Abstract
Background
Neutrophils, the most populous innate immune cell type, are the first responders to sites of infection and inflammation. Neutrophils can release their DNA to form extracellular traps (NETs), webs of DNA and granular proteases that contribute to pathogen clearance and promote thrombus formation. At present, the study of NETs is in part limited to the qualitative analysis of fluorescence microscopy-based images, thus quantification of the interactions between NETs and coagulation factors remains ill-defined.
Aim
Develop a quantitative method to measure the spatial distribution of DNA and colocalization of coagulation factor binding to neutrophils and NETs utilizing fluorescence-based microscopy.
Approach
Human neutrophils were purified from peripheral blood, bound to fibronectin and treated with the PKC-activator phorbol myristate acetate (PMA) to induce neutrophil activation and NETs formation. Samples were incubated with purified coagulation factors or plasma before staining with a DNA-binding dye and coagulation factor-specific antibodies. The spatial distribution of DNA and coagulation factors was imaged via fluorescence microscopy and quantified via a custom-built MATLAB-based image analysis algorithm. The algorithm first established global thresholding parameters on a training set of fluorescence image data and then systematically quantified intensity profiles across treatment conditions. Quantitative comparison of treatment conditions was enabled through the normalization of fluorescent intensities using the number of cells per image to determine the percent and area of DNA and coagulation factor binding per cell.
Results
Upon stimulation with PMA, NETs formation resulted in an increase in the area of DNA per cell. The coagulation factor fibrinogen bound to both the neutrophil cell body as well as NETs, while prothrombin, FX and FVIIa binding was restricted to the neutrophil cell body. The Gla domain of FX was required to mediate FX-neutrophil binding. Activated protein C (APC), but not Gla-less APC, bound to neutrophil cell bodies and NETs in a punctate manner. Neither FXIIa nor FXIa were found to bind either neutrophil cell bodies or NETs. Fibrinogen binding was dependent on extracellular DNA, while FX and APC required phosphatidylserine exposure for binding to activated neutrophils.
Conclusions
We have developed a quantitative measurement platform to define the spatial localization of fluorescently-labeled coagulation factor binding to neutrophils and extracellular DNA during NETosis.
Keywords: Neutrophil extracellular traps, coagulation factors, fluorescence microscopy, biophysical measurement
1. Introduction
Polymorphonuclear leukocytes, or neutrophils, are the most abundant circulating white blood cell type in humans, and play an indispensable role in innate immune host defense. Conversely, in the setting of inflammatory disease, excessive neutrophil activation has been shown to contribute to thrombotic complications. At sites of inflammation, activated neutrophils release intracellular granule proteins and chromatin that together form neutrophil extracellular traps (NETs) in an alternative form of cell death.(Brinkmann et al., 2004; Fuchs et al., 2007) These decondensed chromatin fibers are decorated with antimicrobial proteins and proteases including elastase, cathepsin G and myeloperoxidase (MPO), forming a physical trap to sequester and clear pathogens, considered an additional host defense mechanism.(McDonald et al., 2012; Mócsai, 2013) In addition to these microbial clearance functions, NETs have been shown to promote thrombin generation, fibrin formation and platelet activation in both in vitro and in vivo models to promote thrombus formation.(Gould et al., 2014; Massberg et al., 2010; Semeraro et al., 2011) Previous studies have indicated that fibrinogen and coagulation factors of the intrinsic pathway associate with the DNA, histones or proteases comprising NETs.(Fuchs et al., 2010; Oehmcke et al., 2009; von Brühl et al., 2012) However, it is unclear whether NETs are involved in an active or passive manner in promoting thrombus formation. Specifically, it is unclear whether NETs directly promote activation of coagulation factors, such as factor XII, or act as a scaffold for the assembly and activation of coagulation factors. In contrast, it is possible that NETs serve to bind and sequester active serine proteases, analogous to the binding and inactivation of thrombin on fibrin. These questions are difficult to address experimentally as they require systems that permit quantitative assessment of the binding, assembly and activation of coagulation factors on NETs. Here, we report on the development of a custom-built MATLAB-based image analysis algorithm that coupled with fluorescence microscopy-based images can provide quantitative analysis and spatial localization of coagulation factors binding to neutrophils as they undergo NETosis.
2. Materials and Methods
2.1. Reagents
Activated factor XII(a), factor XIa, factor X, FX-GD, factor VIIa, protein C, APC-GD, prothrombin, fibrinogen, and anti-human mouse antibodies to protein C/APC (AHPC-5071), FVII (AHFVII-5031), FX (AHX-5050), and prothrombin (AHP-5013) were purchased from Hematologic Technologies Inc. (Essex Junction, VT, USA). Activated protein C (APC) was a gift from Dr. András Gruber (Oregon Health & Science University, Portland, OR, USA). Polymorphprep was from Axis-Shield PoC AS (Oslo, Norway). Rabbit polyclonal antibody to fibrinogen was from MP Biomedicals (Santa Ana, CA). Mouse monoclonal antibody to factor XII heavy chain (sc-59517) was from Santa Cruz Biotech (Dallas, TX). The cell-permeable DNA dye Hoechst 33342 was from Invitrogen (Grand Island, NY). Alexa Fluor conjugated anti-mouse antibodies and rabbit polyclonal anti-histone H3 (ab5103) were from Abcam (Cambridge, MA). The antibody 1A6, against the A3 domain of human FXI was generated as described.(Tucker et al., 2009) All other reagents were purchased from Sigma-Aldrich (St. Louis, MO).
2.2. Preparation of human neutrophils
Human neutrophils were purified as previously described.(Itakura and McCarty, 2013) Briefly, human blood was drawn in accordance with an Oregon Health & Science University Institutional Review Board-approved protocol from healthy donors by venipuncture into citrate-phosphate-dextrose (1:7 vol/vol). Blood was layered over an equal volume of Polymorphprep and centrifuged at 500 g for 45 min at 18°C. The lower layer containing neutrophils was subsequently collected and washed with HBSS by centrifugation at 400 g for 10 min. To remove red blood cells from the sample, the pellet was resuspended in sterile H2O for 30 s, followed by immediate addition of 10× PIPES buffer (250 mM PIPES, 1.1 mM CaCl2, and 50 mM KCl, pH 7.4). After centrifugation at 400 g for 10 min, the pellet was resuspended in buffer (HBSS containing 2 mM CaCl2, 2 mM MgCl2 and 1% wt/vol BSA).
