Abstract
Mammalian olfaction depends on the development of specialized olfactory sensory neurons (OSNs) that each express one odorant receptor (OR) protein from a large family of OR genes encoded in the genome. The lysine-specific demethylase-1 (LSD1) protein removes activating H3K4 or silencing H3K9 methylation marks at gene promoters and is required for proper OR regulation. We show that LSD1 protein exhibits variable organization within nuclei of developing OSNs, and tends to consolidate into a single dominant compartment at the edges of chromocenters within nuclei of early post-mitotic cells of the mouse olfactory epithelium (MOE). Using an immortalized cell line derived from developing olfactory placode, we show that consolidation of LSD1 appears to be cell-cycle regulated, with a peak occurrence in early G1. LSD1 co-compartmentalizes with CoREST, a protein known to collaborate with LSD1 to carry out a variety of chromatin-modifying functions. We show that LSD1 compartments co-localize with 1–3 OR loci at the exclusion of most OR genes, and commonly associate with Lhx2, a transcription factor involved in OR regulation. Together, our data suggests that LSD1 is sequestered into a distinct nuclear space that might restrict a histone-modifying function to a narrow developmental time window and/or range of OR gene targets.
Keywords: Olfactory receptor genes, LSD1/KDM1A, neurogenin, p27Kip1, H3K9 methylation, chromocenters, compartmentalization
Introduction
Study of the mouse olfactory epithelium (MOE) is crucial for understanding neurogenesis, as well as neuronal specialization. During development, each olfactory sensory neuron (OSN) expresses only one type of odorant receptor (OR) in a monogenic and monoallelic fashion (Buck and Axel, 1991; Chess et al., 1994; Malnic et al., 1999; Serizawa et al., 2000), selected from among ~1400 OR genes that are clustered at ~50 different chromosomal loci (Sullivan et al., 1996; Zhang et al., 2004a). This singularity in OR expression is the basis of odor discrimination, permitting specialized OSNs to distinguish among an astonishing number of odorant stimuli (Bushdid et al., 2014), as well as the basis for wiring OSNs to distinct targets in the brain (Wang et al., 1998). How this singularity in OR expression is achieved in the first place is still an active area of research and the topic of this study.
OR expression is observed at the onset of OSN differentiation but before axon targeting in embryonic mice (Rodriguez-Gil et al., 2010; Sullivan et al., 1995). In adult mice, OR expression is shown in immature GAP43-positive cells, but OR expression lags the onset of neuronal differentiation (Iwema and Schwob, 2003), and OSN axons target into olfactory bulb before OR expression (Rodriguez-Gil et al., 2015). OR gene switching takes place in the immature OSN layer, which permits reselection in the event that an initial choice is unproductive (Lewcock and Reed, 2004; Serizawa et al., 2003; Shykind et al., 2004). Once a productive OR choice is made, an OR-protein-mediated feedback mechanism enables commitment to that OR selection by preventing additional OR activations (Dalton et al., 2013; Lewcock and Reed, 2004; Serizawa et al., 2003). Driving OR transgene expression in immature OSNs results in broad suppression of endogenous ORs, whereas driving expression in mature OSNs results in the broad suppression of the transgenic OR, suggesting that OR selection occurs in the immature neurons (Nguyen et al., 2007). Therefore, investigation of molecular mechanisms underlying OR selection, switching, and commitment is focused on regulatory proteins that function in the early post-mitotic cells of the OSN lineage.
Recent research indicates critical epigenetic mechanisms that underlie mutually exclusive OR transcription. OR alleles are decorated with the constitutive heterochromatic marks H3K9me3 and H4K20me3(Magklara et al., 2011). These marks are deposited prior to OR choice suggesting that ORs are silenced in their ground state(Magklara et al., 2011). Importantly, these repressive marks are removed on the transcribed allele (Magklara et al., 2011). If deposition of these histone marks is disrupted, expression is biased towards a small OR subset and multigenic expression is observed in some OSNs (Lyons et al., 2014). Another epigenetic layer of regulation is achieved via the organization of OR genes in the nucleus. All but the expressed OR allele are sequestered in heterochromatic chromocenters in mature OSNs, an organization that is necessary for maintaining singularity of OR expression (Clowney et al., 2012). These observations suggest a mechanism whereby only one OR gene per cell is liberated from heterochromatin, thereby restricting access to RNA polymerase for only the de-repressed OR allele.
LSD1, histone lysine specific demethylase (also known as KDM1A), functions in the H3K4 and H3K9 demethylation pathways, whose removal contributes to both gene silencing (Su et al., 2009; Sun et al., 2010; Tsai et al., 2008) and activation (Garcia-Bassets et al., 2007; Metzger et al., 2005; Perillo et al., 2008; Ray et al., 2014), respectively. The various complexes formed by LSD1 dictate which of these two opposing activities will occur in the regulation of a specific target (Wang et al., 2007). Moreover, LSD1 can exhibit both functions on the same gene target, as has been observed in the regulation of androgen receptor genes (Cai et al., 2014). Therefore, LSD1 is a versatile protein utilized in a broad range of developmental contexts. In the MOE, LSD1 is expressed in early cells of the lineage coincident with OR choice and is down-regulated in mature OSNs after OR choice is stabilized (Dalton et al., 2013; Krolewski et al., 2013; Lyons et al., 2013). The deletion of Lsd1 before OR selection results in dramatic decrease in OR expression in the MOE suggesting it is involved in OR regulation (Lyons et al., 2013). The specific hypothesis we explore in this paper is a possible role for LSD1 in achieving mutually exclusive OR transcription via selective H3K9 demethylation during selection and/or H3K4 demethylation on the previously active OR allele during switching. Both of these possible functions for LSD1 in OR regulation predict some mechanism for restricting protein activity to one or a small number of potential OR target gene loci.
We investigated the nuclear organization of LSD1 in the developing OSN lineage, as well as in the OP6 cell line, that represents immature cells of the MOE at a point in development when OR selection has occurred but has not apparently stabilized, in order to gain further insights into a role for LSD1 in OR activation/switching events. We find that LSD1 is compartmentalized into a single compartment per nucleus at the edges of nuclear chromocenters within the earliest, post-mitotic cells of the OSN lineage. In the OP6 cell line, we show that these compartments consist of the LSD1 co-factor CoREST and the OR transcriptional regulator, Lhx2. In both the cell line and in vivo, we show that LSD1 compartments interact with one or a small number of OR genes at the exclusion of the vast majority of OR loci. We speculate that these compartments might form a distinct complex during a narrow developmental window to restrict a chromatin-modifying function during OSN differentiation.
Methods
Mouse OP6 cell preparation and immunofluorescence
The OP6 cell line was cultured under media conditions described previously (Illing et al., 2002; Kilinc et al., 2014; Pathak et al., 2009). Briefly, OP6 cells are grown in Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies) supplemented with 10% fetal bovine serum (FBS, Gibco) at 33°C and differentiated by deactivating the large-T-antigen at 39°C for 4–15 days in DMEM-F12 media (Life Technologies) containing N2 supplement (Life Technologies), 100µM ascorbic acid (Sigma), and 10µM retinoic acid (Sigma). For subsequent immunofluorescence and FISH analysis, cells were seeded on 22cm2 coverslips coated with 0.1% gelatin(Sigma) in a 6 well plate at about 50% confluency and expanded for one day to near confluency. OP6 cells were incubated for 16 hours in 100µM nocodazole for G2/M synchronization, in 0.5mM L-mimosine for G1/S synchronization. Immunofluorescence conditions were modified slightly from procedures described elsewhere (Chaumeil et al., 2004). Briefly, cells were fixed with 3% paraformaldehyde for 10 minutes, permeabilized in 0.5% Triton-X (Sigma) for 10 minutes, and blocked in 1% BSA for 20 minutes at 37°C. The primary and secondary antibody incubations were performed at 37°C for 45 minutes in a humidified chamber. The primary antibodies used in this study were rabbit anti-Lsd1 (Abcam, ab129195, 1:100), mouse anti-Lsd1 (Millipore, 05-939, 1:100), chicken anti-α tubulin (Abcam, ab89984, 1:200), mouse anti-α tubulin (Sigma Aldrich, T5168, 1:1000), mouse anti-8-oxoguanine (Abcam, ab64548, 1:100), rabbit anti-Lhx2 (Millipore, AB5756, 1:200), and mouse anti-CoREST (Millipore, MABN486, 1:250). The secondary antibodies used in this study were donkey anti-mouse-Cy3 (Jackson Immunoresearch, 715-165-150, 1:100), donkey anti-rabbit Alexa 488 (Jackson Immunoresearch, 711-545-152, 1:100), goat anti-rabbit-Cy3 (Millipore, AP132C, 1:800) and goat anti-chicken Alexa 488 (Abcam, ab150173, 1:500). Parallel primary no-antibody controls were used in all experiments to ensure an absence of secondary antibody background staining. In experiments where antibodies were pooled, we tested for secondary antibody cross-reactivity by including controls in which each primary antibody was omitted.
Mitotic shake-off
Five T225 flasks were grown to ~85–90% confluency to obtain OP6 cell populations enriched for M-phase. The flasks were initially washed with 1X PBS to eliminate floating or dead cells. After adding fresh media, flasks were stacked and gently tapped against a hard surface several times to release mitotic cells. The media from all flasks were pooled and centrifuged. The pellet containing released M-phase cells were seeded into 4 well chamber slides and incubated at 33°C. Cells were fixed and immunofluorescence performed at specific time points. 150–200 cells were scored for each slide and two slides were analyzed for each time point.
