Abstract
DNA insulators organize independent gene regulatory domains and can regulate interactions amongst promoter and enhancer elements. They have the potential to be important in genome enhancing and editing technologies because they can mitigate chromosomal position effects on transgenes. The orthologous genes of the Anopheles stephensi putative gypsy‐like insulator protein complex were identified and expression characteristics studied. These genes encode polypeptides with all the expected protein domains (Cysteine 2 Histidine 2 (C2H2) zinc fingers and/or a bric‐a‐brac/poxvirus and zinc finger). The mosquito gypsy transcripts are expressed constitutively and are upregulated in ovaries of blood‐fed females. We have uncovered significant experimental evidence that the gypsy insulator protein complex is widespread in vector mosquitoes.
Keywords: molecular genetics, gene expression, position effects, evolution
Introduction
DNA insulators comprise nucleotide sequences and associated proteins that regulate interactions amongst promoter and enhancer elements and are able to organize independent gene regulatory domains to prevent inappropriate expression. Chromosomal position effects can be attenuated by flanking genes of interest (for example, transgenes) with insulators to block the effects of neighbouring enhancers and silencers as well as encroaching heterochromatin (Bell et al., 2001). A number of different DNA sequences with insulating activity have been identified in both invertebrate and vertebrate species, including specialized chromatin structures, a portion of the gypsy retrotransposon from the fruit fly, Drosophila melanogaster, sites in the sea urchin histone H3 genes silencing nucleoprotein structure, human matrix attachment regions, the chicken β‐globin genes chicken hypersensitive site‐4 element, the Xenopus ribosomal RNA genes, the human T‐cell receptor‐α/δ locus and the CCCTC‐binding factor (CTCF) factor (Udvardy et al., 1985; Geyer & Corces, 1992; Chung et al., 1993; Palla et al., 1997; Robinett et al., 1997; Zhong & Krangel, 1997; Namciu et al., 1998; Moon et al., 2005).
A large part of the current understanding of insulators in insects is derived from studies of the D. melanogaster suppressor of hairy‐wing [Su(Hw)] complex. The Su(Hw) insulator (also known as the gypsy insulator) phenotype results from a nucleotide sequence of ∼350 base pairs (bp) in length derived from the DNA adjacent to the 3′‐end of the 5′‐end long terminal repeat in the gypsy retrotransposon and a number of host‐derived DNA binding proteins (Gerasimova & Corces, 1998; Gerasimova et al., 2000; Byrd & Corces, 2003). The gypsy insulator complex is proposed to regulate gene expression by establishing higher‐order domains of chromatin structure and blocking interference of nearby enhancers or repressors (Gaszner & Felsenfeld, 2006; Markstein et al., 2008; Bushey et al., 2009). Su(Hw) complex insulator function requires the recruitment of three proteins: Su(Hw), modifier of Mobile dispersed genetic element 4 (mdg4)2.2 [Mod(mdg4)2.2] and centrosomal protein of 190 (CP190) (Parkhurst et al., 1988; Georgiev & Gerasimova, 1989; Spana & Corces, 1990; Pai et al., 2004). The Su(Hw) protein has a domain of 12 zinc‐finger (ZF) motifs, some of which are involved in recognizing and binding the gypsy insulator DNA sequence (Harrison et al., 1993; Kim et al., 1996). Mutations in the Su(Hw) gene cause female sterility but do not result in lethality.
Su(Hw) interacts with the other two protein components of the insulator, Mod(mdg4)2.2 and CP190. Mod(mdg4)2.2 is one of a number of isoforms encoded by the mod(mdg4) gene, and the protein is translated following trans‐splicing to join exons from two different primary transcripts (Labrador et al., 2001; Mongelard et al., 2002). The third protein, CP190, was named originally DMAP190 (Drosophila microtubule associated protein of 190) because it was identified first by microtubule affinity chromatography (Jimenez & Goday, 1993). It has three ZF motifs in its central region that are potentially capable of interacting with DNA. However, a consensus DNA binding sequence has not been identified. Whereas CP190 is essential for viability [homozygous mutant fruit flies die during late stages of pupal development (Oegema et al., 1995; Butcher et al., 2004)], mutations in the gene cause no detectable defects in mitosis or centrosome structure. CP190 is essential for Su(Hw) insulator function and interacts with both Su(Hw) and Mod(mdg4)2.2 proteins as well as itself (Pai et al., 2004).
Little is known about insulators in mosquitoes. Gray & Coates (2005) cloned the cDNAs for two putative Aedes aegypti CTCF proteins expressed constitutively in all life stages. We have data that support the presence of Su(Hw) or gypsy‐like insulator complexes in the Asian malaria vector mosquito, Anopheles stephensi (Carballar‐Lejarazú et al., 2013). These data include evidence of both positive and negative influences of exogenously derived gypsy sequences on expression levels of reporter genes. The Ty3/gypsy elements are one of the most abundant and diverse long terminal repeat‐retrotransposon (LTR) families in the An. gambiae genome (Tubio et al., 2011), but LTR‐retrotransposon families represent only ∼0.7% of the An. stephensi genome (Jiang et al., 2014).
It is not known whether the gypsy DNA sequences represent components of a canonical insulating system in insects. The existing data support the hypothesis that the insulator DNA and corresponding interacting proteins most probably originated in ancestral insect genomes and were acquired later by the gypsy retrotransposons. This hypothesis receives additional, although indirect, support from the discovery of putative Su(Hw), mod(mdg4)2.2 and CP190 orthologous genes in the genomes of four mosquito species (Schoborg & Labrador, 2010; R. Carballar‐Lejarazú et al., unpubl. data). We analyse here the mosquito orthologous genes of the putative Su(Hw)‐like insulator complex in An. stephensi. These studies contribute to the molecular tool set needed to explore the extent to which this specific system represents a fundamental property of genome organization, at least in insects, and how it can be used in both basic and applied studies of mosquito gene expression.