2.3. Immunofluorescence microscopy
Purified human neutrophils (2×106/mL) were stimulated with HBSS or PMA (10 nM) for 3 h at 37°C on fibronectin-coated glass coverslips. For initial colocalization experiments, cell samples were washed and treated with vehicle (HBSS buffer), fibrinogen (2.6 mg/mL), prothrombin (FII, 100 µg/mL), FX (10 µg/mL), FX-GD (10 µg/mL), protein C (300 nM), APC (300 nM), APC-GD (300 nM), FVIIa (300nM), FXIa (20 µg/mL), FXIIa (20 µg/mL) in the presence of HK (20 µg/mL) and ZnCl2 (25 µM) was then incubated for 15 min with the cell samples at 37°C. For select experiments, cell samples were washed and treated with vehicle (HBSS buffer), DNase I (10,000U/mL), RGDS (20 µM), Annexin V (10 µg/mL) for 10 min at 37°C. In these experiments, cell samples were then washed and treated with vehicle (HBSS buffer containing BSA), fibrinogen (2.6 mg/mL), FX (10 µg/mL), and APC (300 nM) for 15 min with the cell samples at 37°C. In select experiments cells were otherwise incubated with platelet-poor plasma or vehicle (HEPES containing 2mM CaCl2, 2mM MgCl2 and 0.1% BSA) (1:1) for 15 min at 37°C.
Subsequently, samples were washed with PBS and fixed with 4% PFA followed by incubation with blocking buffer (PBS containing 10% FBS and 5 mg/mL Fraction V BSA). Cells and coagulation factors were stained with anti-FXII (50 µg/mL), 1A6 (anti-FXI, 1:50), anti-PC/APC (100 µg/mL), anti-FVII (100 µg/mL), anti-fibrinogen (1:100), anti-FX (50 µg/mL) or anti-prothrombin (50 µg/mL) in blocking buffer at 4°C overnight. Secondary goat anti-rabbit IgG antibody conjugated with AlexaFluor 488 (1:500) and goat anti-mouse IgG antibody conjugated with AlexaFluor 546 (1:500) and Hoescht 33342 (10 µg/mL) in blocking buffer were added and incubated for 2 h in the dark. Coverslips were mounted onto glass slides and visualized with a Zeiss Axiovert fluorescence microscope.
2.4. Preparation of endothelial cells
Human umbilical endothelial cells (HUVECs) were grown to confluency on glass coverslips in 24-well plates, cells were then washed once with buffer and treated with vehicle (HBSS buffer), fibrinogen (2.6 mg/mL), FX (10 µg/mL), APC (300 nM), FXIa (20 µg/mL), FXIIa (20 µg/mL) in the presence of ZnCl2 (25 µM) for 15 min at 37°C. Cells were then washed again with HBSS buffer and fixed with 4% paraformaldehyde (PFA), then followed previously described antibody labeling as described in Section 2.3.
2.5. Image Analysis
For presentation of data, the fluorescent intensities of each image were adjusted based on signals detected in neutrophil samples in the absence of primary antibodies. Quantification of fluorescent images was performed using a custom algorithm in MATLAB (The Mathworks, Inc., Natick, MA). This algorithm first establishes global thresholding parameters on training set fluorescence image data to then systematically quantify intensity profiles across treatment conditions. Quantitative comparison of treatment conditions was achieved by the normalization of fluorescence intensities to the number of cells per image to determine the percent and area of DNA and coagulation factor binding per cell.
3. Results
3.1 Quantification of coagulation factor binding to neutrophils during NETosis in a purified system
Activation of coagulation factors involved in thrombus formation, including fibrinogen and members of the intrinsic pathway of coagulation, have been reported to be associated with NETs formation in the context of immunothrombosis.(Fuchs et al., 2010; Oehmcke et al., 2009) The overall goal was to design a platform to quantify the binding and spatial localization of coagulation factor-NETs interactions. Neutrophils were treated with PMA to induce NETs, followed by incubation with vehicle (HBSS buffer) or the following coagulation factors: fibrinogen (2.6 mg/mL), prothrombin (FII, 100 µg/mL), FX (10 µg/mL), Gla-less FX (FX-GD; 10 µg/mL), protein C (300 nM), APC (300 nM), Gla-less APC (APC-GD; 300 nM), FVIIa (300nM), and FXIa (20 µg/mL) or FXIIa (20 µg/mL) in the presence of HK (20 µg/mL) and ZnCl2 (25 µM) or ZnCl2 alone. Samples were fixed and stained for DNA, while coagulation factor binding was detected following staining with monoclonal anti-human mouse antibodies followed by secondary labeling with fluorescent anti-mouse antibodies. Coverslips were then mounted onto microscope slides, and at least 4 images were taken per condition. After exportation of images, a MATLAB code was utilized containing an algorithm that uploaded each image, converted each image into greyscale and created a jet scale image of each signal undergoing analysis. The images in greyscale and jet were then adjusted for threshold levels of signal and the signal was quantified for both the sum area and the area per cell, where the area of the total field of view is calculated to be 12510 µm2. The pixel overlap between the signal resulting from fluorescently labeled coagulation factors and DNA was subsequently computed (Pearson’s correlation coefficient, Rr) where the values from −1 to +1 described the overlap between two colored patterns and was independent of the pixel intensity values.(Berny et al., 2010)
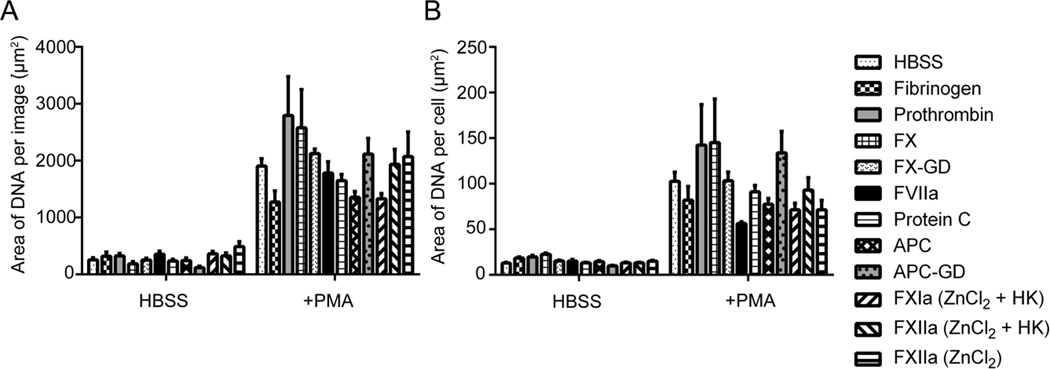
We first validated that this platform was able to detect and quantify NETs formation following stimulation of neutrophils with the PKC activator PMA. As shown in Figure 1, PMA induced robust NETs formation, as observed by the presence of extracellular DNA. Quantification of the surface area coverage of DNA per field of view demonstrated that PMA stimulation promoted an increase in DNA surface area coverage from 297.5 ± 47.1 µm2 for resting neutrophils under basal conditions to 1767.4 ± 201.3 µm2 for neutrophils that had undergone NETosis, as shown in Figure 2A. When quantified on a per cell basis, the results show that PMA stimulation promoted an increase in DNA staining from 13.4 ± 0.9 µm2 per cell to 87.1 ± 10.1 µm2 per cell (Figure 2B). These data are in accord with previous qualitative observations that neutrophils expel their DNA to form extracellular NETs following PMA stimulation, in contrast to DNA fragmentation and condensation observed during apoptosis.(Fuchs et al., 2007)
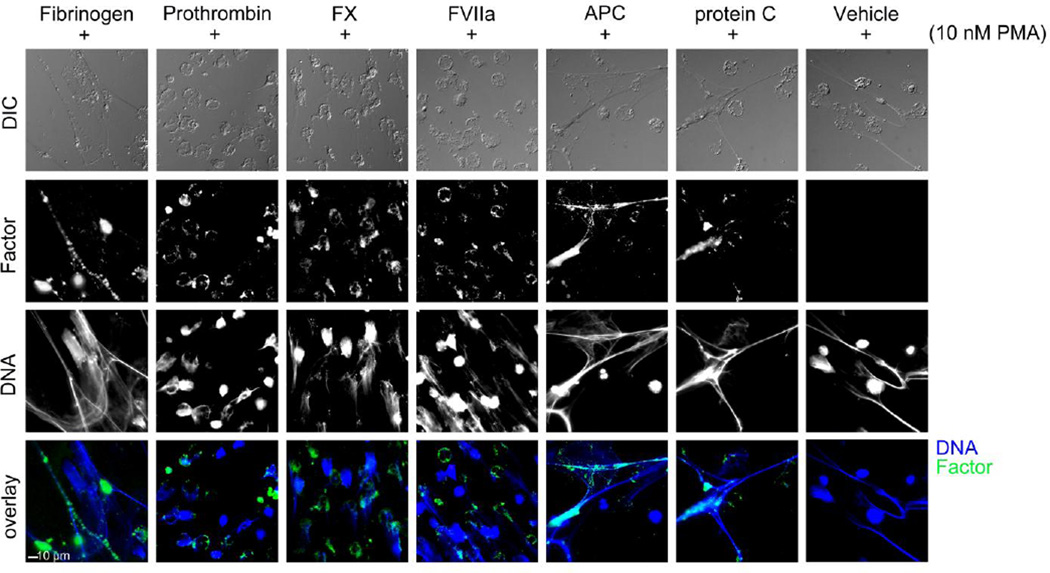
Figure 1. NETs promote binding of select purified coagulation factors.