Mouse olfactory epithelium sections and immunofluorescence
FVBN and B6/129S mice were used for LSD1/neurogenin/p27 and LSD1/CoREST immunofluorescence experiments, respectively. Mice were anesthesized using a triple cocktail of ketamine, xylazine, and acepromazine followed by intracardiac flush with phosphate buffered saline (PBS; pH 7.2) and perfusion with 1% paraformaldehyde-lysine-sodium periodate (for Neurog1-eGFP FVBN animals) or 4% paraformaldehyde. After gross dissection and extraction of the olfactory epithelium, tissue was immersed in fixative under a vacuum for 2–3 hours and subsequently decalcified in saturated ethylenediaminetetraacetic acid (EDTA) overnight. Tissue was then cryoprotected overnight with 30% sucrose and stored in OCT frozen via liquid nitrogen before cryosectioning at 10µm. Immunostaining was performed according to published procedures (Krolewski et al., 2012). Sections were treated with 3% hydrogen peroxide in methanol for 5 minutes, boiled in 0.01M citric acid buffer (pH 6.0) using a commercial food steamer, and incubated in normal donkey block (10% serum + 5% Non fat dry milk + 4% BSA +0.1% Triton X-100) for 15 minutes at room temperature. Sections were incubated with primary antibody diluted in blocking solution (rabbit anti-LSD1, chicken anti-GFP, mouse anti-p27, and mouse anti-CoREST) for 1 hour at room temperature. Visualization of the anti-LSD1 staining was performed using tyramide signal amplification (TSA) with FITC (home-made) or TSA-Plus Cy3 kit (Perkin Elmer, NEL744001KT,1:100). Following TSA, the sections incubated in secondary antibodies conjugated to fluorescein cyanine-3 and Alexa-647 for one hour at room temperature. Nuclei were stained with DAPI for 10 minutes and the sections were sealed with n-propyl gallate. For subsequent DNA FISH experiments, cells were post-fixed in 4% PFA for 1 hour (OP6 preparations) or 10 minutes (MOE preparations) at RT. The following antibodies were used in the staining of mouse MOE sections: rabbit anti-LSD1 (Abcam #ab129195, 1:16,000 for IF (biotin-anti-rabbit, Streptavidin-HRP, TSA-FITC) and 1:1000 for DNA-immunoFISH experiments (anti-rabbit-HRP (4°C, overnight), TSA-Plus-Cy3)), chicken anti-GFP (Abcam, 13970, 1:400), mouse anti-p27 (BD Transductions Laboratories, 610241, 1:25), mouse anti-CoREST (Millipore, MABN486, 1:50), biotin anti-rabbit (Jackson Immuno Research, 711-065-152, 1:100), streptavidin HRP (Jackson Immuno Research, 016-030-084, 1:100), anti-rabbit-HRP (Amersham, NA934, 1:500), donkey anti-chicken-cy3 (Jackson Immuno Research, 703-165-155, 1:100), donkey anti-mouse-647 (Jackson Immuno Research, 715-605-150, 1:100), and donkey anti-mouse-cy3 (Jackson Immuno Research, 715-165-150, 1:100).
DNA fluorescence in situ hybridization (DNA FISH)
DNA FISH was performed on cultured OP6 cells and MOE using BAC clones obtained from BACPAC Resource Center (CHORI); see Tables 1 and 2 below. BAC DNA was nick-translated with DIG or biotin according to manufacture’s instructions (Roche Applied Sciences). Approximately 100ng nick translated probe was mixed with 5µg Cot1-DNA (Invitrogen) and 10mg salmon sperm DNA (Sigma) per reaction. When pooling BACs, ~100ng nick translated template per BAC clone was incubated with 70µg Cot1-DNA and 70mg salmon sperm DNA. For some experiments, a “pan OR” DNA FISH probe was produced by PCR using degenerate primers against well conserved OR sequences. Three different degenerate PCR assays were used on genomic DNA template: 135 (5’ATGGCITAYGAYMGITAYGTIGCIATHTG3’)/P8 (5’RTTICKIARISWRTAIATRAAIGGRTT3’), P26 (5’GCITAYGAYCGITAYGTIGCIATITG3’)/P27 (5’ACIACIGAIAGRTGIGAISCRCAIGT3’), and 5B (5’CCCATGTAYTTBTTYCTCDSYAAYYTRTC3’)/P8. DIG-11-dUTP or Biotin 16-dUTP was incorporated during PCR amplification; degenerate products were digested with MluCI and pooled. This degenerate PCR approach has been used by a number of groups to generate 50–70% of mouse OR templates with high specificity from complex genomic or cDNA templates (Clowney et al., 2012; Dulac and Axel, 1995; Malnic et al., 1999). We tested the specificity of the pan-OR probe by conducting a high-stringency Southern blot with 24 OR-containing BACs and 9 non-OR-containing BACs. As expected, we do not observe any pan-OR hybridization signals against the restriction fragments from the 9 non-OR BACs. We observe 6 hybridization signals against fragments from the 24 OR-containing BACs that do not correspond to annotated OR genes, which may be false-positives or may correspond to non-annotated OR pseudogenes; nevertheless, the overall false-positive rate in these Southern-blot experiments is <1% (a maximum of 6 false-positive fragments in ~2700 total BAC fragments). We observe robust signals against 139 (~61%) of the 227 OR-containing BAC fragments, a result that seems consistent within theoretical expectations (50–70%) given the high-stringency hybridization conditions used. To gain further insights about a false-negative rate for the pan-OR probe in the context of DNA FISH, we conducted two-color experiments with five BAC probes that hybridize to clusters containing 6, 9, 23, 29, and 226 OR genes at their respective chromosomal locations. These five BAC probes produce an average of 3.1 hybridization signals per OP6 nucleus (the OP6 cell line is partially polyploid) (Kilinc et al., 2014). Depending on the probe, the percent co-localization of these signals with the pan-OR probe ranges from 61%-100%, with an average of ~77% co-localization (the pan-OR probe exhibited the lowest false-negative incidence when using BAC probes against large, multi-OR gene clusters). Although this 5-BAC sample is small relative to the full range of OR regions in the genome, these results suggest a modest false-negative rate of <~25% for OR locus recognition using DNA FISH.
Table 1.
Non-OR BAC clones used in this study
| BAC Name | Locus |
|---|---|
| RP24-170L5 (Kif1a) | chr1:94,881,403–95,042,788 |
| RP23-5J14 (actin) | chr5:143,564,278–143,779,524 |
| RP24-274J4 (actin) | chr5:143,523,664–143,689,737 |
| RP24-149A5 (Gusb) | chr5:130,397,230–130,570,162 |
| RP24-358O6 (MyoD) | chr7:53,530,383–53,705,205 |
| RP23-105L18 (Dnm2) | chr9:21,050,795–21,278,503 |
| RP23-220F2 (Ccdc65) | chr15:98,431,217–98,630,847 |
| RP23-155O16 (G-olf) | chr18:67,213,619–67,398,266 |
Table 2.
OR BAC clones used in this study
| BAC Name | Locus |
|---|---|
| RP23-152D9 | chr1:175,023,551–175,239,918 |
| RP23-295H11 | chr2:111,797,452–112,006,004 |
| RP23-291E23 | chr2:36,716,066–36,929,338 |
| RP23-21E22 | chr2:85,524,721–85,759,991 |
| RP23-52M7 | chr2:87,364,531–87,596,132 |
| RP23-35G11 | chr2:89,295,676–89,510,619 |
| RP24-305L5 | chr4:118,319,956–118,530,240 |
| RP23-39C21 | chr6:116,434,892–116,614,239 |
| RP23-109D17 | chr6:42,706,937–42,919,097 |
| RP23-10D18 | chr7:109,634,112–109,873,997 |
| RP23-317I5 | chr7:111,662,557–111,857,606 |
| RP23-299K19 | chr7:114,059,696–114,280,482 |
| RP24-166E20 | chr7:147,273,883–147,436,184 |
| RP23-383P11 | chr7:147,495,086–147,704,126 |
| RP23-415N10 | chr7:6,312,473–6,527,344 |
| RP23-14C11 | chr7:93,425,492–93,664,279 |
| RP23-275I18 | chr9:19,517,332–19,669,581 |
| RP23-289G7 | chr9:38,466,999–38,680,486 |
| RP23-79I22 | chr10:128,951,917–129,202,286 |
| RP23-133O2 | chr10:78,285,813–78,510,630 |
| RP24-82K5 | chr11:73,137,651–73,346,664 |
| RP23-205I24 | chr11:73,587,308–73,813,986 |
| RP23-61D10 | chr13:21,612,336–21,852,385 |
| RP23-345I5 | chr14:51,193,469–51,398,516 |
| RP23-6D17 | chr14:53,012,601–53,243,661 |
| RP24-95L8 | chr16:59,175,390–59,353,402 |
| RP23-68M6 | chr17:38,127,641–38,371,832 |
| RP24-73D4 | chr19:12,301,032–12,519,739 |
For DNA FISH on OP6 cells, cells were fixed, permeabilized and dehydrated in an 80%, 95%, 100% ethanol series, prior to incubation in 50% formamide/2X SSC for 20 minutes at room temperature. Cells were denatured at 85°C for 30 minutes and then hybridized with heat-denatured probes overnight at 37°C. Following hybridization, cells were washed three times with 50% formamide/2X SSC for 5 minutes each and blocked in 4% BSA/4X SSC/0.2% Tween-20 for 20 minutes at 37°C in a humidified chamber. Anti-DIG or avidin antibody incubations in 1% BSA/4X SSC/0.2% Tween-20 were performed for 45 minutes at 37 °C in a humidified chamber. FISH signals were detected with secondary anti-DIG-FITC (Roche, 11207741910), donkey anti-sheep-FITC (Santa Cruz Biotechnology, sc-2476), Avidin-FITC (Vector Labs, A-2011), avidin-rhodamine (Vector Labs, A-2012) or biotinylated anti-avidin (Vector Labs, BA-0300) in various experiments. DNA FISH on MOE sections was performed as described previously (Armelin-Correa et al., 2014).
Microscopy and Image analysis
Images were acquired using a Deltavision RT imaging system (Applied Precision) adapted to an Olympus (IX71) microscope equipped with XYZ motorized stage. Each image was sectioned with 0.5µm intervals to ensure complete coverage of the nucleus. Deconvolved images of MOE sections were generated using Softworx (Applied Precision). Lsd1 compartments were analyzed in FIJI using z-projection of deconvolved images. We defined the LSD1 punctates as an area exhibiting twice the LSD1 immunofluorescence intensity as compared to each of at least three random areas of the nucleus; the mono-punctate cells exhibited only one such area per nucleus, whereas the poly-punctate cells exhibited 2–4 such areas per nucleus. The same criteria were applied when analyzing Co-REST protein consolidation. Lsd1 compartments in MOE sections were also validated using 3D Surface Plot of FIJI software where they match the highest intense regions in nucleus. DNA intensity within as compared to immediately surrounding LSD1 compartments was performed using Radial Profile plug-in of FIJI on deconvolved z-stack projections of MOE sections.
