription start site. An in-house Perl script was used to extract the promoters regions of the selected genes sets. For genes with more than one transcript, the pipeline extracted the promoters for each transcript, and merged overlapping into a single promoter.
Figure 1.
Schematic diagram of the analysis strategy. Obesity-associated gene sets were obtained from GWAS database and genomic coordinates of the promoters were extracted. The promoters were annotated with TFBSs, histone marks, and chromatin segmentation states. Obesity-specific sets of epigenomic elements were identified. All genes were prioritized by the presence of disease-specific epigenomic elements and genes with top scores were validated with association analysis.
Functional annotation
Functional annotation of the genes sets were carried out based on the regulatory annotation files obtained from the UCSC database. The epigenomic elements could be categorized into three groups: transcription factors, histone modifications, and chromatin state segmentation. The data from multiple cell lines were used. As shown in supplementary Table S1, a total of 569 epigenomic elements were used in the analysis.
Enrichment analysis
We first calculated the total number of promoters of obesity associated genes that overlapped with an epigenomic element. If a given promoter overlaps with the same epigenomic element for more than once, it is only counted once. Using the promoters of all genes as a background, according to the promoter number of obesity-associated gene set, we randomly selected the same number of promoters and performed 1000 such random samplings to estimate the average number and variance of random annotation. Compared with random sampling results, we then performed fisher's exact test to identify epigenomic elements that were significantly over-represented or under-represented in obesity related genes. For easier comparison and visualization, P values signifying over-represented epigenomic elements were converted into decimal scale by -log10-transformation while P values signifying under-represented epigenomic elements were converted into decimal scale by log10-transformation. As a positive control, we also tested the promoters of randomly selected genes sets of the same size as the obesity associated genes.
Reverse epigenomic analysis
To identify the promoters of other genes sharing similar epigenomic characters as the promoters of obesity related genes, the promoters of all genes were annotated for the presence of the aforementioned significant epigenomic elements. For each gene, we first calculated the number of times its promoter overlaps with each of the selected epigenomic elements. Then we multiplied the counts of each element by the corresponding transformed P values to prioritize each element by the significance of its association with obesity. Finally, we summed up all counts and the total scores of each gene were obtained.
Gene set enrichment analysis (GSEA)
Genes were ranked according to the scores obtained from the reverse epigenomic analysis. The ranked gene list was supplied to GSEA22 pre-ranked analysis with default parameters and c2 KEGG (curated gene sets from KEGG pathway databases) were used for the analysis.
Validation in GWAS datasets
We used three in-house GWAS datasets to validate the association of the top five genes with obesity related traits. In-house studies were approved by the Institutional Review Boards of the Xi'an Jiaotong University, Creighton University and University of Missouri-Kansas City. Signed informed consent was obtained from all subjects. The three in-house GWAS samples include: 1) Chinese Han subjects with 1627 unrelated healthy adults recruited from Xi'an and Changsha cities; 2) Kansas City sample with 2,286 unrelated US Caucasians of Northern European origin living in Kansas City and its surrounding areas; 3) Omaha sample with 1000 unrelated Caucasian subjects living in Omaha, Nebraska and its surrounding areas. The description of each study has been detailed in our previous studies.23, 24
Phenotype measurement
Body weight and height were recorded and BMI was calculated as body weight (kg) divided by the square of height (m). Body fat mass was measured using Hologic 4500W machines (Hologic Inc., Bedford, MA, USA). Subjects with diseases or conditions that might affect fat metabolism were excluded. The exclusion criteria were detailed in previous studies.25 Age and sex were used as covariates to adjust for the raw BMI and fat mass values. Distribution of the residuals was tested for normality by using Kolmogorov–Smirnov test.
Genotyping and quality control
SNP genotyping was performed using Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA) for the Chinese Han subjects and Kansas City sample. For the Omaha sample, SNP genotyping was performed using the Affymetrix Human Mapping 500K Array (Affymetrix, Santa Clara, CA, USA). The genotyping procedure for each sample has been detailed in our previous studies.23, 24 Quality control was implemented with the following criteria: individual missingness < 5%, SNP call rate > 95%, and Hardy-Weinberg equilibrium (HWE) P-value < 0.0001.
