Abstract
Glucan phosphatases are a recently discovered class of enzymes that dephosphorylate starch and glycogen, thereby regulating energy metabolism. Plant genomes encode for two glucan phosphatases called Starch EXcess4 (SEX4) and Like Sex Four2 (LSF2) that regulate starch metabolism by selectively dephosphorylating glucose moieties within starch glucan chains. Recently, the structures of both SEX4 and LSF2 were determined, with and without phosphoglucan products bound, revealing the mechanism for their unique activities. This review explores the structural and enzymatic features of the plant glucan phosphatases and outlines how they are uniquely adapted for carrying out their cellular functions. We outline the physical mechanisms employed by SEX4 and LSF2 to interact with starch glucans: SEX4 binds glucan chains via a continuous glucan binding platform comprised of its Dual Specificity Phosphatase (DSP) domain and Carbohydrate Binding Module (CBM) while LSF2 utilizes Surface Binding Sites (SBSs). SEX4 and LSF2 both contain a unique network of aromatic residues in their catalytic DSP domains that serve as glucan engagement platforms and are unique to the glucan phosphatases. We also discuss the phosphoglucan substrate specificities inherent to SEX4 and LSF2 and outline structural features within the active site that govern glucan orientation. This review defines the structural mechanism of the plant glucan phosphatases with respect to phosphatases, starch metabolism, and protein-glucan interaction; thereby providing a framework for their applications in both agricultural and industrial settings.
Keywords: Glucan phosphatase, starch, Arabidopsis, reversible phosphorylation, structural biology, Dual Specificity Phosphatase, glucan interactions, enzyme specificity
Introduction
Phosphatases in the Protein Tyrosine Phosphatase (PTP) superfamily are critical regulators of a variety of cellular signaling events via dephosphorylation of specific proteinaceous and non-proteinaceous substrates [1, 2]. Over the last 20 years, many PTP structures have been determined and the relationship between structure and target substrate of both protein and lipid phosphatases has been defined in exquisite detail. The recent discovery of glucan phosphatases, PTPs within the Dual Specificity Phosphatase (DSP) clade that act on phosphorylated carbohydrates, highlights the striking diversity within the PTP family that includes members that dephosphorylate proteins, lipids, nucleic acids, and carbohydrates [3-5]. Recent structural and biochemical studies on the plant glucan phosphatases Starch EXcess4 (SEX4) and Like Sex Four2 (LSF2) have provided detailed insights into the mechanisms by which these PTPs target and dephosphorylate starch glucans [6-8]. This review provides a comparative structural and functional analysis of SEX4 and LSF2 and outlines the novel features unique to the glucan phosphatases. Understanding these unique features will expand our comprehension of the PTP superfamily, allow us to make informed predictions about related phosphatases in different biological systems, and harness their activity for biotechnological purposes.
Glucan Phosphatases in Starch Metabolism
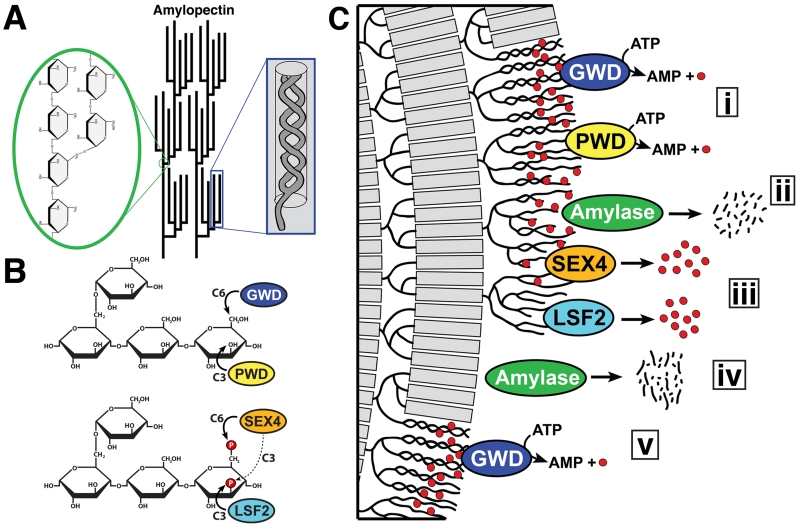
Glucan phosphatases play a critical role in the regulation of transitory leaf starch metabolism during the diurnal photosynthesis cycle [3, 9-11]. Starch is the primary form of glucose storage in plants and permits partitioning of excess glucose produced during photosynthesis for both short- and long-term storage [12, 13]. Starch is composed almost entirely of the glucose polymers amylose and amylopectin, with amylose being the minor constituent (10-30%) and amylopectin the major constituent (70-90%) (Figure 1A). Trace amounts of proteins, lipids, and ions are also present in the starch granule, depending on the source [10]. Both amylose and amylopectin are formed from α-1,4-glycosidic linked glucan chains, but amylopectin also contains α-1,6-branches clustered every 20-25 glucose units [10, 14, 15]. Adjacent amylopectin chains interact to form double helices that organize into crystalline lamellae [16]. This tightly packed arrangement of amylopectin helices results in the expulsion of water and causes starch granules to be semi-crystalline and water-insoluble [10]. Ultimately, this property allows starch granules to be stable, energy dense, and highly effective glucose storage molecules. The cytosolic, starch-like Floridean starch serves an equivalent function in red algae and during the hibernation stage of several eukaryotic protozoans [17-22]. Although starch and glycogen are each synthesized from glucose, and they utilize the same chemical linkages, the biophysical properties of starch differ from glycogen. Glycogen is water-soluble and available for dynamic bursts of metabolic activity [23].
Figure 1. Reversible phosphorylation of the plant starch granule.
A. Amylopectin is a glucose polymer and the main component of starch. It is formed from α-1,4-glycosidic linked glucose chains with α-1,6-branches that are clustered at regular intervals (green inset). Adjacent amylopectin chains form double helices (blue inset) that expel water, contributing to starch being water-insoluble. Modified from [10]. B. Amylopectin glucans are phosphorylated and dephosphorylated by four enzymes. GWD phosphorylates glucans at the C6-position followed by phosphorylation at the C3-position by PWD. SEX4 dephosphorylates the C6- and C3-positions, with a preference for the C6-position, and LSF2 exclusively dephosphorylates the C3-position. C. Reversible phosphorylation in starch degradation. i. GWD and PWD phosphorylate amylopectin helices (gray bars) causing them to unwind. ii. Amylases, particularly β-amylase, are now able to access and degrade the glucan chains, but β-amylase is unable to hydrolyze glucans past a phosphate group (red circles). iii. SEX4 and LSF2 dephosphorylate the exposed glucan chains, iv. allowing further degradation. v. The process continues with phosphorylation on the next layer of amylopectin helices within the starch granular lattice.
Starch is adapted for the predictable diurnal cycles of plants’ photosynthetic-based metabolism [10]. In chloroplasts, transitory starch is continually synthesized each day during the photosynthetic period and the water-insolubility of starch hinders its breakdown via glycolytic enzymes [24]. However, starch is almost completely degraded during the night to facilitate plant growth when photosynthesis is not occurring [25]. Therefore, during the degradative phase hydrolytic enzymes must access the energy cache stored in this insoluble form. To overcome this obstacle, plants utilize a system of reversible phosphorylation that alters the biophysical properties of starch and increases the bioavailability of glucose chains to hydrolytic enzymes, thus permitting transition from starch synthesis to degradation [9, 26-28].
While the presence of phosphate in starch was first reported in the 1890s, the role of phosphate in diurnal starch metabolism came to light via biochemical experiments and the identification of mutant Arabidopsis plants containing a starch excess (SEX) phenotype [29-35]. An excellent history of starch phosphorylation was recently reviewed [36]. SEX mutants contain increased starch content, larger and often malformed starch granules, and a decrease in plant growth [33, 34]. Together these features indicate an inability to efficiently degrade starch granules during non-photosynthetic periods. Biochemical experiments coupled with these mutant lines resulted in the identification of two dikinases called α-Glucan Water Dikinase (GWD/SEX1) and Phosphoglucan Water Dikinase (PWD) [30, 35, 37-40]. GWD exclusively phosphorylates the hydroxyl group at the C6-position of starch glucose, and this event triggers phosphorylation by PWD of hydroxyls at the C3-position (Figure 1B) [30, 37, 39-43]. Multiple studies have proposed that the introduction of covalently bound phosphate groups induces steric hindrance that results in unwinding of amylopectin helices and local solubilization of the outer starch granules (Figure 1C) [26, 42, 44-47]. Thus, the glucan dikinases increase starch surface glucan solubility and allow hydrolytic enzymes access to individual glucan chains. More recently, a role for starch phosphorylation by GWD has been identified during starch synthesis [48]. The importance of these processes is highlighted by the strict conservation of the dikinases in plants and algae [49-51].
While starch phosphorylation is necessary for proper starch metabolism, it also results in a molecular dilemma that became apparent with the identification of an additional SEX mutant called starch excess 4 (sex4) [34]. The identification of the sex4 locus in Arabidopsis resulted in the discovery of a gene encoding a trimodular protein comprised of a chloroplast targeting peptide (cTP), Dual Specificity Phosphatase (DSP) domain, and a Carbohydrate Binding Module (CBM) (Figure 2A) [50, 52, 53]. It was subsequently demonstrated that SEX4 is a glucan phosphatase that dephosphorylates starch-bound phosphate incorporated by GWD and PWD (Figure 1B) [21, 54, 55]. While starch phosphorylation is necessary to solubilize surface glucans, starch dephosphorylation is necessary because starch-bound phosphate groups obstruct the movement of β-amylase, the primary enzyme that degrades starch [56]. β-amylases degrade glucan chains up to a phosphate group, but SEX4 must remove the phosphate before it can proceed and fully degrade the chain (Figure 1C) [55]. Therefore, glucan phosphatase activity essentially resets the cycle and it is an essential component of efficient starch degradation. These studies clearly demonstrated that efficient starch degradation requires the coordinated activity of dikinases, amylases, and phosphatases.
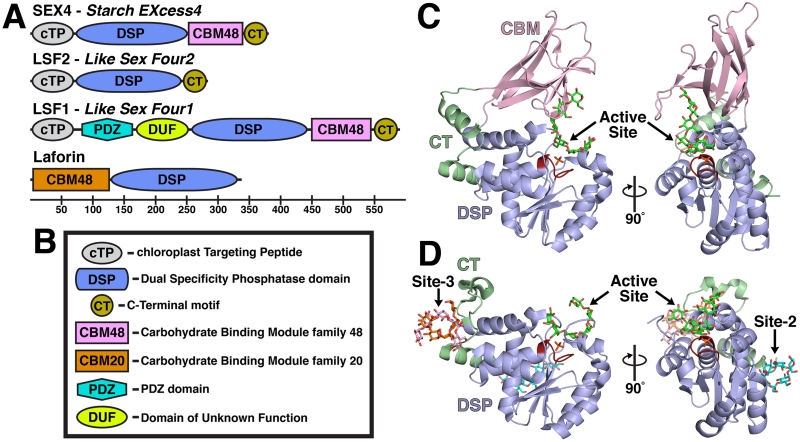
Figure 2. The glucan phosphatase family and structure of the plant glucan phosphatases.
A. Domain outline of the plant proteins SEX4, LSF2 and LSF1 and the human protein laforin with the residue numbers below. B. Domain legend. C. Glucan-bound structure of SEX4 (4PYH [6]) showing the Dual Specificity Phosphatase (DSP) domain (blue) with the Cx5R catalytic site (red), the Carbohydrate Bind Module (CBM, pink), and the CT-motif (CT, green). A single glucan chain (green) and phosphate (orange) were found at the active site. D. Glucan-bound structure of LSF2 (4KYR [7]) showing the DSP domain, Cx5R catalytic site, and CT. A single glucan chain (green) and phosphate (orange) were found at the active site. Three additional glucan chains were found at non-catalytic Secondary Binding Sites (SBSs). A single glucan chain (cyan) was found at SBS Site-2 and two glucan chains (orange and pink) were found at SBS Site-3.
An additional glucan phosphatase was discovered called Like Sex Four2 (LSF2), based on sequence similarity with the SEX4 DSP domain [52, 57, 58]. Enzymatic characterization of LSF2 revealed that it possesses robust glucan phosphatase activity against starch [58]. This discovery was unexpected due to the lack of a Carbohydrate Binding Module (CBM) in LSF2, as a CBM was predicted to be essential for glucan phosphatase activity (Figure 2A) [21, 55, 59]. An additional surprise was that LSF2 exclusively dephosphorylates the C3-position of starch glucose moieties (Figure 1C) [58]. This specificity is in contrast to SEX4, which prefers the C6-position, but can also dephosphorylate the C3-position [6, 54, 60]. The characterization of LSF2 therefore established an elegant two-enzyme model for both phosphorylation and dephosphorylation of starch in plants with the activity of GWD and PWD balanced by SEX4 and LSF2. SEX4 and LSF2 are the only glucan phosphatases in plant genomes based on bioinformatics analyses.
