Abstract
DNA-dependent protein kinase (DNA-PK) is a serine/threonine kinase that plays an essential role in the repair of DNA double-strand breaks (DSBs) in the non-homologous end-joining (NHEJ) pathway. The DNA-PK holoenzyme consists of a catalytic subunit (DNA-PKcs) and DNA-binding subunit (Ku70/80, Ku). Ku is a molecular sensor for double-stranded DNA and once bound to DSB ends it recruits DNA-PKcs to the DSB site. Subsequently, DNA-PKcs is activated and heavily phosphorylated, with these phosphorylations modulating DNA-PKcs. Although phosphorylation of DNA-PKcs is well studied, other post-translational modifications of DNA-PKcs are not. In this study, we aimed to determine if acetylation of DNA-PKcs regulates DNA-PKcs-dependent DSB repair. We report that DNA-PKcs is acetylated in vivo and identified two putative acetylation sites, lysine residues K3241 and K3260. Mutating these sites to block potential acetylation results increased radiosensitive, a slight decrease in DSB repair capacity as assessed by γH2AX resolution, and increased chromosomal aberrations, especially quadriradial chromosomes. Together, our results provide evidence that acetylation potentially regulates DNA-PKcs.
1. Introduction
DNA double-stranded breaks (DSBs) are deleterious DNA lesions that are primarily repaired by two pathways; non-homologous end-joining (NHEJ) and homologous recombination (HR) [1]. NHEJ is the prominent pathway responsible for repairing DSBs in human cells [2]. A central player in NHEJ is the DNA-dependent protein kinase (DNA-PK) (see reviews for more details) [3, 4]. DNA-PK consists of a DNA binding subunit (Ku70/80, Ku) and a catalytic subunit (DNA-PKcs). DNA-PKcs is composed of HEAT (Huntington-elongation-A-subunit-TOR) repeats in its N-terminus, which produce a pincer-shaped structure that forms a central channel and a C-terminal region that contains the PI3 kinase domain, which is flanked by the FAT (FRAP, ATM, TRRAP) domain at its N-terminal side and by the FATC domain at its C-terminal side [5, 6]. Following DSB induction, the Ku heterodimer quickly binds to the DSB ends and recruits DNA-PKcs to the break site, which is mediated by the N-terminal region of DNA-PKcs [7–9]. Upon interacting with the DSB-Ku complex, DNA-PKcs is activated. It is believed that activation of DNA-PKcs is dependent on a conformational change in both the FAT and FATC domains [4]. Once activated, DNA-PKcs phosphorylates a number of substrates with the best characterized being itself.
Following DSB formation, DNA-PKcs is heavily phosphorylated and these phosphorylations are critical for its role in DSB repair [4, 10]. DNA-PKcs autophosphorylates itself and is also phosphorylated by the ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad 3 related (ATR) protein kinases [11, 12]. A large number of the phosphorylation sites are clustered in different regions of DNA-PKcs [13–16]. Two prominent clusters phosphorylated in response to DSB induction are the T2609 [13, 17] and S2056 [11, 18, 19] phosphorylation clusters. S2056 is an autophosphorylation site [18], whereas phosphorylation of the T2609 cluster can be mediated by DNA-PKcs, ATM, or ATR. Blocking phosphorylation of DNA-PKcs at the T2609 phosphorylation cluster by mutating the serines/threonines to alanine results in blocking the release of DNA-PKcs from DNA ends, reduced DSB repair capacity, and increased radiosensitivity [17, 20, 21]. Blocking phosphorylation of the S2056 cluster causes increased radiosensitivity and results in increased DNA end processing, suggesting that it is required for NHEJ [19, 22]. Although, regulation of DNA-PKcs through its phosphorylation has been extensively investigated, other posttranslational modifications of DNA-PKcs are not well studied.
Acetylation of lysine residues is a reversible posttranslational modification, which neutralizes the positive charge of this amino acid and changes the functionality of the protein in diverse ways [23]. There are many individual reports of protein acetylation modulating diverse biological processes, suggesting that lysine acetylation has broad regulatory functions [24]. For example, it plays a key role in the regulation of gene expression through the modification of core histone tails by histone acetyltransferases (HATs) or histone deacetylases (HDACs) [25]. Lysine acetylation has also been implicated to play a role in modulating the activity of DSB repair proteins, including Ku70 [26], ATM [27], and CtIP [28]. A recent proteomic study revealed that there are at least 16 lysine residues acetylated in DNA-PKcs [29], but the biological function of DNA-PKcs acetylation is not characterized. Here, we show that DNA-PKcs is acetylated, and we identified two lysine residues (K3241 and K3260) that are potentially acetylated on DNA-PKcs. Mutating these lysine residues to block acetylation results in increased radiosensitivity and chromosomal aberrations, suggesting that these sites are important for DNA-PKcs dependent DSB repair. Collectively, the data provide initial evidence that acetylation may modulate DNA-PKcs.