Association analysis
For the Chinese Han subjects and Kansas City sample, association analyses assuming an additive inheritance model were carried out with PLINK.26 For the Omaha sample, to facilitate the comparison of association results at the same SNPs, we first used the IMPUTE2 program27 to impute genotypes of SNPs that detected in the Affymetrix SNP 6.0 array but not in the 500K array based on the 1000 genome data (version 3). SNPTEST28 was then used to examine associations in this sample assuming an additive inheritance model. Only SNPs with minor allele frequency (MAF) over 0.05 in all three GWAS samples were used in association analysis. Summary statistics of associations from the three GWAS samples were subjected to Meta-analysis calculations using the METAL software (http://csg.sph.umich.edu/abecasis/Metal/) under the inverse-variance weighted fixed-effect model. Multiple test correction was carried out using the Bonferroni correction method.
We further checked the association with glycemic traits for significant SNPs after multiple testing corrections using data from the MAGIC datasets. The meta-analyses of glucose and insulin-related traits consortium (MAGIC, http://www.magicinvestigators.org/) represents combined data from multiple GWAS to identify loci that impact on glycemic and metabolic traits, which might influence the risk of type 2 diabetes.
RESULTS
Obesity-related genes
With P-value < 5 ×10−8, a total of 413 transcribed genes associated with obesity were extracted from GWAS Catalog, PheGenI database and a recently published GWAS research on BMI.8, 9 All genes were supplied to pathway enrichment analysis with the STRING online tool (http://string-db.org/) and enrichment (false discovery rate (FDR) < 0.05) of six gene sets were found, including neurotrophin signaling pathway, pathways in cancer, MAPK signaling pathway, non-small cell lung cancer, PI3K-Akt signaling pathway and cholinergic synapse (Table 1A).
Table 1A.
KEGG pathway enrichement analysis of the known 413 obesity related genes
| KEGG_id | Description | adjusted p-value |
|---|---|---|
| hsa04722 | Neurotrophin signaling pathway | 6.05 × 10−4 |
| hsa05200 | Pathways in cancer | 2.24 × 10−2 |
| hsa04010 | MAPK signaling pathway | 3.44 × 10−2 |
| hsa05223 | Non-small cell lung cancer | 4.01 × 10−2 |
| hsa04151 | PI3K-Akt signaling pathway | 4.59 × 10−2 |
| hsa04725 | Cholinergic synapse | 4.99 × 10−2 |
Enrichment and depletion analysis
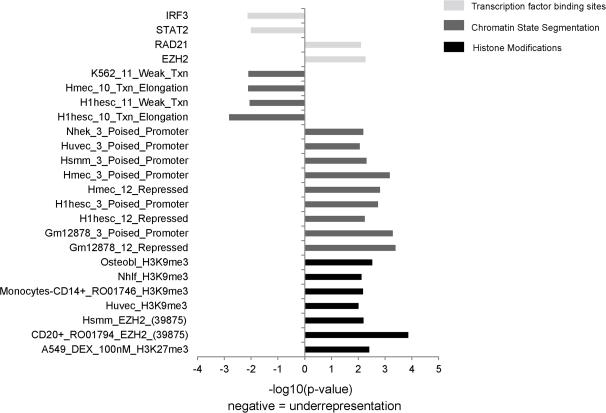
We began by examining whether or not any of the transcription factor binding sites (TFBSs) were enriched or depleted in our gene sets. The results showed that two TFBSs (EZH2 and RAD21, P = 5.41 × 10−3 and 7.85 × 10−3, respectively) enriched in the promoters of obesity susceptibility genes while these genes were depleted in two TFBS: IRF3 and STAT2 (P = 7.36 × 10−3 and 9.77 × 10−3, respectively, Figure 2 and supplementary Table S1).