Co-immunopurification experiments
Nuclear protein was isolated from OP6 cell populations using protocols from the Nuclear Complex Co-IP kit (Active Motif #54001). Immunoprecipitations were performed with 5 µg of antibody at 4°C overnight, followed by incubation in a Protein-G agarose column (Active Motif #53039) for 1 hour at 4°C. Two IP antibodies were used: rabbit anti-LSD1 (Abcam #ab129195) and mouse anti-CoREST (Millipore #MABN486). Retained precipitates were washed and eluted using 2× Reducing Buffer (130 mM Tris pH 6.8, 4% SDS, 100 mM DTT). IP fractions were run on 10% SDS-PAGE gels and blotted on PVDF membrane. Membranes were blocked in 5% BSA, incubated in primary antibodies at 4°C overnight in 5% BSA, and secondary antibodies for 1 hour at room temperature in 5% BSA. Following washing, alkaline-phosphatase signals were visualized with NBT/BCIP per standard protocols. The following antibodies were used in various Western blots: rabbit anti-LSD1 (Abcam #ab129195, 1:1000), mouse anti-CoREST (Millipore #MABN486, 1:1000), donkey anti-rabbit-AP (Abcam #97061, 1:1000), and goat anti-mouse-AP (Sigma #A3562, 1:1000).
Results and Discussion
The OP6 cell line as a model system for OR choice and switching
We used an immortalized cell line (OP6) that is derived from E10 mouse olfactory placode and expresses a transcriptional profile representing post-progenitor immature receptor neurons (Illing et al., 2002; Pathak et al., 2009). These cells can be induced to further differentiate along the lineage by addition of retinoic acid and by preventing re-entry into the cell cycle via deactivation of the temperature-sensitive large-T-antigen (Illing et al., 2002; Pathak et al., 2009). We have previously shown that both undifferentiated and differentiated OP6 cells express OR genes monoallelically and monogenically (Kilinc et al., 2014; Pathak et al., 2009). Since normalized OR expression levels in these cells are significantly lower than has been described for mature OSNs (Iwema and Schwob, 2003; Tietjen et al., 2003; Zhang et al., 2004b), and other markers for mature OSNs are not fully realized even in the differentiated state (Illing et al., 2002; Pathak et al., 2009), we presume that full maturation in the lineage is not occurring in these cultures. Consistent with this perspective, we observe that OR switching occurs in the OP6 cell line (Pathak et al., 2009), suggesting that the feedback inhibition mechanism associated with stabilizing OR choice is not occurring. Moreover, we do not observe complete consolidation of silenced OR genes at the nuclear chromocenters (Kilinc et al., 2014), an organization that is essential for maintaining mutually exclusive OR transcription in mature OSNs (Clowney et al., 2012). Together, our observations suggest that the OP6 cell line represents a differentiated state that is partially advanced along the lineage; i.e., after an initial OR choice is specified yet prior to commitment to that choice during maturation.
The lysine-specific demethylase-1 (LSD1) has been implicated as a factor involved with the mutually exclusive removal of repressive histone-3-lysine-9 (H3K9) methylation marks at OR loci during the selection process (Lyons et al., 2013). LSD1 is down-regulated in mature OSNs after OR choice (Krolewski et al., 2013; Lyons et al., 2013), which could be an essential step to ensure commitment to a single OR choice; i.e., to prevent removal of these repressive marks on additional OR genes after a choice is made (Dalton et al., 2013; Lyons et al., 2013). In OP6 cells, LSD1 is not down-regulated in the undifferentiated or even the more differentiated cells, another indication that OP6 cells do not fully mature in culture (Fig. 1). Therefore, if LSD1 down-regulation is part of the feedback inhibition process required to stabilize OR choice, then its persistence in OP6 cells might partly account for ongoing OR switching in the cell line.
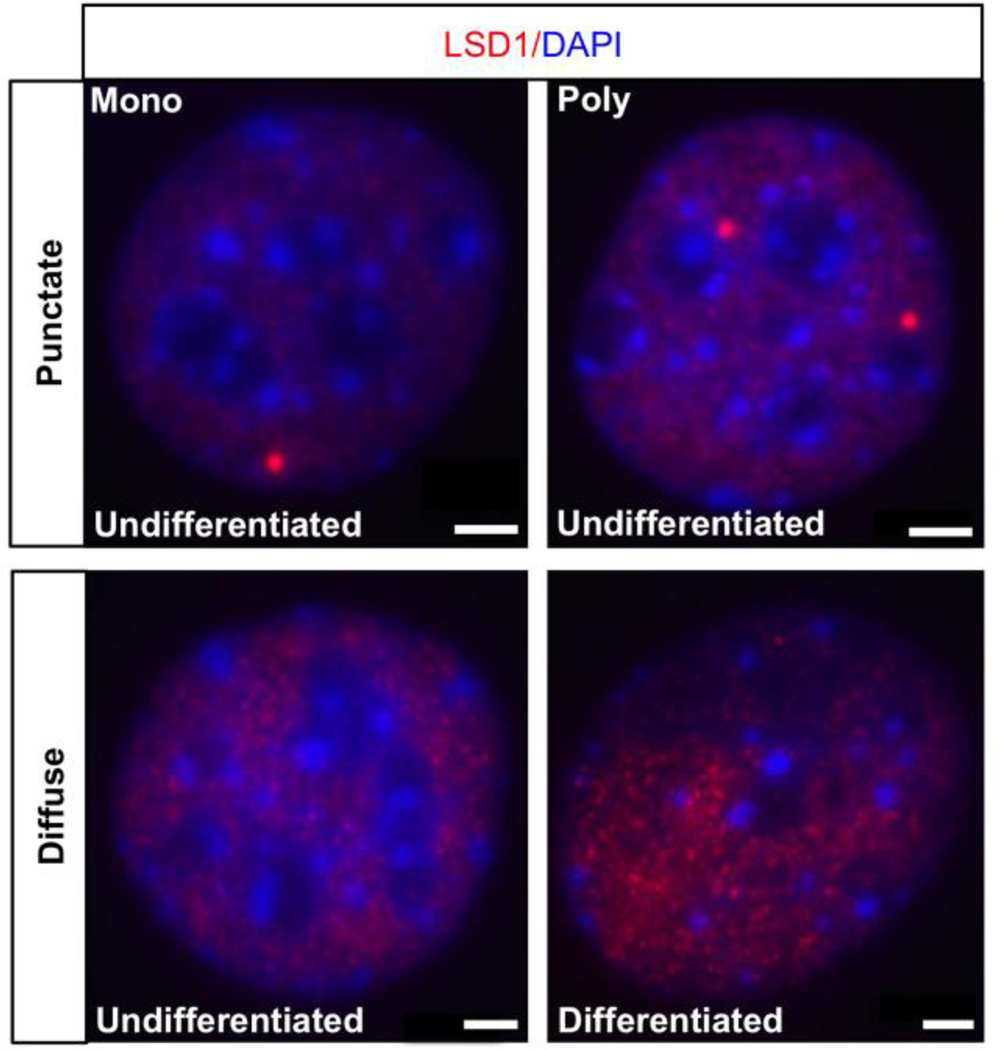
Figure 1. LSD1 exhibits mono and poly punctate compartmentalization in undifferentiated OP6 cells.
LSD1 (red) consolidates into one (“Mono”, top left panel) or a small number (“Poly”, top right panel) of foci in ~15% of undifferentiated OP6 cells. The remaining ~85% of undifferentiated OP6 cells exhibits diffuse LSD1 staining (bottom left panel). Differentiated OP6 cells do not exhibit any evidence of LSD1 compartmentalization (bottom right panel). Cell nuclei are stained with DAPI (blue). Scale bar: 3µm.
LSD1 exhibits a consolidated phenotype in early G1 of OP6 cells
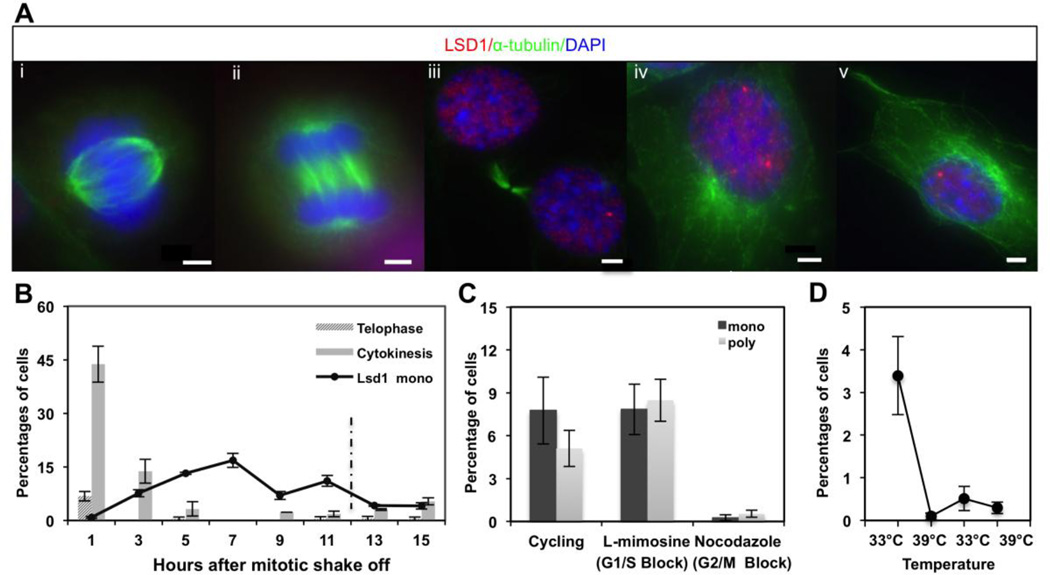
We observe a variable organization of LSD1 protein within nuclei of cycling OP6 cells. The majority of cycling OP6 cells (~85%) exhibits a diffuse nuclear staining of LSD1 protein. LSD1 is lost within nuclei during M-phase of the cell cycle but returns within each daughter cell nucleus in early G1 (Fig. 2A) (Blobel et al., 2009; Nair et al., 2012). Notably, we observe that LSD1 exhibits a consolidated phenotype in ~15% of OP6 cell nuclei, including consolidation into a single, dominant compartment in ~8% of the nuclei (Fig. 1). We blocked the cells at the G2/M boundary by treatment with nocodazole and observe no consolidated LSD1 phenotypes, indicating that LSD1 consolidation is disrupted during S and/or G2 phase (Fig 2). The absence of the consolidated LSD1 phenotype after G2 and during M-phase suggests that LSD1 nuclear compartmentalization is likely occurring during G1. We synchronized OP6 cells using a mitotic shake-off method (see Methods), which enriches for a population of cultured cells at M-phase (Schorl and Sedivy, 2007). The incidence of LSD1 compartmentalization steadily rises during the first 7 hours after mitotic shake-off up to a maximum of ~17%; after this maximum at 7 hours, the incidence decreases significantly up through the point in time when the majority of cells are expected to re-enter the next cell cycle at approximately the 12 hour mark (Fig. 2B). These observations suggest that LSD1 compartmentalization is a transient event that occurs early during G1 after OP6 cells exit the cell cycle. To further investigate this hypothesis, we blocked OP6 cells at the G1/S boundary using L-mimosine (Fig. 2C). We find that ~7.5% of the cells exhibit the single-compartment phenotype in these populations, which is significantly less than the maximum (~17%) observed during the time course, and comparable to the incidence between 11–13 hours after M-phase when a majority of released cells are expected to be re-entering the cell cycle (Fig. 2B). These observations are consistent with an interpretation that LSD1 compartmentalization peaks at some point after cytokinesis and prior to the next G1/S boundary. The overall incidence at any one time point never exceeds 20% of the cells in the population. The lack of a sharp, dominant peak might be explained by imprecise synchrony of the cell population, very transient LSD1 consolidation relative to the temporal resolution in the experiment and/or if only a fraction of cells compartmentalize LSD1 each cell cycle.