SEX4 and LSF2 are critical components of starch catabolism, but the structural mechanism used by these enzymes to target and dephosphorylate glucan substrates was unknown. Recently, we determined the X-ray crystal structures of SEX4 and LSF2 both with and without glucan ligands bound [6-8]. These data allowed us to establish the structural bases of their respective mechanisms, define how the glucan phosphatases incorporate phospho-glucans into their active sites, determine how the glucan chain is oriented to achieve their respective substrate specificities, and hypothesize about the activity of other phosphatases. This new understanding of glucan phosphatase activity expands our comprehension of PTP activity with respect to how substrate targeting is achieved.
Structure of SEX4 and LSF2 – Requirements for Glucan Phosphatase Activity
The first structure of a glucan phosphatase was of SEX4 in the absence of a glucan and we subsequently determined the structure of SEX4 bound to a phosphoglucan product (PDB 3NME and 4PYH) [6, 8]. These structures revealed the interdomain interactions essential to SEX4 activity. SEX4 is 43 kDa protein containing 379 residues and the crystallized construct (residues 90-379) contained the DSP and CBM domains. The non-glucan bound structure provided the first information regarding the fold of SEX4 and the physical relationship between domains.
The DSP domain (residues 90-252) of SEX4 contains a characteristic αβα DSP fold with a central five-stranded β-sheet flanked by eight α-helices (Figure 2C) [8]. The catalytic site Cx5R sequence of SEX4 is located between β5 and α6 at the base of the active site pocket. The SEX4 CBM (residues 253-338) contains six β-strands that fold into a characteristic compact β-sandwich composed of antiparallel sheets. The conserved binding site of the SEX4 CBM domain is positioned between the β-sandwich and an adjacent loop region comprised of residues I323 to N332. Interestingly, the DSP and CBM domains contain an extended and intimate interdomain interaction with 457Å2 of surface area. This interaction is maintained by a previously unrecognized C-Terminal (CT-) motif (residues 338-379) consisting of two α-helices that is connected to the CBM and forms extended contacts with the DSP. As a result of this extended CBM-DSP interaction, SEX4 contains a continuous glucan binding pocket from the CBM binding site through the DSP catalytic site that engages a single glucan chain via both aromatic and hydrophilic residues [6].
Following the determination of the SEX4 structure, the structure of LSF2 was also determined (PDB: 4KYQ and 4KYR) [7]. LSF2 is a 282 amino acid, 32 kDa protein containing a cTP and DSP domain (Figure 2A). Sequence alignments also predicted that LSF2 contained a CT-motif similar to SEX4 [58]. The LSF2 DSP domain (residues 79 to 244) also possesses a characteristic αβα DSP fold consisting of a central five-stranded β-sheet region flanked by eight α-helices (Figure 2D) [7]. The LSF2 CT-motif (residues 245 to 282) consists of a loop region culminating in an α-helix that integrally folds into the DSP domain. Although LSF2 does not contain a CBM, the structure of LSF2 bound to the glucan ligand maltohexaose revealed the presence of two non-catalytic surface binding sites (SBSs) associated with the CT-domain in addition to a glucan ligand located at the DSP catalytic site. These results suggested that LSF2 uses these two SBSs to engage glucan ligands in lieu of a CBM [7].
Interestingly, researchers have identified an additional Arabidopsis protein called Like Sex Four1 (LSF1) that possesses a CBM and DSP, and lsf1 mutant plants exhibit a starch excess phenotype [3, 57]. LSF1 is comprised of a cTP, followed by a protein-protein interaction domain known as a PDZ domain, an extended domain of unknown function (DUF), a DSP domain, a family 48 CBM domain, and CT-motif (Figure 2A) [57]. Since LSF1 has similar domain architecture to SEX4, it was hypothesized that LSF1 is also a glucan phosphatase. However, multiple groups have reported that LSF1 lacks detectable glucan phosphatase activity [57] (personal communication). Despite this lack of activity, LSF1 has a clear role in maintaining proper starch metabolism, but the precise nature of its function is unknown. Therefore, a comparative analysis of SEX4 and LSF2 activity should highlight the features distinct to this family of enzymes as well as provide informed predictions about LSF1.
The SEX4 and LSF2 structures coupled with structure-guided mutagenesis and biochemical data established the major structural requirements for glucan phosphatase activity in these two plant glucan phosphatases. SEX4 requires the CBM-DSP interaction maintained by a previously unrecognized CT-motif and LSF2 requires its DSP glucan-binding site in addition to two previously unrecognized SBSs associated with its CT-motif [6-8, 60]. Thus, both proteins possess a unique glucan binding platform that allows binding and dephosphorylation of the starch surface.
SEX4 and LSF2 Substrate Targeting
Although starch is largely comprised of extended glucose chains, its structure contains multiple complexities that present unique challenges to starch modifying enzymes [10]. The starch granule is a highly condensed, semi-accessible substrate with a low frequency of individual phosphate groups, ~1 per 2000 glucose units in Arabidopsis leaf starch [39]. While this frequency is presumably higher during starch degradation, the difficulty of locating a phosphate group within the starch superstructure requires additional mechanisms to interact with the substrate. Consequentially, glucan phosphatases invariably contain non-catalytic glucan binding motifs, in the form of a CBM or SBSs, to efficiently interact with and dephosphorylate starch [3].
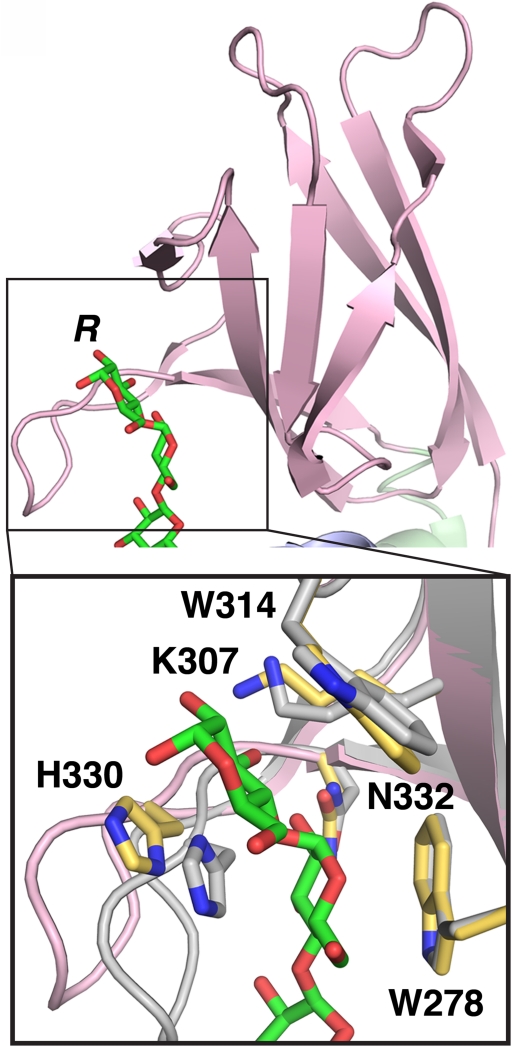
SEX4 contains a single CBM that serves as its glucan binding motif. The maltoheptaose-bound SEX4 structure revealed a tight interaction between the CBM-binding interface and two glucose moieties of the glucan chain (Figure 3A) [6]. The central platform for this interaction is comprised of a dual-tryptophan motif formed from W278 and W314. In addition, hydrogen-bonding interactions are formed between CBM residues H330, N332, and K307 with hydroxyl groups of the glucose moieties. All five of these CBM glucose-interacting residues are highly conserved among SEX4 orthologs. Mutagenesis of these SEX4 CBM residues revealed that the CBM is critical for glucan binding and subsequent dephosphorylation of insoluble starch [6]. Single point mutants of CBM residues resulted in almost total abolishment of phosphatase activity in some cases, which is due to an inability of these mutants to bind. Therefore, the CBM is necessary for substrate targeting in the SEX4 mechanism of activity.
Figure 3. SEX4 CBM-glucan interaction and conservation.
Structure of the glucan-binding site of the SEX4 CBM (pink, 4PYH [6]) showing the bound glucan chain (green). R – reducing end of glucan chain. Inset: Zoom-in of the carbohydrate binding site at the SEX4 CBM showing glucan-interacting side chains. The interacting residues from the glucan bound SEX4 structure (pink, yellow residues) is aligned with the non-glucan bound structure (grey, grey residues, 3NME [8]), highlighting movement of H330.
A CBM domain is a contiguous amino acid sequence with a conserved β-strand-rich tertiary fold that possesses carbohydrate-binding ability and is found within carbohydrate modifying or binding proteins [61]. The most common function of these modalities is to bring the catalytic portion of an enzyme in close and prolonged interaction with a mono-, oligo-, or polysaccharide [62]. CBMs are common in starch-interacting enzymes, but are also found in enzymes that modify cellulose, chitin, β-glucans, glycogen, pullulan, and xylan [62]. The CBM is structured to accommodate its target substrate, with the carbohydrate-binding platform adopting a flat or curved structure to target cellulose or starch, respectively [63]. This positioning is clearly apparent in SEX4, where the dual-tryptophan platform is curved to accommodate the natural shape of the α-glucan substrate [6].
CBMs are categorized according to the Carbohydrate Active enZymes (CAZy) database into 71 different families [64, 65]. The SEX4 CBM belongs to the CBM48 family and the dual-tryptophan platform and K307 represent a conserved functional motif in both CBM48 and the related CBM20 family [61, 66, 67]. The most similar CBM to that of SEX4 is the CBM48 in the human AMP-activated protein kinase β1-subunit (AMPK-β, PDB: 1Z0N) that has a root mean square deviation (RMSD) of 1.4Å compared to the SEX4 CBM (Figure 3B) [68]. AMPK-β contains all the SEX4 CBM glucan- interacting residues except for a threonine where H330 is located in SEX4. A comparison between the maltoheptaose bound and ligand-free SEX4 structures reveals that H330 undergoes a conformational shift upon glucan binding, bringing H330 directly in line with Glc6 and W314 (Figure 3C) [6]. Mutation of the SEX4 H330 to Ala resulted in a >60% decrease in SEX4 starch dephosphorylation [6], highlighting the importance of this residue.
The plant protein LSF1 and the human phosphatase laforin also contain a CBM, and the necessity for CBM functionality has also been demonstrated in laforin [57, 69, 70]. SEX4 and LSF1 both contain CBM48 domains while laforin contains a CBM20 domain [61]. The CBM20 and CBM48 families are closely related based on conserved binding regions, and intermediates between the CBM48 and CBM20 families that contain conserved elements of both families have been identified [61]. Moreover, CBM20 and CBM48 domains do not exclusively target starch or glycogen. Instead, examples of each family has been found in starch-targeting enzymes and glycogen-targeting enzymes. For example, the plant enzyme PWD contains a CBM20 domain although it targets starch, and the human protein AMPK-β contains a CBM48 although it targets glycogen [61]. This overlap between the two families is demonstrated by the ability for SEX4 to dephosphorylate solubilized amylopectin, which is reminiscent of glycogen, and the ability for laforin to rescue some aspects of the sex4 phenotype as well as its ability to dephosphorylate starch [21]. In humans, laforin binds and dephosphorylates glycogen [21, 59, 71, 72]. Mutations in the human gene encoding laforin that decrease its ability to bind and/or dephosphorylate glycogen lead to the accumulation of starch-like, intracellular accumulations called Lafora bodies and result in the fatal neurodegenerative disease called Lafora disease [22, 73, 74].
In addition to differences in family designation, the glucan phosphatases also have differences in the order of the CBM with respect to the catalytic domain. Laforin contains an N-terminal CBM followed by a DSP domain, whereas both SEX4 and LSF1 possess a C-terminal CBM (Figure 2A) [52, 57, 69]. Typically, the CBM precedes the catalytic domain, as in laforin, and the domain order in SEX4 and LSF1 is an exception to the common trend [61]. In addition to similar domain structure, the glucan phosphatases are also similar at the primary amino acid level. The SEX4 and LSF1 CBMs are most similar, sharing 21% identity, whereas the laforin CBM20 only shares 15% sequence identity with SEX4 and LSF1 (Figure 3B); all three CBMs share the conserved glucan binding residues that form a Trp-Lys-Trp motif for glucan interactions, which defines both the CBM20 and CBM48 families [61].