Results and discussion
Primary structures of the genes, transcripts and putative proteins of the An. stephensi Su(Hw)‐like complex components
Heterologous D. melanogaster Su(Hw), mod(mdg4)2.2 and CP190 DNA sequences were used in reciprocal BLAST searches of the whole genome of the An. stephensi Indian strain (Assembly: AsteI2; Vectorbase) to find putative orthologues. Sequence similarity supports designating the genes ASTEI00266, ASTEI03335 and ASTEI00117 as As‐Su(Hw), As‐mod(mdg4) and As‐CP190, respectively. Gene‐specific primers were used in amplification studies [rapid amplification of cDNA ends (RACE) and primer walking] to determine the As‐Su(Hw), As‐mod(mdg4) and As‐CP190 primary sequence structures, the complete sequences of their corresponding transcripts and the putative alternative transcripts for each gene. Predicted amino acid (aa) sequence alignments showed 36.8, 35 and 47.9% identity between An. stephensi Su(Hw), mod(mdg4) and CP190, respectively, with the putative orthologous proteins in D. melanogaster (Fig. S2).
As‐Su(Hw) is a single‐copy gene with five exons (270, 563, 1060, 610, 1998 bp in length), four introns (165, 75, 68, 68 bp), a 348‐ and 246‐bp 5′ and 3′ untranslated region (UTR), respectively, and encodes a single transcript (Figs 1, 2A, S1A, B). A protein of ∼103 kDa was detected in ovaries 48 h post‐bloodmeal (48 hPBM; Fig. 2B). The ZF domain constitutes an ancient DNA‐binding motif present in all eukaryotes and some Archaea (Bouhouche et al., 2000), and the C2H2 ZF in particular is the most common DNA‐binding motif of eukaryotic transcription factors (Clarke & Berg, 1998; Tadepally et al., 2008). The Su(Hw) ZF domains are located in the central portion of the protein (Parkhurst et al., 1988). Nine of the 12 ZFs and a domain of ∼150 aa including the C‐terminal leucine zipper, but not the amino (N)‐ and carboxyl (C)‐terminal acidic regions, are required for enhancer blocking (Harrison et al., 1993; Kim et al., 1996). The An. stephensi Su(Hw) putative protein contains the 12 C2H2 ZF motifs located between exon 3 and exon 4 (Fig. 1) and has the zinc‐coordinating residues (Fig. S3). These ZF domains are conserved amongst all the mosquito species included in the alignment analysis despite the significant differences in the N‐terminal and C‐terminal regions of the protein. However, there was only 56% identity and 38% similarity across all ZF domains.
Figure 1.
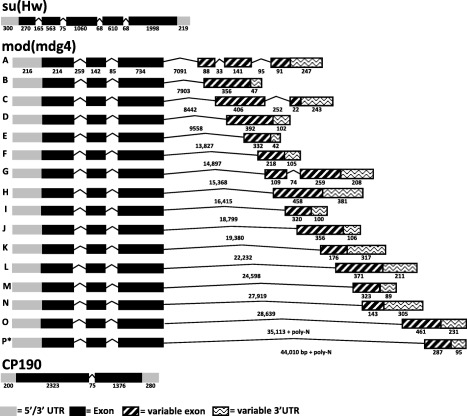
Gene structures of orthologues of the suppressor of hairy‐wing [Su(Hw)] insulator complex in Anopheles stephensi (As). Schematic representations of transcript products for As‐Su(Hw), As‐modifier of mobile dispersed genetic element 4 (mod[mdg4]) and As‐centrosomal protein of 190 (As‐CP190) detected in samples of ovaries 48 h post‐bloodmeal. Exons and introns are represented by boxes and lines, respectively; length in nucleotides is indicated below each. Abbreviation: UTR, untranslated region.
Figure 2.
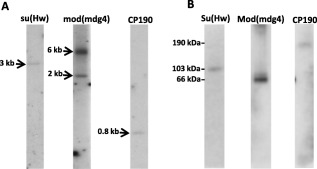
Gene copy number and protein expression of suppressor of hairy‐wing [Su(Hw)], modifier of mobile dispersed genetic element 4 (mod[mdg4]) and centrosomal protein of 190 (CP190) in Anopheles stephensi. (A) Southern blot analyses were used to determine the copy number of each gene. Genomic DNA samples were digested with EcoRI and hybridized with Su(Hw)‐, mod(mdg4)‐ and CP190‐specific probes. Approximate fragment lengths in kb are indicated to the left of each image. (B) Immunoblot analyses of ovaries 48 h post‐bloodmeal extracts were used to determine Su(Hw), Mod(mdg4) and CP190 protein expression. Proteins were detected with Drosophila polyclonal anti‐Su(Hw), anti‐Mod and anti‐CP190 antibodies. Approximate weights in kDa are indicated to the left of the image.
Su(Hw) orthologues are present in all arthropod groups with sequenced genomes; however, their absence in nematodes (sister phylum to arthropods) supports the conclusion that the gene appeared first in a common ancestor at the base of arthropods or close to this base (Heger et al., 2013). As‐Su(Hw) is less complex than the D. melanogaster ortholog, the latter of which transcribes three different RNAs of 3.3, 1.4, and 1.1 kb in length (Parkhurst et al., 1988). Only the 3.3 kb transcript is involved in gypsy insulator function. Furthermore, the D. melanogaster alternative transcripts have nine variable introns, nine alternative exons and a variable 5′ UTR (flybase.org). Functions associated with these alternative forms are unknown.
mod(mdg4), also known as enhancer of position‐effect variegation E(var)3‐93‐D, was first described in D. melanogaster as a modifier of the effects of the insertion of the mdg4 retrotransposon into the yellow locus (Georgiev & Gerasimova, 1989). The fruit fly locus encodes a large number of splice‐variants, each having the same conserved 5′‐end region and a transcript‐specific 3′‐end region (Krauss & Dorn, 2004). Concordantly, the protein isoforms derived from these transcripts are characterized by a common N‐terminus of ∼400 aa, including the broad‐complex, tramtrack, and bric‐a‐brac/poxvirus and zinc finger (BTB/POZ) domains, which facilitate protein‐to‐protein homodimerization (Büchner et al., 2000; Krauss & Dorn, 2004). The C‐termini of the proteins are variable and share little sequence similarity with each other.
As‐mod(mdg4) exhibits some of the same complexity seen with the fruit fly gene and has four exons and three introns (Fig. 1). The conserved 5′‐end region of the An. stephensi transcripts comprises three exons (214, 142 and 734 bp in length) and two introns (259 and 85 bp), and has the same number of exons and introns annotated in the An. gambiae 5′‐end region (three exons: 253, 143 and 730 bp; two introns: 248 and 102 bp). The length of the third intron, which determines the alternative splicing and ultimate identity of the transcript, varies in An. stephensi from 7091 to > 44 010 bp in length and from 2500 to 37 000 bp in An. gambiae.