Figure 2. LSD1 compartmentalization appears to be enriched during early G1 phase of the cell cycle in immortalized OP6 cell populations.
A. LSD1 (red) is excluded from the nucleus during metaphase (i) and anaphase (ii), staged by α-tubulin staining (green) showing mitotic spindle organization at these respective phases of the cell cycle. LSD1 returns to the nucleus in each new daughter cell (iii) and starts to compartmentalize into one or more number of compartments during G1 phase (iv, v). Nuclei are stained with DAPI (blue). Scale bars: 3 µm. B. The average percent distribution of telophase, cytokinesis and mono-punctate LSD1 phenotypes after OP6 cell synchronization at M-phase by mitotic shake-off (see Methods). The approximate time point when cells are expected to re-enter S-phase (12 hours) is indicated with a dashed line. Error bars represents upper and lower limit in two experiments; ~200 cells were scored at each time point per experiment. C. The average percentages of cells exhibiting the “mono-punctate” and “poly-punctate” LSD1 phenotype in cycling OP6 cells, as well as cells blocked at the G1/S and G2/M cell cycle boundaries. Error bars represent standard error of the mean from three independent experiments; ~150 cells were scored in each experiment. D. The average percentage of OP6 cells exhibiting combined poly- and mono-punctate LSD1 organization under initial cycling conditions, followed by deactivation of the large T-antigen at 39°C for days 1–3, followed by reactivation of the large T-antigen at 33°C for days 3–5, followed by a second deactivation of the large T-antigen at 39°C for days 5–7. Error bars reflect standard error of the mean from three independent experiments; ~350 cells were scored in each experiment.
The fact that LSD1 compartmentalization appears to peak shortly after exit from M-phase raises the possibility that this organization is regulated by cell-cycle factors. To further explore this question, we differentiated OP6 cells, a process that involves permanent exit from the cell cycle. We observe no incidence of LSD1 compartmentalization in differentiated OP6 cells; all differentiated OP6 cells exhibit a diffuse distribution of LSD1 protein (Fig. 1). These observations suggest that either developmental progression and/or loss of cell cycling prevent consolidation of LSD1 protein. To distinguish between these two possible interpretations, we conducted a simpler experiment in which the large-T-antigen was deactivated in the absence of any differentiation factors; under these conditions, OP6 cells do not divide or differentiate. After two days without cycling, we again observe a complete absence of LSD1 compartmentalization; when we subsequently reactivate the large-T-antigen in these previously stagnated OP6 populations, we observe a small but appreciable return of LSD1 compartmentalization after two additional days of growth (Fig. 2D). Together, our data suggest that the compartmentalization of LSD1 protein might be dependent on regulatory factors transiently present in cells that have recently undergone mitosis. To dismiss the possibility that this organization within OP6 cells is an artifact of the large-T-antigen itself, we next investigated LSD1 organization within nuclei of developing OSNs of the MOE.
LSD1 protein is compartmentalized in early post-mitotic cells of the OSN lineage
The mouse olfactory epithelium (MOE) is stratified, with globose and horizontal basal cells that give rise to olfactory sensory neurons (OSNs) positioned along the inner basal layers, the post-mitotic immature neuronal layers occupying the middle regions, and the mature OSNs along with supporting non-neuronal sustentacular cells in the outer apical layers. We used anti-eGFP antibodies to identify Neurog1-expressing globose basal cells (GBCs) and immature neurons in Neurog1-eGFP BAC-expressing mice, and an antibody against p27Kip1 (p27) to visualize post-mitotic cells, in order to characterize the expression domain and consolidation phenotypes of LSD1 in vivo. We observe a partial overlap of eGFP- and p27-positive cells in the basal regions of the MOE (Fig. 3). We presume that these double-positive cells represent the most recently specified cells of the OSN lineage where p27 has been activated as a newly post-mitotic cell and GFP persists from previous Neurog1 expression associated with basal cells. Curiously, we find that p27, which regulates re-entry into the cell cycle, does not persist in all post-mitotic cells of the lineage, and is expressed only in a small, inner layer of cells immediately adjacent and partially overlapping with eGFP-positive cells. The majority of the developing neurons in the MOE (>90% of cells) do not express p27, even though these cells are post-mitotic (Fig. 3) (Guo et al., 2010). Therefore, it appears that p27 function is only required at earlier stages of OSN differentiation. We note that although p27 is down-regulated in the outer MOE layers (including mature OSNs), it robustly expresses in non-neuronal sustantacular cells that occupy the outermost layer of the MOE (Fig. 3).
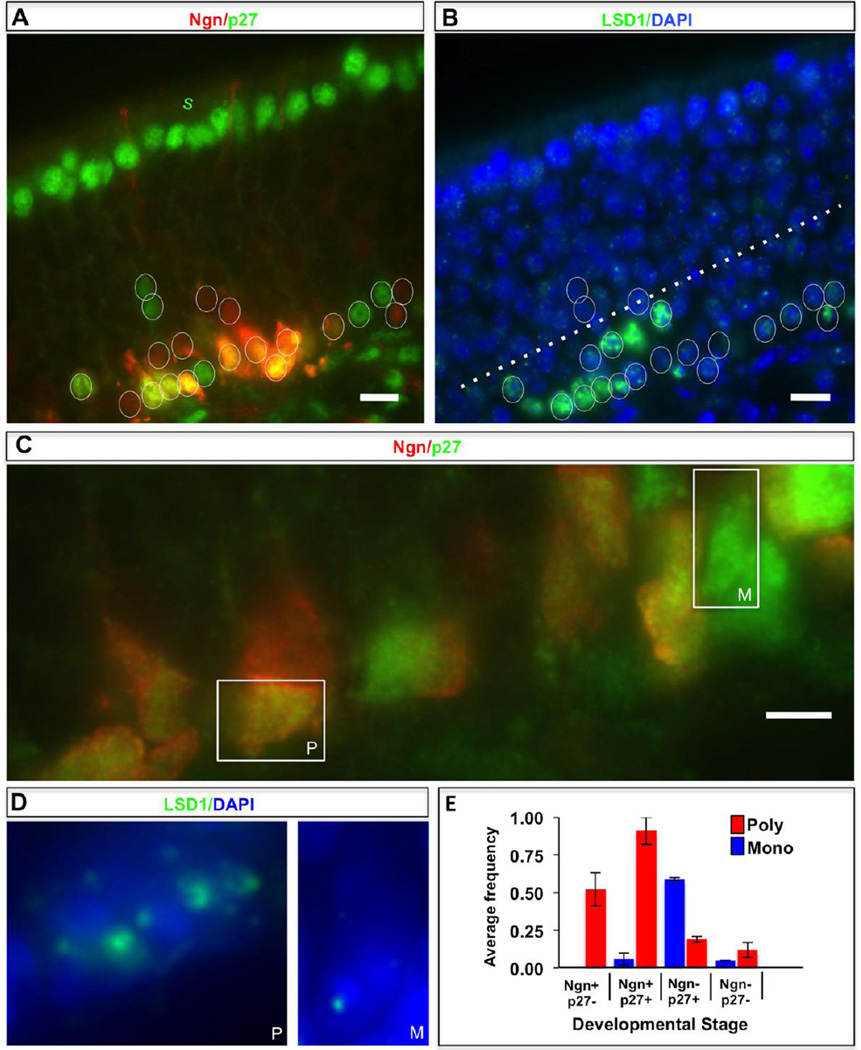
Figure 3. LSD1 expression and compartmentalization in the most immature cells of the MOE.