Substrate targeting via additional domains is a common theme among dual specificity phosphatases. These ancillary domains include slingshot and PDZ domains involved in protein-protein interaction, and PH-GRAM and C2 domains involved in interaction with lipid membranes [1, 2, 75]. There are structures for three DSPs with C2 domains: PTEN (1D5R) [4], Ciona intestinalis voltage sensing phosphatase (Ci-VSP, 3AWE) [76], and Auxilin PTEN-like protein (3N0A) [77], and one structure for a DSP containing a PH-GRAM domain: MTMR2 (1M7R) [78]. SEX4, PTEN, Ci-VSP, and Auxilin PTEN-like proteins all contain ancillary domains C-terminal to the DSP, while MTMR2 contains an N-Terminal PH-GRAM domain and laforin an N-terminal DSP. Moreover, it suggests that the spatial relationship of the two domains has likely consequences on the coupling of the two activities, i.e. substrate interaction and catalysis.
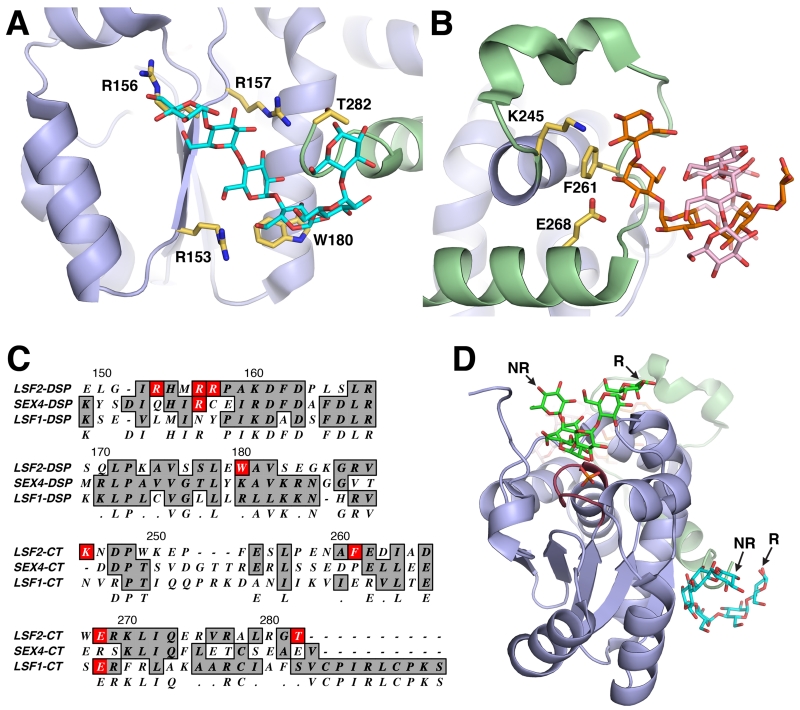
In contrast to the CBM-driven mechanism for glucan targeting found in SEX4, LSF2 possesses a unique DSP active site that incorporates both a glucan binding platform and phosphatase catalytic site [7]. Additionally, LSF2 possesses two non-catalytic SBSs >20Å from the active site that involve the CT-motif to achieve efficient glucan binding [7]. One SBS, Site-2, is located in a binding pocket formed by residues from the DSP domain and the CT-motif (Figure 4A). Specific interactions are formed between DSP residues R153, R157, M155, and W180 with the maltohexaose chain. In addition, the glucan chain wraps around the end of the CT-motif itself. Another SBS, Site-3, is located within the loop region of the CT-motif (Figure 4B). The LSF2 structure revealed two separate glucan chains (Hex-1 and Hex-2) located at Site-3 that formed a helical-like structure. LSF2 primarily interacts with Hex-1, forming hydrogen bonding interactions via K245 and E268 and van der Waals interactions via F261. Interestingly, mutation of residues in the LSF2 SBSs had a nearly identical effect on enzymatic activity and binding compared to the SEX4 CBM [7]. Mutation of Site-2 and Site-3 residues resulted in a dramatic decrease in enzymatic activity that directly correlated with their ability to bind amylopectin.
Figure 4. Non-catalytic surface binding sites (SBSs) in LSF2.
A. LSF2 (4KYR [7]) SBS Site-2, containing a single glucan chain (cyan) that interacts with residues (yellow) located on the DSP (blue) and the c-terminus of the CT (green). B. LSF2 SBS Site-3, containing two glucan chains (orange and pink) that interact with residues (yellow) located on the CT (green). C. Alignment of SBS residues found on LSF2 with SEX4 and LSF1. A portion of the LSF2 DSP (residues 149-189) and the entire CT (residues 245-282) are represented. D. Directional continuity between the glucan chain at the LSF2 active site (green) and Site-2 (cyan). The non-reducing (NR) and reducing (R) ends are labeled.
Similarly to CBMs, SBSs are also found in starch-active enzymes, primarily in glycosyl hydrolases, and have been reported in α-amylase, β-amylase, branching enzyme, xylanase, β-agarase, β-glucosidase, galactosidase, chitinase, and α-glucosidase enzymes [80]. Like CBMs, the necessity for SBSs in starch active enzymes is likely linked to the complex structure of starch granules, which have inherently poor accessibility. The main difference between the CBM and SBSs is that SBSs exist in a fixed position relative to the catalytic site, whereas CBMs are commonly connected to the catalytic domain via a flexible linker.
SBSs are difficult to identify by sequence analysis and instead are commonly identified after co-crystallization with a glucan and structural determination, as was the case with LSF2 [80]. This mode of reverse identification is due to the absence of a unifying structural consensus among different SBSs, despite the number of identified SBSs [80]. Although there are no strictly conserved SBS signatures, aromatic residues typically serve as the basis for glucan interaction in the SBSs. This is also the case with LSF2, with F261 in Site-3 and W180 in Site-2 [7]. The lack of a consensus signature makes a preemptive identification of SBS sites in other glucan phosphatases difficult. Both of the SBSs in LSF2 are associated with the CT-motif, which both SEX4 and LSF1 possess. However, neither SEX4 nor LSF1 contain the conserved residues that comprise the glucan-binding sites in the LSF2 SBSs (Figure 4C). Moreover, the SEX4 CT has the additional function of maintaining the interaction between its CBM and DSP [8]. Therefore, it is likely that the use of SBSs in LSF2 is a unique mechanism in the glucan phosphatase family despite the similar function that the SBSs impart on LSF2 function compared to the CBM in the other glucan phosphatases.
In addition to substrate targeting, both CBMs and SBSs have been postulated to participate in other related functionalities, including substrate disruption, passing on reaction products, allosteric regulation, and substrate guidance into the active site [80]. These additional functionalities have not been tested in SEX4 and LSF2, but the structural data permits insights into their feasibility. Substrate disruption driven by the glucan binding motifs of SEX4 and LSF2 would assist in and increase the efficiency of their enzymatic activity, as the overriding purpose of reversible phosphorylation is indeed substrate disruption. The multiple, disparate binding sites in LSF2 may increase the disorder of the starch granule, with each SBS engaging distinct glucan chains and interrupting glucan helices. This effect is less likely in the single continuous glucan binding site in SEX4. Substrate guiding into the active site via the SEX4 CBM is clearly a factor in its dephosphorylation mechanism, however, the same mechanism in LSF2 will require additional investigation. There may indeed be connectivity between LSF2 SBSs and the active site that was not revealed by co-crystallization with maltohexaose, a relatively short glucan chain. Interestingly, the glucan chains at the LSF2 active site and Site-2 are directionally continuous with respect to reducing and non-reducing ends, indicating these two sites may interact with a single glucan chain and supporting a role for substrate guidance (Figure 4D). The possibility of processivity in the glucan phosphatases has not been explored, but could involve the participation of non-catalytic glucan binding motifs.
Although the mechanisms dramatically differ, the non-catalytic glucan binding modalities in SEX4 and LSF2 are both necessary for starch dephosphorylation. SBSs and CBMs are both found in carbohydrate active enzymes, most relevantly in starch-active glucosyl hydrolases, and function in the same capacity as they do in the plant glucan phosphatases. Although starch-active enzymes generally contain active sites capable of interacting with target glucans, these non-catalytic motifs are necessary to bring the enzyme in direct and prolonged contact with the target substrate.
Glucan Interaction at the SEX4 and LSF2 Active Sites
Based on the functional importance of the CBM and SBSs, it was initially thought that glucan phosphatases could theoretically function via the connection of a CBM or SBSs to any phosphatase domain [21]. This theoretical model would only require the binding domain to bring the phosphatase domain in close contact with the polyglucan substrate. However, early studies with SEX4 and laforin indicated that they contain necessary glucan-interacting elements within the DSP domain itself. A fusion protein of the SEX4 CBM with the DSP domain of the human protein phosphatase VHR possessed generic phosphatase activity, but this fusion protein could not dephosphorylate glucan substrates [59]. Indeed, specific elements unique to SEX4 and LSF2 were found within the DSP active site itself that contribute towards glucan interaction at the active site [6, 7, 60].
Both SEX4 and LSF2 DSP domains contain a typical αβα PTP fold that is highly conserved among DSPs despite a low level of sequence identity [7, 8]. Not surprisingly, the DSP structures of SEX4 and LSF2 are most similar to each other with a RMSD of 1.1Å. With respect to other proteins, the SEX4 DSP is most structurally similar to the thermophilic Archaea Sulfolobus solfataricus-PTP (Ss-PTP, 2I6O) [81], and the human phosphatases STYX (2R0B) [82], KAP (1FPZ) [83], VHZ (2IMG) [84], and DUSP27 (2Y96) [85]. LSF2 is most structurally similar to human PTPMT1 (3RGQ) [86], VHZ, Ss-PTP, KAP, and STYX. The average RMSD between the glucan phosphatases and these other DSPs is 2.5Å, although sequence identities are only 10-18%. Despite their structural similarity with the glucan phosphatases, these structural homologs cover a strikingly diverse array of target substrates. PTPMT1 is a lipid phosphatase that dephosphorylates phosphatidylgycerol phosphate [86], VHZ and DUSP27 are true DSPs that dephosphorylate both p-Tyr and p-Ser/Thr protein residues [84, 85], Ss-PTP is a p-Tyr phosphatase [81], KAP is a p-Thr phosphatase [83], and STYX is a pseudophosphatase that that binds phospho-proteins yet has no phosphatase activity [87]. A structural commonality linking SEX4 and LSF2 with most of these diverse phosphatases is the presence of an α-helical Variable (V-) loop. The V-loop is a substrate-determining PTP subdomain that is a loop in most phosphatases [2], but the glucan phosphatases and their structural homologs instead possess an α-helix. Despite this commonality, the global structure of the SEX4 and LSF2 DSPs are not indicative of their unique target substrate. Therefore, the glucan phosphatase DSP domains require specific features that govern their glucan-interacting ability.
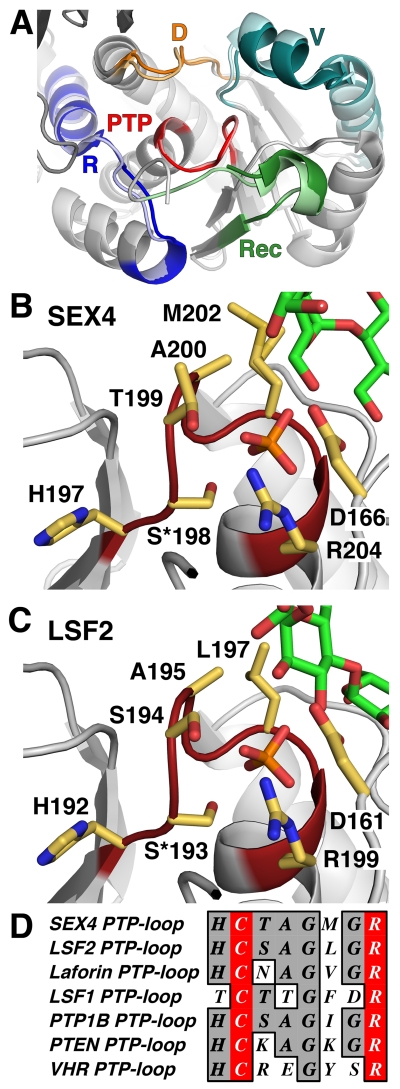
Based on previous structures, a number of PTP subdomains, including the V-loop, have been identified that function to target specific substrates at the catalytic site (Figure 5A) [2]. The PTP-loop is the most notable subdomain, comprising the catalytic core of all members of the PTP superfamily [1, 5]. The PTP-loop is characterized by an HCxxGxxRA/T (Cx5R) motif with the conserved catalytic cysteine generating a nucleophilic attack on the phosphate group and the conserved arginine necessary for integrating the phosphate within the catalytic pocket [5]. The catalytic cysteine has been shown to be sensitive to redox regulation in several PTPs [88]. Indeed, previous studies indicate the catalytic cysteine of SEX4 (C198) is rendered inactive via formation of a disulfide bond with adjacent C130 [89]. Subsequent reduction via thioredoxins likely allows SEX4 activity to be regulated within the chloroplast in a diurnal pattern without changes in protein concentration [89].
Figure 5. DSP subdomains and PTP-loop catalytic site.