Alternative splicing of the As‐mod(mdg4) product results in 16 different transcripts (Fig. 1). Thirteen of the 16 have a single exon that constitutes its transcript‐specific 3′‐end region, whereas 17 of the 20 mod(mdg4) transcripts in An. gambiae annotated in Vectorbase have the same attribute. These similarities support a high extent of conservation between the An. stephensi and An. gambiae mod(mdg4) genes. Finally the As‐mod(mdg4) 5′‐end UTR is 216 bp in length and is conserved amongst the 16 transcripts, similar to the ∼140 bp 5′‐end UTR seen in An. gambiae. Despite all of the transcripts having a uniform 5′‐end UTR, all of the As‐mod(mdg4) transcripts have a variable 3′‐end UTR that ranges from 47 to 380 bp (Fig. S1A, B).
As‐mod(mdg4) encodes transcripts in the size range of 0.5 to 1.8 kb and these are enriched in ovaries from blood‐fed females (Fig. 3). All alternative transcripts are located on the An. stephensi supercontig APCG01002702.1, where the annotated As‐mod(mdg4) gene maps. These results confirm the existence of alternative cis‐splicing in An. stephensi and the consequent existence of multiple splice‐variants.
Figure 3.
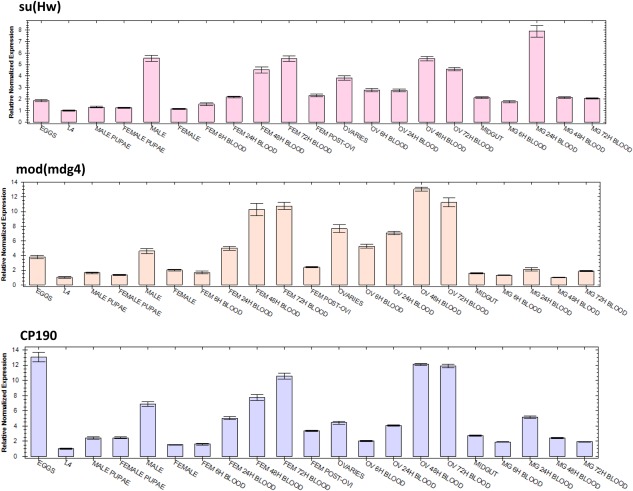
Abundance profiles of Anopheles stephensi suppressor of hairy‐wing [As‐Su(Hw)], As‐modifier of mobile dispersed genetic element 4 (mod[mdg4]) and As‐centrosomal protein of 190 (As‐CP190) transcripts during development. Each histogram represents data of three biological replicates normalized using ribosomal gene S7 transcript abundance as a reference (average ± SEM). Eggs were collected 2 h following oviposition. Sugar‐fed adult females and males were collected 5 days post‐eclosion. Abbreviations: BLOOD, hours after bloodmeal; L4, fourth‐instar larvae; FEM, female; FEM POST‐OVI, females post egg laying; OV, ovaries; MG, midgut.
The sequence and length of the introns and exons corresponding to each splice‐variant were aligned with the conserved region to recreate the exact sequence of each complete As‐mod(mdg4) transcript (Table S2). All of the transcripts were analysed for stop codons and polyadenylation signals, allowing for reconstruction of the gene models (Fig. 1, Table S2). All of the identified transcripts have the conserved exons 1 and 2 in which the BTB/POZ domain is located (Figs 1, S3). A 5′ RACE PCR using a reverse primer designed to anneal to the conserved region of the transcripts was performed to confirm the lack of any splice‐variation in the 5′‐end region of the transcripts. Only one 5′ UTR sequence was obtained, confirming that there is no splice‐variation within the 5′‐end region of the As‐mod(mdg4) locus (data not shown).
Interestingly, Southern‐blot analysis supports a duplication of the As‐mod(mdg4) gene in An. stephensi (Fig. 2A). There is no evidence of a duplication of this locus in Drosophila or other insects (Krauss & Dorn, 2004). Further BLAST analysis using the Southern‐blot probe sequence showed no additional complementary DNA sequences with e‐values ≤ 0.13, supporting the interpretation that the lower molecular‐weight species observed in the Southern‐blot analysis is the result of nonspecific annealing. The conserved region of the As‐mod(mdg4) gene sequence was determined by a sequencing primer‐walking strategy, and this allowed the reconstruction of a 9.4 kb genomic DNA fragment for the shortest isoform [As‐mod(mdg4)‐A] and up to a 46 kb genomic DNA fragment for the longest isoform [As‐mod(mdg4)‐P].
A single Mod(Mdg4) polypeptide of ∼66 kDa was detected in ovaries using Drosophila antibodies from blood‐fed females (48 hPBM; Fig. 2B). Mod(Mdg4) protein isoforms are characterized by a common N‐terminus of ∼400 aa, including the BTB/POZ‐domain that facilitates protein‐to‐protein homodimerization (Büchner et al., 2000). Anopheles stephensi Mod isoform C was used for protein domains conservation analysis amongst vector mosquitoes (Fig. S3). A BTB/POZ‐domain 97 aa in length is located at aa 32 to 124 in all the analysed sequences, and has 85.6% identity amongst all the mosquito sequences.
Analyses in silico support the presence of 41 and 31 transcript splice variants in An. gambiae and D. melanogaster, respectively (Krauss & Dorn, 2004), whereas 3′ RACE PCR verified 16 for As‐mod(mdg4) in our study (Fig. S1A, B). These differences could be the result of experimental limitations of the isolation of the transcripts via the 3′ RACE PCR or reflect a lower number of As‐mod(mdg4) transcripts. Eight of the 16 transcript‐specific regions had significant sequence similarity to An. gambiae transcript‐specific regions and this supports the conclusion that although the general structure of the mod(mdg4) loci are conserved between the two species, there are a number of unique transcripts present in An. stephensi that are absent in the closely related species. Furthermore, little is known of the function of the products of these splice‐variants. Given the conservation of the locus and its large number of transcripts across millions of years of evolution, it is likely that these proteins play an important role in mosquito biology, but only one of these, the Mod(mdg4) protein, is present in the Su(Hw) insulator.
