Abstract
Different types of autophagy co-exist in most mammalian cells and each of them fulfill very specific tasks in intracellular degradation. Some of this autophagic pathways contribute to cellular metabolism by directly hydrolyzing intracellular lipid stores and glycogen. Chaperone-mediated autophagy (CMA), in contrast, is a selective form of autophagy that can only target proteins for lysosomal degradation. Consequently, it was anticipated that the only possible contribution of this pathway to cellular metabolism was by providing free amino acids resulting from protein breakdown. However, recent studies have demonstrated that disturbance in CMA leads to important alterations in glucose and lipid metabolism and in overall organism energetics. Here, we describe the unique mechanisms by which CMA contributes to the regulation of cellular metabolism and discuss the possible implications that these previously unknown functions of CMA could have in the pathogenesis of common metabolic diseases.
Keywords: chaperones, diabetes, fatty liver, glucose metabolism, lipid metabolism, lysosomes, proteases, proteolysis
Introduction
Cells recycle cytosolic components through autophagy, the process that mediates degradation of proteins and organelles in lysosomes [1]. The two final steps of autophagy, breakdown and recycling of degradation products, occur in lysosomes, a cellular compartment shared by all types of autophagy [2]. However, the mechanisms that deliver the cytosolic components to be degraded (cargo) toward lysosomes are different for each of the three main types of autophagy (Fig. 1) [3]. In the case of macroautophagy, cargo is first identified by soluble receptors and adaptor proteins and sequestered inside double membrane vesicles known as autophagosomes. Lysosomal delivery occurs through direct fusion of autophagosomes with lysosomes [1]. Cells sequester cytosolic cargo in microautophagy through invaginations that form directly in the surface of lysosomes or late endosomes [4, 5]. These invaginations seal and pinch-off from the membrane into the lumen in the form of small vesicles that undergo degradation along with the trapped cargo (Fig. 1). In chaperone-mediated autophagy (CMA) cargo delivery also occurs at lysosomes but it does not require formation of vesicles; instead the substrate proteins for this autophagic pathway cross the lysosomal membrane through a protein-translocation complex [6, 7].
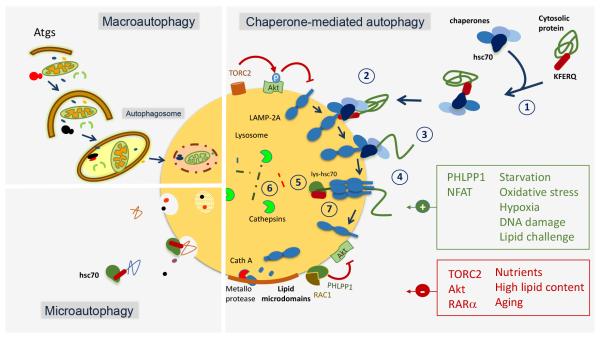
Figure 1. Schematic model of the different types of autophagy in mammals.
The three autophagic pathways that normally co-exist in most mammalian cells are depicted: Macroautophagy requires sequestration of cytosolic cargo by a limiting membrane that forms through conjugation of autophagy-related proteins (Atgs) and seals into a double membrane vesicle (autophagosome). Degradation occurs upon fusion of the autophagosome with the lysosome. In microautophagy, cytosolic cargo is trapped in small vesicles that form by invagination of the lysosomal membrane. A selective form of microautophagy is also depicted in which cytosolic proteins are selectively targeted by hsc70 to the microvesicles for internalization and degradation in the lumen. Details are shown of the individual steps that mediate degradation of cytosolic proteins via chaperone-mediated autophagy: 1) substrate recognition by hsc70; 2) binding to the cytosolic tail of LAMP-2A; 3) unfolding of the substrate protein by the chaperone complex; 4) multimerization of LAMP-2A to form the translocation complex; 5) substrate translocation with the assistance of the lysosome resident hsc70 (lys-hsc70); 6) substrate degradation and 7) disassembly of LAMP-2A from the translocation complex to initiate a new round of substrate binding and translocation. Regulation of CMA through the TORC2-Akt-PHLPP1 axis and through the degradation of LAMP-2A in lipid membrane microdomains through the combined action of a membrane metalloprotease and cathepsin A is shown. Activators and inhibitors of CMA are depicted in green and red, respectively. Abbreviations: Atg: autophagy-related proteins; Akt: protein kinase B; cath: cathepsin; hsc: heat shock cognate protein; LAMP: lysosome-associated membrane protein; NFAT: nuclear factor of activated T cells; PHLPP1: PH domain and leucine rich repeat protein phosphatase 1; RAC1: ras related C3 botulinum toxin substrate1; RARα: retinoic acid receptor alpha; TORC: target of rapamycin complex.
Due to this unique mechanism of accessing lysosomes, only proteins can be targeted for lysosomal degradation by CMA, in contrast with macroautophagy and microautophagy that can deliver any kind of macromolecules (proteins, lipids, glycogen), organelles and even pathogens. However, recent studies support that, being restricted only to protein degradation is not a limitation for the number and variety of intracellular functions that CMA contributes to modulate. Besides the role in protein quality control [8] (resulting from its ability to selectively target damaged or no-longer functional proteins for degradation), the diversity of the subproteome degraded by CMA has linked CMA to the regulation of transcriptional programs [9], cell death and cell survival mechanisms [10-13], DNA repair and cell cycle progression [14] as well as a variety of intracellular processes related to the control of cellular energetics [15-18]. In this minireview, we focus on this last function because recent studies have provided a new understanding of the role of CMA in the regulation of cellular metabolism in response to different nutritional challenges.