A. Structural alignment of SEX4 (darker shade, 4PYH [6]) and LSF2 (lighter shade, 4KYR [7]) highlighting the DSP subdomains. The PTP-loop (PTP, red) is the catalytic site and is located centrally, surrounded by the D-loop (D, orange), V-loop (V, teal), Recognition Motif (Rec, green), and R-motif (R, blue). B. PTP-loop of SEX4 (red) showing the glucan chain (green) and phosphate (orange) with PTP-loop residues in yellow. S*198 (mutated from cysteine in crystallized construct), R204, and D166 comprise the catalytic triad. C. PTP-loop of LSF2 (red) showing the glucan chain (green) and phosphate (orange) with PTP-loop residues in yellow. S*193, R199, and D161 comprise the catalytic triad. D. Sequence alignment of the PTP-loop of SEX4, LSF2, laforin, LSF1, and human PTP1B, PTEN, and VHR. The catalytic residues are highlighted in red.
In both the glucan-bound SEX4 and LSF2 structures, a phosphate group is located within the PTP-loop directly below the glucan chain (Figure 5B,C). Structural data from multiple DSPs have shown that the primary sequence of the PTP-loop corresponds with its target substrate [5]. For instance, PTPMT1 was identified as a lipid phosphatase due to homologous basic residues within its PTP-loop that are also present in the PTP-loop of the lipid phosphatase PTEN [86]. SEX4 and LSF2 both have very similar PTP-loop sequences of HCTAGMGR and HCSAGLGR, respectively (Figure 5D). Defining the catalytic cysteine as residue 0, both glucan phosphatases have a short-chain hydrophilic residue at the +1 position, followed by an alanine and glycine, then a non-aromatic hydrophobic residue at the +4 position, and a glycine before the Cx5R arginine. This motif is significantly distinct compared to other DSPs that target protein or lipid substrates. Additionally, laforin has a similar PTP-loop consistent with SEX4 and LSF2, yielding a glucan phosphatase signature motif of CζAGΨGR where ζ is a hydrophilic residue and Ψ a long chain aliphatic [60].
Conversely, LSF1 has a very different PTP-loop sequence of TCTTGFDR (Figure 5D). It has been noted that the residue at the −1 position in LSF1 is a threonine instead of the conserved histidine found in most PTPs [57]. This histidine residue is hypothesized to be integral for catalysis by decreasing the pKa of the adjacent catalytic cysteine and facilitating nucleophilic attack [5]. This amino acid change has been postulated to account for LSF1 inactivity [3, 57]. There are several examples of inactive phosphatases that have regulatory functions, including STYX and members of the myotubularin family, but these inactive phosphatases contain substitutions in catalytic triad residues (Dx20Cx5R) [87, 90, 91]. Loss of activity solely due to a mutated histidine has not been reported previously. Subsequent analyses of LSF1 demonstrated that residues within the LSF1 PTP-loop sequence are also highly divergent from those found in the other glucan phosphatases [60]. LSF1 contains Thr and Glu at positions 2 and 5, respectively, instead of a shorter chain Ala or Gly found in the glucan phosphatases. Additionally, position 4 is an aromatic residue in LSF1 and a long chain aliphatic residue in the three glucan phosphatases. Therefore, based on the signature motif identified in SEX4, LSF2 and laforin of CζAGΨGR, the LSF1 PTP-loop cannot accommodate phosphoglucan substrates [60].
Surrounding the PTP-loop are four additional subdomains that interact with the target substrate and integrate it into the catalytic site; the D-loop, V-loop, R-motif, and recognition motif (Figure 5A) [2]. The combined structure of these subdomains comprise the overall active site of the phosphatases and the shape and chemical composition dictate the substrates that can be targeted and accommodated into the catalytic site for dephosphorylation.
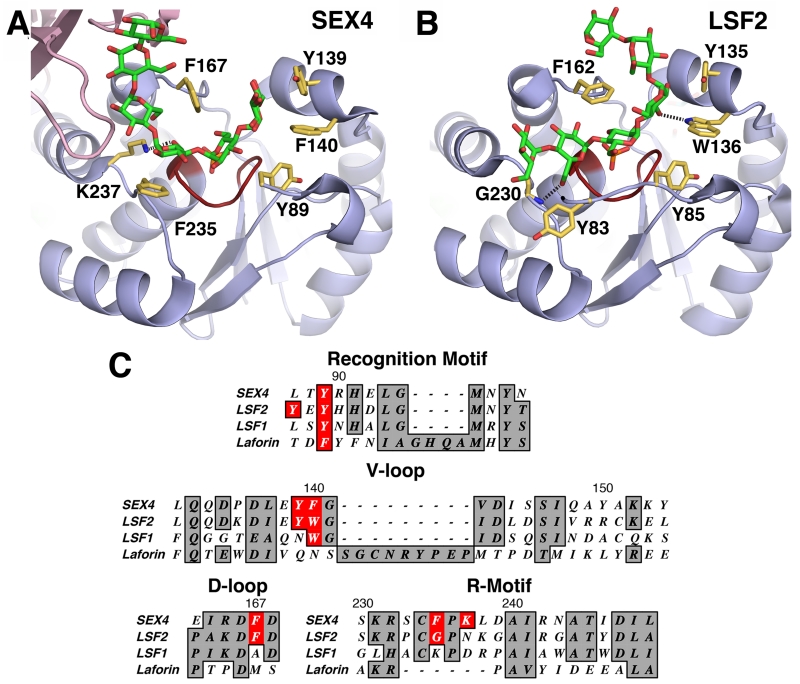
The glucan-bound structures of SEX4 and LSF2 illustrate features found in the DSP subdomains that contribute specifically to glucan targeting (Figure 6A,B) [6-8]. The D-loop is notable for containing a conserved aspartate that is part of the catalytic triad and operates in catalysis as a general acid/base to form the phospho-enzyme intermediate followed by expulsion of the phosphate from the active site [92, 93]. At the +1 position from the catalytic aspartate, the D-loops of SEX4 and LSF2 both contain a phenylalanine residue (SEX4-F167, LSF2-F162) that interacts with the glucan chain directly over the active site. This residue caps the targeted phosphoglucose moiety and was observed to be flexible when comparing the non-glucan bound structure with the glucan bound SEX4 and LSF2 structures. C-terminal from the active site, the R-motif of SEX4 contains phenylalanine and lysine residues (F235 and K237) that interact with the glucan chain via van der Waals interaction and hydrogen bonding, respectively. Conversely, LSF2 contains a glycine residue (G230) in its R-motif that forms a hydrogen bond with the glucan chain upstream of the active site moiety. N-terminal of the active site, the V-loop of SEX4 contains tyrosine and phenylalanine residues (Y139 and F140) that interact with the glucan moiety. The LSF2 V-loop contains similar tyrosine and tryptophan residues (Y135 and W136) that make equivalent glucan contacts. Finally, the recognition motifs of SEX4 and LSF2 both contain a tyrosine residue (SEX4-Y89, LSF2-Y85) that interacts with the glucan chain at the active site, and LSF2 possesses an additional tyrosine residue (Y83) that is not present in SEX4, which makes additional glucan contacts. The DSP subdomains found in SEX4 and LSF2 participate extensively with the interaction between the catalytic domain and the glucan chain.
Figure 6. Glucan phosphatase DSP interaction with the glucan chain.
A. SEX4 DSP (blue, 4PYH [6]) with the glucan chain (green) and phosphate (orange) at the catalytic site (red). Interacting residues are shown in yellow with hydrogen bond between K237 and the glucan chain shown. B. LSF2 DSP (blue, 4KYR [7]) with the glucan chain (green) and phosphate (orange) at the catalytic site (red). Interacting residues are shown in yellow with hydrogen bond between the glucan chain and G230 and W136 shown. C. Sequence alignment of DSP subdomains between SEX4, LSF2, LSF1, and laforin. Sequence number of SEX4 residues are shown. Glucan interaction residues in SEX4 and LSF2 are highlighted with conserved residues in LSF1 and laforin.
The most obvious and functionally significant feature of the DSP subdomains in SEX4 and LSF2 is the large population of aromatic residues that interact with the glucan chain (Figure 6) [6, 7]. These aromatic residues form a network that provides an extended glucan-binding platform throughout the entire active site. Aromatic residues are essential for carbohydrate interaction in a variety of contexts via van der Waals interactions between the aromatic residues and glucose rings [61, 63, 80]. Indeed, most classes of CBMs rely heavily on aromatic residues to engage various carbohydrate substrates [63]. The active sites of multiple glycosyl hydrolases contain similar networks of aromatic residues, although they are typically less concentrated than is observed in the glucan phosphatase active sites [94, 95]. An analysis of the sequences of 34 additional DSPs reveals that the aromatic network found in the glucan phosphatases are novel and not conserved among DSPs. This network of aromatic residues in SEX4 and LSF2 is therefore a characteristic and defining feature of the plant glucan phosphatases.
Interestingly, a DSP alignment between SEX4, LSF2, and LSF1 suggests that this aromatic network is not conserved in LSF1 (Figure 6C). LSF1 lacks many of the aromatic residues found in SEX4 and LSF2 and these differences likely contribute to the absence of its glucan phosphatase activity (Figure 6C). Determination of the LSF1 structure and enzymatic analysis using strategic mutagenesis would shed further light on this hypothesis.
As expected, mutation of glucan-interacting residues in the SEX4 and LSF2 DSP subdomains had a clear effect on each enzyme’s ability to dephosphorylate glucan substrates [6, 7]. However, the extent to which these mutations hindered activity differed notably. In SEX4, mutation of DSP residues decreased activity 10-80% with an average decrease of 38% [6]. DSP mutation in LSF2 had a more notable affect activity. Single point mutations of the LSF2 DSP resulted in a decrease in activity ranging from 40-95% with an average decrease of 66% [7]. These differences appear to stem from the compensatory activity of the SEX4 CBM, and the presence of hydrogen-bonding residues in the LSF2 DSP. In SEX4, the CBM and DSP share a continuous binding pocket; therefore the effect of DSP mutations has a muted effect due to glucan binding and proximity of the CBM platform [6]. Furthermore, LSF2 contains two residues (W136 and G230) that form hydrogen-bonding interactions with the glucan chain that are not present in SEX4 (Figure 6) [7]. Their presence likely balances the loss of active site interaction that could be envisioned given the distance between the active site and SBSs. Therefore, the DSP subdomains of SEX4 and LSF2, despite having a common aromatic signature, contain distinct affinities for glucans in isolation of additional glucan binding domains or motifs.
Conformation of Glucan Chains
The glucan-bound SEX4 and LSF2 structures permitted a clear study of DSP residues that interact with the glucan chain. However, α-glucan chains are directional molecules that may be engaged from multiple orientations. Considering the orientation of the glucan chain at the SEX4 and LSF2 active site is particularly relevant to determining the structural basis of their observed substrate specificity. SEX4 preferentially dephosphorylates the C6-position of starch glucans whereas LSF2 exclusively dephosphorylates the C3-position. Understanding the structural basis of this difference will provide possible means to manipulate glucan phosphatase substrate specificity for biotechnological purposes.
In both the SEX4 and LSF2 structures, six glucose moieties of the glucan chain were observed in the active site [6, 7]. In SEX4, this chain spanned both the CBM and the DSP in a continuous binding pocket, whereas in LSF2 the entire chain was present at the DSP active site.
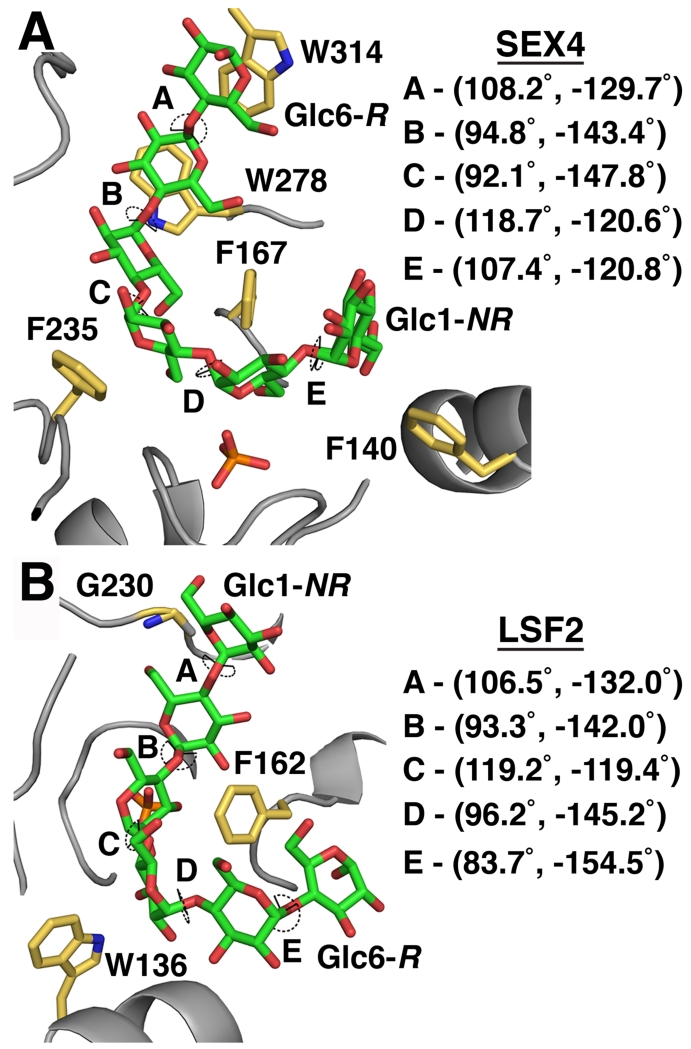
Both enzymes also bound the glucan chains in a manner conducive to the twisting/helical geometry of the substrate, which is likely significant for optimal affinity. However, the enzymes also distorted this helical conformation in interestingly consistent ways. Previous studies have determined that the α-1,4-glycosidic dihedral torsion in helical amylopectin chains have values of (91.8°, −153.2°), (85.7°, −145.3°), and (91.8°, −151.3°) [96]. The inequality of the two angles results in torsion consistent with a helical geometry. Conversely, equality in the glycosidic bond torsion angle, i.e. (119.3°, −119.3°), would indicate a lack of torsion and linearity.