As‐CP190 is a single‐copy gene comprising two exons (1376 and 2323 bp) and one intron (75 bp), and encodes a single transcript of 3 kb, including a 200‐bp 5′‐ and 280‐bp 3′‐end UTR (Figs 1, 2 S1A, B). This transcript is translated into a 190 kDa protein detected in ovaries 48 hPBM (Fig. 2B). Although CP190 was identified and characterized initially in D. melanogaster as a result of its association with centrosomes and microtubules, later studies showed it to be localized in the nucleus and binding to specific sites on polytene chromosomes, consistent with a role in the interphase nuclei (Whitfield et al., 1995). Although CP190 does not bind directly to DNA, it is required for Su(Hw)‐dependent gypsy and Drosophila CTCF (dCTCF)‐dependent Frontabdominal‐8 (Fab‐8) insulator function (Pai et al., 2004; Mohan et al., 2007).
Analysis of the As‐CP190 conceptual translation product shows that the BTB/POZ domain comprises position 33–130 aa, with an identity of 88.7% amongst all the mosquito species (Fig. S3). Interestingly, four C2H2 ZF motifs were predicted in three of the mosquito species (An. stephensi, An. gambiae, Ae. aegypti). Culex quinquefasciatus, which has only three ZF motifs, has all of the zinc‐coordinating residues and 85% identity and 15% similarity with the other species. The D. melanogaster CP190 protein has BTB/POZ, D‐rich and E‐rich domains essential for its association with insulator subclasses and insulator function (Ahanger et al., 2013). Furthermore, the E‐rich region is important for the disassociation of CP190 from the chromosome during heat‐shock, which may provide a mechanism for regulating insulator function (Oliver et al., 2010).
Stage‐ and tissue‐specific transcript abundance
The differences in mRNA abundance levels for As‐Su(Hw), As‐mod(mdg4) and As‐CP190 were detected and measured by quantitative real‐time PCR (qPCR) at embryonic, larval, pupal and adult developmental stages, and in ovaries and midguts of adult females at different times (6, 24, 48 and 72 h) after a bloodmeal (Fig. 3, Table S3). In general, all three genes are upregulated in adult females and ovaries following a bloodmeal when compared with adult sugar‐fed females. Interestingly, the three transcripts are expressed four, two and fivefold higher for As‐Su(Hw), As‐mod(mdg4) and As‐CP190, respectively, in adult males than in adult females (sugar‐fed) (P = 0.001, 0.814, 0.000 respectively); and As‐Su(Hw) is the only gene with transcripts that are upregulated in the midgut 24 hPBM (P = 0001) (Fig. 3, Table S3).
As‐Su(Hw) transcript accumulation in adult females was fourfold higher at 72 hPBM when compared with sugar‐fed females (P = 0.001) and a similar trend was observed in ovaries, twofold higher at 48 hPBM (P = 0.028) (Fig. 3, Table S3). This expression profile is consistent with Drosophila microarray data that show a high expression of Su(Hw) products in the ovaries; however, we did not find the high expression in embryos as reported in the fruit fly (http://flybase.org/reports/FBgn0003567.html). The function of Su(Hw) in female germline development is not well understood, although new evidence supports distinct roles for germline function and insulator activity. Soshnev et al. (2012) demonstrated that Su(Hw) binding is constitutive during development, supporting the hypothesis that its function in ovaries is not tissue‐specific. Based on these data, they propose that Su(Hw) may not play a global architectural role in establishing genome regulation important for oogenesis but that it may be required generally for establishment of domain boundaries that permit appropriate gene expression.
Quantitative transcript accumulation for As‐mod(mdg4) using specific primers for the modC isoform [BLAST analysis supports the conclusion that this isoform is equivalent to the mod(mdg4) isoform involved in insulator function in D. melanogaster; however functional experiments are needed to confirm this] showed that it is most abundant in ovaries 48 hPBM (sixfold higher than ovaries from sugar‐fed females, P = 0.000) and in adult females at 24 and 48 hPBM (eightfold higher than sugar‐fed females, P = 0.371, 0.000 respectively) (Fig. 3, Table S3). mod(mdg4) is expressed at high levels during Drosophila oogenesis (peak transcript accumulation is observed within 0–12 h post egg deposition during the embryonic stages), and its accumulation in all larval and adult organs/tissues ranges from moderate to undetectable, with the exception of the ovaries, adult male accessory gland and larval salivary glands (http://flybase.org/reports/FBgn0002781.html). However, we were not able to detect a high abundance of the As‐mod(mdg4) transcripts in embryos. In addition to the qPCR data, Reverse transcriptase‐PCR (RT‐PCR) performed with primers specific for each isoform determined the relative abundance of all the 16 As‐mod(mdg4) isoforms (Fig. 4). Accumulation of all isoforms ranged from low to high in ovaries 48 hPBM, and low to undetectable in the midgut, with the exception of the MP54 isoform, which is the only isoform that accumulates highly in the midgut before and after blood feeding. Isoform A showed a stage‐specific high accumulation in female pupae and was low to undetectable in all the remaining stages or tissues. It is clear that all the mod(mdg4) isoforms have characteristic expression profiles in different developmental stages and tissues, and functional experiments are needed to determine the biological role of each and their associated phenotypes.
Figure 4.
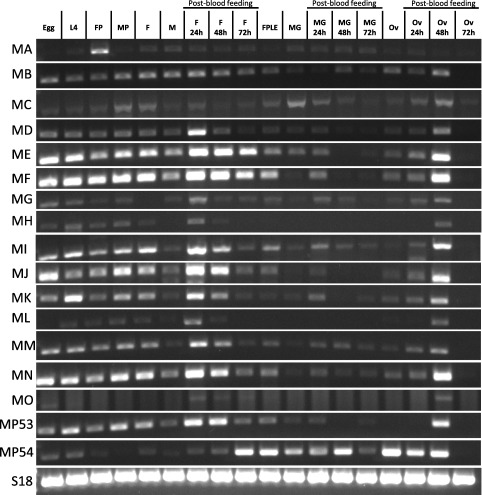
Developmental expression profiles of As‐mod(mdg4) isoforms in Anopheles stephensi. The abundances of As‐modifier of mobile dispersed genetic element 4 (mod[mdg4]) isoforms were analysed using RNA isolated from multiple individuals at each of the indicated stages: L4, fourth‐instar larvae; FP, female pupae; MP, male pupae; F, adult female; M, male; FPLE, females post egg laying; MG, midgut; Ov, ovaries. Adult females and tissues (ovaries and midguts) were analysed at different time points after a bloodmeal (24, 48 and 72 h). The small ribosomal protein gene 18 (S18), was used as loading control.