2. Materials and methods
2.1. Cell culture and transfections
Chinese hamster ovary (CHO) DNA-PKcs-deficient (V3) cell line [30] and V3 cells stably expressing YFP-tagged DNA-PKcs were cultured in Hyclone MEM media containing 10% Fetal Bovine Serum and Newborn Calf Serum (1:1 mixture), 100 U/ml penicillin and 100 U/ml streptomycin. The cells were incubated at 37°C in a humidified incubator with 5% CO2. For generation of stable cell lines, cells were transfected with the linearized expression plasmid using Lipofectamine® 2000 transfection reagent (Invitrogen) according to the manufacturer’s procedures. Stable cell lines expressing YFP-tagged DNA-PKcs were maintained with 500 μg/ml of G418.
2.2. Fluorescent immunostaining, western blotting and antibodies
Fluorescence immunostaining was performed as described previously [31]. Nuclear extraction, immunoprecipitation and western blotting were performed as described previously [22]. Anti-γH2AX (07–164; mouse monoclonal, Millipore), anti-pS2056 (ab124918; rabbit monoclonal, abcam), anti-acetyl-lysine (SA615; rabbit polyclonal, Enzo Life Sciences) are commercially available.
2.3. Cell survival and chromosome analysis
Cell survival curves were obtained by measuring the colony forming abilities of cell populations irradiated with varying doses of irradiation as previously described [32]. Chromosome analysis was performed as described previously [33]. Briefly, exponentially growing cells were irradiated, and cultured in a presence of colcemid (1 μg/mL), starting 30 min post-irradiation for 4 hours. Mitotic cells were harvested by trypsinization and then treated with a hypotonic solution. Cells were fixed in methanol:acetic acid (3:1) and chromosomes were spread by air drying. After the slides were stained with Giemsa, chromosome aberrations were scored.
2.4. Live cell imaging and laser micro-irradiation
Live cell imaging combined with laser micro-irradiation was performed as described previously. Fluorescence signal of YFP-DNA-PKcs was monitored by using an Axiovert 200M microscope (Carl Zeiss, Inc), with a Plan-Apochromat 63X/NA 1.40 oil immersion objective (Carl Zeiss, Inc) [20]. A 365-nm pulsed nitrogen laser (Spectra Physics) was directly coupled to the epifluorescence path of the microscope and used to generate DSBs in a defined area of the nucleus. Analysis of acquired images was done as previously described [34]. Briefly, fluorescence intensity (IN) of each time point was based on pre-laser background intensity using the formula: IN(t) = Idt/Ibt×IbpreIR [Idt: the difference between the accumulation spot intensity and the undamaged site background intensity of each time point; Ibt: the background intensity of each time point; IbpreIR: the background intensity before irradiation. Relative fluorescence intensity (RF) was calculated using the formula: RF(t) = (INt−INpreIR)/(INmax−INpreIR) [INpreIR: IN of the micro-irradiated area before laser damage; INmax: the maximum IN in the micro-irradiated area of all time points]. Each data point is the average of 10 independent measurements.
2.5. Statistics
Statistical analysis was performed utilizing the student’s t-test (paired, 1-sided). We refer to statistically significant as p<0.05. Each point represents the mean ± SD of three independent experiments unless otherwise stated.