The glycosidic bonds of glucans between the SEX4 CBM and DSP binding sites (Glc3-Glc4 and Glc4-Glc5) have dihedral torsion angles consistent with a helical conformation (Figure 7) [6, 7]. In contrast, the glycosidic bonds of glucans at the DSP active site (Glc1-Glc2 and Glc2-Glc3) and the CBM binding site (Glc5-Glc6) have more linear dihedral torsion angles. The same trend is found in LSF2 (Figure 7). The glycosidic bonds of glucans at the DSP active site in LSF2 (Glc1-Glc2 and Glc3-Glc4) have more linear dihedral torsion angles, whereas the glycosidic bonds of glucans outside of the LSF2 active site (Glc4-Glc5 and Glc5-Glc6) contain more helical dihedral torsion angles. The only outlier is the helical glycosidic bond of LSF2 Glc2-Glc3, which appears to be influenced by hydrogen bonding contributions from G230. Taken together, these results indicate that hotspots of glucan interaction in the glucan phosphatases (DSP active site, CBM) tend towards linearization of the glucan chain, and regions that bind more loosely inherently accommodate the helical substrate. These results suggest an interesting interplay in the glucan phosphatases between accommodating the natural conformation of the substrate and modifying this conformation for enzymatic purposes.
Figure 7. Directionality and torsion angles of glucans at the SEX4/LSF2 active site.
A. Dihedral torsion angles of the glucan chain (green) at the SEX4 active site (4PYH [6]). Interacting residues are labeled in yellow. R – reducing end of the glucan chain, NR – non-reducing end. B. Dihedral torsion angles of the glucan chain (green) at the LSF2 active site (4KYR [7]). Interacting residues are labeled in yellow.
Although engagement of the helical α-glucan chains is similar in SEX4 and LSF2, the chains are engaged with opposing directionality (Figure 7). In SEX4, the non-reducing end of the glucan chain is positioned at the V-loop and the reducing end is positioned at the R-motif [6]. In LSF2, the opposite is observed; the non-reducing end is positioned at the R-motif and the reducing end is positioned at the V-loop [7]. Starch granules are polarized, with the reducing end towards the interior of the granule and the non-reducing ends towards the outside [10, 97]. Thus, the structure data suggests that SEX4 and LSF2 may engage glucans from different ends of the chain within starch granules.
Glucan Phosphatase Substrate Specificity
One of the most significant aspects of SEX4 and LSF2 glucan chain engagement is the orientation of the glucan moiety at the catalytic site. This orientation is important to substrate specificity, as the C6-hydroxyl and the C3-hydroxyl groups are on opposite facets of the glucan chain. Therefore, the substrate specificity of SEX4 and LSF2 must arise from integrating the glucan chain in a C6- or C3-specific orientation.
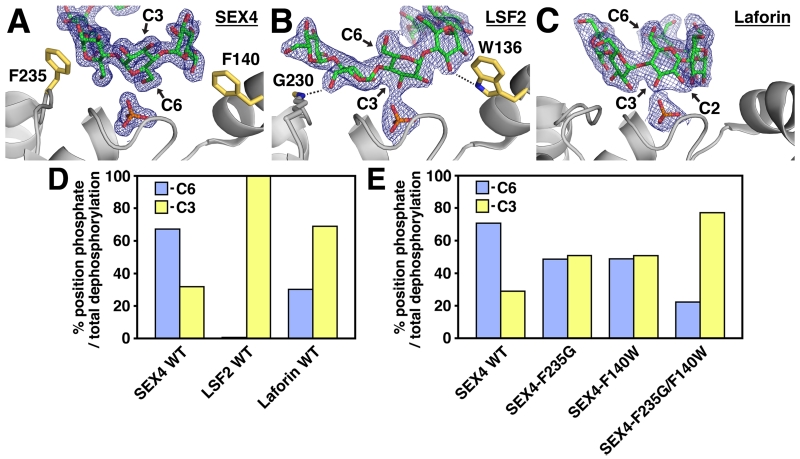
The SEX4 structure shows that the maltoheptaose chain is clearly positioned in a C6-specific orientation at the catalytic site (Figure 8A) [6]. The O6 group of Glc2 interacts with the phosphate in the catalytic site at a distance of 2.6Å, compared with 7.1Å for the O3 group. In addition, the glucose moieties upstream (Glc1) and downstream (Glc3) of Glc2 are also oriented with the O6 group pointed towards the catalytic site. Furthermore, the orientation of the PTP-catalytic triad (S(C)198, R204, and D166) is proximal to the Glc2 O6 and phosphate, thus poised for catalysis. Interestingly, this orientation is reversed at the SEX4 CBM, where the C3-hydroxyl is positioned towards the body of the protein and the C6-hydroxyl is pointed towards the solvent. Therefore, between the CBM and DSP, SEX4 is structured to interact with opposite facets of the glucan chain. The C6-oriented glucan-ligand within the SEX4 structure reaffirms the substrate specificity indicated by the enzymatic data.
Figure 8. Substrate Specificity in the Plant Glucan Phosphatases.
Active site of (A) SEX4 (4PYH [6]), (B) LSF2 (4KYR [7]), and (C) Laforin (4RKK [79]) showing 2Fo-Fc electron density (1.0 σ, blue mesh) of the glucan chain (green) and phosphate (orange). The orientation of the glucan moiety at the catalytic site (C6 and C3 position) is indicated. Residues contributing to substrate specificity in SEX4 and LSF2 are shown in yellow. D. Relative specific activity of SEX4, LSF2, and Laforin at the C6- (blue) and C3- (yellow) positions of Arabidopsis starch represented as the percentage of total phosphate position dephosphorylation per minute per µg of protein. Modified from [60]. E. Relative specific activity of SEX4 active site mutants at the C6- (blue) and C3- (yellow) positions of Arabidopsis starch represented as the percentage of total phosphate position dephosphorylation per minute per µg of protein. Modified from [6].
The opposite is found in the LSF2 structure, in which the maltohexaose chain is clearly positioned in a C3-specific orientation at the catalytic site (Figure 8B) [7]. The O3 group of Glc3 interacts with the phosphate in the active site at a distance of 2.4Å, compared with 7.0Å for the O6 group. Once again, the glucose moieties upstream (Glc4) and downstream (Glc2) are also oriented with the O3 group towards the catalytic site. We found that the glucan-bound LSF2 PTP-catalytic triad is also proximal to the Glc3 O3 group and phosphate and is therefore poised for catalysis. As with SEX4, the LSF2 structure also provides a biophysical confirmation to the enzymatic data indicating exclusive C3-specific activity. Additional work by the Zeeman lab has demonstrated increased C3-bound phosphate in lsf2 plants [58]. Thus, in planta, biochemical, and structural data all converge to define the activity of the glucan phosphatases.
Laforin was also co-crystallized in the presence of maltohexaose and a bound maltohexaose chain and phosphate was integrated in the DSP domain active site [79]. As predicted, the laforin DSP domain is structurally most similar to SEX4 and LSF2 with a RMSD of 2.0 and 2.1Å, respectively. The O2 and O3 groups of Glc3 are positioned 3.8Å and 2.7Å from the phosphate bound in the active site, while the O6 position is 7.7Å away and pointed towards the solvent (Figure 8C). Utilizing the position specific assay, we determined that laforin preferentially dephosphorylates C3 hydroxyls and possess substantial activity towards the C6 position, nearly the exact opposite of the SEX4 specificity data (Figure 8D).
The C6-oriented glucan ligand within the SEX4 structure and the C3-oriented glucan ligand within the LSF2 structure provided an opportunity for structural analysis to determine the underlying basis for the differences in substrate specificity in the glucan phosphatases. The most obvious hypothesis regarding substrate specificity would be the fundamental difference in glucan-binding mechanism used by SEX4 and LSF2. SEX4 uses a CBM to interact with glucan chains and LSF2 uses SBSs, therefore these external factors (with respect to the active site) could drive substrate specificity. However, all subsequent experiments demonstrated that the CBM and SBSs have no effect on substrate specificity [60]. Mutation of the CBM or SBSs, or removal or the CBM altogether, did not affect the ratio between C6- and C3- dephosphorylation in SEX4 and LSF2 [60].
If the binding mechanism used by the glucan phosphatases is not responsible for glucan orientation, then the DSP active site itself must define substrate specificity. The SEX4 and LSF2 glucan phosphatase active sites must have distinct structural differences that serve to position the glucan chain in a C6- or C3-specific orientation. In the previous section, we outlined similar features of the glucan phosphatase DSP that account for its unique ability to bind glucans, namely its network of aromatic residues and wide/shallow topology. However, to unravel the differences in substrate specificity differences between the active site of SEX4 and LSF2 were investigated.
Indeed, SEX4 and LSF2 have different residues comprising the active site boundaries that serve to tailor the binding capabilities and shape of the active site to accommodate their respective specificities. In SEX4, F235 and F140 make van der Waals interactions with the glucose moieties upstream and downstream of the active site moiety, respectively (Figure 8A) [6]. Both of these residues are altered in LSF2, where G230 and W136 make hydrogen-bonding interactions with the O3 groups of the glucan chain (Figure 8B) [7]. In addition, the residue differences affect the active site topology. The absence of a side chain at G230 in LSF2 results in a small pocket, whereas the β-carbon of F235 in SEX4 forms a distinct ridge in the same area (Figure 9) [6].
Figure 9. Active Site Topology of PTP superfamily members.
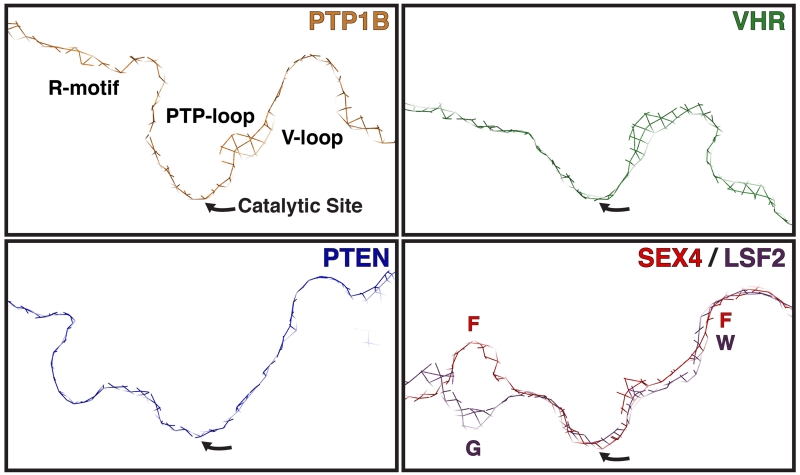
The active site topology of representative PTP superfamily members as represented by the cross section of the active site formed by the R-motif, PTP-loop, and V-loop surface (general positions are labeled in PTP1B panel). The catalytic site is located at the lowest trough of the active site pocket. PTP1B (orange, 1EEN [104]) is a canonical Protein Tyrosine Phosphatase (PTP) that dephosphorylates p-Tyr exclusively and contains a narrow, deep active site topology. VHR (green, 1VHR, [5]) is a Dual Specificity Phosphatase (DSP) that dephosphorylates p-Tyr and p-Ser/Thr residues and has a wide, shallow active site topology. PTEN (blue, 1D5R [4]) is a DSP that dephosphorylates lipid phosphatidylinositol-3,4,5-triphosphate (PI3P) and has a wide, deep active site topology. The glucan phosphatases SEX4 (red, 4PYH [6]) and LSF2 (purple, 4KYR [7]) have wide, shallow active site topologies compared to the other PTP superfamily members. The SEX4 and LSF2 active sites are relatively uniform with the exception of the R-motif region, which has a distinct ridge in SEX4 and a pocket in LSF2. The position of residues that contribute to R-motif (SEX4-F235 (F)/LSF2-G230 (G)) and V-loop (SEX-F140 (F)/LSF2-W136 (W)) topology are indicated.