Figure 2.
Enrichment/depletion of the 24 epigenomic elements in the promoters of obesity-associated genes.
Since the epigenomic landscape of cells derived from different tissue sources or levels of pluropotency can vary considerably, contributing to distinct gene expression programs and biological functions,29, 30 we then analyzed cell type-specific epigenomic factors that control the accessibility of chromatin, including chromatin states and histone marks for obesity genes. As shown in Figure 2 and supplementary Table S1, among the chromatin state segmentation types, the “poised promoter” chromatin region was significantly enriched in the promoters of obesity genes and this signature was shared in 6 cell lines, including Nhek (normal human epidermal keratinocytes, P = 6.53 × 10−3), Huvec (human umbilical vein endothelial cells, P = 8.79 × 10−3), Hsmm (human skeletal muscle myoblasts, P = 4.90 × 10−3), Hmec (human mammary epithelial cells, P = 6.65 × 10−4), H1hesc (human embryonic stem cells, P = 1.83 × 10−3), and Gm12878 (B-lymphoblastoid, normal karyotype, European Caucasian, Epstein-Barr Virus transformed, P = 5.15 × 10−4). Significant enrichment of the “repressed” chromatin region was also found in three cell lines: Hmec (P = 1.56 × 10−3), H1hesc (P = 5.78 × 10−3), and Gm12878 (P = 4.07 × 10−4). Depletion of “txn elongation” was found in H1hesc (P = 1.5 × 10−3) and Hemc (P = 7.81 × 10−3). Depletion of “weak txn” was found in both K562 (P = 7.99 × 10−3) and H1hesc (P = 8.82 × 10−3).
Among all of the histone marks, we did not detect any depletion in the promoters of the obesity genes. As shown in Figure 2 and supplementary Table S1, Enrichment of the H3K9me3 repressive mark was found in four cell lines, including osteoblasts (P = 2.99 × 10−3), Nhlf (normal human lung fibroblast, P = 7.57 × 10−3), monocytes (P = 6.7 × 10−3), and Huvec (P = 9.89 × 10−3). Enrichment of EZH2 (H3K27me3) was found in CD20+ (P = 1.36 × 10−4), Hsmm (P = 6.42 × 10−3), and A549 (P = 3.89 × 10−3).
In summary, non-cell type-specific enrichment of chromatin states and histone marks was detected in the promoters of obesity genes, which might reflect the fact that obesity is a complex disease involving many pathophysiological systems.
Reverse epigenomic analysis suggested novel obesity related genes and miRNAs
The analysis results indicated that a set of epigenomic elements tended to be enriched or depleted in obesity gene promoters. We hypothesized if these signatures indeed reflect obesity-relevant regulatory factors; other genes with similar features in their promoters would tend to be related to obesity as well. Thus, prioritizing the promoters of all genes according to the enrichment or depletion of the selected epigenomic elements might detect novel genes associated with obesity. We searched for the significantly enriched or depleted epigenomic elements within the promoters of all genes, weighted each element by the significance of their associations with obesity related genes, and ranked the genes by the resulting scores. This ranking scheme expectedly identified known obesity associated genes, with ELAVL4, BDNF, and SOX5 being among the top 20 (Table 2). However, not all obesity associated genes scored high, with many other genes demonstrating higher scores. To investigate whether genes identified by reverse epigenomic analysis may provide novel candidate genes relevant to obesity, we performed GSEA on all genes prioritized by the total scores. KEGG enrichment analysis resulted similar pathways to the original gene sets. As shown in Table 1B, neuroactive ligand receptor interaction and pathways related to general growth and patterning were detected.
Table 2.