A. The mouse olfactory epithelium (MOE) is organized with the inner basal layers consisting of globose basal cells, a subset of which are neurogenin-eGFP (Neurog1)-positive (red), intermediate layers with immature neurons at various stages of differentiation, and the outer apical layers consisting of mature olfactory sensory neurons, as well as supporting sustentacular cells (denoted by s). Expression of the p27Kip1 protein (green-false colored), required during differentiation to prevent further entry into the cell cycle, is restricted to the earliest post-mitotic cells immediately adjacent to the basal cells, as well as the outermost sustentacular cell (s). Individual nuclei of immature cells of the OSN lineage that are eGFP-positive, p27-positive, or positive for both markers are circled. Scale bar:10 µm. B. The identical MOE section as shown in Panel A (with the same circled cells) exhibiting DNA (blue) and Lsd1 protein (green) staining. LSD1 is expressed in the basal cell layer, as well as the p27-positive cells in the inner ~25% of the MOE (below the dotted line). Scale bar: 10 µm. C. A larger-scale image (scale bar: 5 µm) of a portion of the inner MOE layers with a boxed eGFP-p27 double-positive cell (yellow) and p27-positive cell (green-false colored), which exemplify the poly-punctate (P) and mono-punctate (M) LSD1 organization, respectively, as shown in Panel D. D. LSD1 staining within the same two boxed nuclei from Panel C showing a “poly-punctate” (P, left) and “mono-punctate” (M, right) organization, respectively. E. Histogram estimating the average frequency for two MOE sections (error bars = one standard deviation) of the “poly-punctate” (red) and “mono-punctate” (blue) LSD1 phenotypes in MOE cell populations scored for neurogenin (Ngn+) and p27Kip1 (p27+) expression.
LSD1 is expressed in the late GBC layer (eGFP-positive cells), as well as in the innermost layers containing the least differentiated cells of the OSN lineage; e.g., in p27-positive cells (Fig. 3). As expected, LSD1 is increasingly down-regulated towards the outer layers of the MOE, including mature OSNs, confirming results published elsewhere (Krolewski et al., 2013; Lyons et al., 2013). Therefore, LSD1 is expressed in both the dividing basal cells and in the early post-mitotic cells of the OSN lineage; these are the cell populations where OR selection is presumably occurring (Iwema and Schwob, 2003; Nguyen et al., 2007; Shykind et al., 2004). Importantly, we observe that the LSD1 protein exhibits a variable organization within the nuclei of p27-positive cells, just as was observed in OP6 cell populations. In ~67% of p27-positive cells, LSD1 protein is consolidated into a small number of foci, a phenotype we refer to as “poly-punctate”, and LSD1 protein is consolidated into a single compartment in ~23% of p27-positive cells, a phenotype we refer to as “mono-punctate” (Fig. 3). The remaining ~10% of p27-positive cells exhibit a more diffuse organization of LSD1 protein. Of note, in most cells, the single LSD1 compartment (“mono-punctate”) is positioned on the edge of a nuclear chromocenter where OR genes are generally located (Fig. 5A, 5C) (Clowney et al., 2012).
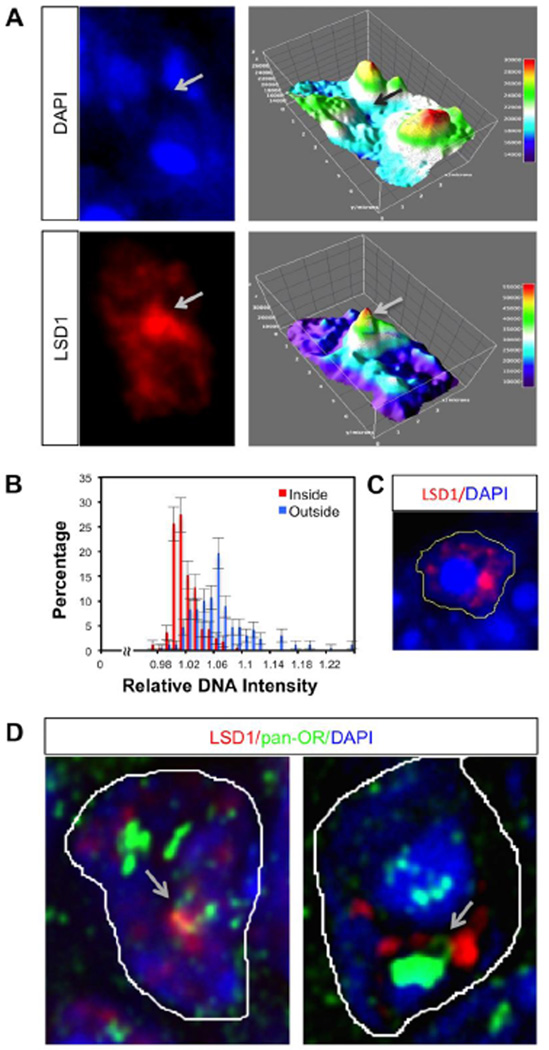
Figure 5. LSD1 compartments show decreased DNA staining relative to surrounding regions of the nucleus.
A. An immature OSN nucleus with DAPI and LSD1 staining (left panels) and their corresponding Thermal LUT 3D Surface plots generated using ImageJ (right panels) Signal intensity is color coded (representing low values with blue and high values with red) and the luminance of the image is interpreted as height for the plot. B. A histogram showing the percentage frequency of relative DNA intensities inside (originating from the center of the compartment (0) throughout the edge of the compartment (R)) and outside (from the edge of the compartment (R) out to one radial distance (2R)) of the compartment is constructed using the radial profile plug-in of ImageJ. The integrated intensities along this distance (0–2R) are normalized to the center (0) of each compartment. The frequency distributions of 0–2R distance of 30 different cells are depicted. Error bars represent counting errors, which were estimated as one standard deviation of total measurements of intensities, calculated from the binomial distribution C. A representative image showing the positioning of an LSD1 compartment at the edge of a nuclear chromocenter, as depicted by intense DAPI staining. Yellow line represents the boundary of the nucleus. D. Two representative images illustrating DNA loops (arrows) for OR loci that interact with LSD1 compartments.
LSD1 consolidation in the MOE appears to occur in a stepwise manner. Although both poly- and mono-punctate LSD1 phenotypes are prevalent in p27-positive cells, their respective occurrence correlates with the presence or absence of neurogenin in this population (Fig. 3E). Within the p27-positive subpopulation, most (>90%) of the poly-punctate cells are neurogenin (eGFP)-positive, whereas most (>85%) of the mono-punctate cells are neurogenin (eGFP)-negative. Therefore, the poly-punctate organization appears to precede the mono-punctate organization during developmental progression in the MOE. Together, our data indicates that LSD1 protein becomes increasingly consolidated (poly- to mono-punctate) in vivo, and that the consolidation into a single nuclear compartment is a common organization (>60% incidence) during the narrow developmental window in p27-positive cells that have recently lost the neurogenin basal cell marker. Both the OP6 cell line data and these observations from the MOE hint that LSD1 compartmentalization is associated with recent exit from the cell cycle, as might be predicted if this organization was involved regulating target genes (e.g., OR genes) within a newly differentiated daughter cell.
LSD1 compartments exhibit restricted associations with OR gene loci
We conducted DNA FISH experiments to investigate whether OR gene loci interact with LSD1 compartments in both OP6 cells and in the context of the MOE. We used BAC-sized probes each containing portions of various OR gene clusters. We find that individual OR probes co-localize with LSD1 compartments, however the incidence for each probe tested was very low in the overall population (Fig. 4A). We conducted 22 DNA FISH experiments on OP6 cells using individual OR-containing BAC probes, imaging ~30–50 nuclei with compartmentalized LSD1 in each experiment. This was not an adequate sample size, since most BAC probes exhibited <3 co-localizations per experiment; tentatively, we estimate that the average frequency was ~1% (or less) per probe. One possible explanation for this rarity is that few OR genes are permitted access to LSD1 compartments per cell, perhaps as part of a mechanism that ensures only one or a small number of OR genes are targeted by LSD1 at a time.
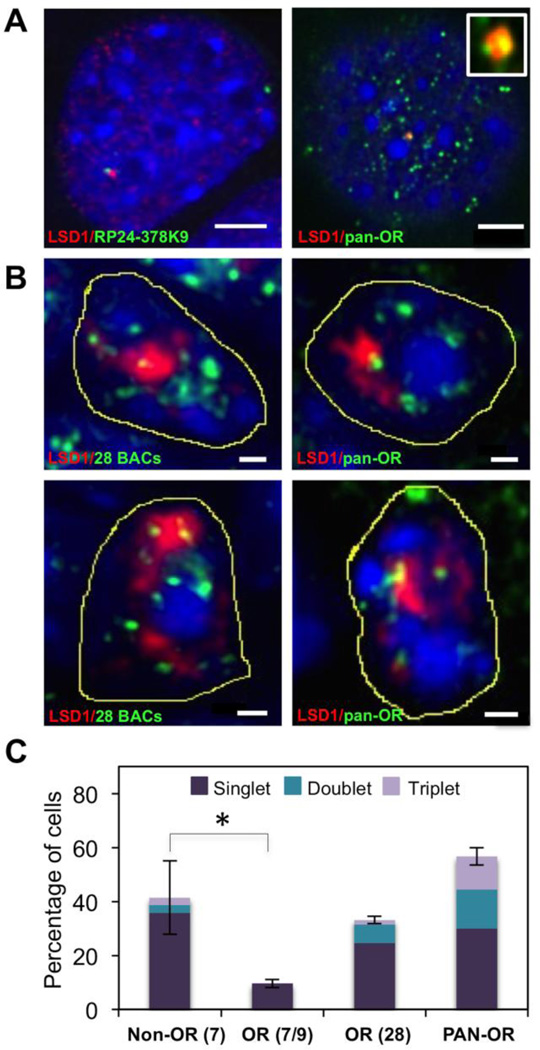
Figure 4. LSD1 compartments co-localize with OR gene loci in OP6 cells and in the MOE.