These differences create distinct shapes and were found to contribute to substrate specificity in the plant glucan phosphatases (Figure 8E) [6]. Single mutations of F235 or F140 in SEX4 to the corresponding residues found in LSF2 abolished substrate specificity with C6- and C3-phosphates removed at equal rates. In addition, the double mutation of both residues resulted in a complete reversal of substrate specificity in SEX4 from the C6- to the C3-position, with the C3-position dephosphorylated at an approximately 3-fold higher rate than the C6-position. These results indicated that the substrate specificity of the glucan phosphatases is influenced by elements in the DSP active site in both enzymes. Indeed, we recently utilized soluble and insoluble glucan substrates with a series of SEX4, LSF2, and laforin mutants and chimeras to address this hypothesis. We found that the glucan phosphatase DSP domain is sufficient for dephosphorylation of soluble polyglucans [60]. Conversely, the ancillary CBM domain and SBSs are required for efficient dephosphorylation of insoluble polyglucans [60].
In addition to determining the chemistry of the active site, the PTP subdomains also compose the overall topology or shape of the phosphatase active site. It has previously been established that the active site topology of PTPs and DSPs are tailored to their respective target substrates in terms of width and depth [4]. This topology is normally defined by the surface of the enzyme from the R-motif to the V-loop, with the catalytic PTP-loop directly in the center of these two subdomains [4]. Thus, p-Tyr specific PTPs such as PTP1B contain a narrow and deep active site that permits access to p-Tyr residues, but excludes shorter p-Thr/Ser residues (Figure 9) [98]. In contrast, dual specificity phosphatases that dephosphorylate p-Tyr, p-Thr, and p-Ser such as VHR generally have a shallower and wider active site that permits catalytic access to both short and long side chains [5]. The lipid phosphatase PTEN, which engages a large tri-phosphorylated phosphatidylinositol head group, contains a wide and deep active site compared to the proteinaceous phosphatases [4]. Therefore, the characteristics of the target substrate closely match the topology of the phosphatases in the classical lock-and-key manner.
Comparing the active sites of SEX4 and LSF2, we found that the active sites of SEX4 and LSF2 are wide and shallow, allowing them to engage multiple glucose moieties of a long glucan chain (Figure 9) [6]. The active site of SEX4 is most similar to PTEN, and LSF2 is even more shallow and wide. These results indicate that the non-proteinaceous DSPs typically contain an active site that can engage an extended substrate, rather than target individual protein side chains. Furthermore, the glucan phosphatases must engage multiple glucose units simultaneously and possibly cycle glucose moieties through the active site to identify a phosphate group. The active site topology of SEX4 and LSF2 is consistent with these enzymatic requirements.
An additional feature of glucan phosphatase activity that has not been explored is the influence of the LSF2 SBSs on the position of the glucan chain at the active site. It is very plausible that the three binding sites in LSF2 function cooperatively when binding complex carbohydrate substrates. The extended length of glucan chains in amylopectin and their close spatial relationship would fit well into a model whereby two binding sites interact with a single glucan chain or two separate glucan chains simultaneously. Evidence for their relationship is found in the consistent directionality, i.e. position of reducing and non-reducing ends, between the glucan chains at the active site and Site-2 (Figure 4D). This consistency implies that these two sites are structured to bind a single glucan chain. Furthermore, two glucan chains were found at Site-3 complexed into a helical conformation (Figure 4B). This leaves open the possibility that Site-3 is capable of binding helical portions of the amylopectin superstructure that have yet to be phosphorylated or solubilized. More dynamic biophysical investigations into the LSF2 interaction with complex glucans will shed light on the more complex and interconnected nature of its structural mechanism of activity.
Lastly, there is currently no structural or biochemical data regarding how SEX4 and LSF2 accommodate additional features of the starch granule, such as linear phosphoglucan chains on the granule surface, branches, adjacent glucan chains, residual helical structures, and the radial orientation of the starch granule which is formed from reducing to non-reducing end. The position of phosphate with respect to branch points is also unknown, but correlation between them can easily be imagined, possibly as a mechanism for specific phosphate positioning by GWD and/or PWD. It is likely that SEX4 and LSF2 are equipped to either accommodate branches or to circumvent them to more efficiently locate phosphate groups in the starch granule. Investigation into this area will provide a more global view of how SEX4 and LSF2 interact with some of the more complex elements of the starch granule superstructure.
In summary, the plant glucan phosphatases SEX4 and LSF2 engage glucans in a manner conducive to their natural helical conformation and each bind the glucan chain in opposite directions and orientations. The basis for opposing orientations of the glucan chain at the active site of SEX4 and LSF2 is due to different residues present in the DSP active site. This difference in active site structure accounts for the difference in substrate specificity between the two glucan phosphatases.
Summary and Future Directions
Glucan phosphatases represent a new subset of PTPs that are essential for complex carbohydrate metabolism. Recent structural and enzymatic characterization of SEX4 and LSF2 provides detailed information on the physical basis for starch interaction and dephosphorylation. SEX4 binds glucans via an extended CBM-DSP interface that couples strong glucan binding at the active site with phosphoglucan integration into the catalytic site by the DSP. Conversely, LSF2 contains SBSs that non-catalytically interact with glucan chains and a stronger binding DSP active site that integrates the glucans into the catalytic site. A comparison of SEX4 and LSF2 with other DSPs indicates that the glucan phosphatase DSP contains a unique network of aromatic residues that function as glucan platforms and a wide, shallow active site to accommodate three glucan moieties of a longer glucan chain. Lastly, the substrate specificity inherent in SEX4 and LSF2 activity and essential for proper starch degradation is based upon discreet elements within the DSP domain and is able to be manipulated by simple mutagenesis in SEX4. This information provides a complete characterization of the fundamental enzymology of the plant glucan phosphatases. Although SEX4, LSF2, laforin, and LSF1 are the only known members of the glucan phosphatase family, identification of future members will likely reveal variations on the structural themes described.
This initial characterization of the plant glucan phosphatases provides a framework for future research regarding finer details of their interaction with the complex starch granule. Additional insights into SEX4 and LSF2 interaction with branches, adjacent glucan chains, and helical features of the starch granule will be necessary to more fully understand how these enzymes contend with a complex and semi-accessible substrate. Starch, as an enzymatic target, contains multiple microenvironments that complicate binding and understanding how glucan phosphatases are structured for efficient starch interaction may assist in the development of more efficient starch hydrolyzing enzymes. This will also require an investigation into a possible processive mechanism used by the glucan phosphatases, which is currently not understood.
The current review defines the structural mechanisms of SEX4 and LSF2 and summarizes our current understanding of LSF1. While LSF1 is undoubtedly involved in starch metabolism its precise role is currently unclear. What is interesting is that while LSF1 resembles the glucan phosphatases SEX4, LSF1 lacks the glucan phosphatase signature motif and glucan phosphatase activity. The determination of the structural mechanism of SEX4 and LSF2 activity revealed that LSF1 is missing several key elements important to their function, including a PTP-loop consensus sequence, some of the aromatic network of residues in the DSP, and key specificity determining residues. In addition, LSF1 contains elements not present in SEX4 and LSF2, namely a PDZ domain and DUF. Determining the structure of LSF1 and further studies on its interactions with starch, and enzymes involved in starch metabolism, will likely reveal the exact nature of this enigmatic enzyme and possibly provide more biotechnological tools to further exert molecular control over starch metabolism.
As critical regulators of starch metabolism, the plant glucan phosphatases are promising targets to harness plant energy metabolism to both increase starch yield in crops and improve the efficiency of starch processing for industrial purposes. Due to the recent identification of the plant glucan phosphatases, their genetic manipulation has not yet been fully explored in crops. However, silencing of GWD has recently been shown to positively impact starch yield and biomass in wheat plants [99], thereby providing a powerful proof of principle for the glucan phosphatases. In addition, studies on the expression, conservation, and function of SEX4 in barley [100], rice [101], maize [102], and chestnut [103] have been performed. However, the role of glucan phosphatases in storage starch metabolism or any link to plant pathology is currently unclear. Understanding the precise structural mechanism of plant glucan phosphatase interaction with starch may grant an even greater potential for control as biotechnological tools advance in the coming years. In addition, the multitude of non-food applications of starch often requires diverse chemical properties that require extensive processing. Using the glucan phosphatases to enzymatically alter the phosphorylation pattern on starch or to function in starch breakdown may yield industry-wide improvements.
Acknowledgements
This work was supported by National Institutes of Health Grants R01NS070899; KSEF grants KSEF-2268RDE-014 and KSEF-2971-RDE-017; Mitzutani Foundation for Glycoscience Award; NSF Grants IIA-1355438 and MCB-1252345. M.S.G. and C.W.V.K. are founders of OptiMol Enzymes LLC.
Abbreviations
- SEX4
Starch EXcess4
- LSF2
Like Sex Four2
- LSF1
Like Sex Four1
- GWD
α-Glucan Water Dikinase
- PWD
Phosphoglucan Water Dikinase
- DSP
Dual Specificity Phosphatase (domain)
- CBM
Carbohydrate Binding Module
- CT
C-Terminal motif
- SBS
Surface Binding Site
- PTP
Protein Tyrosine Phosphatase
- DUF
Domain of Unknown Function
Footnotes
Author Contributions
D.A.M., C.W.V.K., and M.S.G. wrote the paper.