Top twenty genes with the largest number of epigenomic elements enriched in the promoters of obesity-associated genes set.
| Gene | Description | Total Score |
|---|---|---|
| MIR7641-2 | MicroRNA 7641-2 | 197.17 |
| ESRRG | Estrogen-related receptor gamma | 140.16 |
| PDE4D | Phosphodiesterase 4D | 109.97 |
| PDE4DIP | Phosphodiesterase 4D interacting protein | 101.93 |
| LINC00461 | Long intergenic non-protein coding RNA 461 | 95.15 |
| ELAVL4 | ELAV like neuron-specific RNA binding protein 4 | 90.27 |
| GATA4 | GATA binding protein 4 | 87.04 |
| CHN2 | Chimerin 2 | 85.62 |
| PHACTR3 | Phosphatase and actin regulator 3 | 85.20 |
| TFAP2A | Transcription factor AP-2 alpha | 81.81 |
| BDNF | Brain-derived neurotrophic factor | 80.24 |
| ONECUT1 | One cut homeobox 1 | 80.14 |
| PROX1-AS1 | PROX1 antisense RNA 1 | 80.09 |
| RTN1 | Reticulon 1 | 77.88 |
| SATB2 | SATB homeobox 2 | 77.68 |
| SOX5 | SRY (sex determining region Y)-box 5 | 77.65 |
| TRIM36 | Tripartite motif containing 36 | 76.66 |
| KCNK10 | Potassium channel, two pore domain subfamily K, member 10 | 76.45 |
| RGMA | Repulsive guidance molecule family member a | 76.25 |
| PDE4B | Phosphodiesterase 4B | 75.54 |
Table 1B.
KEGG pathway enrichment analysis results on all genes prioritized by the obesity weighted total numbers of epigenomic elements.
| KEGG_id | Term | Size | ES | NES | adjusted p-value |
|---|---|---|---|---|---|
| hsa04950 | Maturity onset diabetes of the young | 25 | 0.80 | 1.72 | 1.00 × 10−5 |
| hsa04340 | Hedgehog signaling pathway | 56 | 0.76 | 1.70 | 1.00 × 10−3 |
| hsa05217 | Basal cell carcinoma | 55 | 0.74 | 1.66 | 1.00 × 10−3 |
| hsa04080 | Neuroactive ligand receptor interaction | 270 | 0.68 | 1.61 | 1.10 × 10−2 |
| hsa04020 | Calcium signaling pathway | 176 | 0.68 | 1.60 | 1.80 × 10−2 |
| hsa05412 | Arrhythmogenic right ventricular cardiomyopathy | 74 | 0.69 | 1.57 | 3.60 × 10−2 |
| hsa04730 | Long term depression | 70 | 0.69 | 1.56 | 3.70 × 10−2 |
Note: Size: Number of genes in the gene set; ES: Enrichment Score; NES: Normalized Enrichment Score.
Validation in GWAS datasets
To further confirm the association between the novel genes identified by reverse epigenomic analysis and obesity, the top five genes were subjected to association analysis in our three GWAS samples. Basic characteristics of the samples are listed in supplementary Table S2. As shown in supplementary Table S3, a total of 677 SNPs located within and near the selected five genes were included for association analyses, including 4 SNPs for PDE4DIP, 267 SNPs for ESRRG, 16 SNPs for MIR7641-2, 371 SNPs for PDE4D and 19 SNPs for LINC00461.
For BMI, after combining results from these three sample sets, we listed all nominally significant association results with meta-analysis P values < 0.05 in supplementary Table S4. The results showed that 54 SNPs in four genes (ESRRG, MIR7641-2, PDE4D and LINC00461) were nominally significantly associated with BMI (P < 0.05). Among them, the SNP rs7522101 in ESRRG, remained significantly associated with BMI after multiple testing corrections (Table 3, combined P = 7.25 × 10−5). As shown in Table 3, the minor allele “C” of this SNP was negatively associated with BMI.
Table 3.
Significant association results identified for BMI.
| SNP | Region | Gene | Allelea | Omaha | Kansas | Chinese Han | Combine | MAGIC consortium β-cell function | Fasting Glucose | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | P | Beta | P | Beta | P | P | Beta | P | Beta | P | ||||
| rs7522101 | intronic | ESRRG | G/C | 0.1192 | 5.62 × 10−2 | 0.6581 | 3.40 × 10−3 | 0.1824 | 5.53 × 10−2 | 7.25 × 10−5 | 0.0140 | 1.99 × 10−3 | −0.0089 | 3.32 × 10−2 |
Note:
The second allele is the risk allele. SNPs associated with BMI in the combined three data sets after multiple testing corrections are shown.