3. Results
3.1. Mutating 8 potential acetylation sites of DNA-PKcs results in radiosensitivity
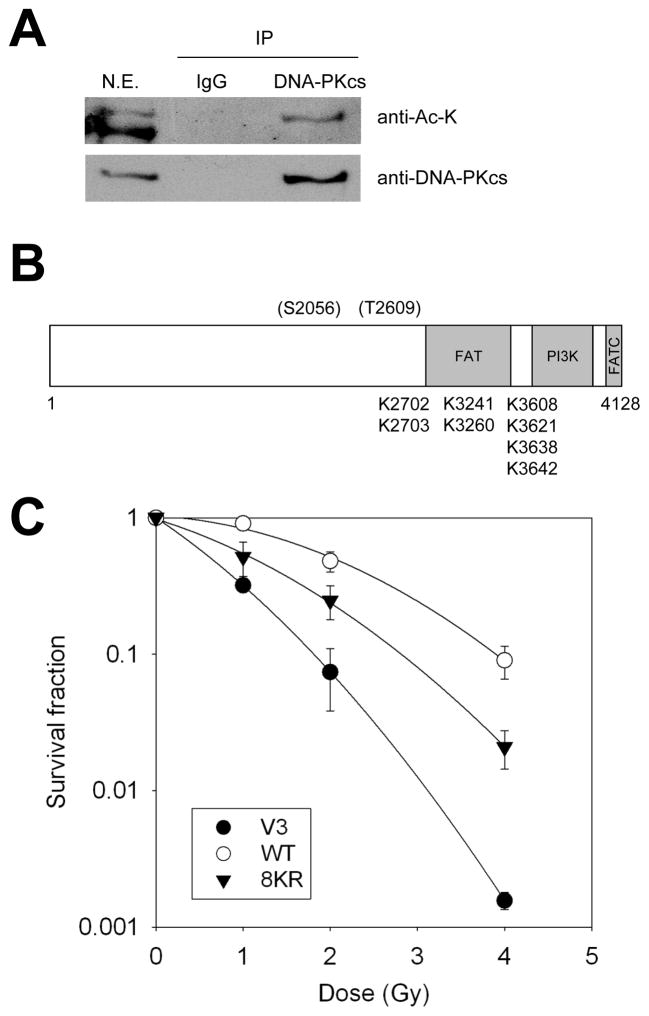
A proteomic study identified 16 potential lysine residues acetylated in DNA-PKcs; therefore, we postulated that acetylation modulates DNA-PKcs activity [29]. To test this hypothesis, we first examined whether DNA-PKcs is acetylated in vivo. Immunoprecipitated DNA-PKcs from HeLa nuclear extract was probed with anti-pan-acetyl-lysine antibodies, and we found that DNA-PKcs is acetylated in normal cycling cells (Fig. 1A). Next, we aimed to identify acetylation sites that may modulate the activity of DNA-PKcs. From the 16 lysine residues identified in the proteomic study [29], we initially focused on eight lysines located around the FAT and kinase domains of DNA-PKcs, as we speculated that acetylation at these sites may affect the activity of DNA-PKcs. Approximate position of each lysine is illustrated in Figure 1B. To examine if these lysines modulate DNA-PKcs function, we blocked potential acetylation at these sites by replacing the lysines (K) with arginines (R), hereafter termed as 8KR. The 8KR mutant was stably expressed in the DNA-PKcs-deficient Chinese hamster ovary (CHO) cell line, V3, and we assessed if mutating these putative acetylation sites affected the repair of DNA damage by monitoring cell survival to varying doses of IR. As shown in Figure 1C, DNA-PKcs deficient V3 cells are extremely radiosensitive (D10 = 1.8 Gy). V3 cells complemented with 8KR are a moderately radiosensitive (D10 = 2.8 Gy) compared to V3 cells completed with wild-type DNA-PKcs (V3-WT) (D10 = 4.0 Gy), indicating that possible acetylation at these lysine residues plays a role in DNA-PKcs-dependent repair of DNA damage.
Figure 1.
DNA-PKcs is acetylated and blocking acetylation causes radiosensitivity. (A) Immunoprecipitated DNA-PKcs from HeLa nuclear extract was detected by pan-acetyl-Ab. (B) Summary of sites of K-to-R mutations in DNA-PKcs. (C) V3 cells complemented with wild-type or 8KR mutant DNA-PKcs were subjected to clonogenic survival analysis following irradiation with indicated doses.