References
- 1.Tonks NK. Protein tyrosine phosphatases--from housekeeping enzymes to master regulators of signal transduction. FEBS J. 2013;280:346–78. doi: 10.1111/febs.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso A, Rojas A, Godzik A, Mustelin T. The Dual-Specific Protein Tyrosine Phosphatse Family. Springer; Berlin: 2003. [Google Scholar]
- 3.Silver DM, Kotting O, Moorhead GB. Phosphoglucan phosphatase function sheds light on starch degradation. Trends Plant Sci. 2014 doi: 10.1016/j.tplants.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P, Pavletich NP. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–34. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 5.Yuvaniyama J, Denu JM, Dixon JE, Saper MA. Crystal structure of the dual specificity protein phosphatase VHR. Science. 1996;272:1328–31. doi: 10.1126/science.272.5266.1328. [DOI] [PubMed] [Google Scholar]
- 6.Meekins DA, Raththagala M, Husodo S, White CJ, Guo HF, Kotting O, Vander Kooi CW, Gentry MS. Phosphoglucan-bound structure of starch phosphatase Starch Excess4 reveals the mechanism for C6 specificity. Proc Natl Acad Sci U S A. 2014;111:7272–7. doi: 10.1073/pnas.1400757111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meekins DA, Guo HF, Husodo S, Paasch BC, Bridges TM, Santelia D, Kotting O, Vander Kooi CW, Gentry MS. Structure of the Arabidopsis glucan phosphatase like sex four2 reveals a unique mechanism for starch dephosphorylation. Plant Cell. 2013;25:2302–14. doi: 10.1105/tpc.113.112706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vander Kooi CW, Taylor AO, Pace RM, Meekins DA, Guo HF, Kim Y, Gentry MS. Structural basis for the glucan phosphatase activity of Starch Excess4. Proc Natl Acad Sci U S A. 2010;107:15379–15384. doi: 10.1073/pnas.1009386107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smirnova J, Fernie AR, Steup M. Starch degradation. In: Nakamura Y, editor. Starch: Metabolism and Structure. Spinger Japan; 2015. pp. 239–290. [Google Scholar]
- 10.Streb S, Zeeman SC. Starch metabolism in Arabidopsis. Arabidopsis Book. 2012;10:e0160. doi: 10.1199/tab.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hejazi M, Fettke J, Steup M. Starch phosphorylation and dephosphorylation: the consequtive action of starch-related dikinases and phosphatases. In: Tetlow IJ, editor. Starch: Origins, Structure and Metabolism. SEB; London: 2012. pp. 279–308. [Google Scholar]
- 12.Zeeman SC, Kossmann J, Smith AM. Starch: Its Metabolism, Evolution, and Biotechnological Modification in Plants. Annu Rev Plant Biol. 2010;61:209–234. doi: 10.1146/annurev-arplant-042809-112301. [DOI] [PubMed] [Google Scholar]
- 13.Fettke J, Fernie AR, Steup M. Transitory starch and its degradation in higher plant cells. In: Tetlow IJ, editor. Starch: Origins Structure and Metabolism. SEB; London: 2012. pp. 309–372. [Google Scholar]
- 14.Tester RF, Karkalas J, Qi X. Starch - Composition, fine structure and architecture. J Cereal Sci. 2004;39:151–165. [Google Scholar]
- 15.Bertoft E, Seetharaman K. Starch Structure. In: Tetlow IJ, editor. Starch: Origins, Structure, and Metabolism. SEB; Lodon: 2012. pp. 1–28. [Google Scholar]
- 16.Buleon A, Colonna P, Planchot V, Ball S. Starch granules: structure and biosynthesis. Int J Biol Macromol. 1998;23:85–112. doi: 10.1016/s0141-8130(98)00040-3. [DOI] [PubMed] [Google Scholar]
- 17.Ball SG, Morell MK. From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu Rev Plant Biol. 2003;54:207–33. doi: 10.1146/annurev.arplant.54.031902.134927. [DOI] [PubMed] [Google Scholar]
- 18.Coppin A, Varre JS, Lienard L, Dauvillee D, Guerardel Y, Soyer-Gobillard MO, Buleon A, Ball S, Tomavo S. Evolution of plant-like crystalline storage polysaccharide in the protozoan parasite Toxoplasma gondii argues for a red alga ancestry. J Mol Evol. 2005;60:257–67. doi: 10.1007/s00239-004-0185-6. [DOI] [PubMed] [Google Scholar]
- 19.Meeuse BJD, Andries M, Wood JA. Floridean Starch. J Exp Bot. 1960;11:129–140. [Google Scholar]
- 20.Viola R, Nyvall P, Pedersen M. The unique features of starch metabolism in red algae. Proc Biol Sci. 2001;268:1417–22. doi: 10.1098/rspb.2001.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gentry MS, Dowen RH, 3rd, Worby CA, Mattoo S, Ecker JR, Dixon JE. The phosphatase laforin crosses evolutionary boundaries and links carbohydrate metabolism to neuronal disease. J Cell Biol. 2007;178:477–88. doi: 10.1083/jcb.200704094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gentry MS, Dixon JE, Worby CA. Lafora disease: insights into neurodegeneration from plant metabolism. Trends Biochem Sci. 2009;34:628–39. doi: 10.1016/j.tibs.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roach PJ, Depaoli-Roach AA, Hurley TD, Tagliabracci VS. Glycogen and its metabolism: some new developments and old themes. Biochem J. 2012;441:763–87. doi: 10.1042/BJ20111416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeeman SC, Tiessen A, Pilling E, Kato KL, Donald AM, Smith AM. Starch synthesis in Arabidopsis. Granule synthesis, composition, and structure. Plant Physiol. 2002;129:516–29. doi: 10.1104/pp.003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith AM, Zeeman SC, Smith SM. Starch degradation. Annu Rev Plant Biol. 2005;56:73–98. doi: 10.1146/annurev.arplant.56.032604.144257. [DOI] [PubMed] [Google Scholar]
- 26.Blennow A, Nielsen TH, Baunsgaard L, Mikkelsen R, Engelsen SB. Starch phosphorylation: a new front line in starch research. Trends Plant Sci. 2002;7:445–50. doi: 10.1016/s1360-1385(02)02332-4. [DOI] [PubMed] [Google Scholar]
- 27.Kotting O, Kossmann J, Zeeman SC, Lloyd JR. Regulation of starch metabolism: the age of enlightenment? Curr Opin Plant Biol. 2010;13:321–9. doi: 10.1016/j.pbi.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Blennow A, Engelsen SB. Helix-breaking news: fighting crystalline starch energy deposits in the cell. Trends Plant Sci. 2010;15:236–40. doi: 10.1016/j.tplants.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 29.Fernbach A. Quelques observations sur la composition de l’amidon de pommes de terre. C R Acad Sci. 1904:428–430. [Google Scholar]
- 30.Ritte G, Lloyd JR, Eckermann N, Rottmann A, Kossmann J, Steup M. The starch-related R1 protein is an alpha -glucan, water dikinase. Proc Natl Acad Sci U S A. 2002;99:7166–71. doi: 10.1073/pnas.062053099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorberth R, Ritte G, Willmitzer L, Kossmann J. Inhibition of a starch-granule-bound protein leads to modified starch and repression of cold sweetening. Nat Biotechnol. 1998;16:473–7. doi: 10.1038/nbt0598-473. [DOI] [PubMed] [Google Scholar]
- 32.Cohen A. Über elektrische Wanderung von Kolloiden. Z Elektrochem. 1897:63–67. [Google Scholar]
- 33.Caspar T, Lin TP, Kakefuda G, Benbow L, Preiss J, Somerville C. Mutants of Arabidopsis with altered regulation of starch degradation. Plant Physiol. 1991;95:1181–8. doi: 10.1104/pp.95.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeeman SC, Northrop F, Smith AM, Rees T. A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolysing enzyme. Plant J. 1998;15:357–65. doi: 10.1046/j.1365-313x.1998.00213.x. [DOI] [PubMed] [Google Scholar]
- 35.Yu TS, Kofler H, Hausler RE, Hille D, Flugge UI, Zeeman SC, Smith AM, Kossmann J, Lloyd J, Ritte G, Steup M, Lue WL, Chen J, Weber A. The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell. 2001;13:1907–18. doi: 10.1105/TPC.010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blennow A. Phosphorylation of the starch granule. In: Nakamura Y, editor. Starch: Metabolism and Structure. Springer Japan; 2015. pp. 399–424. [Google Scholar]
- 37.Baunsgaard L, Lutken H, Mikkelsen R, Glaring MA, Pham TT, Blennow A. A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated alpha-glucans and is involved in starch degradation in Arabidopsis. Plant J. 2005;41:595–605. doi: 10.1111/j.1365-313X.2004.02322.x. [DOI] [PubMed] [Google Scholar]
- 38.Ritte G, Steup M, Kossmann J, Lloyd JR. Determination of the starch-phosphorylating enzyme activity in plant extracts. Planta. 2003;216:798–801. doi: 10.1007/s00425-002-0931-1. [DOI] [PubMed] [Google Scholar]
- 39.Ritte G, Heydenreich M, Mahlow S, Haebel S, Kotting O, Steup M. Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS Lett. 2006;580:4872–6. doi: 10.1016/j.febslet.2006.07.085. [DOI] [PubMed] [Google Scholar]
- 40.Kotting O, Pusch K, Tiessen A, Geigenberger P, Steup M, Ritte G. Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves. The phosphoglucan, water dikinase. Plant Physiol. 2005;137:242–52. doi: 10.1104/pp.104.055954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skeffington AW, Graf A, Duxbury Z, Gruissem W, Smith AM. Glucan, Water Dikinase Exerts Little Control over Starch Degradation in Arabidopsis Leaves at Night. Plant Physiol. 2014;165:866–879. doi: 10.1104/pp.114.237016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahlow S, Hejazi M, Kuhnert F, Garz A, Brust H, Baumann O, Fettke J. Phosphorylation of transitory starch by alpha-glucan, water dikinase during starch turnover affects the surface properties and morphology of starch granules. New Phytol. 2014;203:495–507. doi: 10.1111/nph.12801. [DOI] [PubMed] [Google Scholar]
- 43.Hejazi M, Fettke J, Paris O, Steup M. The two plastidial starch-related dikinases sequentially phosphorylate glucosyl residues at the surface of both the A- and B-type allomorphs of crystallized maltodextrins but the mode of action differs. Plant Physiol. 2009;150:962–76. doi: 10.1104/pp.109.138750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansen PI, Larsen FH, Motawia SM, Blennow A, Spraul M, Dvortsak P, Engelsen SB. Structure and hydration of the amylopectin trisaccharide building blocks--Synthesis, NMR, and molecular dynamics. Biopolymers. 2008;89:1179–93. doi: 10.1002/bip.21075. [DOI] [PubMed] [Google Scholar]
- 45.Hansen PI, Spraul M, Dvortsak P, Larsen FH, Blennow A, Motawia MS, Engelsen SB. Starch phosphorylation--maltosidic restrains upon 3′- and 6′-phosphorylation investigated by chemical synthesis, molecular dynamics and NMR spectroscopy. Biopolymers. 2009;91:179–93. doi: 10.1002/bip.21111. [DOI] [PubMed] [Google Scholar]
- 46.Edner C, Li J, Albrecht T, Mahlow S, Hejazi M, Hussain H, Kaplan F, Guy C, Smith SM, Steup M, Ritte G. Glucan, water dikinase activity stimulates breakdown of starch granules by plastidial beta-amylases. Plant Physiol. 2007;145:17–28. doi: 10.1104/pp.107.104224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hejazi M, Fettke J, Haebel S, Edner C, Paris O, Frohberg C, Steup M, Ritte G. Glucan, water dikinase phosphorylates crystalline maltodextrins and thereby initiates solubilization. Plant J. 2008;55:323–34. doi: 10.1111/j.1365-313x.2008.03513.x. [DOI] [PubMed] [Google Scholar]
- 48.Hejazi M, Mahlow S, Fettke J. The glucan phosphorylation mediated by alpha-glucan, water dikinase (GWD) is also essential in the light phase for a functional transitory starch turn-over. Plant Signal Behav. 2014;9 doi: 10.4161/psb.28892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gentry MS, Pace RM. Conservation of the glucan phosphatase laforin is linked to rates of molecular evolution and the glucan metabolism of the organism. BMC Evol Biol. 2009;9:138. doi: 10.1186/1471-2148-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kerk D, Conley TR, Rodriguez FA, Tran HT, Nimick M, Muench DG, Moorhead GB. A chloroplast-localized dual-specificity protein phosphatase in Arabidopsis contains a phylogenetically dispersed and ancient carbohydrate-binding domain, which binds the polysaccharide starch. Plant J. 2006;46:400–13. doi: 10.1111/j.1365-313X.2006.02704.x. [DOI] [PubMed] [Google Scholar]
- 51.Moorhead GB, De Wever V, Templeton G, Kerk D. Evolution of protein phosphatases in plants and animals. Biochem J. 2009;417:401–9. doi: 10.1042/BJ20081986. [DOI] [PubMed] [Google Scholar]
- 52.Fordham-Skelton AP, Chilley P, Lumbreras V, Reignoux S, Fenton TR, Dahm CC, Pages M, Gatehouse JA. A novel higher plant protein tyrosine phosphatase interacts with SNF1-related protein kinases via a KIS (kinase interaction sequence) domain. The Plant Journal. 2002;29:705–715. doi: 10.1046/j.1365-313x.2002.01250.x. [DOI] [PubMed] [Google Scholar]
- 53.Niittyla T, Comparot-Moss S, Lue W-L, Messerli G, Trevisan M, Seymour MDJ, Gatehouse JA, Villadsen D, Smith SM, Chen J, Zeeman SC, Smith AM. Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J Biol Chem. 2006;281:11815–11818. doi: 10.1074/jbc.M600519200. [DOI] [PubMed] [Google Scholar]