For the fat mass, as shown in supplementary Table S5, after combining results from the three sample sets, 30 SNPs in four genes (ESRRG, MIR7641-2, PDE4D and LINC00461) were nominally significantly associated with fat mass (combined P < 0.05). The SNP rs7522101 in ESRRG was nominally significantly negatively associated with fat mass (combined P = 3.46 × 10−3). However, none of these SNPs remained significant after multiple testing corrections.
For rs7522101 (Table 3), we also check its association with glycemic traits in the MAGIC datasets. It was associated with indices of β-cell function (P = 1.99 × 10−3, beta = 0.0140)31 and fasting glucose level (P = 3.32 × 10−2, beta = -0.0089).31, 32 These findings are particularly important since obesity is associated with increased risk of diabetes, which might be influenced by glycemic and metabolic traits.
DISCUSSION
GWASs have identified many genes associated with obesity. However, there remains limited understanding of experimentally identified epigenomic regions involved in regulation of these genes. Here we illustrated the distinct epigenomic characters of genes genetically associated with obesity.
In addition, the reverse epigenomic analysis and subsequent association analysis suggested that ESRRG may be a novel candidate gene for obesity. TFBSs analysis identified the overrepresentation of two TFBSs: RAD21 and EZH2. RAD21 is mainly involved in general growth and patterning (cell cycle), while the histone H3K27 methyltransferase EZH2 functioning upstream of Wnt genes. A previous study has indicated that EZH2 could facilitate adipogenesis through directly repressing Wnt genes.33 Corroborated with the enrichment of EZH2, overrepresentation of the H3K27me3 and EZH2_ (39875) histone marks were also detected. STAT2 was significantly depleted in obesity gene promoters. This transcription factor may be involved in the regulation of obesity related biological processes,34 including lipid metabolism, fatty acid transport, lipid transport and generation of precursor metabolites and energy. Depletion of STAT2 may indicate lack of active transcription in general. IRF3 was also significantly depleted in obesity gene promoters. IRF3 encodes a member of the interferon regulatory transcription factor (IRF) family. Consistent with our results, previous study observed that IRF3 expression was markedly decreased in the liver in obese mice.35 Moreover, IRF3 protects against obesity-related insulin resistance and hepatic steatosis through inhibiting the IKKβ/NF-κB signaling pathway.35
Chromatin state segmentation analysis revealed a common feature of inactive transcription status, since “poised promoter” and “repressed” were overrepresented in multiple cell lines. In addition, enrichment of H3K9me3, a mark of transcriptional repression36 was also detected in the histone modification marks analysis. These results suggested that the progression of obesity may be due to the depression of most obesity related genes.
GSEA of genes prioritized by the epigenomic elements revealed obesity related disease pathways, including diabetes and cardiac diseases. In addition, compared with the original obesity gene sets, similar enriched pathways were detected (Table 1), including neuroactive ligand receptor interaction and pathways related to general growth and patterning. This suggests that selecting novel candidate genes associated with obesity according to the ranking list is workable.