Chaperone-mediated autophagy: the basics
All substrates for CMA contain in their amino acid sequence a pentapeptide motif [19] that when recognized by hsc70, a constitutive cytosolic chaperone, targets them to the lysosomal surface [20]. There, the substrate protein/chaperone complex binds the lysosome-associated membrane protein type 2A (LAMP-2A) [21], and this promotes multimerization of LAMP-2A into a translocation complex [22, 23] (Fig. 1). The substrate undergoes unfolding [24] and its translocation into the lysosomal lumen is completed by a luminal resident chaperone (lys-hsc70) [25, 26]. The substrate protein is then rapidly degraded by lysosomal proteases and LAMP-2A disassembles from the translocation complex to allow for a new cycle of substrate binding and translocation [27]. CMA activity is determined by the levels of LAMP-2A at the lysosomal membrane [28] and by the efficiency of assembly and disassembly of LAMP-2A in this compartment [27].
CMA is active in most cells under basal conditions but it is maximally activated in response to stressors such as oxidative stress [29], hypoxia [12, 13], DNA damage [14] and prolonged starvation [30]. The signaling mechanisms that regulate CMA activity are in general poorly understood. The nuclear receptor, retinoic acid receptor alpha is an endogenous inhibitor of CMA [31], whereas the calcineurin-NFAT pathway, described in the context of T cell response, was the first CMA-activating signaling pathway identified [9]. Recent studies have demonstrated the regulation of CMA directly on the surface of lysosomes by the mTOR-Akt-PHLPP pathway [32]. Phosphorylation of Akt by mTOR complex 2 (TORC2) sustains the inhibitory effect of lysosomal Akt over CMA and is responsible for the CMA basal tone. Maximal activation of CMA is attained by the active recruitment of the phosphatase PHLPP towards the lysosomal membrane in a Rac-1-dependent manner, and the subsequent dephosphorylation of Akt [32] (Fig. 1).
CMA activity is coordinated with the activity of other autophagic pathways and even with the ubiquitin/proteasome system. Thus, cells respond to blockage of CMA both in vitro and in vivo by upregulating macroautophagy and the ubiquitin/proteasome system [16, 33, 34]. Similarly, inhibition of macroautophagy or the proteasome leads to increased CMA activity [35, 36]. This compensation among proteolytic systems allows maintenance of basal protein homeostasis even when one of the proteolytic systems fails. However, the absence of the defective system becomes evident once the cells are exposed to stress, because although these pathways can compensate for each other they are not redundant.
Chaperone-mediated autophagy in the control of cellular energetics
Cellular energetics have been linked to CMA since its discovery, when it became evident that removal of serum in cultured cells was a very efficient way to maximally induce degradation of cytosolic proteins by CMA [37, 38]. A similar response of CMA to nutritional deprivation was also confirmed in vivo in different organs from rats and mice [30, 39]. Interestingly, the kinetics of maximal activation of CMA by nutrient deprivation are different to those of macroautophagy, also activated in response to starvation. Studies in cultured fibroblasts and in mouse liver demonstrate that activation of macroautophagy occurs as early as 30 min of starvation (in animals) or of the removal of nutrients from the culture media (in cells) [33, 40, 41]. Maximal protein breakdown through macroautophagy is reached by 4-6 hours of starvation and is then followed by a gradual decline. Later studies have revealed that although protein breakdown through macroautophagy decreases in organs such a liver when starvation persists, the process of macroautophagy remains active at this late time but switches toward preferential degradation of lipids [42]. Degradation of proteins at late starvation times is in part sustained through CMA. Although basal CMA is detectable in most cell types, the activity of this pathway undergoes an almost exponential increase at 8-10 hours of nutrient deprivation, both in mouse liver and in cultured fibroblasts [33, 36]. CMA activity peaks at 16-18 hours into starvation and persists elevated for days [30].
Activation of CMA in absence of nutrients was interpreted as a response activated by cells to replenish the intracellular pool of free amino acids and, in this way, sustain protein synthesis and cellular energetics. The selectivity that characterizes CMA would allow triaging essential from non-essential proteins, and using the latter ones as a source of amino acids upon their degradation by CMA. The findings in support of this role were the fact that starvation enhanced CMA activity [30], that blockage of CMA in vitro reduced intracellular ATP levels [33, 43] and that restoration of normal CMA levels in old mice increased their liver ATP content [44]. However, recent findings have demonstrated that the changes in cellular energetics as a result of CMA activation may, to a large extent, be due to the function of the proteins degraded by CMA in glucose and lipid metabolism rather than the mere contribution of amino acids.
Regulation of glucose metabolism by CMA
It is perplexing that although glycolytic enzymes, such as glyceraldehyde 3-phosphate dehydrogenase and aldolase, were identified as bona fide CMA substrates since very early in the discovery of this autophagic pathway [45] and have been extensively used to track CMA activity, it is only recently that a connection between CMA and glucose metabolism has been established (Fig. 2).