A. DNA FISH using probes made from individual OR-containing BAC clones co-localize with “mono-punctate” LSD1 compartments (red) at a very low incidence in OP6 cell populations (left panel). A “pan-OR” DNA FISH probe, constructed from a degenerate PCR product from multiple OR templates encoded in mouse genomic DNA, hybridizes to ~100 chromosomal regions per nucleus. This “panOR” probe exhibits exclusive associations (~one per nucleus) with the LSD1 compartment (right panel); we observe some examples in which more than one “pan-OR” association occurs within a single compartment (right inset). Scale bars: 3 µm. B. DNA FISH using pool of 28 OR-containing BAC clones or the “pan-OR” probe co-localize with “mono-punctate” compartments (top panels) in MOE nuclei. Some compartments exhibit more than one OR signal per compartment (lower panels). Scale bar:1 µm. Condensed DNA is stained with DAPI (blue) in all of the above images. C. The average percentage (error bars indicate minimum and maximum values of two independent experiments) of co-localized DNA FISH signals (fully embedded) is shown for a pool of 7 non-OR BAC probes, a pool of 7 or 9 OR BAC probes, a pool of 28 OR BAC probes, and the “pan-OR” probe. The fraction of singlet, doublet and triplet co-localizations per compartment is denoted by indicated colors. Approximately 30–50 MOE cell nuclei with “mono-punctate” LSD1 organization were analyzed per experiment. A two-tailed unpaired Student’s t-test comparing non-OR(7) and OR BACs (7/9) demonstrates a statistically significant difference (p=0.004), as denoted by the asterisk.
To explore whether OR access to an LSD1 compartment is exclusive, we next conducted DNA FISH experiments on MOE sections using pooled OR-containing BAC probes, as well as with a “pan-OR” probe made from degenerate PCR on gDNA templates (see Methods). We scored two different co-localization categories: an “embedded” signal is an OR locus that is fully encompassed by the LSD1 compartment, whereas a “touching” signal is an OR locus that is at the very edge of the LSD1 compartment (not enclosed, but no intervening space is evident). When using large 28-BAC pools or the pan-OR probe, we find that almost all of the DNA FISH signals reside outside of the LSD1 compartment (Fig. 4). This is especially evident in OP6 cells, in which OR genes are less consolidated than they are in vivo (Kilinc et al., 2014), where we identify ~110 (+/− ~30) distinct spots per nucleus using the pan-OR probe, with only ~one signal (and at most three signals) located within a compartment. (Fig. 4A) The total number of pan-OR FISH signals per nucleus presumably under-estimates the number of OR genes being detected (since all genes within a single OR cluster will not be resolved), but is likely to minimally identify a majority of OR-containing chromosomal loci. Our results indicate that the vast majority of OR genes/gene loci are excluded from the LSD1 compartment when it forms.
While the vast majority of OR loci are not interacting with an LSD1 compartment in any given cell, it appears that each LSD1 compartment in the population might interact with at least one OR locus. When examining large 28-BAC pools on MOE sections, we observe ~33% of the mono-punctate LSD1 compartments (24/73 compartments) in the MOE contain an “embedded” OR signal. This percentage increases to ~57% (47/83) of the mono-punctate LSD1 compartments with an “embedded” OR signal when using the pan-OR probe; if the “touching” phenotype is included, ~87% of the mono-punctate compartments are associated with a pan-OR signal (Fig. 4B). Assuming an approximate false-negative rate of ~30% for the pan-OR probe (see Methods), these observations suggest that a very large fraction, if not all, of the LSD1 compartments interact with an OR. Moreover, the observed strong correlation between the percentage of LSD1 compartments exhibiting an associated DNA FISH signal and the size of the DNA FISH pools used in the experiment (Fig. 4C) is consistent with the hypothesis that OR associations are common from cell to cell, but exclusive to one or a small number of ORs per cell.
While we infer a degree of “exclusivity” from these experiments, our data do not support a hypothesis in which LSD1 compartment associations are “mutually exclusive” to a single OR gene. First, we note that BAC-sized probes contain a cluster of multiple OR genes; given the DNA FISH signals are commonly punctate and fully enclosed within an LSD1 compartment, we presume that multiple OR genes from the cluster are present therein. Second, when scoring large 28-BAC probe pools, as well as the pan-OR probe, we identify some LSD1 compartments in the MOE containing more than one DNA FISH signal (Fig 4B). However, regardless of probe pool size, we never observe more than 3 signals per compartment, suggesting an upper limit of OR-LSD1 compartment interactions per cell. Nevertheless, the fact that the compartment is not strictly “mutually exclusive” does not rule out a role for these compartments in contributing to the exclusive activation of only one OR gene per cell; e.g., if the compartments serve to gather one or a small number of eligible loci, yet only one gene of one cluster might be positioned in the complex so that it is subsequently processed and delivered to a Pol2 factory.
Finally, we wondered if the LSD1 compartment is exclusive to OR loci, or whether other non-OR loci associate with the compartment. We tested 7 non-OR containing BAC probes (see Methods) that contain both housekeeping and developmental genes. When tested individually, we observed a much higher incidence of co-localization with the LSD1 compartment than we ever observed with any OR-containing BAC probe. None of these randomly selected non-OR loci appear to be fully excluded from the compartment. When we pooled these 7 non-OR BAC probes, we observed ~4-fold higher incidence of overlap as compared to the pool of 7 OR-containing BAC probes, and approximately the same incidence of overlap as observed for a much larger pool of 28 OR-containing BAC probes (Fig. 4C). Although we do not yet know anything about the specific activities of the LSD1 compartment, this observation raises the possibility that these compartments could serve a broader (i.e., non-OR-specific) function. A more generic role for these compartments might also predict a similar consolidated LSD1 organization in early post-mitotic cells of other, non-OSN lineages. Yet, the apparent lack of discrimination among a random set of non-OR loci could also mean that these non-OR associations are simply unregulated and inconsequential. If this is the case, and non-OR associations indeed represent the “random” or “unbiased” incidence, then it would seem that OR associations are under-represented (t-test p<0.01) as compared to the expected level of chance interactions. Such a conclusion would suggest that OR loci are actively excluded from the LSD1 compartments, perhaps as part of a mechanism to restrict widespread LSD1 activity at OR loci.
Decondensation and demethylation activity within LSD1 compartments
We make two interesting observations hinting that chromatin might be decondensed when associated with LSD1 compartments. First, we observe a depression of DAPI stain correlated with the space within as compared to immediately surrounding LSD1 compartments (Fig 5A). This observation is consistent with the fact that LSD1 compartments tend to reside at the edges of chromocenters, where DNA is highly condensed (an example is illustrated in Fig. 5C). The surface plot in Figure 5A suggest that LSD1 compartments are a local minimum for DAPI stain, and among the least stained regions of the nucleus at large. Regions with low DAPI stain are typically associated with euchromatic spaces, such as the interchromatin compartments where transcriptional factories reside (Schneider and Grosschedl, 2007). A second observation hinting that chromatin might tend to be more decondensed while associated with LSD1 compartments is made when visualizing probe arrays across an OR locus. Both in OP6 cells and on MOE sections, we observe several striking examples of “DNA loops” on the allele associated with the LSD1 compartment, but not on other alleles in the same cell (Fig 5C). Interestingly, the effect of Lsd1 on intergenic looping was also observed for E2-stimulated bcl-2 gene activation (Ombra et al., 2013). Such loops may be a prerequisite for interacting with the compartment or may form as a consequence of this interaction.
With the latter possibility in mind, we next asked whether LSD1 compartments are high-throughput hubs of H3K9 demethylation, a process expected to be associated with chromatin decondensation (Perillo et al., 2008). To investigate this question in OP6 cells, we used an antibody against oxidative DNA damage (8-oxo-guanine, or 8-oxodG), a mark that results on DNA in the vicinity of LSD1-mediated histone demethylation reactions (Amente et al., 2010; Perillo et al., 2008). This approach has been used previously as a means to monitor recent (not yet repaired) oxidative damage by LSD1-mediated demethylation events at OR loci, since it is presumed that LSD1-OR interactions are transient in nature (Lyons et al., 2013). Here, we presume some degree of false-negative (under-representing actual LSD1 demethylation events), nevertheless, the antibody robustly stains at mitochondria where oxidative DNA damage is known to be abundant. If the compartment was a focus of hyperactive LSD1 demethylation, we would predict an enrichment of oxidative DNA damage here as compared to random regions of the nucleus. Perhaps surprisingly, the majority of LSD1 compartments (>80%) do not contain any evidence of 8-oxodG-damaged DNA, nor is there evidence of enrichment of 8-oxodG foci within or immediately surrounding the compartment as compared to other random areas of the nucleus (not shown). Moreover, even those compartments containing some evidence of 8-oxodG DNA damage, exhibit isolated foci within a very small portion of the overall compartment, suggesting that most of the compartmentalized LSD1 protein is inactive at any given moment. Therefore, if 8-oxodG represents a reasonable proxy for recent LSD1-mediated demethylation activity and assuming the damaged DNA has some reasonable probability of detection with this assay before eviction/repair, it would appear that the LSD1 compartments are not high-throughput hubs of H3K9 or H3K4 demethylation activity. We speculate that these compartments might instead by designed to mostly limit LSD1-mediated demethylation, a possibly advantageous scenario in the context of mutually exclusive OR activation, where LSD1 activity might need to be extremely rate-limiting.
Other protein components of the LSD1 compartment
A well-characterized partner in various LSD1 regulatory complexes is the CoREST protein, where the catalytic domain of LSD1 makes direct contacts with the CoREST SANT2 domain, which facilitates target DNA interactions required for efficient LSD1-mediated H3K4 demethylation and silencing of target genes (Shi et al., 2005). In another context, LSD1 and CoREST collaborate in a de-repression complex that demethylates H3K9 during NeuroD1-mediated activation of target genes (Ray et al., 2014). Thus, it seems that LSD1 and CoREST collaborate in a variety of developmental contexts and outcomes. With this precedence in mind, we next wondered if CoREST might partner with LSD1 in this context.