We choose the top five genes for further validation and the results confirmed the association of ESRRG with obesity. The selected SNP in ESRRG was also confirmed to be associated with glycemic traits. ESRRG (estrogen-related receptor gamma) encodes a member of the estrogen receptor-related receptor family. It has been reported this gene encodes a transcriptional activator of DNA cytosine-5-methyltransferases 1 expression, modulating cell proliferation and estrogen signaling in breast cancer.37 Consistent with our results, previous study reported that it also regulates energy metabolism through modulating gene expression involved in the processes of oxidative metabolism and mitochondrial biogenesis.38 Due to early post-natal lethality of global Esrrg knockout (Esrrg−/−) mice because of severe renal, gastric and cardiac dysfunction, Esrrg−/− mice didn't appear to have obvious obesity-related phenotypes.39, 40 However, a previous in vivo study41 indicated that mice lacking one copy of ESRRG in muscle exhibited decreased exercise capacity and muscle mitochondrial function. The relationship between mitochondrial dysfunction in muscle and type 2 diabetes or obesity has been reported by many studies.42, 43 There is an impaired bioenergetic capacity of skeletal muscle mitochondria in T2D and obesity subjects.42 Reduced mitochondrial function may predispose patients to intramyocellular lipid accumulation, leading to the interruption of insulin-stimulated glucose transport activity and decreased insulin-stimulated muscle glycogen synthesis, which is a major risk factor in the pathogenesis of T2D.43 ESRRG plays important roles in causing a shift toward slow twitch muscle type and activation of this gene in muscle provides a potential node for therapeutic intervention for obesity.41
Only nominal associations of miR-7641, PDE4D, and LINC00461 with obesity were detected in our results. However, previous studies have suggested their potential relationship with obesity. For example, miR-7641 is a regulator of CXCL1,44 and increased serum CXCL1 was linked to obesity.45 Nominal association signals between PDE4D and BMI have been detected in a previous study,46 and PDE4D is critical in regulating insulin secretion from β-cells which might affect the regular energy metabolism.47 For LINC00461, the SNP rs6893807 with nominal association (supplementary Table S4-S5) in our results was associated with BMI at near genome-wide significance level (P = 1.81 × 10−7) in a previous meta-analysis study in east Asian-ancestry populations.48 The transcript encoded by LINC00461 has been conserved in both sequence and brain expression across diverse mammals.49 Therefore, these genes might also be candidate risk genes for obesity. Further studies are needed to confirm their associations with obesity. Taking together, our results suggested that epigenomic features could be used to predict susceptibility genes for obesity.
Limitations of this study should be addressed. We only focused on the epigenomic features for the promoter of the susceptibility genes considering its important roles in regulating gene expression. Since other genetic regions may also be involved in the regulation of gene expression, further investigations on exploring epigenomic features for the whole gene region are needed.
In summary, using epigenomic data from the ENCODE project, we identified a set of epigenomic elements enriched in obesity related genes. Our results demonstrated the existence of a set of epigenomic elements as a regulatory basis for the functions of genes genetically associated with obesity. Through prioritizing all genes based on the selected epigenomic elements, we suggested that ESRRG might be a novel susceptibility gene associated with obesity.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by National Natural Science Foundation of China [grant numbers 31371278, 31471188, 81573241, 31511140285]; China Postdoctoral Science Foundation [2015M570820]; Natural Science Basic Research Program Shaanxi Province [2015JQ3089], and the Fundamental Research Funds for the Central Universities; and grants from National Institutes of Health [grant numbers P50AR055081, R01AG026564, R01AR050496, and R01AR057049].
Footnotes
Conflict of Interest statement
The authors have no financial conflicts of interest.