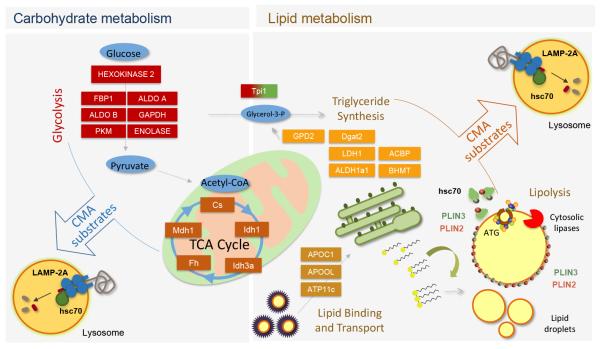
Figure 2. Contribution of CMA to regulation of glucose and lipid metabolism.
Left: CMA participates in the regulation of glycolysis by controlling levels of glycolytic enzymes and enzymes involved in the TCA cycle. Right: CMA modulates rates of lipid uptake and lipogenesis through the degradation of proteins involved in lipid binding and transport and enzymes of the triglyceride synthesis pathway. In addition, CMA determines rates of lipolysis through the selective removal of perilipins (PLIN), proteins that cover the surface of lipid droplets. Removal of perilipins is a pre-requisite to allow access to lipid droplets of cytosolic lipases and autophagy-related proteins (ATG) that initiate lipophagy.
Studies with a transgenic mouse model with selective knock-out of LAMP-2A in hepatocytes, and consequently defective for CMA in liver, have revealed a pronounced metabolic phenotype already manifested at 4-6 months of age [16]. These mice are initially lean, show reduced adiposity both of white and brown adipose tissue, and a negative energetic balance (higher energy expenditure than littermate controls with similar food intake and ambulatory activity). Analysis of the respiratory exchange ratio in these animals revealed a marked preference for carbohydrate consumption as a source of energy. This pronounced usage of carbohydrates is behind, at least in part, their higher glucose tolerance and insulin responsiveness in these animals at young age [16]. Altered glucose metabolism is also noticeable directly in their hepatocytes, that show unusually low levels of glycogen stores, due to reduced neoglucogenesis, and very high rates of glycolysis. In fact, glycolysis persists elevated in these animals even under starvation conditions, when the liver normally shuts off its own glycolysis and becomes the energetic support for the rest of the organs [16].
At the molecular level, the elevated rates of glycolysis are due to the inability of the CMA-incompetent mice to timely degrade in lysosomes the enzymes that participate in glucose catabolic pathways and thus accommodate to the metabolic switch imposed by starvation [16] (Fig. 2). Along with the previously known transcriptional and allosteric enzymatic regulation, selective degradation by CMA reveals as an additional mechanisms utilized by cells to regulate flux though metabolic pathways. Interestingly, some of the enzymes of the glycolytic and TCA cycle undergo some level of CMA degradation even under normal nutritional conditions, suggesting that this autophagic pathway may also contribute to the regulation of homeostatic glucose metabolism [16] (Fig. 2).
Cells respond to CMA blockage by upregulating other proteolytic pathways, and similar activation of macroautophagy and the ubiquitin/proteasome system was also observed in vivo in the liver of CMA-incompetent mice [16, 34]. This compensatory activity seems sufficient to replace for the role of CMA in protein quality control because, contrary to studies in cultured cells, hepatic blockage of CMA in vivo did not result in accumulation of oxidized, misfolded or aggregated proteins in this organ [16]. However, metabolic alterations were noticeable despite activation of these other proteolytic systems, suggesting that CMA-dependent remodeling of the hepatic enzymatic content cannot be attained, or at least not in the same way, through the other proteolytic pathways. It is possible that the inability to identify single proteins for degradation by macroautophagy or the different kinetics of activation of macroautophagy and the proteasome system, when compared with CMA, are responsible for the failure to compensate for CMA.
Recent studies have demonstrated that the ability of CMA to degrade key glycolytic enzymes, such as, hexokinase-2 can be detrimental for cellular energetics in conditions with abnormally enhanced CMA activity, such as individuals with autosomal recessive mutations in the tripeptidyl peptidase II complex (TPPII) [46] (Fig. 3). TPPII-deficiency leads to reduced intracellular amino acid levels and cells respond to this decrease by increasing lysosomes and CMA activity. This elevated CMA activity beyond normal levels reduces hexokinase-2 content and glycolysis and interferes in this way with a normal immune response and neurodevelopment [46].
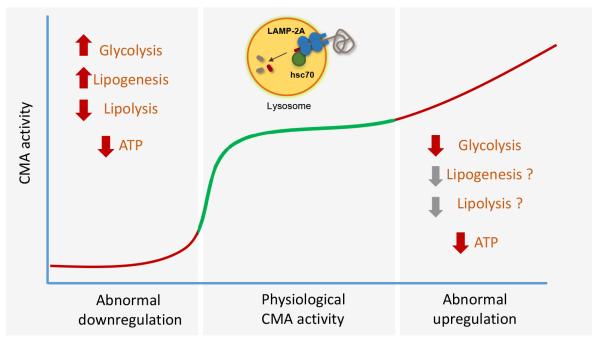
Figure 3. Consequences of CMA malfunctioning in cellular energetics.
Blockage of CMA activity in liver and fibroblasts in culture results in abnormally increased glycolytic flux and lipogenesis. However, the energetic balance is negative and the ATP low, due to the inability to mobilize the stored lipid through lipolysis. Abnormal upregulation of CMA has also shown to alter cellular energetics in two different pathological conditions. In these cases, high rates of degradation of a key glycolytic enzyme by CMA reduce glycolytic flux and cellular energetics. Although there is no information on lipid metabolism under these conditions it is anticipated that reduced lipogenesis may also contribute to inefficient lipid handling in this disease context.