3.2. Mutating K3241 and K3260 of DNA-PKcs results in radiosensitivity
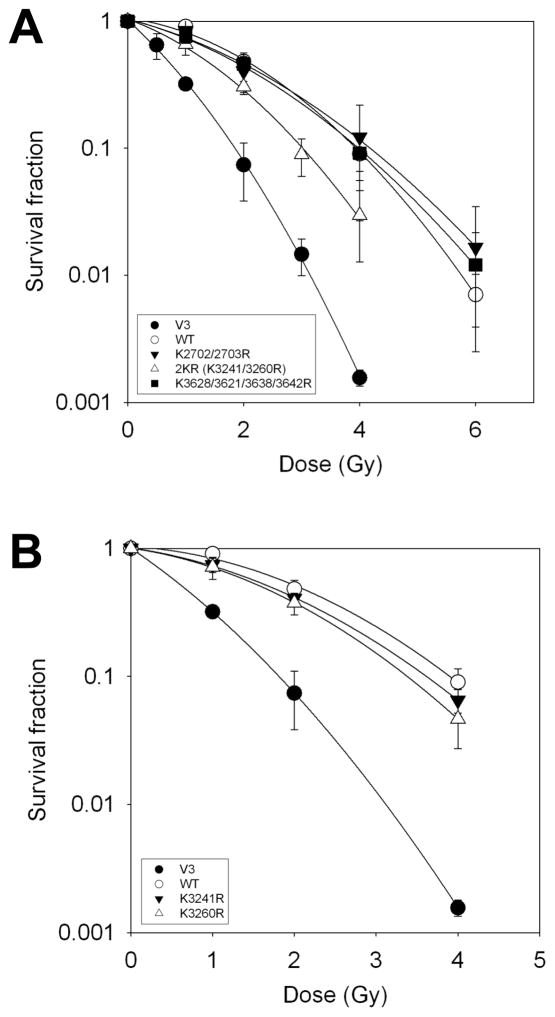
To delineate which of the eight lysine residues are required for the repair of IR-induced DNA damage, we created three groups of K-to-R mutants: K2702/2703R, K3241/3260R (termed as 2KR), K3608/3621/3638/3642R. As shown in Figure 2A, V3 cells complemented with 2KR are moderately sensitive (D10 = 3.2 Gy) compared to V3-WT cell line and show similar radiosensitivity as the V3-8KR cells. On the other hand, V3 cells complemented with K2702/2703R (D10 = 4.0 Gy) or K3608/3621/3638/3642R (D10 = 4.0 Gy) showed radioresistance similar to the V3-WT cell line (Fig. 2A). V3 cells complemented with K3241R (D10 = 3.6 Gy) and K3260R (D10 = 3.3 Gy) also showed less resistance than V3-WT cell line, but not as similar as 8KR or 2KR (Fig. 2B). Collectively, the results suggest that both K3241 and K3260 play an important role in the DNA-PKcs-dependent repair of IR-generated DSBs.
Figure 2.
K3241/K3260 have more impact among 8 acetylation sites in C-term of DNA-PKcs. (A–B) V3 cells complemented with wild-type or 2KR (K3241/3260R), K2702/2703R, K3608/3621/3638/3642R (A), or K3241R, K3260R (B) mutant DNA-PKcs were subjected to clonogenic survival analysis following irradiation with indicated doses.
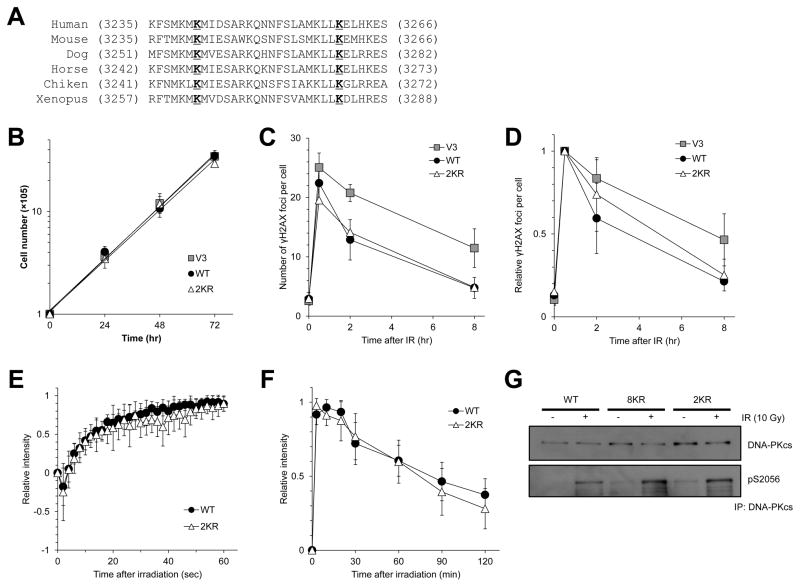
Lysines 3241 and 3260 are conserved in all known DNA-PKcs homologues (Fig. 3A), suggesting that these two lysine residues may be required for DNA-PKcs function. To check if the phenotypes we observed with the K-to-R mutations on K3241 and K3260 were simply due to a general defect in growth, we examined cell proliferation in V3 cells and V3 cells complemented with WT and 2KR. As shown in Figure 3B, 2KR did not lead to growth defects. To determine if radiosensitivity of 2KR is caused by defective DSB repair, we monitored DSB repair via IR-induced γH2AX foci resolution. As shown in Figure 3C, absolute number of induced γH2AX foci at 30 min after IR (1 Gy) was quite different among the different cell lines (25.0±2.5 for V3 cells, 22.4±4.5 for WT, and 19.5±2.8 for 2KR); therefore, we used relative number of γH2AX foci to the ones at 30 min after IR for our analyses. Relative remaining γH2AX foci number at 2 hrs after IR was slightly higher in 2KR than WT (0.84±0.11 for V3 cells, 0.59±0.21 for WT, and 0.74±0.22 for 2KR) (Fig. 3D). Although the difference between WT and 2KR was not significant (p=0.069) at 2 hrs post-IR, the trend does that blocking potential acetylation at these two sites mildly impairs DSB repair.