Acid-washed glass coverslips were coated with 20 µg/mL fibronectin and then blocked with denatured BSA (5 mg/mL). Purified human neutrophils (2×106/mL) were plated on the coverslips, and were treated with HBSS or PMA (10 nM) for 3 hours at 37°C. Cell samples were washed and treated with vehicle (HBSS buffer), fibrinogen (2.6 mg/mL), prothrombin (FII, 100 µg/mL), FX (10 µg/mL), FVIIa (300nM), APC (300 nM), protein C (300 nM) was then incubated for 15 minutes with the cell samples at 37°C. Samples were then fixed with 4% PFA. (A) Samples were incubated overnight with primary antibodies. Samples were then incubated with Hoechst 33342 (1:1000) and secondary antibodies Alexa Fluor 488 goat anti-rabbit and Alexa Fluor 546 goat anti-mouse (Invitrogen, 1:500). Images were normalized to secondary antibody alone images and quantified in a custom MATLAB program to quantify each pixel positive signal, shown above are representative images of coagulation factors with positive cell staining.
Figure 2. DNA was quantified as area per image and per cell.
Images were normalized to secondary antibody alone images and quantified in a custom MATLAB program to quantify each pixel positive signal as (A) the area of DNA per image and (B) area DNA per cell. Data are mean±SEM n=3.
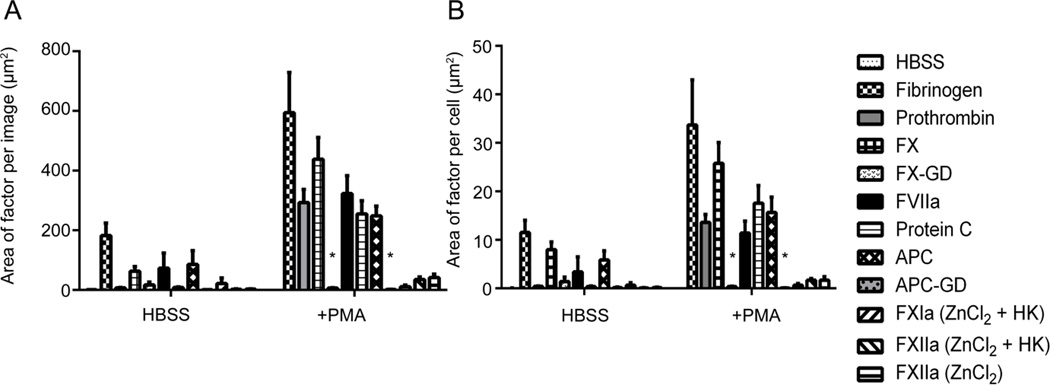
We next utilized the platform to quantify the binding and spatial localization of coagulation factor-NETs interactions. Resting or PMA-stimulated neutrophils were incubated with the coagulation factor fibrinogen in a purified system. The results show that purified fibrinogen bound to resting adherent neutrophils, as evidenced by an area of 182.57 ± 41.73 µm2 for fibrinogen labeling per field of view. A dramatic increase in fibrinogen binding was observed following PMA-induced NETosis, as evidenced by a greater than 3-fold increase in fibrinogen staining, when calculated as area coverage (Figure 3A). A similar trend was observed when the signal for fibrinogen binding was normalized per cell, as shown in Figure 3B. Moreover, the correlation coefficient between fibrinogen binding and DNA staining after NETs formation increased to 0.82, reflecting the observation that fibrinogen colocalized with NETs as seen in Figure 1.
Figure 3. Purified coagulation factor proteins quantified as area per image and per cell.
Acid-washed glass coverslips were coated with 20 µg/mL fibronectin and then blocked with denatured BSA (5 mg/mL). Purified human neutrophils (2×106/mL) were plated on the coverslips, and were treated with HBSS or PMA (10 nM) for 3 hours at 37°C. Cell samples were washed and treated with vehicle (HBSS buffer), fibrinogen (2.6 mg/mL), prothrombin (FII, 100 µg/mL), FX (10 µg/mL), FX-GD (10 µg/mL), protein C (300 nM), APC (300 nM), APC-GD (300 nM), FVIIa (300nM), FXIa (20 µg/mL), FXIIa (20 µg/mL) in the presence of HK (20 µg/mL) and ZnCl2 (25 µM) was then incubated for 15 minutes with the cell samples at 37°C. Samples were then fixed with 4% PFA. (A) Samples were incubated overnight with primary antibodies. Samples were then incubated with Hoechst 33342 (1:1000) and secondary antibodies Alexa Fluor 488 goat anti-rabbit and Alexa Fluor 546 goat anti-mouse (Invitrogen, 1:500). Images were normalized to secondary antibody alone images and quantified in a custom MATLAB program to quantify each pixel positive signal as (A) the area of factor per image and (B) area of signal per cell. Data are mean±SEM n=3.