Regulation of lipid metabolism by CMA
Blockage of hepatic CMA in vivo also had an associated phenotype of hepatosteatosis. In contrast with the reduced size of peripheral adipose stores in these animals, lipids accumulated inside hepatocytes and failed to be mobilized for distribution to other organs when nutrients are scarce [16]. Differential proteomics of lysosomes revealed as bona fide CMA substrates a considerable number of enzymes involved in lipogenesis (triglyceride synthesis) and in hepatic lipid uptake [16] (Fig. 2). Failure to timely turnover these proteins through CMA degradation, is behind their higher abundance in CMA-incompetent livers and the increase internalization and storage of lipids. However, increased lipid biogenesis alone could not explain the massive accumulation of lipids in this organ in conditions such as starvation, when hepatic fat stores are usually mobilized for energetic support of other organs. The preferred usage by the CMA-incompetent animals of carbohydrates as energetic source was a clue to identify that part of the hepatic lipid accumulation originates from the inability of hepatocytes defective in CMA to activate lipolysis [18] (Fig. 3).
Mobilization of intracellular lipids through breakdown can take place by cytosolic lipases (conventional lipolysis) or through delivery of lipid droplets to lysosomes via a specialized form of macroautophagy named lipophagy [42, 47] (Fig. 2). Both forms of lipolysis are markedly reduced in CMA defective cells, despite the observed increase of other forms of macroautophagy, because the failure occurs at the level of cargo recognition [18]. Lipid droplets are normally covered by proteins of the perilipin family that prevent exposure of the hydrophobic lipids of these organelles to the cytosol [47]. Removal of perilipins in specific areas of the lipid droplet is required for cytosolic lipases to access the core lipids and for the macroautophagy machinery to assemble on the surface of lipid droplets before sequestration into autophagosomes. Recent studies have demonstrated that degradation of lipid droplet-bound perilipins via CMA contributes to their removal, a pre-requisite for initiation of lipolysis [18] (Fig. 2). Hsc70 binds directly the CMA-targeting motif of these novel substrates and a second AMPK-mediated phosphorylation event on the perilipins triggers their detachment from the droplet and targeting to lysosomes for degradation [48]. When CMA is blocked, rather than accumulating in the cytosol, the perilipin/hsc70 complex persists bound to the surface of the lipid droplets blocking the access of lipases and lipophagy proteins (Fig. 2).
These in vivo studies support that CMA contributes to lipid metabolism at least by these two different mechanisms, degradation of lipogenic enzymes and selective removal of lipid droplet proteins. However, it is possible that additional CMA-related functions could also have an impact on cellular metabolism. Although in the case of CMA-incompetent livers, changes at the transcriptional level were discrete [16], it is possible that CMA may also contribute to regulate metabolic programs at the transcriptional level by modulating degradation of transcription factors or their modulators.
Effect of aging on the role of CMA in cellular metabolism
The enhanced hepatic glycolytic flux that forces higher energetic expenditure in mice with CMA-incompetent livers may appear advantageous, as these animals maintain low adiposity and good response to insulin. However, as they reach middle age, they develop pronounced glucose intolerance and insulin resistance and problems with hepatic protein quality control become evident [34]. A similar metabolic and quality control catastrophe can be precipitated even in young liver-specific CMA-incompetent animals upon exposure to additional stressors such as lipid challenges or oxidative stress [34]. Part of the metabolic failure could be a direct consequence of the severe hepatosteatosis developed by these animals that often progresses to fibrosis and to hepatic tumors [34]. In addition, the accumulation of protein damage, due in part to the gradual loss with age of the compensatory activation of macroautophagy and the proteasome, could also contribute to the observed increase in oxidative stress in these livers and further aggravate the metabolic insufficiency [34].
CMA activity decreases with age in multiple organs and tissues due to a decrease in lysosomal levels of LAMP-2A [49, 50]. Considering the metabolic changes observed in CMA-deficient mice, it is possible that reduced CMA in aging is one of the factors that contributes to the high incidence of the metabolic syndrome in elders.
Reciprocal interplay between CMA and cellular metabolic pathways
CMA is activated in response to multiple stressors including both excess and lack of nutrients. The reasons for activation of CMA in the context of a lipid challenge are now easier to understand in light of the described role of this pathway in the regulation of lipid synthesis and lipid store mobilization [16]. Cells exposed to elevated concentrations of free fatty acids and animals on a high fat diet show an initial increase in CMA activity [18, 51] (Fig. 1). Similarly, increased levels of LAMP-2A have also been described in a rodent model of nonalcoholic steatohepatitis [52]. However, when the lipid challenge is intense or prolonged, CMA activity decreases to levels below those observed under normal conditions. Inhibition of CMA occurs, at least in part, due to shortening of the half-life of LAMP-2A in lysosomes [51]. LAMP-2A levels at the lysosomal membrane are tightly regulated through LAMP-2A degradation in discrete lipid microdomains in this membrane by a pair of two proteases, cathepsin A and a membrane metalloprotease [53] (Fig. 1). Lipidomic analysis of the membrane of lysosomes from animals exposed to diets with high lipid content have demonstrated an expansion of the lipid microdomains and subsequent higher rates of LAMP-2A degradation [51]. Interestingly, comparative lipidomics revealed that lysosomes from old mice, even when maintained under normal diet, show similar membrane lipid changes with age to those observed in young animals in high fat diets [51]. These findings provide an explanation to the reduced stability of LAMP-2A in lysosomes during aging and also highlight that high fat consumption has an accelerating effect on aging of CMA.