As‐CP190 transcripts accumulated to their highest levels in the embryos (Fig. 3, Table S3). Ovaries from 48 and 72 hPBM had eightfold more transcript accumulation than those from sugar‐fed females (P = 0.431, 0.186 respectively). Adult females after 72 hPBM had a ninefold increase of AsCP190 transcripts when compared with sugar‐fed females (P = 0.003; Fig. 3, Table S3). CP190 in D. melanogaster has its peak accumulation in embryonic stages and in the adult ovary, and nearly all larvae and adult tissues/organs, including adult midgut, accumulate it at moderate levels (flybase.org/reports/FBgn0000283.html).
The presence of large amounts of Su(Hw), Mod(mdg4) and CP190 proteins in D. melanogaster in all stages of oogenesis and early embryogenesis supports a strong maternal component. Their presence in both nurse and follicle cell nuclei, and their proposed roles as general transcriptional regulators, support the hypothesis that they are required for control of maternal genes during oogenesis. Mod(mdg4) protein does not become localized in nuclei in the embryo until cleavage cycle 9, further arguing against a function in chromatin organization during early cleavage cycles (Büchner et al., 2000). The An. stephensi transcription profiles are consistent with these data and their interpretation.
The gypsy transposon insulator DNA contains 12 degenerate binding sites for the Su(Hw) protein, with each site comprising a 12‐bp core sequence (5′‐YRYTGCATAYYY‐3′; Smith & Corces, 1995; Parnell et al., 2006). Furthermore, Parnell et al. (2006) demonstrated that the Su(Hw) protein associates with > 500 non‐gypsy transposon‐associated genomic sites on Drosophila polytene chromosomes, and other studies have shown that clusters of Su(Hw)‐binding sites are located mostly in intergenic regions or in introns of large genes (eg Ramos et al., 2006). Furthermore, the DNA‐binding protein Su(Hw) shows unique distribution patterns with respect to the location and expression level of genes, and moreover, Su(Hw) and CP190 have cell line‐specific localization sites (Bushey et al., 2009). At present, there is no information about the presence or minimum sequence requirements for endogenous Su(Hw) binding sites or any other insulator sequence in mosquitoes. Future experiments will focus on determining the chromosome and genome binding pattern of these insulator proteins to map the landscape of insulator genomic interactions in vector mosquitoes.
Experimental procedures
Mosquito rearing and maintenance
The Indian strain of An. stephensi (Jiang et al., 2014) bred in our insectary for > 7 years was used in the experiments. The mosquitoes were maintained at 26 ± 1°C with 77% humidity and 12 h day/night, 30 min dusk/dawn lighting cycle. Larvae were fed a diet of powdered fish food (Tetramin, Blacksburg, VA, USA) mixed with yeast. Adults were provided ad libitum with water and a 10% w/v sucrose solution.
Isolation of DNA, RNA and preparation of cDNA by reverse‐transcription
RNA was extracted from mosquito samples (10 adult females, 15 adult males, 30 ovaries, 30 midguts and 20 larvae) using a RNA Clean and Concentrator kit (Zymo Research, Irvine, CA, USA). A total of 0.5 μg of RNA was used for reverse transcription in reaction volumes of 20 μl using qScript cDNA SuperMix (Quanta Biosciences, Gaithersburg, MD, USA). DNA extraction was performed on samples of 10 adult sugar‐fed females using a Wizard Genomic DNA Extraction kit (Promega, Madison, WI, USA).
Quantitative RT‐PCR analysis
mRNA quantification was performed on a CFX96 Real‐Time PCR Detection System (Bio‐Rad, Hercules, CA, USA). Oligonucleotide primers (Table S1) were designed using Prime3 software, Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi), GraphPad Prism (GraphPad Software, Inc; La Jolla, CA, USA). Values were normalized to mRNA abundance levels of the An. stephensi Small ribosomal 7 (S7) gene with primers that amplify an 84‐bp product (Brown et al., 2003). Gene expression was quantified using gene‐specific primers in a 20 μl final reaction volume containing 10 μl SoFast EvaGreen SuperMix (Bio‐Rad), 150 nM each of the forward and reverse primers, and 5.0 μl cDNA sample. The amplification protocol consisted of 30 s at 95°C, followed by 40 cycles of amplification (95°C for 5 s, 63°C for 5 s), the plate read for SYBR Green I fluorescence, after which a melting‐curve reaction was conducted from 65 to 95°C with plate readings every 0.5°C. Measurements of mRNA abundance were taken in triplicate and their mean used for further analyses. A negative control (no cDNA) and an RNA sample without a reverse transcriptase step (to determine genomic DNA contamination) were included in each run. GraphPad Prism software was used to calculate statistical significance using t‐tests. P‐values ≤ 0.05 were considered significant.
Primer‐walking sequencing of As‐Su(Hw), As‐mod(mdg4) and As‐CP190
Reciprocal BLAST was performed with the Drosophila Su(Hw) complex genes against the An. stephensi genome annotated in Vectorbase (www.vectorbase.org). Primers for genomic and RT‐PCR amplification of segments of the An. stephensi genes were designed to anneal to regions with high sequence identity to the An. gambiae orthologues (Table S1). These primers were used in a walking strategy to sequence the gene structure and open reading frame (ORF) for all three genes. Sequencing of the amplification products was performed by Laguna Scientific (Irvine, CA, USA) using the M13F and M13R universal primers, and reconstruction of the complete gene structure and ORF of each gene was carried out using SeqMan software (www.DNASTAR.com).