We next evaluated whether FX and prothrombin bound to neutrophils following NETosis, as FX and prothrombin have been shown to assemble on surfaces of blood cells, such as platelets and endothelial cells as part of the prothrombinase complex.(Hamilton et al., 1990; Rodgers and Shuman, 1983; Tracy et al., 1985) The results show that while FX binding to neutrophils was minimal under resting conditions, FX bound to neutrophil cell bodies following PMA stimulation (Figure 1). Quantification of FX labeling demonstrated that the mean surface area of FX binding to neutrophils increased from 62.9 ± 15.5 µm2 for resting cells to 438.4 ± 72.9 µm2 after PMA stimulation (Figure 3A). On a per cell basis, FX labeling was shown to increase from 7.96 ± 1.57 µm2 on resting cells to 25.8 ± 4.3 µm2 after PMA stimulation as seen in Figure 3B. The correlation coefficient between FX and DNA from NETs formation was 0.13, indicative of the fact that FX bound in a punctate manner to the neutrophil cell body, rather than to the DNA-rich NETs. Based on the colocalization pattern, we hypothesized the γ-carboxyglutamic acid (Gla)-rich domain of FX could promote the binding to neutrophils underwent NETosis. To test this, a Gla domain-less derivative of FX was utilized to determine whether the Gla-domain was required for FX-NETs binding. After quantification, the results demonstrated that FX-GD labeling accounted for less than <20 µm2 for either resting or PMA-activated neutrophils, which is approximately a 14-fold reduction in binding as compared to wild-type FX (Figure 3A & B). Overall these data suggesting that the Gla domain of FX plays a critical role in mediating the binding of FX to neutrophils during NETosis. Next, prothrombin was incubated with neutrophils under resting conditions and following NETs formation with PMA. The results show that binding of prothrombin to the cell body increased after PMA stimulation to form NETs (Figure 1). Upon quantification, the mean area of prothrombin on resting cells increased from 6.8 ± 1.5 µm2 to 292.9 ± 43.8 µm2 following PMA stimulation (Figure 3A). Prothrombin labeling per cell was determined on resting cells to be 0.4 ± 0.06 µm2 and increased after PMA stimulation to 13.62 ± 1.6 µm2 (Figure 3B). After NETs formation, the correlation coefficient between prothrombin and DNA was found to be 0.25, again indicative of the fact that prothrombin bound in a punctate manner to the neutrophil cell body rather than to NETs.
Neutrophils are known to contain and, upon activation, express tissue factor, the initiator of the extrinsic coagulation pathway.(Kambas et al., 2012) However, it is unknown whether neutrophil tissue factor can complex with FVII to activate FX. The results show that after incubation with FVIIa, the mean area coverage of FVIIa labeling was 73.3 ± 50.2 µm2 for resting cells and increased to 323.3 ± 59.8 µm2 after PMA stimulation (Figure 3A). After normalization per cell, the mean area of FVIIa labeling was 3.4 ± 3.1 µm2 for resting cells and again increased to 11.4 ± 2.4 µm2 after PMA stimulation (Figure 3B).
Following the initiation and propagation of the coagulation cascade to generate thrombin, thrombin itself feeds back to downregulate thrombin generation through the generation of APC. Thrombin in complex with thrombomodulin cleaves protein C to generate APC, an endogenous anticoagulant which inactivates FVIIIa and FVa to dampen thrombin activation. Moreover, APC has been demonstrated to cleave histones, the most abundant protein associated with DNA. (Xu et al., 2009) Our group has previously shown that APC binding to leukocytes inhibits endotoxin-induced tissue factor procoagulant activity.(Yang et al., 2009) Experiments were designed to determine whether APC bound to neutrophils during NETosis. Thus, resting neutrophils and neutrophils that had undergone NETosis were incubated with APC as well as the zymogen, protein C in a purified system. These data show that the mean area of APC labeling was 85.5 ± 46.3 µm2 for resting neutrophils, while induction of NETs resulted in a 3-fold increase in APC binding, to 248.5 ± 32.1 µm2 after PMA stimulation (Figure 3A). After normalization, the mean area of APC binding per cell was 5.9 ± 1.9 µm2 for resting neutrophils as compared to 15.63 ± 3.16 µm2 per cell after NETs formation (Figure 3B). The correlation coefficient between labeling for APC and DNA after NETs induction was 0.54, reflective of the binding of APC to both the cell body and DNA-rich NETs as observed in Figure 1. In the presence of protein C, a mean area of 8.1 ± 2.2 µm2 was found for protein C-labeling of neutrophils. Following PMA stimulation to form NETs, the area increased to 255.3 ± 43.5 µm2 for protein C. Following normalization per cell, the pattern of increased area after NETs formation in response to PMA was again observed. The correlation coefficient between PC and DNA after NETs induction was calculated to be 0.45, again reflective of the binding of PC to NETs as observed in Figure 1.
We have previously shown that APC binding to leukocytes is mediated in part by the endothelial protein C receptor (EPCR).(Yang et al., 2009) Based on the fact that the N-terminus of APC also contains a Gla-rich domain which mediates binding to EPCR,(Sturn et al., 2003) a Gla domain-less derivative of APC was utilized to determine whether the Gla-domain was required for APC-NETs binding. The results show that incubation of neutrophils with Gla-less APC only resulted in a mean area of <2 µm2 for either resting or PMA-activated (Figure 4A). After normalization per cell, Gla-less APC labeling was quantified as less than 0.2 µm2 for resting or PMA-stimulated neutrophils stimulation (Figure 4B), a nearly 40-fold reduction in labeling as compared to wild-type APC. This data suggests that the Gla domain of APC plays a critical role in mediated APC binding to activated neutrophils and NETs.