Figure 3.
Effects of mutation at K3241 and K3260 of DNA-PKcs on growth, DSB repair, recruitment/dissociation, and autophosphorylation. (A) Alignment of human DNA-PKcs K3241 and K3260 with DNA-PKcs sequences. (B) Growth curves corresponding to the indicated cell lines. (C, D) Cells were subjected to 1 Gy of γ-rays and γH2AX immunostaining. DSB repair kinetics by γH2AX foci formation was analyzed; absolute number (C) and relative number (D). (E) Initial accumulation kinetics of WT and 2KR (K3241/3260R) at laser-generated DSBs. (F) 2-hour kinetics of WT and 2KR (K3241/3260R) at laser-generated DSBs. (G) V3 cells complemented with WT, 8KR, or 2KR (K3241/3260R) DNA-PKcs were subjected to 10 Gy of γ-rays and recovered for 30 min. IR-induced autophosphorylation of DNA-PKcs was detected by anti-pS2056 antibody.
Next, we aimed to determine if mutating these two lysines modulates the activity of DNA-PKcs. It was previously shown that impairment of either the kinase activity or clustered phosphorylation of DNA-PKcs leads to sustained retention time at laser-induced DSBs [20, 35]. To check if these two residues play a role in regulating the dynamics of DNA-PKcs at DSBs, we examined the kinetics of 2KR at laser-generated DSBs. The 2KR-mutation did not cause any defects on the recruitment and retention of DNA-PKcs at DSBs in cycling cells when assayed using live cell imaging combined with micro-irradiation (Fig. 3E,F). Next, we assessed if mutating these sites affects the kinase activity of DNA-PKcs, as they lie in the FAT domain of DNA-PKcs, which has previously been implicated to be required for DNA-PKcs activity [36–38]. Surprisingly, 8KR and 2KR have similar kinase activity as WT, as assessed via radiation induced auto-phosphorylation of DNA-PKcs at S2056 (Fig. 3G).
3.3. K3241 and K3260 of DNA-PKcs are important for genomic stability
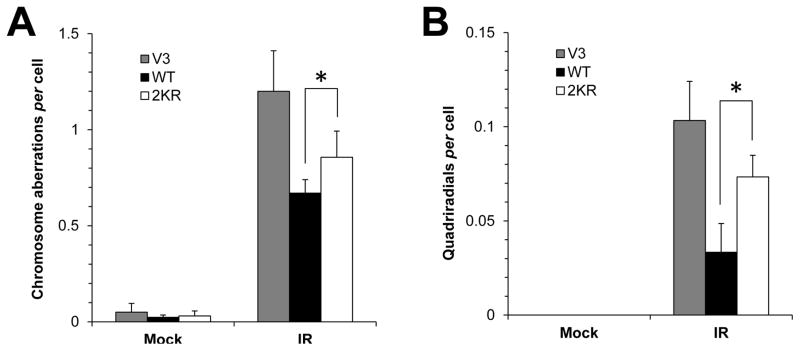
Finally, in order to test if mutation of DNA-PKcs at K3241 and K3260 has any effect on genomic stability, we measured IR-induced chromosome aberrations. As shown in Figure 4A, IR-induced chromosomal aberrations were significantly higher in 2KR cells (0.86±0.14 per cell) compared with WT cells (0.67±0.07 per cell) (p=0.031). Furthermore, as shown in Figure 4B, quadriradial chromosomes [39] were detected significantly more readily in 2KR cells compared to WT cells (p=0.029). The frequency of IR-induced quadriradial chromosome aberrations was 0.033±0.015 per cell in WT cells and 0.073±0.012 per cell in 2KR cells.
Figure 4.