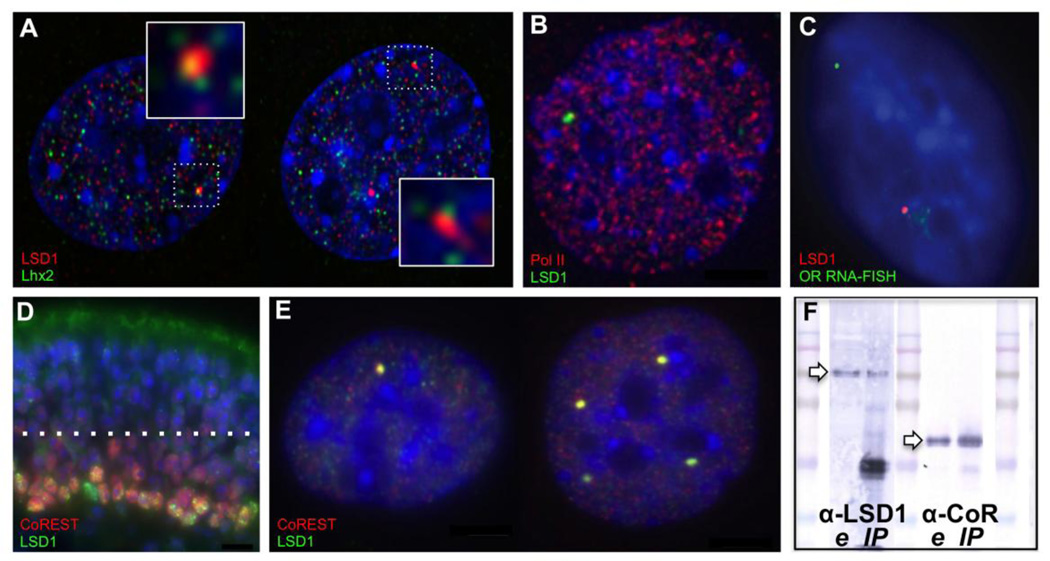
CoREST exhibits striking co-consolidation with mono- and poly-punctate LSD1 compartments in OP6 cells (Fig. 6E). Subsequent co-IP experiments confirm an interaction between LSD1 and CoREST proteins isolated from OP6 cell nuclei (Fig. 6F). Surprisingly, we do not observe the same striking degree of CoREST-LSD1 co-consolidation in the MOE, nevertheless, the CoREST protein is expressed in overlapping cell subpopulations of the MOE (Fig. 6D). CoREST is a known partner in LSD1 complexes in other contexts (Cai et al., 2014; Lee et al., 2005; Ray et al., 2014; Shi et al., 2005).
Figure 6. LSD1 compartments associate with Lhx2 and CoREST in OP6 cell nuclei.
A. During early G1 phase, Lhx2 protein (green) is distributed in a broad, punctate pattern in OP6 cell nuclei, including frequent association at the edges of both mono- (left) and poly- (right) punctate LSD1 compartments (red). B. LSD1 compartments do not contain active RNA polymerase 2, indicating that they are probably not transcriptional factories. C. Accordingly, OR RNA FISH signals are not associated with LSD1 compartments, indicating that these compartments are not sites of active OR transcription. D. LSD1 (green) and CoREST (red) staining in the immature layers of the mouse MOE. Several mono- and poly-punctate LSD1 nuclei are evident with CoREST co-localization (yellow spots). E. LSD1 (green) and CoREST (red) co-consolidate in mono- (left) and poly- (right) punctate compartments in OP6 cells, as well as OP27 cells (31, not shown). Condensed DNA is stained with DAPI (blue) in all panels. F. Western blots showing the presence of LSD1 (~100 kD, left arrow) using an anti-LSD1 antibody (α-LSD1, left lanes) in nuclear extract (e) and in the LSD1 immunoprecipitated fraction (IP), as well as the presence of CoREST (~48 kD, right arrow) using an anti-CoREST antibody (α -CoR, right lanes) in nuclear extract (e) and in the LSD1 immunoprecipitated fraction (IP).
The Lhx2 transcription factor has been previously implicated in OR regulation. It interacts with OR gene promoters and is expressed strongly in the basal cells of the MOE where OR gene expression is initiated (Hirota and Mombaerts, 2004; Hirota et al., 2007). Although we have not yet identified a robust immunofluorescence protocol for Lhx2 study on MOE tissue sections, we have conducted preliminary Lhx2 immunofluorescence experiments in OP6 cells. Lhx2 expression in OP6 cells is non-uniform, with numerous distinct spots evident per nucleus. Approximately half of LSD1 compartments contain an Lhx2 signal; we rarely, if ever, observe more than one Lhx2 signal per compartment (Fig. 6A). We are currently developing three-color immunofluorescence strategies to investigate whether an OR locus, whenever it is associated with an LSD1 compartment, is also associated with Lhx2, and whether Lhx2 is associated with one or both OR loci when two ORs are present in the same compartment. These studies might provide insights into a role for Lhx2 in either the delivery of eligible ORs to the LSD1 compartment, which predicts all/any ORs present within the compartment will be associated with Lhx2, or in the selection or Pol2 delivery step, which predicts only one of multiple ORs present within the compartment will be associated with Lhx2.
Finally, we have investigated whether the LSD1 compartment is co-localized at a Pol2 factory. Using immunofluorescence, we identify numerous Pol2 factories per nucleus, consistent with observations published previously (Osborne et al., 2004). We never observe overlap of a Pol2 factory with an LSD1 compartment (Fig 6B). Therefore, the LSD1 compartment is probably not an OR transcriptional hub. Noting that visualized Pol2 factories contain many consolidated molecules of Pol2 protein, it remains possible that a single or very small number of undetectable or inactive Pol2 protein molecules might exist within the LSD1 compartment. To investigate this further, we conducted OR RNA FISH experiments and never observed co-localization of the transcribed OR locus with the LSD1 compartment (Fig. 6C); moreover, LSD1 is not required for ongoing OR transcription in mature OSNs (Krolewski et al., 2013; Lyons et al., 2013). Together, these observations seem to argue against the hypothesis that LSD1 compartments are foci for OR transcription. Rather, it seems more plausible that LSD1 compartments, if they function in OR regulation at all, might be involved at an earlier selection stage, perhaps to prepare the selected OR for subsequent delivery to a Pol2 factory elsewhere in the nucleus.
Conclusions
We have described the regulation of LSD1 protein into discrete compartments at the edges of nuclear chromocenters in early post-mitotic cells of the OSN lineage. Nuclear chromocenters are compacted chromosomal regions that contain silenced genes and repetitive DNA enriched in constitutive heterochromatic marks, such as H3K9 trimethylation (Almouzni and Probst, 2011; Guenatri et al., 2004; Maison et al., 2010). The positioning of LSD1 compartments at chromocenters hints that these foci might function either in the delivery of previously active genes (e.g., silenced by H3K4 demethylation) and/or liberation of previously silenced genes (e.g., de-repressed by H3K9 demethylation) to/from the chromocenters.
Why does LSD1 consolidate in such an extreme way during a narrow time window in OSN development? Several hypotheses seem feasible, including two opposing ideas: (1) LSD1 might compartmentalize to tightly control histone demethylation; e.g., to limit throughput or prevent widespread LSD1 activity; or (2) LSD1 compartments might assemble multi-protein complexes for efficient, high-throughput processing of gene targets along a chromatin regulatory pathway; e.g., to efficiently traffic gene loci between chromatin states during cell differentiation or following cell division. The first hypothesis differs from the latter in the predicted level of LSD1 activity that might be observed within these compartments. In an extreme version of the first hypothesis, LSD1 might be sequestered into a mostly inactive state, which might result in very low (or no) histone demethylation activity at any given time. In contrast, extreme versions of the latter hypothesis might predict hyperactive hubs of histone demethylation. We describe preliminary immunofluorescence experiments using an antibody against oxidative DNA damage (8-oxodG) that do not seem to support the latter “hub” hypothesis. Instead, it would appear that the LSD1 within these compartments might only rarely be active, as might be predicted if compartmentalization functioned to restrict widespread LSD1 activity. We speculate that sequestration of LSD1 in a mostly idle form might be important for slowing down the de-repression process; e.g., to ensure a low probability of activating a second OR gene prior to the completion of the selection and feedback process (Dalton et al., 2013; Lewcock and Reed, 2004; Serizawa et al., 2003; Tan et al., 2013). Interestingly, a recently published theoretical model suggests that OR singularity could be achieved by a combination of a slow H3K9 demethylation process followed by a rapid feedback response to efficiently prevent additional demethylation events. (Tan et al., 2013)
Our interest in LSD1 organization in differentiating OSN populations is based on the hypothesis that LSD1 functions in the regulation of mutually exclusive OR transcription. The timing of LSD1 consolidation in vivo, occurring in the early p27-positive cells of the OSN lineage is coincident with the timing of the establishment of monogenic OR transcription during OSN development (Nguyen et al., 2007; Shykind et al., 2004). Using DNA FISH, we show that at least one OR locus is probably associated with each of the consolidated LSD1 compartments at the exclusion of most other OR loci in that cell. We also show evidence of a putative OR regulator, Lhx2, that is present in about half of these compartments in a cell population at a given time. These observations provide momentum for the hypothesis that the consolidation of LSD1 into discrete compartments in developing OSNs is important in the context of OR gene regulation. Genetic perturbation of LSD1 and its impact on OR selection/switching and detailed study of chromatin states at OR loci when interacting with LSD1 compartments, as well as characterization of LSD1 activity, post-translational modifications, and associated proteins in the complex, will be important for elucidating what role, if any, these compartments play in mutually exclusive OR expression.
Highlights.