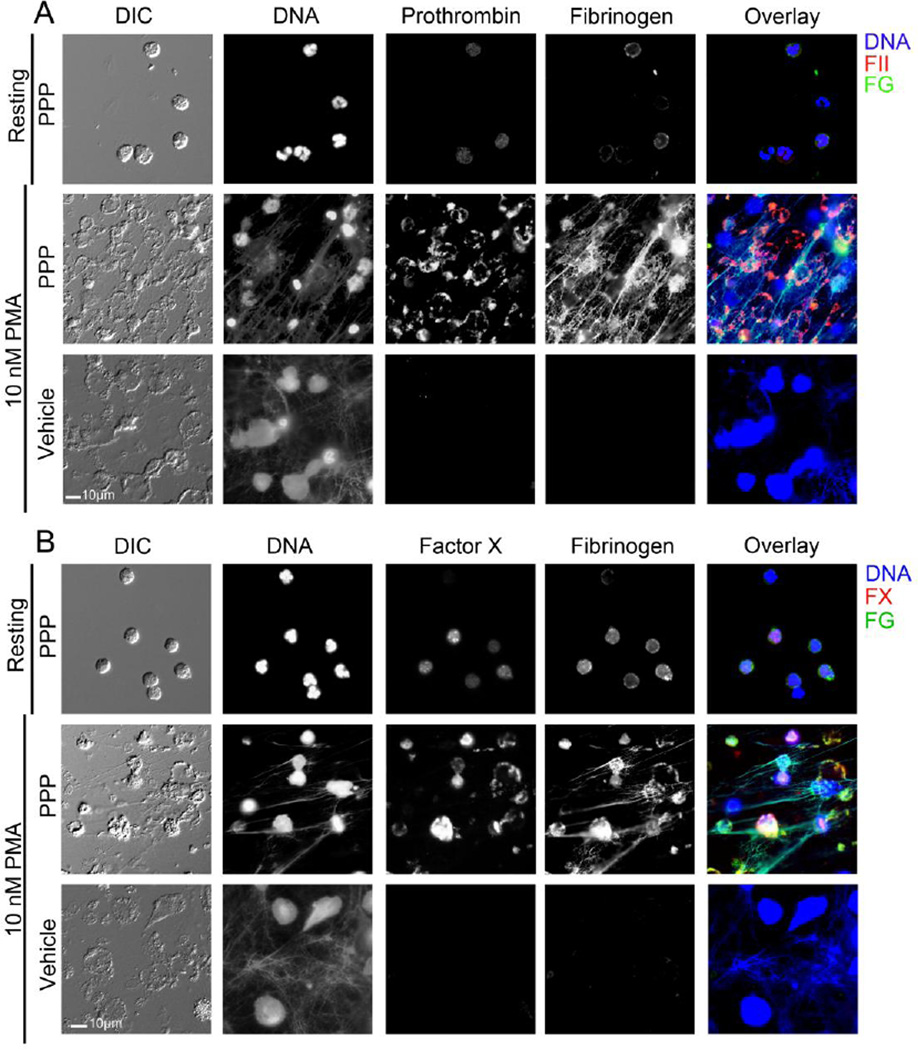
Figure 4. NETs promotes localization of plasma prothrombin, FX and fibrinogen.
Representative images of neutrophils and NETs treated with buffer or plasma and stained for DNA, fibrinogen and either (A) prothrombin or (B) FX.
To generate thrombin, the serine protease essential to formation of fibrin, coagulation can be activated and propagated through two main pathways, namely the extrinsic or the intrinsic pathways of coagulation. Activation of the intrinsic pathway of coagulation is initiated when the coagulation factor XII comes into contact with negatively charged surfaces, causing FXII to undergo autoactivation resulting in the generation of FXIIa, which subsequently proteolytically activates FXI to form the serine protease, FXIa. As DNA is a highly charged negative molecule, it has been hypothesized that NETs, by being comprised primarily of DNA, can promote activation of the intrinsic pathway of coagulation, specifically through activation of FXII.(Kannemeier et al., 2007; von Brühl et al., 2012) Thus, experiments were designed to test detection of labeled NETs with either FXIa or FXIIa. Our quantitative analysis failed to show labeling of either neutrophils or NETs with FXIIa or FXIa, either alone or in the presence of the cofactors HK and/or ZnCl2 (Figure 4A & B), suggesting that under these experimental conditions, NETs may not be able to support the binding of the coagulation factors of the intrinsic pathway.
Conversely, as a positive control we tested if we could detect FXIIa and FXIa binding to cultured endothelial cells (Supplemental Figure 1A & B).(Mahdi et al., 2003, 2002) Quantification of FXIIa labeling of endothelial cells in the presence of ZnCl2 demonstrated a mean surface area of 1001.2 ± 68.5 µm2 that was normalized to 37.4 ± 3.3 µm2 per cell. The correlation coefficient between FXIIa and DNA of endothelial cells was 0.55, reflecting the colocalization and strong signal of FXIIa observed on the endothelium. Similarly, incubation of endothelial cells with FXIa in the presence of 25µM ZnCl2 resulted in a surface area of 977.3 ± 44.5 µm2 that after normalization per cell was found to be 35.8 ± 1.7 µm2. The correlation coefficient was calculated to be 0.36 for FXIa and DNA found in endothelial cells, reflective of the more diffuse staining pattern of FXIa to the endothelium in these studies. Our quantitative platform was also able to detect the binding of fibrinogen, FX and APC to endothelial cells (Supplemental Figure 1A & B). These results demonstrate that our quantitative analysis platform is able to detect purified coagulation factor binding to different cell types, such as endothelial cells.
3.2. Quantification of coagulation factor binding to neutrophils during NETosis in plasma
We next designed experiments to utilize platelet-poor plasma in this assay in order to quantify endogenous coagulation factor binding to neutrophils during NETosis. As shown in Figure 5, neutrophils were observed to form DNA-rich NETs following stimulation with PMA. First, results show that resting neutrophil DNA area was 316.8 ± 78.0 µm2, which after normalization per cell corresponded to a mean area of DNA/cell of 19.8 ± 1.8 µm2. After NETs formation, the total DNA area increased to 2234.6 ± 129.3 µm2, which after normalization per cell corresponded to a mean area of DNA/cell of 116.5 ± 14.1 µm2. These results in the presence of plasma were similar to the extent of NETosis observed in the presence of purified coagulation factors.
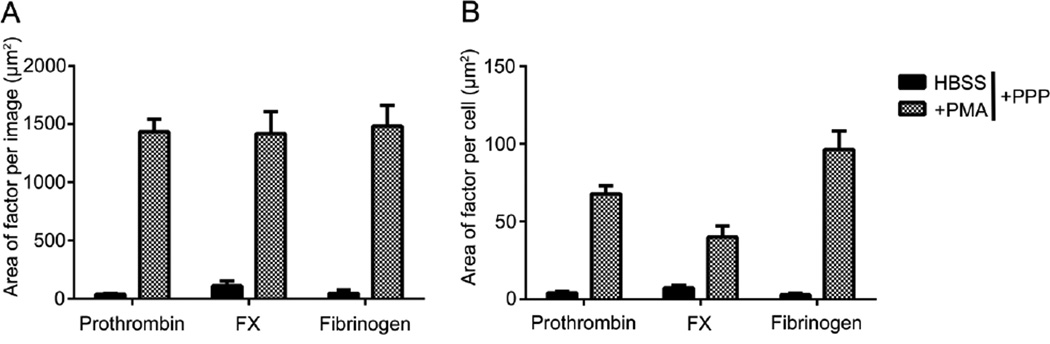
Figure 5. Coagulation factors in platelet poor plasma colocalize to NETs, quantified as area per image and per cell.
Acid-washed glass coverslips were coated with 20 µg/mL fibronectin and then blocked with denatured BSA (5 mg/mL). Purified human neutrophils (2×106/mL) were plated on the coverslips, and were treated with HBSS or PMA (10 nM) for 3 hours at 37°C. Cell samples were washed and incubated with platelet-poor plasma or vehicle (HEPES containing 2mM CaCl2, 2mM MgCl2 and 0.1% BSA) (1:1) for 15 min at 37°C. Samples were incubated overnight with primary antibodies. Samples were then incubated with Hoechst 33342 (1:1000) and secondary antibodies Alexa Fluor 488 goat anti-rabbit and Alexa Fluor 546 goat anti-mouse (Invitrogen, 1:500). Images were normalized to secondary antibody alone images and quantified in a custom MATLAB program to quantify each pixel positive signal as (A) the area per image and (B) area per cell. Data are mean±SEM n=3.