Chromosome analysis of 2KR cells compared with WT and V3 cells. (A, B) Cells were subjected to 0.5 Gy of γ-rays and mitotic cells were analyzed for chromosomal aberrations; (A) total chromosome aberrations and (B) quadriradial-type chromosome aberrations. (*p<0.05)
4. Discussion
For the past decade, extensive work has been performed to identify the regulatory mechanisms governing the activity of DNA-PKcs. A number of groups have revealed that phosphorylation of DNA-PKcs is a key post-translational modification that modulates DNA-PKcs [4, 10]. In this study, we aimed to provide evidence that acetylation regulates DNA-PKcs. We showed that DNA-PKcs is acetylated in vivo in normal cycling cells. Furthermore, blocking potential acetylation at two sites previously identified to be acetylated in a proteomic screen in the C-terminus of DNA-PKcs results in moderate radiosensitivity, a small decrease in DSB repair capacity, and increased chromosomal aberrations. This data shows that mutating the potential acetylation lysines results in attenuated DNA-PKcs dependent DSB repair.
Interacting with the Ku-DNA complex results in a conformational change in the FAT and FATC domains of DNA-PKcs, which is believed to result in the alteration of the catalytic groups and/or the ATP binding pocket of DNA-PKcs, leading to full activation of its kinase activity [36–38]. As lysines 3241 and 3260 are in the FAT domain of DNA-PKcs, we postulated that acetylation of DNA-PKcs at these sites would directly modulate the kinase activity of DNA-PKcs. However, ablating potential acetylation of K3241 and K3260 did not significantly alter the kinase activity of DNA-PKcs as monitored by autophosphorylation of DNA-PKcs. Furthermore, we did not find that mutating these sites results in a defect in the dynamics of DNA-PKcs at laser-generated DSBs. Therefore, enhanced radiosensitivity and reduced DSB repair capacity by mutation at these sites are not due to alterations in the two primary functions of DNA-PKcs.
As lysines K3214 and K3260 do not affect the two primary functions of DNA-PKcs, we postulate that they are required for a secondary function of DNA-PKcs. We hypothesize that they may be required for the ability of DNA-PKcs to modulate HR and/or DSB repair pathway choice. Previously, the phosphorylation status of DNA-PKcs was shown to play a role in regulating the HR pathway [40]. In addition, the role of DNA-PKcs in controlling HR pathway was reported to be dependent on the T2609 phosphorylation cluster [41, 42]. Moreover, one of the pro-HR factors BRCA1 blocks autophosphorylation of DNA-PKcs at the serine 2056 phosphorylation cluster and promotes HR pathway [22]. DSB repair analysis by γH2AX resolution shows that there is a minor difference between 2KR and WT cells in repairing DSBs. However, chromosome analysis in cells exposed to IR showed that 2KR cells had increased chromosome aberrations compared to WT cells, in particular a significant increase in quadriradial-type aberrations was observed in the 2KR cells. The quadriradial-type of chromosome aberration is a chromatid-type aberration, which is frequently observed in cells with HR deficiency [43]. The HR pathway helps to maintain a high level of genome stability, even though immediate cell survival may be only minimally affected in cells with HR deficiency [44]. If 2KR cells have deficiency in regulating HR pathway, the intermediate radiosensitivity and quadriradial chromosomes in 2KR cells may explain why radiosensitivity of 2KR cells are mild compared with NHEJ-deficient V3 cells.
How lysines K3241 and K3260 affect DNA-PKcs-dependent DSB repair remains unclear. It is possible that these two sites are required for the ability of DNA-PKcs to interact with a specific NHEJ/HR factor, required for a subset of NHEJ/HR, or influence the kinase activity of DNA-PKcs moderately. Furthermore, it is possible that these sites are required for DNA-PKcs function that is required for the cellular response to DSBs that is independent of DSB repair. It is also indicated that secondary replication-associated DSBs formed following exposure to IR are major substrates for IR-induced HR repair [45]. DNA-PKcs regulates the replication stress response through Chk1-Claspin pathway [46] and chromosome stability through the Chk2-BRCA1 pathway [47]. Thus, it is possible that the quadriradial chromosomes observed in 2KR cells might be arising from these secondary replication-associated DSBs after IR combined with its deficiency in appropriate control of HR pathway. Altogether, our data suggests that mutating lysines K3241 and K3260 to arginines to potentially blocking acetylation at these residues leads to a mild defect in DSB repair and decreased genomic stability. Undoubtedly, further investigation is required to elucidate the role of these lysine residues and potentially acetylation at these sites in regulating DNA-PKcs.
Supplementary Material
Highlights.
DNA-PKcs is acetylated in cells.
Mutating K3241 and K3260 of DNA-PKcs results in increased radiosensitivity.