Although further studies are required, it is likely that high levels of glucose may have a similar inhibitory effect on CMA activity. Studies in experimental mouse models of diabetes reported reduced CMA activity in kidney and attributed it to the lower abundance of lysosomal LAMP-2A [54]. In fact, CMA failure may be behind the hypertrophy of the diabetic kidney because of its inability to downregulate levels of the growth factor Pax7, identified as a bona fide, CMA substrate [55].
Little is known on the impact that changes in amino acid metabolism could have on CMA, other than the well-known activation of this pathway in response to amino acid depletion. However, it is interesting that defective CMA activity has been recently reported in cystinosis, a lysosomal storage disease where defect in the cystine transporter leads to lysosomal accumulation of cystine [56]. Although defective CMA may be a final common feature of all lysosomal disorders due to the disturbances of the lysosomal compartment, it is interesting that in cystinosis, reduced CMA activity originates from mislocalization of LAMP-2A [56].
Unique use of CMA in cancer cells for regulation of cellular energetics
Studies of the changes in CMA activity in oncogenic processes and cancer cells have also revealed a tight connection between CMA and cellular energetics but, interestingly, the mechanisms by which CMA contributes to regulate metabolic flux in cancer cells are different and the consequences of CMA manipulation are opposed to those described in non-transformed cells (Fig. 4).
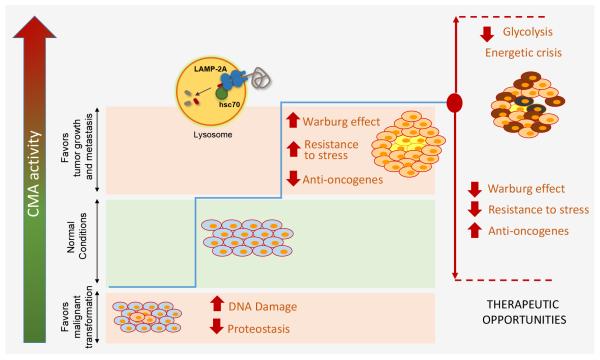
Figure 4. Complex interplay between CMA and cancer biology.
Both reduced and increased CMA activity can be pro-oncogenic depending on the cellular context. Reduced CMA has been shown to increase DNA damage and reduce proteostasis providing thus an environment favorable for malignant transformation. However, once cells have undergone transformation, they upregulate CMA and utilize it to their advantage to sustain the Warburg effect and increase their resistance to cytotoxic stressors. The high dependence of many cancer cells on CMA to regulate their energetic balance highlights the possible therapeutic value of modulation of CMA in cancer. Blockage of CMA in cancer cells reduces tumor growth and metastasis by abrogating part of the Warburg effect and increasing the tumor susceptibility to cytotoxic agents. Abnormally high CMA has been proposed to contribute to the energetic crisis induced in certain cancers by blockage of other autophagic pathways.
A large number of cancer cell lines display abnormally high levels of basal CMA activity when compared with their non-transformed counterpart cell types [43, 57, 58]. This upregulation of CMA is also noticeable during experimental cell transformation and in many types and stages of human tumors [43, 57]. Furthermore, genetic blockage of CMA in cancer cell lines, through interference against the limiting factor LAMP-2A, reduced their tumorigenic ability and CMA blockage in pre-formed xenographic tumors induced tumor shrinkage and lowered metastasis number [43]. Reduced proliferative activity upon CMA blockage, at least in the case of lung cancer cells and melanoma, has been directly linked to glucose metabolism [15, 43] (Fig. 4). Thus, many cancer cells need functional CMA to sustain the Warburg effect (aerobic glycolysis) [15, 43], extensively reported to be required for cancer progression. The contribution of CMA to this effect may occur through combination of different mechanisms. Contrary to normal cells, where CMA reduces levels of glycolytic enzymes through degradation, in different types of cancer cells, CMA contributes to sustain glycolysis by reducing p53 levels and thus neutralizing its transcriptional repression over glycolytic enzymes such as GAPDH and aldolase [43]. In addition, CMA selectively degrades an acetylated form of embryonic M2 isoform of pyruvate kinase, more common in cancer cells. Reduced levels of this enzyme via the enhanced CMA in tumors, favors accumulation of glycolytic intermediates that promote cell proliferation and cell growth [15]. In both scenarios, blockage of CMA reduces tumor growth by reducing glycolytic flux and/or the accumulation of glycolytic intermediates [15, 43] (Fig. 4).
However, the advantages of upregulated CMA for cancer cells may be multiple and not all of them limited to cellular energetics. Augmented CMA may also make cancer cells more resistant to different stressors such as oxidative stress, hypoxia or even DNA damaging agents. LAMP-2A knock-down in breast cancer cells associates with increase oxidative stress, induction of apoptosis and increased sensitivity to chemotherapeutic drugs [57]. Recent studies using a hepatocellular carcinoma cell line infected with hepatitis C virus have demonstrated a marked increase in LAMP-2A levels upon challenge with free fatty acids [59]. Although further studies directly measuring CMA activity in these conditions are needed, the observed upregulation of LAMP-2A is suggestive of a conserved role of CMA in the defense against lipotoxicity in cancer cells, similar to the one described in non-transformed cells [51]. Lastly, CMA upregulation may also favor cancer transformation and progression through elimination of endogenous anti-oncogenic mechanisms. CMA accelerates degradation of the tumor-suppressor mammalian STE20-like kinase 1 (MST1), contributing to rapid breast cancer growth [58], and also promotes a misbalance between the pro-oncogenic and anti-oncogenic forms of PEA-15 [60] (Fig. 4).