The binding of plasma coagulation factors to neutrophils during NETosis was then quantified. In the presence of plasma, no prothrombin, FX nor fibrinogen labeling of resting neutrophils was observed. However, as shown in Figure 4, the coagulation factors prothrombin, FX and fibrinogen bound to PMA-stimulated neutrophils. The mean area of prothrombin binding to PMA-stimulated neutrophils was 1434.1 ± 107.9 µm2, which upon normalization per cell was quantified to be 67.9 ± 5.0 µm2 (Figure 5A & B). The mean area of FX binding to PMA-stimulated neutrophils was 1417.5 ± 188.4 µm2, which upon normalization per cell was quantified to be 40.0 ± 7.4 µm2 (Figure 5A & B). The mean area of fibrinogen binding to PMA-stimulated neutrophils was 1483.9 ± 174.5 µm2, which upon normalization per cell was quantified to be 96.4 ± 11.8 µm2. The correlation coefficient between fibrinogen, prothrombin and FX binding and DNA staining after NETs formation was quantified to be 0.75, 0.65 and 0.49, respectively, reflective of the observation that fibrinogen colocalized with NETs, whereas both prothrombin and FX were observed to bind to the neutrophil cell body (Figure 4).
3.3. Quantification of coagulation factor binding to DNA and phospholipids comprising NETs
We next designed experiments to determine the role of DNA and phospholipids that respectively comprise NETs, may play in promoting the binding of coagulation factors.(Berny et al., 2010; Fuchs et al., 2010; Plow et al., 2000) To first validate our platform and determine the role of DNA, we quantified the DNA extruded after NETs were pretreated with 10,000U/mL DNase. After imaging and analysis the surface area of DNA was significantly reduced to 329.2 ± 57.9 µm2 after pretreatment with DNase, and the DNA area was normalized to 16.8 ± 2.4 µm2 per cell.
After validating that DNase treatment could significantly reduce the degree of extracellular DNA in our system, we incubated DNase-treated NETs with fibrinogen, FX and APC. First, fibrinogen binding was found to be significantly reduced in absence of extracellular DNA, where the total surface area was 298.65 ± 24.9 µm2 and normalized to 14.0 ± 1.5 µm2 per cell, similar to the result for resting neutrophil-fibrinogen binding (Figure 6A & B). Similarly, the surface area of FX was reduced to 200.7 ± 19.9 µm2 which normalized to 8.6 ± 1.0 µm2 per cell. Lastly, APC labeling was significantly reduced in the presence of DNase-treatment, with an area of 149.6 ± 23.5 µm2, which normalized to 5.9 ± 0.9 µm2 per cell (Figure 6A & B). Overall, these results confirm the role of extracellular DNA in supporting the binding of select coagulation factors during NETosis.
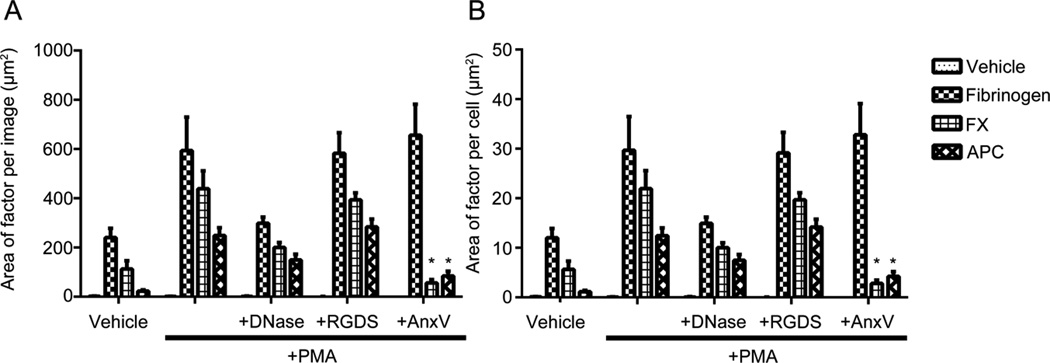
Figure 6. Coagulation factors bind neutrophils and neutrophil extracellular traps in a DNA- and phospholipid-dependent manner quantified as area per image and per cell.
Acid-washed glass coverslips were coated with 20 µg/mL fibronectin and then blocked with denatured BSA (5 mg/mL). Purified human neutrophils (2×106/mL) were plated on the coverslips, and were treated with HBSS or PMA (10 nM) for 3 hours at 37°C. Cell samples were washed and treated with vehicle (HBSS buffer), DNase I (10,000U/mL), RGDS (20 µM), Annexin V (10 µg/mL) for 10 minutes at 37°C. Cell samples were then washed and treated with vehicle (HBSS buffer), fibrinogen (2.6 mg/mL), FX (10 µg/mL), and APC (300 nM) was then incubated for 15 minutes with the cell samples at 37°C. Samples were then fixed with 4% PFA. (A) Samples were incubated overnight with primary antibodies. Samples were then incubated with Hoechst 33342 (1:1000) and secondary antibodies Alexa Fluor 488 goat anti-rabbit and Alexa Fluor 546 goat anti-mouse (Invitrogen, 1:500). Images were normalized to secondary antibody alone images and quantified in a custom MATLAB program to quantify each pixel positive signal as (A) the area of factor per image and (B) area of signal per cell. Data are mean±SEM n=3.
Following activation, the cell membrane of blood cells ‘flip’ to expose phosphatidylserine to the extracellular environment. Moreover, phospholipid exposure is considered a marker of cell activation and enhancer of coagulation factor activation, in part through binding of calcium ions and the Gla-domain of vitamin K-dependent coagulation factors.(Huang et al., 2003) As our results demonstrated a critical role for the Gla domain of FX and APC in supporting the binding of these coagulation factors to activated neutrophils, we designed experiments to determine the role of the exposure of phosphatidylserine by activated neutrophils in mediating FX and APC binding. In order to test this hypothesis, activated neutrophils were pretreated with purified Annexin V (AnxV) in order to competitively inhibit binding of coagulation factors to phosphatidylserine. Our results demonstrate that fibrinogen binding to NETs was relatively unchanged in the presence of AnxV, where the total surface area was 655.9 ± 126.3 µm2 and normalized to 37.7 ± 6.8 µm2 per cell (Figure 6A & B). Next, after inhibition of activated neutrophils with Annexin V, the surface area of FX labeling of activated neutrophils was significantly reduced to 56.5 ± 13.4 µm2 which normalized to 3.4 ± 0.9 µm2 per cell. Lastly, APC labeling of activated neutrophils was significantly reduced in the presence of AnxV, with an area of 84.9 ± 18.4 µm2, which normalized to 3.7 ± 1.0 µm2 per cell (Figure 6A & B). Overall, these results demonstrate additional evidence that the Gla domains in FX and APC may be essential to mediate binding to the cell body during NETosis in a phosphatidylserine-dependent manner.