Mutating K3241 and K3260 of DNA-PKcs results in genomic instability.
Acknowledgments
The work was supported by the National Institute of Health [CA50519 and CA13499 to DJC] and the Cancer Prevention Research Institute of Texas [RP110465 to DJC].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 2.Davis AJ, Chen DJ. DNA double strand break repair via non-homologous end-joining. Transl Cancer Res. 2013;2:130–143. doi: 10.3978/j.issn.2218-676X.2013.04.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis AJ, Chen BP, Chen DJ. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst) 2014;17:21–29. doi: 10.1016/j.dnarep.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartley KO, Gell D, Smith GC, Zhang H, Divecha N, Connelly MA, Admon A, Lees-Miller SP, Anderson CW, Jackson SP. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 6.Perry J, Kleckner N. The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell. 2003;112:151–155. doi: 10.1016/s0092-8674(03)00033-3. [DOI] [PubMed] [Google Scholar]
- 7.Williams DR, Lee KJ, Shi J, Chen DJ, Stewart PL. Cryo-EM structure of the DNA-dependent protein kinase catalytic subunit at subnanometer resolution reveals alpha helices and insight into DNA binding. Structure. 2008;16:468–477. doi: 10.1016/j.str.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2010;463:118–121. doi: 10.1038/nature08648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis AJ, Lee KJ, Chen DJ. The N-terminal region of the DNA-dependent protein kinase catalytic subunit is required for its DNA double-stranded break-mediated activation. J Biol Chem. 2013;288:7037–7046. doi: 10.1074/jbc.M112.434498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobbs TA, Tainer JA, Lees-Miller SP. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst) 2010;9:1307–1314. doi: 10.1016/j.dnarep.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen BP, Uematsu N, Kobayashi J, Lerenthal Y, Krempler A, Yajima H, Lobrich M, Shiloh Y, Chen DJ. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J Biol Chem. 2007;282:6582–6587. doi: 10.1074/jbc.M611605200. [DOI] [PubMed] [Google Scholar]
- 12.Yajima H, Lee KJ, Chen BP. ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol Cell Biol. 2006;26:7520–7528. doi: 10.1128/MCB.00048-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merkle D, Douglas P, Moorhead GB, Leonenko Z, Yu Y, Cramb D, Bazett-Jones DP, Lees-Miller SP. The DNA-dependent protein kinase interacts with DNA to form a protein-DNA complex that is disrupted by phosphorylation. Biochemistry. 2002;41:12706–12714. doi: 10.1021/bi0263558. [DOI] [PubMed] [Google Scholar]
- 14.Douglas P, Cui X, Block WD, Yu Y, Gupta S, Ding Q, Ye R, Morrice N, Lees-Miller SP, Meek K. The DNA-dependent protein kinase catalytic subunit is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Mol Cell Biol. 2007;27:1581–1591. doi: 10.1128/MCB.01962-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 16.Ma Y, Pannicke U, Lu H, Niewolik D, Schwarz K, Lieber MR. The DNA-dependent protein kinase catalytic subunit phosphorylation sites in human Artemis. J Biol Chem. 2005;280:33839–33846. doi: 10.1074/jbc.M507113200. [DOI] [PubMed] [Google Scholar]
- 17.Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, Chen DJ. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, Botvinick E, Qin J, Chen DJ. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J Biol Chem. 2005;280:14709–14715. doi: 10.1074/jbc.M408827200. [DOI] [PubMed] [Google Scholar]
- 19.Cui X, Yu Y, Gupta S, Cho YM, Lees-Miller SP, Meek K. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol Cell Biol. 2005;25:10842–10852. doi: 10.1128/MCB.25.24.10842-10852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, Rambo RP, Hura GL, Pelikan M, So S, Abolfath RM, Chen DJ, Lees-Miller SP, Tainer JA. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem. 2010;285:1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis AJ, Chi L, So S, Lee KJ, Mori E, Fattah K, Yang J, Chen DJ. BRCA1 modulates the autophosphorylation status of DNA-PKcs in S phase of the cell cycle. Nucleic Acids Res. 2014;42:11487–11501. doi: 10.1093/nar/gku824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang XJ. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays. 2004;26:1076–1087. doi: 10.1002/bies.20104. [DOI] [PubMed] [Google Scholar]