In light of most studies showing marked upregulation of CMA in cancer cells and detrimental effects on tumor growth, metastasis and resistance to cytotoxic drugs when this pathway is inhibited, blockage of CMA is currently being explored as a possible anti-oncogenic intervention. However, as in the case of macroautophagy, the effect of modulating CMA on cancer biology may be context dependent (Fig. 4). For example, simultaneous treatment with inhibitors of the tyrosine kinase FLT3 and of macroautophagy in non-acute myeloid leukemia leads to metabolic stress, and marked activation of CMA has been shown to contribute in part to this stress. CMA may reduce glycolysis through depletion of hexokinase-2 [17] and also sensitize cancer cells to metabolic challenges through degradation of mutant (but not wild-type) p53 [61]. Although future studies are required to determine if upregulation of CMA by itself, in the absence of the other two treatments would be equally effective to induce a metabolic crisis, these results offer support to the important role of CMA in the regulation of the cellular energetic balance in cancer cells.
Pending questions and concluding remarks
Better understanding of the multiple connections between CMA and cellular metabolism has been a consequence of the realization that this autophagic process does not merely contribute amino acids through protein breakdown, but that instead, part of its metabolic function is due to CMA’s ability to control levels of key players in cellular metabolism. Studies modulating CMA in vivo have also helped to expand CMA function beyond amino acid and protein catabolism to include lipid and glucose metabolism. As it is the case in other forms of autophagy, the interplay of CMA with cellular metabolic processes is bi-directional, whereby CMA activity is also under the control of these processes and can even be negatively impacted by them. The inhibitory effect of some of these metabolic challenges on CMA along with the gradual decrease in the activity of this autophagic pathway with age may underlie the bases for the increased incidence of the metabolic syndrome in elders.
In need of further investigation are the signaling mechanisms that could contribute to regulate the bi-directional interplay between CMA and the cellular energetic balance. Besides the recent discovery of the regulatory role of TORC2-Akt-PHLPP1 axis on CMA, it is anticipated that other signaling pathways may contribute to fine tuning the adaptation of CMA activity to cellular energetics. Predominant contribution of CMA to quality control or cellular energetics could be cell- or tissue-dependent, making necessary the study of the consequences of CMA blockage in a tissue-specific manner in the future. In addition, ongoing studies in mouse models with regulatable systemic blockage of CMA should provide more information on the consequences of the observed global malfunction of CMA in aging to organismal energetics. Similar studies but with models that allow for organ-specific or systemic upregulation of CMA will further provide information of the possible therapeutic value of CMA modulation in metabolic disorders.
ACKNOWLEDGEMENTS
Work in our laboratory is supported by National Institutes of Health grants AG021904, AG031782 and DK098408 and the generous support of R&R Belfer.
Abbreviations
- Akt
protein kinase B
- Atg
autophagy-related proteins
- cath
cathepsin
- CMA
chaperone-mediated autophagy
- hsc
heat shock cognate protein
- LAMP
lysosome-associated membrane protein
- NFAT
nuclear factor of activated T cells
- PHLPP1
PH domain and leucine rich repeat protein phosphatase 1
- PLIN
perilipin
- RAC1
ras related C3 botulinum toxin substrate1
- RARα
retinoic acid receptor alpha
- TORC
target of rapamycin complex
- TPPI
tripeptidyl peptidase II complex
- MST1
mammalian STE20-like kinase 1
Footnotes
AUTHOR’S CONTRIBUTION: IT performed bibliographic search, summarized and analyzed the content of the selected manuscripts, assisted with draft of the review and figures and revised the edited version; AMC contributed the design of sections and organization of the original outline, edited and added to the original draft and review the final version of the manuscript.