4. Discussion
Fluorescence- and brightfield-based microscopy imaging modalities have been extensively utilized to visualize and study the mechanisms of NETs formation. Herein, we present a quantitative platform to study neutrophil-coagulation factor binding to neutrophils during NETosis.
Using this custom-built MATLAB-based image analysis algorithm, it was first validated that NETs support the binding of fibrinogen in both purified and plasma-based systems, in line with previous observations.(Fuchs et al., 2010) The fibrinogen-NETs interaction was found to occur in an extracellular DNA-dependent manner. These findings show that FVIIa, FX and prothrombin bound to the neutrophil cell body during NETosis, suggesting that the surface of activated neutrophils may serve as a site to assemble the prothrombinase complex and promote thrombin generation at sites of inflammation. FX binding could be abrogated with the engineered loss of its Gla domain, as well as after inhibition of phosphatidylserine exposed on the neutrophil cell membrane during NETosis. Using cultured endothelial cells, we observed binding of fibrinogen, FX and APC, as well as the intrinsic factors FXIIa and FXIa in a ZnCl2-dependent manner. Conversely, we failed to observe a measurable direct interaction of either FXII or FXI with neutrophils during NETosis. As several reports have suggested that NETs activate the intrinsic pathway of coagulation,(Oehmcke et al., 2009; von Brühl et al., 2012) perhaps indirect pathways may be involved in the activation of FXII by NETs rather than direct binding and autoactivation of FXII by DNA.
As coagulation is activated and propagated, protein C is cleaved to become APC, whereupon the active serine protease exerts its anticoagulant function to dampen the amplification of thrombin generation. The proteolytic activity of APC as an essential endogenous mechanism to downregulate coagulation, and together with its activity as an anti-inflammatory protease APC plays a critical role in dampening thromboinflammation associated with disease states such as sepsis. Neutrophil activation leading to NETs formation has been implicated to promote pathologic thrombus formation at sites of inflammation. Thus, uncovering the molecular mechanisms by which APC binds to neutrophils during NETosis, and determining whether this interaction downregulates NETs formation may provide clues for the development of therapies targeted to safely reduce thromboinflammation. These results demonstrated that APC and protein C binds to DNA-rich NETs and the cell membrane, which support previous studies suggesting that histone degradation is associated with the proteolytic activity of APC, and supports the hypothesis that APC may regulate neutrophil cell death.(Iba and Nagakari, 2015; Xu et al., 2009)
In conclusion, we have developed and utilized a quantitative fluorescence-based imaging platform to study the spatial distribution of DNA and coagulation factors binding to neutrophils during NETosis. We demonstrated an increase in fibrinogen binding to NETs that is dependent upon extracellular DNA, while the coagulation factors prothrombin, FX and FVIIa bind to the neutrophil cell bodies following NETosis in both purified systems and in plasma. We found that the Gla domain of FX played an essential role in mediating binding of FX to activated neutrophils during NETosis, and that this is dependent upon the exposure of anionic phospholipids on the activated neutrophil cell membrane. Conversely, we were not able to measure the binding of FXIIa or FXIa to neutrophils during NETosis, but were able to measure the binding of FXIIa and FXIa to endothelial cells. We also found that the zymogen protein C, as well as APC, both bound DNA-rich NETs, and identified a critical role for the Gla domain of APC in mediating binding to the neutrophil cell body during NETosis, mediated by phosphatidylserine exposure. Overall, this study suggests that NETs may serve as a scaffold for the assembly of select coagulation factors, providing rationale for the targeted inhibition of NETosis to reduce thromboinflammation.
Highlights.
The coagulation factor fibrinogen binds DNA-rich NETs.
Coagulation factors prothrombin, FX and FVIIa bind activated neutrophils during NETosis.
The anticoagulant activated protein C (APC) binds activated neutrophils and DNA-rich NETs.
The binding of FX and APC to activated neutrophils is dependent upon the coagulation factor Gla-domain and exposure of neutrophil phosphatidylserine.
Acknowledgments
We thank the volunteers who kindly donated their blood for this study. We would like to thank Dr. András Gruber for stimulating discussions and for the generous gift of APC.
Sources of funding
This work was supported by grants from the National Institutes of Health (R01HL101972 and R01GM116184 to O.J.T.M. and T32AI007472 to L.D.H.). O.J.T. McCarty is an American Heart Association (AHA) Established Investigator (13EIA12630000). C.P. is an AHA Fellow (14POST18180011). O.J.T.M and E.E.G received funding from the Australian Academy of Science (AAS) in collaboration with the U.S. Air Force Office of Scientific Research (AFOSR).
Abbreviations
- NETs
neutrophil extracellular traps
- FXII
factor XII
- FXI
factor XI
- FX
factor X
- FX-GD
Gla-less FX
- FVIIa
factor VIIa
- APC
Activated protein C
- APC-GD
Gla-less APC
- PMA
phorbol 12-myristate 13-acetate
- PKC
protein kinase C
- Gla-domain
γ-carboxyglutamic acid domain
- HK
high molecular weight kininogen
- AnxV
Annexin V
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authorship contributions
Conceived and designed experiments: L.D.H., A.I., O.J.T.M. Performed experiments: L.D.H., C.P., T.C., A.I., D.K.R., A.B., E.E.G. MATLAB development: L.D.H., A.B., K.G.P. Analyzed the data: L.D.H., T.C., A.B. Wrote and edited the manuscript: L.D.H., C.P., A.I., K.G.P., E.E.G., O.J.TM.
Disclosures
The authors have no conflicts of interest to declare.