- 54.Hejazi M, Fettke J, Kotting O, Zeeman SC, Steup M. The Laforin-like dual-specificity phosphatase SEX4 from Arabidopsis hydrolyzes both C6- and C3-phosphate esters introduced by starch-related dikinases and thereby affects phase transition of alpha-glucans. Plant Physiol. 2010;152:711–22. doi: 10.1104/pp.109.149914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kotting O, Santelia D, Edner C, Eicke S, Marthaler T, Gentry MS, Comparot-Moss S, Chen J, Smith AM, Steup M, Ritte G, Zeeman SC. STARCH-EXCESS4 is a laforin-like Phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana. Plant Cell. 2009;21:334–46. doi: 10.1105/tpc.108.064360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takeda Y, Hizukuri S. Studies on starch phosphate. Part 5. Reexamination of the action of sweet-potato beta-amylase on phophorylated (1->4)-a-D-glucan. Carbohydr Res. 1981;39:151–165. [Google Scholar]
- 57.Comparot-Moss S, Kotting O, Stettler M, Edner C, Graf A, Weise SE, Streb S, Lue WL, MacLean D, Mahlow S, Ritte G, Steup M, Chen J, Zeeman SC, Smith AM. A putative phosphatase, LSF1, is required for normal starch turnover in Arabidopsis leaves. Plant Physiol. 2010;152:685–97. doi: 10.1104/pp.109.148981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santelia D, Kotting O, Seung D, Schubert M, Thalmann M, Bischof S, Meekins DA, Lutz A, Patron N, Gentry MS, Allain FH, Zeeman SC. The phosphoglucan phosphatase like sex four2 dephosphorylates starch at the c3-position in Arabidopsis. Plant Cell. 2011;23:4096–111. doi: 10.1105/tpc.111.092155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Worby CA, Gentry MS, Dixon JE. Laforin, a dual specificity phosphatase that dephosphorylates complex carbohydrates. J Biol Chem. 2006;281:30412–8. doi: 10.1074/jbc.M606117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meekins DA, Raththagala M, Auger KD, Turner BD, Santelia D, Kotting O, Gentry MS, Vander Kooi CW. Mechanistic Insights into Glucan Phosphatase Activity Against Polyglucan Substrates. J Biol Chem. 2015 doi: 10.1074/jbc.M115.658203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Janecek S, Svensson B, MacGregor EA. Structural and evolutionary aspects of two families of non-catalytic domains present in starch and glycogen binding proteins from microbes, plants and animals. Enzyme Microb Technol. 2011;49:429–40. doi: 10.1016/j.enzmictec.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 62.Guillen D, Sanchez S, Rodriguez-Sanoja R. Carbohydrate-binding domains: multiplicity of biological roles. Appl Microbiol Biotechnol. 2010;85:1241–9. doi: 10.1007/s00253-009-2331-y. [DOI] [PubMed] [Google Scholar]
- 63.Abbott DW, van Bueren AL. Using structure to inform carbohydrate binding module function. Curr Opin Struct Biol. 2014;28:32–40. doi: 10.1016/j.sbi.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 64.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–8. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2013;42:D490–5. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Christiansen C, Hachem MA, Glaring MA, Vikso-Nielsen A, Sigurskjold BW, Svensson B, Blennow A. A CBM20 low-affinity starch-binding domain from glucan, water dikinase. FEBS Lett. 2009;583:1159–63. doi: 10.1016/j.febslet.2009.02.045. [DOI] [PubMed] [Google Scholar]
- 67.Machovic M, Janecek S. Starch-binding domains in the post-genome era. Cell Mol Life Sci. 2006;63:2710–24. doi: 10.1007/s00018-006-6246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Polekhina G, Gupta A, van Denderen BJ, Feil SC, Kemp BE, Stapleton D, Parker MW. Structural basis for glycogen recognition by AMP-activated protein kinase. Structure. 2005;13:1453–62. doi: 10.1016/j.str.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 69.Minassian BA, Ianzano L, Meloche M, Andermann E, Rouleau GA, Delgado-Escueta AV, Scherer SW. Mutation spectrum and predicted function of laforin in Lafora’s progressive myoclonus epilepsy. Neurology. 2000;55:341–6. doi: 10.1212/wnl.55.3.341. [DOI] [PubMed] [Google Scholar]
- 70.Ganesh S, Tsurutani N, Suzuki T, Hoshii Y, Ishihara T, Delgado-Escueta AV, Yamakawa K. The carbohydrate-binding domain of Lafora disease protein targets Lafora polyglucosan bodies. Biochem Biophys Res Commun. 2004;313:1101–9. doi: 10.1016/j.bbrc.2003.12.043. [DOI] [PubMed] [Google Scholar]
- 71.Tagliabracci VS, Turnbull J, Wang W, Girard JM, Zhao X, Skurat AV, Delgado-Escueta AV, Minassian BA, Depaoli-Roach AA, Roach PJ. Laforin is a glycogen phosphatase, deficiency of which leads to elevated phosphorylation of glycogen in vivo. Proc Natl Acad Sci U S A. 2007;104:19262–6. doi: 10.1073/pnas.0707952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tagliabracci VS, Girard JM, Segvich D, Meyer C, Turnbull J, Zhao X, Minassian BA, Depaoli-Roach AA, Roach PJ. Abnormal metabolism of glycogen phosphate as a cause for Lafora disease. J Biol Chem. 2008;283:33816–25. doi: 10.1074/jbc.M807428200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gentry MS, Roma-Mateo C, Sanz P. Laforin, a protein with many faces: glucan phosphatase, adapter protein, et alii. FEBS J. 2013;280:525–37. doi: 10.1111/j.1742-4658.2012.08549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roach PJ. Glycogen phosphorylation and Lafora disease. Mol Aspects Med. 2015 doi: 10.1016/j.mam.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–46. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 76.Matsuda M, Takeshita K, Kurokawa T, Sakata S, Suzuki M, Yamashita E, Okamura Y, Nakagawa A. Crystal structure of the cytoplasmic phosphatase and tensin homolog (PTEN)-like region of Ciona intestinalis voltage-sensing phosphatase provides insight into substrate specificity and redox regulation of the phosphoinositide phosphatase activity. J Biol Chem. 2011;286:23368–77. doi: 10.1074/jbc.M110.214361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guan R, Dai H, Harrison SC, Kirchhausen T. Structure of the PTEN-like region of auxilin, a detector of clathrin-coated vesicle budding. Structure. 2010;18:1191–8. doi: 10.1016/j.str.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Begley MJ, Taylor GS, Kim SA, Veine DM, Dixon JE, Stuckey JA. Crystal structure of a phosphoinositide phosphatase, MTMR2: insights into myotubular myopathy and Charcot-Marie-Tooth syndrome. Mol Cell. 2003;12:1391–402. doi: 10.1016/s1097-2765(03)00486-6. [DOI] [PubMed] [Google Scholar]
- 79.Raththagala M, Brewer MK, Parker MW, Sherwood AR, Wong BK, Hsu S, Bridges TM, Paasch BC, Hellman LM, Husodo S, Meekins DA, Taylor AO, Turner BD, Auger KD, Dukhande VV, Chakravarthy S, Sanz P, Woods VL, Jr., Li S, Vander Kooi CW, Gentry MS. Structural mechanism of laforin function in glycogen dephosphorylation and lafora disease. Mol Cell. 2015;57:261–72. doi: 10.1016/j.molcel.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cuyvers S, Dornez E, Delcour JA, Courtin CM. Occurrence and functional significance of secondary carbohydrate binding sites in glycoside hydrolases. Crit Rev Biotechnol. 2012 doi: 10.3109/07388551.2011.561537. [DOI] [PubMed] [Google Scholar]
- 81.Chu HM, Wang AH. Enzyme-substrate interactions revealed by the crystal structures of the archaeal Sulfolobus PTP-fold phosphatase and its phosphopeptide complexes. Proteins. 2007;66:996–1003. doi: 10.1002/prot.21262. [DOI] [PubMed] [Google Scholar]
- 82.Almo SC, Bonanno JB, Sauder JM, Emtage S, Dilorenzo TP, Malashkevich V, Wasserman SR, Swaminathan S, Eswaramoorthy S, Agarwal R, Kumaran D, Madegowda M, Ragumani S, Patskovsky Y, Alvarado J, Ramagopal UA, Faber-Barata J, Chance MR, Sali A, Fiser A, Zhang ZY, Lawrence DS, Burley SK. Structural genomics of protein phosphatases. J Struct Funct Genomics. 2007;8:121–40. doi: 10.1007/s10969-007-9036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song H, Hanlon N, Brown NR, Noble ME, Johnson LN, Barford D. Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2. Mol Cell. 2001;7:615–26. doi: 10.1016/s1097-2765(01)00208-8. [DOI] [PubMed] [Google Scholar]
- 84.Agarwal R, Burley SK, Swaminathan S. Structure of human dual specificity protein phosphatase 23, VHZ, enzyme-substrate/product complex. J Biol Chem. 2008;283:8946–53. doi: 10.1074/jbc.M708945200. [DOI] [PubMed] [Google Scholar]
- 85.Lountos GT, Tropea JE, Waugh DS. Structure of human dual-specificity phosphatase 27 at 2.38 A resolution. Acta Crystallogr D Biol Crystallogr. 2011;67:471–9. doi: 10.1107/S090744491100970X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiao J, Engel JL, Zhang J, Chen MJ, Manning G, Dixon JE. Structural and functional analysis of PTPMT1, a phosphatase required for cardiolipin synthesis. Proc Natl Acad Sci U S A. 2011;108:11860–5. doi: 10.1073/pnas.1109290108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wishart MJ, Dixon JE. Gathering STYX: phosphatase-like form predicts functions for unique protein-interaction domains. Trends Biochem Sci. 1998;23:301–6. doi: 10.1016/s0968-0004(98)01241-9. [DOI] [PubMed] [Google Scholar]
- 88.Salmeen A, Barford D. Functions and mechanisms of redox regulation of cysteine-based phosphatases. Antioxid Redox Signal. 2005;7:560–77. doi: 10.1089/ars.2005.7.560. [DOI] [PubMed] [Google Scholar]
- 89.Silver DM, Silva LP, Issakidis-Bourguet E, Glaring MA, Schriemer DC, Moorhead GB. Insight into the redox regulation of the phosphoglucan phosphatase SEX4 involved in starch degradation. FEBS J. 2013;280:538–48. doi: 10.1111/j.1742-4658.2012.08546.x. [DOI] [PubMed] [Google Scholar]
- 90.Amoasii L, Hnia K, Laporte J. Myotubularin phosphoinositide phosphatases in human diseases. Curr Top Microbiol Immunol. 2012;362:209–33. doi: 10.1007/978-94-007-5025-8_10. [DOI] [PubMed] [Google Scholar]
- 91.Hsu F, Mao Y. The structure of phosphoinositide phosphatases: Insights into substrate specificity and catalysis. Biochim Biophys Acta. 2015;1851:698–710. doi: 10.1016/j.bbalip.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cho H, Ramer SE, Itoh M, Kitas E, Bannwarth W, Burn P, Saito H, Walsh CT. Catalytic domains of the LAR and CD45 protein tyrosine phosphatases from Escherichia coli expression systems: purification and characterization for specificity and mechanism. Biochemistry. 1992;31:133–8. doi: 10.1021/bi00116a019. [DOI] [PubMed] [Google Scholar]
- 93.Guan KL, Broyles SS, Dixon JE. A Tyr/Ser protein phosphatase encoded by vaccinia virus. Nature. 1991;350:359–62. doi: 10.1038/350359a0. [DOI] [PubMed] [Google Scholar]
- 94.Robert X, Haser R, Mori H, Svensson B, Aghajari N. Oligosaccharide binding to barley alpha-amylase 1. J Biol Chem. 2005;280:32968–78. doi: 10.1074/jbc.M505515200. [DOI] [PubMed] [Google Scholar]
- 95.Koropatkin NM, Smith TJ. SusG: a unique cell-membrane-associated alpha-amylase from a prominent human gut symbiont targets complex starch molecules. Structure. 18:200–15. doi: 10.1016/j.str.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 96.Imberty A, Chanzy H, Perez S, Buleon A, Tran V. The double-helical nature of the crystalline part of A-starch. J Mol Biol. 1988;201:365–78. doi: 10.1016/0022-2836(88)90144-1. [DOI] [PubMed] [Google Scholar]
- 97.Recondo E, Leloir LF. Adenosine diphosphate glucose and starch synthesis. Biochem Biophys Res Commun. 1961;6:85–8. doi: 10.1016/0006-291x(61)90389-8. [DOI] [PubMed] [Google Scholar]
- 98.Barford D, Flint AJ, Tonks NK. Crystal structure of human protein tyrosine phosphatase 1B. Science. 1994;263:1397–404. [PubMed] [Google Scholar]
- 99.Ral JP, Bowerman AF, Li Z, Sirault X, Furbank R, Pritchard JR, Bloemsma M, Cavanagh CR, Howitt CA, Morell MK. Down-regulation of Glucan, Water-Dikinase activity in wheat endosperm increases vegetative biomass and yield. Plant Biotechnol J. 2012;10:871–82. doi: 10.1111/j.1467-7652.2012.00711.x. [DOI] [PubMed] [Google Scholar]
- 100.Ma J, Jiang QT, Wei L, Yang Q, Zhang XW, Peng YY, Chen GY, Wei YM, Liu C, Zheng YL. Conserved structure and varied expression reveal key roles of phosphoglucan phosphatase gene starch excess 4 in barley. Planta. 2014;240:1179–90. doi: 10.1007/s00425-014-2140-0. [DOI] [PubMed] [Google Scholar]
- 101.Nunez-Lopez L, Aguirre-Cruz A, Barrera-Figueroa BE, Pena-Castro JM. Improvement of enzymatic saccharification yield in Arabidopsis thaliana by ectopic expression of the rice SUB1A-1 transcription factor. PeerJ. 2015;3:e817. doi: 10.7717/peerj.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Weise SE, Aung K, Jarou ZJ, Mehrshahi P, Li Z, Hardy AC, Carr DJ, Sharkey TD. Engineering starch accumulation by manipulation of phosphate metabolism of starch. Plant Biotechnol J. 2012;10:545–54. doi: 10.1111/j.1467-7652.2012.00684.x. [DOI] [PubMed] [Google Scholar]
- 103.Berrocal-Lobo M, Ibanez C, Acebo P, Ramos A, Perez-Solis E, Collada C, Casado R, Aragoncillo C, Allona I. Identification of a homolog of Arabidopsis DSP4 (SEX4) in chestnut: its induction and accumulation in stem amyloplasts during winter or in response to the cold. Plant Cell Environ. 2011;34:1693–704. doi: 10.1111/j.1365-3040.2011.02365.x. [DOI] [PubMed] [Google Scholar]
- 104.Sarmiento M, Puius YA, Vetter SW, Keng YF, Wu L, Zhao Y, Lawrence DS, Almo SC, Zhang ZY. Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition. Biochemistry. 2000;39:8171–9. doi: 10.1021/bi000319w. [DOI] [PubMed] [Google Scholar]