References
- 1.Zhang H, Baehrecke EH. Eaten alive: novel insights into autophagy from multicellular model systems. Trends Cell Biol. 2015;25:376–87. doi: 10.1016/j.tcb.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandez AF, Lopez-Otin C. The functional and pathologic relevance of autophagy proteases. J Clin Invest. 2015;125:33–41. doi: 10.1172/JCI73940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, Santambrogio L. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20:131–9. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell. 2005;19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 6.Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 2014;24:92–104. doi: 10.1038/cr.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–17. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arias E, Cuervo AM. Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol. 2011;23:184–9. doi: 10.1016/j.ceb.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valdor R, Mocholi E, Botbol Y, Guerrero-Ros I, Chandra D, Koga H, Gravekamp C, Cuervo AM, Macian F. Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation. Nat Immunol. 2014;15:1046–54. doi: 10.1038/ni.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Q, She H, Gearing M, Colla E, Lee M, Shacka JJ, Mao Z. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 2009;323:124–7. doi: 10.1126/science.1166088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie W, Zhang L, Jiao H, Guan L, Zha J, Li X, Wu M, Wang Z, Han J, You H. Chaperone-mediated autophagy prevents apoptosis by degrading BBC3/PUMA. Autophagy. 2015;11:1623–35. doi: 10.1080/15548627.2015.1075688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hubbi ME, Hu H, Kshitiz, Ahmed I, Levchenko A, Semenza GL. Chaperone-mediated Autophagy Targets Hypoxia-inducible Factor-1alpha (HIF-1alpha) for Lysosomal Degradation. J Biol Chem. 2013;288:10703–14. doi: 10.1074/jbc.M112.414771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreira JV, Fofo H, Bejarano E, Bento CF, Ramalho JS, Girao H, Pereira P. STUB1/CHIP is required for HIF1A degradation by chaperone-mediated autophagy. Autophagy. 2013;9:1349–66. doi: 10.4161/auto.25190. [DOI] [PubMed] [Google Scholar]
- 14.Park C, Suh Y, Cuervo AM. Regulated degradation of Chk1 by chaperone-mediated autophagy in response to DNA damage. Nat Commun. 2015;6:6823. doi: 10.1038/ncomms7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H, Zha Z, Liu Y, Li Z, Xu Y, Wang G, Huang Y, Xiong Y, Guan KL, Lei QY. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719–30. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider JL, Suh Y, Cuervo AM. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab. 2014;20:417–32. doi: 10.1016/j.cmet.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia HG, Najafov A, Geng J, Galan-Acosta L, Han X, Guo Y, Shan B, Zhang Y, Norberg E, Zhang T, Pan L, Liu J, Coloff JL, Ofengeim D, Zhu H, Wu K, Cai Y, Yates JR, Zhu Z, Yuan J, Vakifahmetoglu-Norberg H. Degradation of HK2 by chaperone-mediated autophagy promotes metabolic catastrophe and cell death. J Cell Biol. 2015;210:705–16. doi: 10.1083/jcb.201503044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol. 2015;17:759–70. doi: 10.1038/ncb3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–9. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 20.Chiang H, Terlecky S, Plant C, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382–5. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- 21.Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–3. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 22.Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28:5747–63. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rout AK, Strub MP, Piszczek G, Tjandra N. Structure of transmembrane domain of lysosome-associated membrane protein type 2a (LAMP-2A) reveals key features for substrate specificity in chaperone-mediated autophagy. J Biol Chem. 2014;289:35111–23. doi: 10.1074/jbc.M114.609446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salvador N, Aguado C, Horst M, Knecht E. Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. Journal of Biological Chemistry. 2000;275:27447–56. doi: 10.1074/jbc.M001394200. [DOI] [PubMed] [Google Scholar]
- 25.Agarraberes F, Dice JF. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J Cell Sci. 2001;114:2491–9. doi: 10.1242/jcs.114.13.2491. [DOI] [PubMed] [Google Scholar]
- 26.Cuervo AM, Dice JF, Knecht E. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J Biol Chem. 1997;272:5606–15. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- 27.Bandyopadhyay U, Sridhar S, Kaushik S, Kiffin R, Cuervo AM. Identification of regulators of chaperone-mediated autophagy. Mol Cell. 2010;39:535–47. doi: 10.1016/j.molcel.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuervo AM, Dice JF. Regulation of lamp2a levels in the lysosomal membrane. Traffic. 2000;1:570–83. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- 29.Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–40. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuervo AM, Knecht E, Terlecky SR, Dice JF. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- 31.Anguiano J, Garner TP, Mahalingam M, Das BC, Gavathiotis E, Cuervo AM. Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat Chem Biol. 2013;9:374–82. doi: 10.1038/nchembio.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arias E, Koga H, Diaz A, Mocholi E, Patel B, Cuervo AM. Lysosomal mTORC2/PHLPP1/Akt Regulate Chaperone-Mediated Autophagy. Mol Cell. 2015;59:270–84. doi: 10.1016/j.molcel.2015.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Nat Acad Sci USA. 2006;103:5905–10. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider JL, Villarroya J, Diaz-Carretero A, Patel B, Urbanska AM, Thi MM, Villarroya F, Santambrogio L, Cuervo AM. Loss of hepatic chaperone-mediated autophagy accelerates proteostasis failure in aging. Aging Cell. 2015;14:249–64. doi: 10.1111/acel.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaushik S, Massey A, Mizushima N, Cuervo AM. Constitutive Activation of Chaperone-mediated Autophagy in Cells with Impaired Macroautophagy. Mol Biol Cell. 2008;19:2179–92. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koga H, Martinez-Vicente M, Macian F, Verkhusha VV, Cuervo AM. A photoconvertible fluorescent reporter to track chaperone-mediated autophagy. Nat Commun. 2011;2:386. doi: 10.1038/ncomms1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Backer JM, Bourret L, Dice JF. Regulation of catabolism of microinjected ribonuclease A requires the amino-terminal 20 amino acids. Proc Natl Acad Sci USA. 1983;80:2166–70. doi: 10.1073/pnas.80.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berger JJ, Dice JF. Effect of serum deprivation and replacement on proteolysis in cultured human fibroblasts. Prog Clin Biol Res. 1985;180:479–81. [PubMed] [Google Scholar]