RACE in An. stephensi
RACE was performed using the Clontech protocol and a SMARTer RACE kit (Clontech, Mountain View, CA, USA). RACE‐ready cDNA was performed using the Clontech reagents and RNA collected from ovaries dissected 48 hPBM. RACE reactions were performed using Phusion High Fidelity Master Mix from New England Biosystems (Beverly, MA, USA). Nested RACE reactions were performed with touchdown PCR cycles on separate preparations of 5′‐ and 3′‐end cDNA templates as described in the Clontech protocol. Primers were designed using Prime3 software (Table S1). RACE and nested RACE products were run on agarose gels, and fragments extracted and cloned into the pCR‐Blunt II TOPO plasmid (ThermoFisher Scientific, Grand Island, NY, USA). Plasmids were amplified in chemically competent TOP10 Escherichia coli and analysed by sequencing using M13 forward and reverse primers.
Southern blot analyses
Standard Southern blotting and hybridization techniques to detect gene copy number included digesting genomic DNA samples with EcoRI, membrane transfer, and hybridization with a 32P‐labelled probe complementary to Su(Hw)‐, mod‐ or CP190‐encoding DNA.
Immunoblot analyses
Mosquito ovaries dissected at 48 hPBM were lysed in ice‐cold buffer [50 mM Tris, pH 7.8; 150 mM NaCl; 1% IGEPAL CA360 (Sigma, St Louis, MO, USA)] with Complete Protease Inhibitor Cocktail (Roche, Indianapolis, IN, USA) and 1 mM phenylmethylsulfonyl fluoride (PMSF). Total cell lysates were separated by 10% sodium dodecyl sulphate polyacrylamide gel electrophoresis and the gel electroblotted to a polyvinylidene fluoride (PVDF) membrane in 1× Towbin buffer (Bio‐Rad, Hercules, CA, USA) buffer. Following protein transfer, the membrane was washed twice for 10 min in 1× Tris‐buffered saline (TBS) buffer (10 mM Tris‐HCl, pH 7.5; 150 mM NaCl), and incubated in blocking buffer (5% non‐fat dry milk, 1× TBS, 0.05% Tween‐20 [St. Louis, MO, USA]) overnight at 4°C. The membrane was washed twice for 15 min in 1× TBST (1× TBS, 0.05% Tween‐20) and incubated for 1 h at room temperature on an orbital shaker with Drosophila polyclonal antisera (provided by Dr Victor Corces, Emory University, Atlanta, GA, USA) diluted 1:2000 [Su(Hw)], 1:1000 (Mod) and 1:10 000 (CP190) in blocking buffer. After antibody binding, the blot was washed three times in 1× TBST for 15 min. Anti‐rabbit Immunoglobulin G (IgG), fragment crystallizable region (Fc region), horseradish peroxidase (HRP) conjugate (Promega) was diluted 1:10 000 in blocking buffer and the blot incubated for 1 h at room temperature on an orbital shaker. The blot was washed five times for 10 min each in 1× TBST, incubated for 5 min in SuperSignal West Pico Chemiluminescent (ECL) HRP substrate (ThermoFisher Scientific, Grand Island, NY, USA) and exposed to BioMax maximum resolution (MR) film (KODAK, Rochester, NY, USA) for 1–20 min.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's web‐site:
Figure S1. Rapid amplification of cDNA ends (RACE) products and reconstructed suppressor of hairy‐wing [Su(Hw)] insulator transcripts. (A) RACE amplification products for Su(Hw), modifier of mobile dispersed genetic element 4 (mod[mdg4]) and centrosomal protein of 190 (CP190). All genes had a single transcript for the 5′ and 3′ RACE reactions, with the exception of mod(mdg4) 3′ RACE, which had nine groups with varying molecular weights. All groups were cloned and sequenced. The following Anopheles stephensi mod(mdg4) isoforms were obtained for each group: group 1: isoform C; group 2: isoform H; group 3: isoform O; group 4: isoforms A, D, L; group 5: isoform G; group 6: isoforms K, N; group 7: isoforms I, M; group 8: isoforms E, J; group 9: isoforms F, P. (B) RACE and walking primer sequencing data from Su(Hw), mod(mdg4) and CP190 from An. stephensi. The 5′ and 3′ untranslated region sequences are highlighted in yellow and bold letters indicate exon–exon junctions.
Figure S2. Alignment of the suppressor of hairy‐wing [Su(Hw)] protein complex. Alignment of predicted amino acid sequences encoding Su(Hw), Modifier of mobile dispersed genetic element 4, Mod(mdg4) and centrosomal protein of 190 (CP190) from Anopheles stephensi and Drosophila melanogaster. The red colour indicates highly conserved residues and the blue indicates less conservation as determined by alignment using Cobalt (http://www.st‐va.ncbi.nlm.nih.gov/tools/cobalt).
Figure S3. Predicted protein domains encoded by the gene orthologues of the suppressor of hairy‐wing [Su(Hw)] insulator complex in mosquitoes. Protein sequences were aligned using the ClustalW algorithm. Identical and highly conserved residues are highlighted in grey. The zinc‐coordinating residues are indicated in red. Abbreviations: STEPHE, Anopheles stephensi; GAMBIAE, Anopheles gambiae; AEDES, Aedes aegypti; CULEX, Culex quinquefasciatus.
Table S1. Oligonucleotide primers.
Table S2. Detailed analysis of the transcript‐specific sequences of the Anopheles stephensi mod(mdg4) isoforms.
Table S3. Transcript abundance delta‐delta quantification cycle (ΔΔCq) and statistical analyses of differences in Anopheles stephensi suppressor of hairy‐wing [As‐Su(Hw)], As‐modifier of mobile dispersed genetic element 4 (mod[mdg4]) and As‐centrosomal protein of 190 (As‐CP190) mRNA abundance in embryonic, larval, pupal and adult developmental stages, and in ovaries and midguts of adult females.
Acknowledgements
We thank Aniko Fazekas for assistance with mosquito rearing. This research was supported in part by a grant from the National Institutes of Health (NIH), National Institute of Allergy and Infectious diseases (NIAID) AI29746.