References
- Berny MA, Munnix ICA, Auger JM, Schols SEM, Cosemans JMEM, Panizzi P, Bock PE, Watson SP, McCarty OJT, Heemskerk JWM. Spatial Distribution of Factor Xa,, Thrombin, and Fibrin(ogen) on Thrombi at Venous Shear. PLoS ONE. 2010;5:e10415. doi: 10.1371/journal.pone.0010415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, Weinrauch Y, Brinkmann V, Zychlinsky A. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 2007;176:231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs TA, Brill A, Duerschmied D, Schatzberg D, Monestier M, Myers DD, Wrobleski SK, Wakefield TW, Hartwig JH, Wagner DD. Extracellular DNA traps promote thrombosis. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15880–15885. doi: 10.1073/pnas.1005743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould TJ, Vu TT, Swystun LL, Dwivedi DJ, Mai SHC, Weitz JI, Liaw PC. Neutrophil extracellular traps promote thrombin generation through platelet-dependent and platelet-independent mechanisms. Arterioscler. Thromb. Vasc. Biol. 2014;34:1977–1984. doi: 10.1161/ATVBAHA.114.304114. [DOI] [PubMed] [Google Scholar]
- Hamilton KK, Hattori R, Esmon CT, Sims PJ. Complement proteins C5b-9 induce vesiculation of the endothelial plasma membrane and expose catalytic surface for assembly of the prothrombinase enzyme complex. J. Biol. Chem. 1990;265:3809–3814. [PubMed] [Google Scholar]
- Huang M, Rigby AC, Morelli X, Grant MA, Huang G, Furie B, Seaton B, Furie BC. Structural basis of membrane binding by Gla domains of vitamin K–dependent proteins. Nat. Struct. Mol. Biol. 2003;10:751–756. doi: 10.1038/nsb971. [DOI] [PubMed] [Google Scholar]
- Iba T, Nagakari K. The effect of plasma-derived activated protein C on leukocyte cell-death and vascular endothelial damage. Thromb. Res. 2015;135:963–969. doi: 10.1016/j.thromres.2015.03.012. [DOI] [PubMed] [Google Scholar]
- Itakura A, McCarty OJT. Pivotal role for the mTOR pathway in the formation of neutrophil extracellular traps via regulation of autophagy. Am. J. Physiol. Cell Physiol. 2013;305:C348–C354. doi: 10.1152/ajpcell.00108.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambas K, Mitroulis I, Apostolidou E, Girod A, Chrysanthopoulou A, Pneumatikos I, Skendros P, Kourtzelis I, Koffa M, Kotsianidis I, Ritis K. Autophagy Mediates the Delivery of Thrombogenic Tissue Factor to Neutrophil Extracellular Traps in Human Sepsis. PLoS ONE. 2012;7:e45427. doi: 10.1371/journal.pone.0045427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannemeier C, Shibamiya A, Nakazawa F, Trusheim H, Ruppert C, Markart P, Song Y, Tzima E, Kennerknecht E, Niepmann M, Bruehl M-L, von, Sedding D, Massberg S, Günther A, Engelmann B, Preissner KT. Extracellular RNA constitutes a natural procoagulant cofactor in blood coagulation. Proc. Natl. Acad. Sci. 2007;104:6388–6393. doi: 10.1073/pnas.0608647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdi F, Madar ZS, Figueroa CD, Schmaier AH. Factor XII interacts with the multiprotein assembly of urokinase plasminogen activator receptor, gC1qR, and cytokeratin 1 on endothelial cell membranes. Blood. 2002;99:3585–3596. doi: 10.1182/blood.v99.10.3585. [DOI] [PubMed] [Google Scholar]
- Mahdi F, Shariat-Madar Z, Schmaier AH. The relative priority of prekallikrein and factors XI/XIa assembly on cultured endothelial cells. J. Biol. Chem. 2003;278:43983–43990. doi: 10.1074/jbc.M304239200. [DOI] [PubMed] [Google Scholar]
- Massberg S, Grahl L, von Bruehl M-L, Manukyan D, Pfeiler S, Goosmann C, Brinkmann V, Lorenz M, Bidzhekov K, Khandagale AB, Konrad I, Kennerknecht E, Reges K, Holdenrieder S, Braun S, Reinhardt C, Spannagl M, Preissner KT, Engelmann B. Reciprocal coupling of coagulation and innate immunity via neutrophil serine proteases. Nat. Med. 2010;16:887–896. doi: 10.1038/nm.2184. [DOI] [PubMed] [Google Scholar]
- McDonald B, Urrutia R, Yipp BG, Jenne CN, Kubes P. Intravascular Neutrophil Extracellular Traps Capture Bacteria from the Bloodstream during Sepsis. Cell Host Microbe. 2012;12:324–333. doi: 10.1016/j.chom.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Mócsai A. Diverse novel functions of neutrophils in immunity, inflammation, and beyond. J. Exp. Med. 2013;210:1283–1299. doi: 10.1084/jem.20122220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehmcke S, Mörgelin M, Herwald H. Activation of the human contact system on neutrophil extracellular traps. J. Innate Immun. 2009;1:225–230. doi: 10.1159/000203700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plow EF, Haas TA, Zhang L, Loftus J, Smith JW. Ligand Binding to Integrins. J. Biol. Chem. 2000;275:21785–21788. doi: 10.1074/jbc.R000003200. [DOI] [PubMed] [Google Scholar]
- Rodgers GM, Shuman MA. Prothrombin is activated on vascular endothelial cells by factor Xa and calcium. Proc. Natl. Acad. Sci. U. S. A. 1983;80:7001–7005. doi: 10.1073/pnas.80.22.7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semeraro F, Ammollo CT, Morrissey JH, Dale GL, Friese P, Esmon NL, Esmon CT. Extracellular histones promote thrombin generation through platelet-dependent mechanisms: involvement of platelet TLR2 and TLR4. Blood. 2011;118:1952–1961. doi: 10.1182/blood-2011-03-343061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturn DH, Kaneider NC, Feistritzer C, Djanani A, Fukudome K, Wiedermann CJ. Expression and function of the endothelial protein C receptor in human neutrophils. Blood. 2003;102:1499–1505. doi: 10.1182/blood-2002-12-3880. [DOI] [PubMed] [Google Scholar]
- Tracy PB, Eide LL, Mann KG. Human prothrombinase complex assembly and function on isolated peripheral blood cell populations. J. Biol. Chem. 1985;260:2119–2124. [PubMed] [Google Scholar]
- Tucker EI, Marzec UM, White TC, Hurst S, Rugonyi S, McCarty OJT, Gailani D, Gruber A, Hanson SR. Prevention of vascular graft occlusion and thrombus-associated thrombin generation by inhibition of factor XI. Blood. 2009;113:936–944. doi: 10.1182/blood-2008-06-163675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Brühl M-L, Stark K, Steinhart A, Chandraratne S, Konrad I, Lorenz M, Khandoga A, Tirniceriu A, Coletti R, Köllnberger M, Byrne RA, Laitinen I, Walch A, Brill A, Pfeiler S, Manukyan D, Braun S, Lange P, Riegger J, Ware J, Eckart A, Haidari S, Rudelius M, Schulz C, Echtler K, Brinkmann V, Schwaiger M, Preissner KT, Wagner DD, Mackman N, Engelmann B, Massberg S. Monocytes, neutrophils, and platelets cooperate to initiate and propagate venous thrombosis in mice in vivo. J. Exp. Med. 2012;209:819–835. doi: 10.1084/jem.20112322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang X, Pelayo R, Monestier M, Ammollo CT, Semeraro F, Taylor FB, Esmon NL, Lupu F, Esmon CT. Extracellular histones are major mediators of death in sepsis. Nat. Med. 2009;15:1318–1321. doi: 10.1038/nm.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XV, Banerjee Y, Fernández JA, Deguchi H, Xu X, Mosnier LO, Urbanus RT, Groot PGde, White-Adams TC, McCarty OJT, Griffin JH. Activated protein C ligation of ApoER2 (LRP8) causes Dab1-dependent signaling in U937 cells. Proc. Natl. Acad. Sci. 2009;106:274–279. doi: 10.1073/pnas.0807594106. [DOI] [PMC free article] [PubMed] [Google Scholar]