LSD1 forms a single compartment in early post-mitotic cells of the OSN lineage
LSD1 complexes with CoREST in early G1 of an immortalized olfactory cell line
One or a small number of OR gene loci interact with the LSD1 compartment per cell
OR loci appear decondensed but are not transcribed from within LSD1 compartments
Histone demethylation activity appears to be limited within LSD1 compartments
Acknowledgments
This work was supported by National Science Foundation Grant MCB-0842868 and National Institutes of Health Grants R01-DC006267 and R01-DC002167. The authors wish to thank Jane Roskams for providing us the OP6 cell line, Amy MacQueen for use of her microscope and imaging software, Rutesh Vyas for contributing a result from his RNA FISH experiments, Brian Lin for his advice and help with mouse immunofluorescence experiments, Joyce Noble for her help with Western blot experiments, Bettina Malnic for providing insights for DNA-FISH experiments on MOE sections, and Diane Meredith for her support in all aspects of this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almouzni G, Probst AV. Heterochromatin maintenance and establishment: lessons from the mouse pericentromere. Nucleus. 2011;2:332–338. doi: 10.4161/nucl.2.5.17707. [DOI] [PubMed] [Google Scholar]
- Amente S, Bertoni A, Morano A, Lania L, Avvedimento EV, Majello B. LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene. 2010;29:3691–3702. doi: 10.1038/onc.2010.120. [DOI] [PubMed] [Google Scholar]
- Armelin-Correa LM, Gutiyama LM, Brandt DY, Malnic B. Nuclear compartmentalization of odorant receptor genes. Proc. Natl. Acad. Sci. U.S.A. 2014;111:2782–2787. doi: 10.1073/pnas.1317036111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel GA, Kadauke S, Wang E, Lau AW, Zuber J, Chou MM, Vakoc CR. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol. Cell. 2009;36:970–983. doi: 10.1016/j.molcel.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Bushdid C, Magnasco MO, Vosshall LB, Keller A. Humans can discriminate more than 1 trillion olfactory stimuli. Science. 2014;343:1370–1372. doi: 10.1126/science.1249168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C, He HH, Gao S, Chen S, Yu Z, Gao Y, Chen S, Chen MW, Zhang J, Ahmed M, Wang Y, Metzger E, Schule R, Liu XS, Brown M, Balk SP. Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell Rep. 2014;9:1618–1627. doi: 10.1016/j.celrep.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Okamoto I, Heard E. X-chromosome inactivation in mouse embryonic stem cells: analysis of histone modifications and transcriptional activity using immunofluorescence and FISH. Meth. Enzymol. 2004;376:405–419. doi: 10.1016/S0076-6879(03)76027-3. [DOI] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton RP, Lyons DB, Lomvardas S. Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell. 2013;155:321–332. doi: 10.1016/j.cell.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L, Rose DW, Glass CK, Fu XD, Rosenfeld MG. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenatri M, Bailly D, Maison C, Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 2004;166:493–505. doi: 10.1083/jcb.200403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Packard A, Krolewski RC, Harris MT, Manglapus GL, Schwob JE. Expression of pax6 and sox2 in adult olfactory epithelium. J. Comp. Neurol. 2010;518:4395–4418. doi: 10.1002/cne.22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota J, Mombaerts P. The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8751–8755. doi: 10.1073/pnas.0400940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota J, Omura M, Mombaerts P. Differential impact of Lhx2 deficiency on expression of class I and class II odorant receptor genes in mouse. Mol. Cell. Neurosci. 2007;34:679–688. doi: 10.1016/j.mcn.2007.01.014. [DOI] [PubMed] [Google Scholar]
- Illing N, Boolay S, Siwoski JS, Casper D, Lucero MT, Roskams AJ. Conditionally immortalized clonal cell lines from the mouse olfactory placode differentiate into olfactory receptor neurons. Mol. Cell. Neurosci. 2002;20:225–243. doi: 10.1006/mcne.2002.1106. [DOI] [PubMed] [Google Scholar]
- Iwema CL, Schwob JE. Odorant receptor expression as a function of neuronal maturity in the adult rodent olfactory system. J. Comp. Neurol. 2003;459:209–222. doi: 10.1002/cne.10583. [DOI] [PubMed] [Google Scholar]
- Kilinc S, Meredith DT, Lane RP. Sequestration within nuclear chromocenters is not a requirement for silencing olfactory receptor transcription in a placode-derived cell line. Nucleus. 2014;5 doi: 10.4161/nucl.29343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krolewski RC, Packard A, Jang W, Wildner H, Schwob JE. Ascl1 (Mash1) knockout perturbs differentiation of nonneuronal cells in olfactory epithelium. PloS One. 2012;7:e51737. doi: 10.1371/journal.pone.0051737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krolewski RC, Packard A, Schwob JE. Global expression profiling of globose basal cells and neurogenic progression within the olfactory epithelium. J. Comp. Neurol. 2013;521:833–859. doi: 10.1002/cne.23204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. Proc. Natl. Acad. Sci. U.S.A. 2004;101:1069–1074. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DB, Allen WE, Goh T, Tsai L, Barnea G, Lomvardas S. An epigenetic trap stabilizes singular olfactory receptor expression. Cell. 2013;154:325–336. doi: 10.1016/j.cell.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DB, Magklara A, Goh T, Sampath SC, Schaefer A, Schotta G, Lomvardas S. Heterochromatin-mediated gene silencing facilitates the diversification of olfactory neurons. Cell Rep. 2014;9:884–892. doi: 10.1016/j.celrep.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Evans ZA, Kheradpour P, Mountoufaris G, Carey C, Barnea G, Kellis M, Lomvardas S. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145:555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C, Quivy JP, Probst AV, Almouzni G. Heterochromatin at mouse pericentromeres: a model for de novo heterochromatin formation and duplication during replication. Cold Spring Harb. Symp. Quant. Biol. 2010;75:155–165. doi: 10.1101/sqb.2010.75.013. [DOI] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Nair VD, Ge Y, Balasubramaniyan N, Kim J, Okawa Y, Chikina M, Troyanskaya O, Sealfon SC. Involvement of histone demethylase LSD1 in short-time-scale gene expression changes during cell cycle progression in embryonic stem cells. Mol. Cell. Biol. 2012;32:4861–4876. doi: 10.1128/MCB.00816-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MQ, Zhou Z, Marks CA, Ryba NJ, Belluscio L. Prominent roles for odorant receptor coding sequences in allelic exclusion. Cell. 2007;131:1009–1017. doi: 10.1016/j.cell.2007.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ombra MN, Di Santi A, Abbondanza C, Migliaccio A, Avvedimento EV, Perillo B. Retinoic acid impairs estrogen signaling in breast cancer cells by interfering with activation of LSD1 via PKA. Biochim. Biophys. Acta. 2013;1829:480–486. doi: 10.1016/j.bbagrm.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Pathak N, Johnson P, Getman M, Lane RP. Odorant receptor (OR) gene choice is biased and non-clonal in two olfactory placode cell lines, and OR RNA is nuclear prior to differentiation of these lines. J. Neurochem. 2009;108:486–497. doi: 10.1111/j.1471-4159.2008.05780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- Ray SK, Li HJ, Metzger E, Schule R, Leiter AB. CtBP and associated LSD1 are required for transcriptional activation by NeuroD1 in gastrointestinal endocrine cells. Mol. Cell. Biol. 2014;34:2308–2317. doi: 10.1128/MCB.01600-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Gil DJ, Bartel DL, Jaspers AW, Mobley AS, Imamura F, Greer CA. Odorant receptors regulate the final glomerular coalescence of olfactory sensory neuron axons. Proc. Natl. Acad. Sci. U.S.A. 2015;112:5821–5826. doi: 10.1073/pnas.1417955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Gil DJ, Treloar HB, Zhang X, Miller AM, Two A, Iwema C, Firestein SJ, Greer CA. Chromosomal location-dependent nonstochastic onset of odor receptor expression. J. Neurosci. 2010;30:10067–10075. doi: 10.1523/JNEUROSCI.1776-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007;21:3027–3043. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- Schorl C, Sedivy JM. Analysis of cell cycle phases and progression in cultured mammalian cells. Methods. 2007;41:143–150. doi: 10.1016/j.ymeth.2006.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa S, Ishii T, Nakatani H, Tsuboi A, Nagawa F, Asano M, Sudo K, Sakagami J, Sakano H, Ijiri T, Matsuda Y, Suzuki M, Yamamori T, Iwakura Y, Sakano H. Mutually exclusive expression of odorant receptor transgenes. Nat. Neurosci. 2000;3:687–693. doi: 10.1038/76641. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Shykind BM, Rohani SC, O'Donnell S, Nemes A, Mendelsohn M, Sun Y, Axel R, Barnea G. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117:801–815. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Su ST, Ying HY, Chiu YK, Lin FR, Chen MY, Lin KI. Involvement of histone demethylase LSD1 in Blimp-1-mediated gene repression during plasma cell differentiation. Mol. Cell. Biol. 2009;29:1421–1431. doi: 10.1128/MCB.01158-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SL, Adamson MC, Ressler KJ, Kozak CA, Buck LB. The chromosomal distribution of mouse odorant receptor genes. Proc. Natl. Acad. Sci. U.S.A. 1996;93:884–888. doi: 10.1073/pnas.93.2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SL, Bohm S, Ressler KJ, Horowitz LF, Buck LB. Target-independent pattern specification in the olfactory epithelium. Neuron. 1995;15:779–789. doi: 10.1016/0896-6273(95)90170-1. [DOI] [PubMed] [Google Scholar]
- Sun G, Alzayady K, Stewart R, Ye P, Yang S, Li W, Shi Y. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol. Cell. Biol. 2010;30:1997–2005. doi: 10.1128/MCB.01116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Zong C, Xie XS. Rare event of histone demethylation can initiate singular gene expression of olfactory receptors. Proc. Natl. Acad. Sci. U.S.A. 2013;110:21148–21152. doi: 10.1073/pnas.1321511111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietjen I, Rihel JM, Cao Y, Koentges G, Zakhary L, Dulac C. Single-cell transcriptional analysis of neuronal progenitors. Neuron. 2003;38:161–175. doi: 10.1016/s0896-6273(03)00229-0. [DOI] [PubMed] [Google Scholar]
- Tsai WW, Nguyen TT, Shi Y, Barton MC. p53-targeted LSD1 functions in repression of chromatin structure and transcription in vivo. Mol. Cell. Biol. 2008;28:5139–5146. doi: 10.1128/MCB.00287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Nemes A, Mendelsohn M, Axel R. Odorant receptors govern the formation of a precise topographic map. Cell. 1998;93:47–60. doi: 10.1016/s0092-8674(00)81145-9. [DOI] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, Liu F, Taylor H, Lozach J, Jayes FL, Korach KS, Glass CK, Fu XD, Rosenfeld MG. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rodriguez I, Mombaerts P, Firestein S. Odorant and vomeronasal receptor genes in two mouse genome assemblies. Genomics. 2004a;83:802–811. doi: 10.1016/j.ygeno.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rogers M, Tian H, Zhang X, Zou DJ, Liu J, Ma M, Shepherd GM, Firestein SJ. High-throughput microarray detection of olfactory receptor gene expression in the mouse. Proc. Natl. Acad. Sci. U.S.A. 2004b;101:14168–14173. doi: 10.1073/pnas.0405350101. [DOI] [PMC free article] [PubMed] [Google Scholar]