References
- Ahanger, S.H. , Shouche, Y.S. and Mishra, R.K. (2013) Functional sub‐division of the Drosophila genome via chromatin looping. Nucleus 4: 115–122. doi: 10.4161/nucl.23389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, A.C. , West, A.G. and Felsenfeld, G. (2001) Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science 291: 447–450. [DOI] [PubMed] [Google Scholar]
- Bouhouche, N. , Syvanen, M. and Kado, C.I. (2000) The origin of prokaryotic C2H2 zinc finger regulators. Trends Microbiol 8: 77–81. [DOI] [PubMed] [Google Scholar]
- Brown, A.E. , Bugeon, L. , Crisanti, A. and Catteruccia, F. (2003) Stable and heritable gene silencing in the malaria vector Anopheles stephensi . Nucleic Acids Res 31: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büchner, K. , Roth, P. , Schotta, G. , Krauss, V. , Saumweber, H. , Reuter, G. et al (2000) Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila . Genetics 155: 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey, A.M. , Ramos, E. and Corces, V.G. (2009) Three subclasses of a Drosophila insulator show distinct and cell type‐specific genomic distributions. Genes Dev 23: 1338–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher, R.D. , Chodagam, S. , Basto, R. , Wakefield, J.G. , Henderson, D.S. , Raff, J.W. et al (2004) The Drosophila centrosome‐associated protein CP190 is essential for viability but not for cell division. J Cell Sci 117: 1191–1199. [DOI] [PubMed] [Google Scholar]
- Byrd, K. and Corces, V.G. (2003) Visualization of chromatin domains created by the gypsy insulator of Drosophila . J Cell Biol 162: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballar‐Lejarazú, R. , Jasinskiene, N. and James, A.A. (2013) Exogenous gypsy insulator sequences modulate transgene expression in the malaria vector mosquito, Anopheles stephensi . Proc Natl Acad Sci USA 110: 7176–7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, J.H. , Whiteley, M. and Felsenfeld, G. (1993) A 5′ element of the chicken beta‐globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila . Cell 74: 505–514. [DOI] [PubMed] [Google Scholar]
- Clarke, N.D. and Berg, J.M. (1998) Zinc fingers in Caenorhabditis elegans: finding families and probing pathways. Science 282: 2018–2022. [DOI] [PubMed] [Google Scholar]
- Gaszner, M. and Felsenfeld, G. (2006) Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 7: 703–713. [DOI] [PubMed] [Google Scholar]
- Georgiev, P.G. and Gerasimova, T.I. (1989) Novel genes influencing the expression of the yellow locus and mgd4 (gypsy) in Drosophila melanogaster . Mol Gen Genet 220: 121–126. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T.I. and Corces, V.G. (1998) Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell 92: 511–521. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T.I. , Byrd, K. and Corces, V.G. (2000) A chromatin insulator determines the nuclear localization of DNA. Mol Cell 6: 1025–1035. [DOI] [PubMed] [Google Scholar]
- Geyer, P.K. and Corces, V.G. (1992) DNA position‐specific repression of transcription by a Drosophila zinc finger protein. Genes Dev 6: 1865–1873. [DOI] [PubMed] [Google Scholar]
- Gray, C.E. and Coates, C.J. (2005) Cloning and characterization of cDNAs encoding putative CTCFs in the mosquitoes, Aedes aegypti and Anopheles gambiae . BMC Mol Biol 6: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, D.A. , Gdula, D.A. , Coyne, R.S. and Corces, V.G. (1993) A leucine zipper domain of the suppressor of Hairy‐wing protein mediates its repressive effect on enhancer function. Genes Dev 7: 1966–1978. [DOI] [PubMed] [Google Scholar]
- Heger, P. , George, R. and Wiehe, T. (2013) Successive gain of insulator proteins in arthropod evolution. Evolution 67: 2945–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, X. , Peery, A. , Hall, B.A. , Sharma, A. , Chen, X.G. , Waterhouse, R.M. et al (2014) Genome analysis of a major urban malaria vector mosquito, Anopheles stephensi . Genome Biol 15: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez, M. and Goday, C. (1993) A centrosome‐associated antibody from Drosophila melanogaster reveals a new microtubule‐dependent structure in the equatorial zone of Parascaris univalens embryos. J Cell Sci 106: 719–730. [DOI] [PubMed] [Google Scholar]
- Kim, J. , Shen, B. , Rosen, C. and Dorsett, D. (1996) The DNA‐binding and enhancer‐blocking domains of the Drosophila suppressor of Hairy‐wing protein. Mol Cell Biol 16: 3381–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss, V. and Dorn, R. (2004) Evolution of the trans‐splicing Drosophila locus mod(mdg4) in several species of Diptera and Lepidoptera. Gene 331: 165–176. [DOI] [PubMed] [Google Scholar]
- Labrador, M. , Mongelard, F. , Plata‐Rengifo, P. , Baxter, E.M. , Corces, V.G. and Gerasimova, T.I. (2001) Protein encoding by both DNA strands. Nature 409: 1000. [DOI] [PubMed] [Google Scholar]
- Markstein, M. , Pitsouli, C. , Villalta, C. , Celniker, S.E. and Perrimon, N. (2008) Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet 40: 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan, M. , Bartkuhn, M. , Herold, M. , Philippen, A. , Heinl, N. and Bardenhagen, I. (2007) The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J 26: 4203–4214. doi: 10.1038/sj.emboj.7601851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongelard, F. , Labrador, M. , Baxter, E.M. , Gerasimova, Y.I. and Corces, V.G. (2002) Trans‐splicing as a novel mechanism to explain interallelic complementation in Drosophila . Genetics 160: 1481–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, H. , Filippova, G. , Loukinov, D. , Pugacheva, E. , Chen, Q. , Smith, S.T. et al (2005) CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab‐8 insulator. EMBO Rep 6: 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namciu, S.J. , Blochlinger, K.B. and Fournier, R.E. (1998) Human matrix attachment regions insulate transgene expression from chromosomal position effects in Drosophila melanogaster . Mol Cell Biol 18: 2382–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema, K. , Whitfield, W.G. and Alberts, B. (1995) The cell cycle‐dependent localization of the CP190 centrosomal protein is determined by the coordinate action of two separable domains. J Cell Biol 131: 1261–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, D. , Sheehan, B. , South, H. , Akbari, O. and Pai, C.Y. (2010) The chromosomal association/dissociation of the chromatin insulator protein Cp190 of Drosophila melanogaster is mediated by the BTB/POZ domain and two acidic regions. BMC Cell Biol 11: 101. doi: 10.1186/1471-2121-11-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai, C.Y. , Lei, E.P. , Ghosh, D. and Corces, V.G. (2004) The centrosomal protein CP190 is a component of the chromatin insulator. Mol Cell 16: 737–748. [DOI] [PubMed] [Google Scholar]