- 39.Wing SS, Chiang HL, Goldberg AL, Dice JF. Proteins containing peptide sequences related to KFERQ are selectively depleted in liver and heart, but not skeletal muscle, of fasted rats. Biochem J. 1991;275:165–9. doi: 10.1042/bj2750165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abeliovich H, Dunn WA, Jr., Kim J, Klionsky DJ. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol. 2000;151:1025–34. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Molecular Biology of the Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–5. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kon M, Kiffin R, Koga H, Chapochnick J, Macian F, Varticovski L, Cuervo AM. Chaperone-mediated autophagy is required for tumor growth. Science translational medicine. 2011;3:109ra117. doi: 10.1126/scitranslmed.3003182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–65. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cuervo AM, Terlecky SR, Dice JF, Knecht E. Selective binding and uptake of ribonuclease A and glyceraldehyde-3-phosphate dehydrogenase by isolated rat liver lysosomes. J Biol Chem. 1994;269:26374–80. [PubMed] [Google Scholar]
- 46.Lu W, Zhang Y, McDonald DO, Jing H, Carroll B, Robertson N, Zhang Q, Griffin H, Sanderson S, Lakey JH, Morgan NV, Reynard LN, Zheng L, Murdock HM, Turvey SE, Hackett SJ, Prestidge T, Hall JM, Cant AJ, Matthews HF, Koref MF, Simon AK, Korolchuk VI, Lenardo MJ, Hambleton S, Su HC. Dual proteolytic pathways govern glycolysis and immune competence. Cell. 2014;159:1578–90. doi: 10.1016/j.cell.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.chweiger M, Zechner R. Breaking the Barrier--Chaperone-Mediated Autophagy of Perilipins Regulates the Lipolytic Degradation of Fat. Cell Metab. 2015;22:60–1. doi: 10.1016/j.cmet.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 48.Kaushik S, Cuervo AM. AMPK-dependent phosphorylation of lipid droplet protein PLIN2 triggers its degradation by CMA. Autophagy. 2016 doi: 10.1080/15548627.2015.1124226. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–13. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 50.Kiffin R, Kaushik S, Zeng M, Bandyopadhyay U, Zhang C, Massey AC, Martinez-Vicente M, Cuervo AM. Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J Cell Sci. 2007;120:782–91. doi: 10.1242/jcs.001073. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez-Navarro JA, Kaushik S, Koga H, Dall'armi C, Shui G, Wenk MR, Di Paolo G, Cuervo AM. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2012;109:E705–14. doi: 10.1073/pnas.1113036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Das S, Seth RK, Kumar A, Kadiiska MB, Michelotti G, Diehl AM, Chatterjee S. Purinergic receptor X7 is a key modulator of metabolic oxidative stress-mediated autophagy and inflammation in experimental nonalcoholic steatohepatitis. Am J Physiol Gastrointest Liver Physiol. 2013;305:G950–63. doi: 10.1152/ajpgi.00235.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaushik S, Massey AC, Cuervo AM. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 2006;25:3921–33. doi: 10.1038/sj.emboj.7601283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sooparb S, Price SR, Shaoguang J, Franch HA. Suppression of chaperone-mediated autophagy in the renal cortex during acute diabetes mellitus. Kidney Int. 2004;65:2135–44. doi: 10.1111/j.1523-1755.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- 55.Franch H, Sooparb S, Du J, Brown N. A mechanism regulating proteolysis of specific proteins during renal tubular cell growth. J Biol Chem. 2001;276:19126–31. doi: 10.1074/jbc.M101777200. [DOI] [PubMed] [Google Scholar]
- 56.Napolitano G, Johnson JL, He J, Rocca CJ, Monfregola J, Pestonjamasp K, Cherqui S, Catz SD. Impairment of chaperone-mediated autophagy leads to selective lysosomal degradation defects in the lysosomal storage disease cystinosis. EMBO molecular medicine. 2015;7:158–74. doi: 10.15252/emmm.201404223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saha T. LAMP2A overexpression in breast tumors promotes cancer cell survival via chaperone-mediated autophagy. Autophagy. 2012;8:1643–56. doi: 10.4161/auto.21654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li L, Fang R, Liu B, Shi H, Wang Y, Zhang W, Zhang X, Ye L. Deacetylation of tumor-suppressor MST1 in Hippo pathway induces its degradation through HBXIP-elevated HDAC6 in promotion of breast cancer growth. Oncogene. 2015 doi: 10.1038/onc.2015.476. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 59.Kubota C, Torii S, Hou N, Saito N, Yoshimoto Y, Imai H, Takeuchi T. Constitutive reactive oxygen species generation from autophagosome/lysosome in neuronal oxidative toxicity. J Biol Chem. 2010;285:667–74. doi: 10.1074/jbc.M109.053058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quintavalle C, Di Costanzo S, Zanca C, Tasset I, Fraldi A, Incoronato M, Mirabelli P, Monti M, Ballabio A, Pucci P, Cuervo AM, Condorelli G. Phosphorylation-regulated degradation of the tumor-suppressor form of PED by chaperone-mediated autophagy in lung cancer cells. J Cell Physiol. 2014;229:1359–68. doi: 10.1002/jcp.24569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vakifahmetoglu-Norberg H, Kim M, Xia HG, Iwanicki MP, Ofengeim D, Coloff JL, Pan L, Ince TA, Kroemer G, Brugge JS, Yuan J. Chaperone-mediated autophagy degrades mutant p53. Genes Dev. 2013;27:1718–30. doi: 10.1101/gad.220897.113. [DOI] [PMC free article] [PubMed] [Google Scholar]