REFERENCES
- 1.Kelly T, Yang W, Chen CS, Reynolds K, He J. Global burden of obesity in 2005 and projections to 2030. Int. J. Obes. (Lond.) 2008;32(9):1431–7. doi: 10.1038/ijo.2008.102. [DOI] [PubMed] [Google Scholar]
- 2.Stunkard AJ, Foch TT, Hrubec Z. A twin study of human obesity. JAMA. 1986;256(1):51–4. [PubMed] [Google Scholar]
- 3.Turula M, Kaprio J, Rissanen A, Koskenvuo M. Body weight in the Finnish Twin Cohort. Diabetes Res. Clin. Pract. 1990;10(Suppl 1):S33–6. doi: 10.1016/0168-8227(90)90137-i. [DOI] [PubMed] [Google Scholar]
- 4.Selby JV, Newman B, Quesenberry CP, Jr., Fabsitz RR, King MC, Meaney FJ. Evidence of genetic influence on central body fat in middle-aged twins. Hum. Biol. 1989;61(2):179–94. [PubMed] [Google Scholar]
- 5.Rose KM, Newman B, Mayer-Davis EJ, Selby JV. Genetic and behavioral determinants of waist-hip ratio and waist circumference in women twins. Obes. Res. 1998;6(6):383–92. doi: 10.1002/j.1550-8528.1998.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 6.Malis C, Rasmussen EL, Poulsen P, Petersen I, Christensen K, Beck-Nielsen H, et al. Total and regional fat distribution is strongly influenced by genetic factors in young and elderly twins. Obes. Res. 2005;13(12):2139–45. doi: 10.1038/oby.2005.265. [DOI] [PubMed] [Google Scholar]
- 7.Fall T, Ingelsson E. Genome-wide association studies of obesity and metabolic syndrome. Mol. Cell. Endocrinol. 2014;382(1):740–57. doi: 10.1016/j.mce.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 8.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Magi R, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518(7538):187–96. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet. 2011;88(3):294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chadwick LH. The NIH Roadmap Epigenomics Program data resource. Epigenomics. 2012;4(3):317–24. doi: 10.2217/epi.12.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41(Database issue):D56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.American Association for Cancer Research Human Epigenome Task F, European Union NoESAB Moving AHEAD with an international human epigenome project. Nature. 2008;454(7205):711–5. doi: 10.1038/454711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Consortium EP The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306(5696):636–40. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 16.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrera BM, Keildson S, Lindgren CM. Genetics and epigenetics of obesity. Maturitas. 2011;69(1):41–9. doi: 10.1016/j.maturitas.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoger R. Epigenetics and obesity. Pharmacogenomics. 2008;9(12):1851–60. doi: 10.2217/14622416.9.12.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arora P. Obesity genetics and epigenetics: dissecting causality. Circ. Cardiovasc. Genet. 2014;7(3):395–6. doi: 10.1161/CIRCGENETICS.114.000698. [DOI] [PubMed] [Google Scholar]
- 20.Dozmorov MG, Wren JD, Alarcon-Riquelme ME. Epigenomic elements enriched in the promoters of autoimmunity susceptibility genes. Epigenetics. 2014;9(2):276–85. doi: 10.4161/epi.27021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(Database issue):D1001–6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang TL, Guo Y, Li SM, Li SK, Tian Q, Liu YJ, et al. Ethnic differentiation of copy number variation on chromosome 16p12.3 for association with obesity phenotypes in European and Chinese populations. Int. J. Obes. (Lond.) 2013;37(2):188–90. doi: 10.1038/ijo.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang TL, Guo Y, Li J, Zhang L, Shen H, Li SM, et al. Gene-gene interaction between RBMS3 and ZNF516 influences bone mineral density. J. Bone Miner. Res. 2013;28(4):828–37. doi: 10.1002/jbmr.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng HW, Deng H, Liu YJ, Liu YZ, Xu FH, Shen H, et al. A genomewide linkage scan for quantitative-trait loci for obesity phenotypes. Am. J. Hum. Genet. 2002;70(5):1138–51. doi: 10.1086/339934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39(7):906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 29.Rivera CM, Ren B. Mapping human epigenomes. Cell. 2013;155(1):39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–9. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42(2):105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 2012;44(6):659–69. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Jin Q, Lee JE, Su IH, Ge K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc. Natl. Acad. Sci. U. S. A. 2010;107(16):7317–22. doi: 10.1073/pnas.1000031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Huang J, Jiang M, Lin H. Signal transducer and activator of transcription 2 (STAT2) metabolism coupling postmitotic outgrowth to visual and sound perception network in human left cerebrum by biocomputation. J. Mol. Neurosci. 2012;47(3):649–58. doi: 10.1007/s12031-011-9702-4. [DOI] [PubMed] [Google Scholar]