- 24.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 26.Cohen HY, Lavu S, Bitterman KJ, Hekking B, Imahiyerobo TA, Miller C, Frye R, Ploegh H, Kessler BM, Sinclair DA. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 27.Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol Cell Biol. 2007;27:8502–8509. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 30.Whitmore GF, Varghese AJ, Gulyas S. Cell cycle responses of two X-ray sensitive mutants defective in DNA repair. Int J Radiat Biol. 1989;56:657–665. doi: 10.1080/09553008914551881. [DOI] [PubMed] [Google Scholar]
- 31.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 32.Kurimasa A, Kumano S, Boubnov NV, Story MD, Tung CS, Peterson SR, Chen DJ. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol Cell Biol. 1999;19:3877–3884. doi: 10.1128/mcb.19.5.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagasawa H, Little JB, Lin YF, So S, Kurimasa A, Peng Y, Brogan JR, Chen DJ, Bedford JS, Chen BP. Differential role of DNA-PKcs phosphorylations and kinase activity in radiosensitivity and chromosomal instability. Radiat Res. 2011;175:83–89. doi: 10.1667/RR2092.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao Z, Davis AJ, Fattah KR, So S, Sun J, Lee KJ, Harrison L, Yang J, Chen DJ. Persistently bound Ku at DNA ends attenuates DNA end resection and homologous recombination. DNA Repair (Amst) 2012;11:310–316. doi: 10.1016/j.dnarep.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis AJ, So S, Chen DJ. Dynamics of the PI3K-like protein kinase members ATM and DNA-PKcs at DNA double strand breaks. Cell Cycle. 2010;9:2529–2536. doi: 10.4161/cc.9.13.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rivera-Calzada A, Maman JD, Spagnolo L, Pearl LH, Llorca O. Three-dimensional structure and regulation of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) Structure. 2005;13:243–255. doi: 10.1016/j.str.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 37.Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell. 2006;22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 38.Rivera-Calzada A, Spagnolo L, Pearl LH, Llorca O. Structural model of full-length human Ku70-Ku80 heterodimer and its recognition of DNA and DNA-PKcs. EMBO Rep. 2007;8:56–62. doi: 10.1038/sj.embor.7400847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.German J. Bloom syndrome: a mendelian prototype of somatic mutational disease. Medicine (Baltimore) 1993;72:393–406. [PubMed] [Google Scholar]
- 40.Neal JA, Dang V, Douglas P, Wold MS, Lees-Miller SP, Meek K. Inhibition of homologous recombination by DNA-dependent protein kinase requires kinase activity, is titratable, and is modulated by autophosphorylation. Mol Cell Biol. 2011;31:1719–1733. doi: 10.1128/MCB.01298-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang S, Yajima H, Huynh H, Zheng J, Callen E, Chen HT, Wong N, Bunting S, Lin YF, Li M, Lee KJ, Story M, Gapud E, Sleckman BP, Nussenzweig A, Zhang CC, Chen DJ, Chen BP. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J Cell Biol. 2011;193:295–305. doi: 10.1083/jcb.201009074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin YF, Nagasawa H, Little JB, Kato TA, Shih HY, Xie XJ, Wilson PF, Jr, Brogan JR, Kurimasa A, Chen DJ, Bedford JS, Chen BP. Differential radiosensitivity phenotypes of DNA-PKcs mutations affecting NHEJ and HRR systems following irradiation with gamma-rays or very low fluences of alpha particles. PLoS One. 2014;9:e93579. doi: 10.1371/journal.pone.0093579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scully R, Puget N, Vlasakova K. DNA polymerase stalling, sister chromatid recombination and the BRCA genes. Oncogene. 2000;19:6176–6183. doi: 10.1038/sj.onc.1203971. [DOI] [PubMed] [Google Scholar]
- 44.Lowndes NF, Toh GW. DNA repair: the importance of phosphorylating histone H2AX. Curr Biol. 2005;15:R99–R102. doi: 10.1016/j.cub.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 45.Groth P, Orta ML, Elvers I, Majumder MM, Lagerqvist A, Helleday T. Homologous recombination repairs secondary replication induced DNA double-strand breaks after ionizing radiation. Nucleic Acids Res. 2012;40:6585–6594. doi: 10.1093/nar/gks315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin YF, Shih HY, Shang Z, Matsunaga S, Chen BP. DNA-PKcs is required to maintain stability of Chk1 and Claspin for optimal replication stress response. Nucleic Acids Res. 2014;42:4463–4473. doi: 10.1093/nar/gku116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shang Z, Yu L, Lin YF, Matsunaga S, Shen CY, Chen BP. DNA-PKcs activates the Chk2-Brca1 pathway during mitosis to ensure chromosomal stability. Oncogenesis. 2014;3:e85. doi: 10.1038/oncsis.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.