- Palla, F. , Melfi, R. , Anello, L. , Di Bernardo, M. and Spinelli, G. (1997) Enhancer blocking activity located near the 3′ end of the sea urchin early H2A histone gene. Proc Natl Acad Sci USA 94: 2272–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst, S.M. , Harrison, D.A. , Remington, M.P. , Spana, C. , Kelley, R.L. , Coyne, R.S. et al (1988) The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA‐binding protein. Genes Dev 2: 1205–1215. [DOI] [PubMed] [Google Scholar]
- Parnell, T.J. , Kuhn, E.J. , Gilmore, B.L. , Helou, C. , Wold, M.S. and Geyer, P.K. (2006) Identification of genomic sites that bind the Drosophila suppressor of Hairy‐wing insulator protein. Mol Cell Biol 26: 5983–5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, E. , Ghosh, D. , Baxter, E. and Corces, V. (2006) Genomic organization of gypsy chromatin insulators in Drosophila melanogaster . Genetics 172: 2337–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett, C.C. , O'Connor, A. and Dunaway, M. (1997) The repeat organizer, a specialized insulator element within the intergenic spacer of the Xenopus rRNA genes. Mol Cell Biol 17: 2866–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoborg, T.A. and Labrador, M. (2010) The phylogenetic distribution of non‐CTCF insulator proteins is limited to insects and reveals that BEAF‐32 is Drosophila lineage Specific. J Mol Evol 70: 74–84. [DOI] [PubMed] [Google Scholar]
- Smith, P.A. and Corces, V. (1995) The suppressor of Hairy‐wing protein regulates the tissue‐specific expression of the Drosophila gypsy retrotransposon. Genetics 139: 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnev, A.A. , He, B. , Baxley, R.M. , Jiang, N. , Hart, C.M. , Tan, K. et al (2012) Genome‐wide studies of the multi‐zinc finger Drosophila Suppressor of Hairy‐wing in the ovary. Nucleic Acids Res 40: 5415–5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana, C. and Corces, V.G. (1990) DNA bending is a determinant of binding specificity for a Drosophila zinc finger protein. Genes Dev 4: 1505–1515. [DOI] [PubMed] [Google Scholar]
- Tadepally, H.D. , Burger, G. and Aubry, M. (2008) Evolution of C2H2‐zinc finger genes and subfamilies in mammals: species‐specific duplication and loss of clusters, genes and effector domains. BMC Evol Biol 8: 176. doi: 10.1186/1471-2148-8-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubio, J.M. , Tojo, M. , Bassaganyas, L. , Escaramis, G. , Sharakhov, I.V. , Sharakhova, M.V. et al (2011) Evolutionary dynamics of the Ty3/gypsy LTR retrotransposons in the genome of Anopheles gambiae . PLoS ONE 6: e16328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardy, A. , Maine, E. and Schedl, P. (1985) The 87A7 chromomere: identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol 185: 341–358. [DOI] [PubMed] [Google Scholar]
- Whitfield, W.G. , Chaplin, M.A. , Oegema, K. , Parry, H. and Glover, D.M. (1995) The 190 kDa centrosome‐associated protein of Drosophila melanogaster contains four zinc finger motifs and binds to specific sites on polytene chromosomes. J Cell Sci 108: 3377–3387. [DOI] [PubMed] [Google Scholar]
- Zhong, X.P. and Krangel, M.S. (1997) An enhancer‐blocking element between alpha and delta gene segments within the human T cell receptor alpha/delta locus. Proc Natl Acad Sci USA 94: 5219–5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's web‐site:
Figure S1. Rapid amplification of cDNA ends (RACE) products and reconstructed suppressor of hairy‐wing [Su(Hw)] insulator transcripts. (A) RACE amplification products for Su(Hw), modifier of mobile dispersed genetic element 4 (mod[mdg4]) and centrosomal protein of 190 (CP190). All genes had a single transcript for the 5′ and 3′ RACE reactions, with the exception of mod(mdg4) 3′ RACE, which had nine groups with varying molecular weights. All groups were cloned and sequenced. The following Anopheles stephensi mod(mdg4) isoforms were obtained for each group: group 1: isoform C; group 2: isoform H; group 3: isoform O; group 4: isoforms A, D, L; group 5: isoform G; group 6: isoforms K, N; group 7: isoforms I, M; group 8: isoforms E, J; group 9: isoforms F, P. (B) RACE and walking primer sequencing data from Su(Hw), mod(mdg4) and CP190 from An. stephensi. The 5′ and 3′ untranslated region sequences are highlighted in yellow and bold letters indicate exon–exon junctions.
Figure S2. Alignment of the suppressor of hairy‐wing [Su(Hw)] protein complex. Alignment of predicted amino acid sequences encoding Su(Hw), Modifier of mobile dispersed genetic element 4, Mod(mdg4) and centrosomal protein of 190 (CP190) from Anopheles stephensi and Drosophila melanogaster. The red colour indicates highly conserved residues and the blue indicates less conservation as determined by alignment using Cobalt (http://www.st‐va.ncbi.nlm.nih.gov/tools/cobalt).
Figure S3. Predicted protein domains encoded by the gene orthologues of the suppressor of hairy‐wing [Su(Hw)] insulator complex in mosquitoes. Protein sequences were aligned using the ClustalW algorithm. Identical and highly conserved residues are highlighted in grey. The zinc‐coordinating residues are indicated in red. Abbreviations: STEPHE, Anopheles stephensi; GAMBIAE, Anopheles gambiae; AEDES, Aedes aegypti; CULEX, Culex quinquefasciatus.
Table S1. Oligonucleotide primers.
Table S2. Detailed analysis of the transcript‐specific sequences of the Anopheles stephensi mod(mdg4) isoforms.
Table S3. Transcript abundance delta‐delta quantification cycle (ΔΔCq) and statistical analyses of differences in Anopheles stephensi suppressor of hairy‐wing [As‐Su(Hw)], As‐modifier of mobile dispersed genetic element 4 (mod[mdg4]) and As‐centrosomal protein of 190 (As‐CP190) mRNA abundance in embryonic, larval, pupal and adult developmental stages, and in ovaries and midguts of adult females.