- 35.Wang XA, Zhang R, She ZG, Zhang XF, Jiang DS, Wang T, et al. Interferon regulatory factor 3 constrains IKKbeta/NF-kappaB signaling to alleviate hepatic steatosis and insulin resistance. Hepatology. 2014;59(3):870–85. doi: 10.1002/hep.26751. [DOI] [PubMed] [Google Scholar]
- 36.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 2010;28(8):817–25. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Q, Li N, Hu X, Li J, Du Z, Chen L, et al. Genome-wide mapping of DNA methylation in chicken. PloS one. 2011;6(5):e19428. doi: 10.1371/journal.pone.0019428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kubo M, Ijichi N, Ikeda K, Horie-Inoue K, Takeda S, Inoue S. Modulation of adipogenesis-related gene expression by estrogen-related receptor gamma during adipocytic differentiation. Biochim. Biophys. Acta. 2009;1789(2):71–7. doi: 10.1016/j.bbagrm.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 39.Alaynick WA, Kondo RP, Xie W, He W, Dufour CR, Downes M, et al. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell metabolism. 2007;6(1):13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 40.Alaynick WA, Way JM, Wilson SA, Benson WG, Pei L, Downes M, et al. ERRgamma regulates cardiac, gastric, and renal potassium homeostasis. Mol. Endocrinol. 2010;24(2):299–309. doi: 10.1210/me.2009-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rangwala SM, Wang X, Calvo JA, Lindsley L, Zhang Y, Deyneko G, et al. Estrogen-related receptor gamma is a key regulator of muscle mitochondrial activity and oxidative capacity. J. Biol. Chem. 2010;285(29):22619–29. doi: 10.1074/jbc.M110.125401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51(10):2944–50. doi: 10.2337/diabetes.51.10.2944. [DOI] [PubMed] [Google Scholar]
- 43.Morino K, Petersen KF, Shulman GI. Molecular mechanisms of insulin resistance in humans and their potential links with mitochondrial dysfunction. Diabetes. 2006;55(Suppl 2):S9–S15. doi: 10.2337/db06-S002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoo JK, Jung HY, Kim CH, Son WS, Kim JK. miR-7641 modulates the expression of CXCL1 during endothelial differentiation derived from human embryonic stem cells. Arch. Pharm. Res. 2013;36(3):353–8. doi: 10.1007/s12272-013-0067-9. [DOI] [PubMed] [Google Scholar]
- 45.Nunemaker CS, Chung HG, Verrilli GM, Corbin KL, Upadhye A, Sharma PR. Increased serum CXCL1 and CXCL5 are linked to obesity, hyperglycemia, and impaired islet function. J. Endocrinol. 2014;222(2):267–76. doi: 10.1530/JOE-14-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melen E, Himes BE, Brehm JM, Boutaoui N, Klanderman BJ, Sylvia JS, et al. Analyses of shared genetic factors between asthma and obesity in children. J. Allergy Clin. Immunol. 2010;126(3):631–7 e1-8. doi: 10.1016/j.jaci.2010.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim MJ, Park SK, Lee JH, Jung CY, Sung DJ, Park JH, et al. Salt-Inducible Kinase 1 Terminates cAMP Signaling by an Evolutionarily Conserved Negative-Feedback Loop in beta-Cells. Diabetes. 2015;64(9):3189–202. doi: 10.2337/db14-1240. [DOI] [PubMed] [Google Scholar]
- 48.Wen W, Zheng W, Okada Y, Takeuchi F, Tabara Y, Hwang JY, et al. Meta-analysis of genome-wide association studies in East Asian-ancestry populations identifies four new loci for body mass index. Human molecular genetics. 2014;23(20):5492–504. doi: 10.1093/hmg/ddu248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oliver PL, Chodroff RA, Gosal A, Edwards B, Cheung AF, Gomez-Rodriguez J, et al. Disruption of Visc-2, a Brain-Expressed Conserved Long Noncoding RNA, Does Not Elicit an Overt Anatomical or Behavioral Phenotype. Cereb. Cortex. 2015;25(10):3572–85. doi: 10.1093/cercor/bhu196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.