Abstract
Neural tissue engineering aims at developing novel approaches for the treatment of diseases of the nervous system, by providing a permissive environment for the growth and differentiation of neural cells. Three-dimensional (3D) cell culture systems provide a closer biomimetic environment, and promote better cell differentiation and improved cell function, than could be achieved by conventional two-dimensional (2D) culture systems. With the recent advances in the discovery and introduction of different types of stem cells for tissue engineering, microfluidic platforms have provided an improved microenvironment for the 3D-culture of stem cells. Microfluidic systems can provide more precise control over the spatiotemporal distribution of chemical and physical cues at the cellular level compared to traditional systems. Various microsystems have been designed and fabricated for the purpose of neural tissue engineering. Enhanced neural migration and differentiation, and monitoring of these processes, as well as understanding the behavior of stem cells and their microenvironment have been obtained through application of different microfluidic-based stem cell culture and tissue engineering techniques. As the technology advances it may be possible to construct a “brain-on-a-chip”. In this review, we describe the basics of stem cells and tissue engineering as well as microfluidics-based tissue engineering approaches. We review recent testing of various microfluidic approaches for stem cell-based neural tissue engineering.
Keywords: Microfluidics, stem cells, neural tissue engineering, 3D culture, stem cell niche, brain on a chip
Graphical abstract
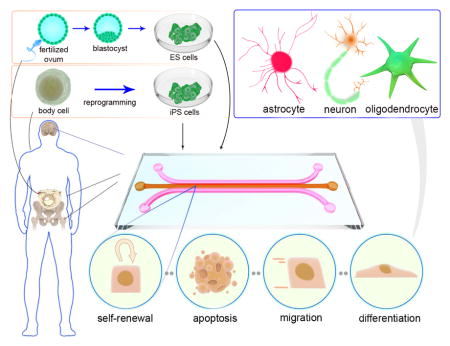
Overall process of stem cell derivation and isolation, as well as microfluidic stem cell culture and neural tissue engineering
1. Introduction
Neural tissue engineering has attracted increasing attention in recent years; however the complexity of the biology of the nervous system, and the intrinsic biological blocks that interfere with its regeneration require innovative approaches 1–4. Scientists, physicians and engineers have applied various approaches and tools for enhancing neural regeneration and tissue engineering by better mimicking of the in vivo environment. These approaches include the development of bioactive scaffolds that imitate the extracellular matrix (ECM), improved monolayer technique using surface patterns with micro- and nano-topographies, nano-enabled materials and emerging technologies such as three-dimensional (3D) printing, microtechnology and microfluidics 1, 3, 5–8. Conventional cell culture and tissue engineering methods suffer from limitations including limited distribution of biomolecules by diffusion, and lack of natural interactions between the ECM and the cells themselves. To overcome these limitations, several 3D-based cell culture approaches have been created such as gel-based and spheroid-based systems, and also various porous structures 5, 9–12. Although theses 3D techniques provide a better biomimetic microenvironment for cells and stem cells than do two-dimensional (2D) cultures, they still have limitations in spatiotemporal control of specific cell culture parameters, and have encouraged researchers to further develop optimized methods.
In addition to the substantial impact of microfluidic technology on biomedical research 13–15, microfluidics has shown much promise in the field of tissue engineering. Microfluidic systems offer advantages over conventional well-plate systems including the ability to: 1) control the spatiotemporal distribution of physico-chemical signals at the cellular level, 2) to analyze cell differentiation and function using fewer cells and smaller quantities of reagents, and 3) perform multiple assays simultaneously 9, 16, 17. Moreover, microfluidic devices can overcome the disadvantages of typical cell/stem cell culture methods and tissue engineering approaches by improved mimicking of in vivo interactions between ECM and cells, and providing opportunities for high-resolution in situ imaging 16–18. In this regard neuroscience research and neural tissue engineering have benefited from different potential applications of microdevices, including improved neuronal culture, better in vitro disease modeling, new methods of cell isolation, and stem cell research 19–21.
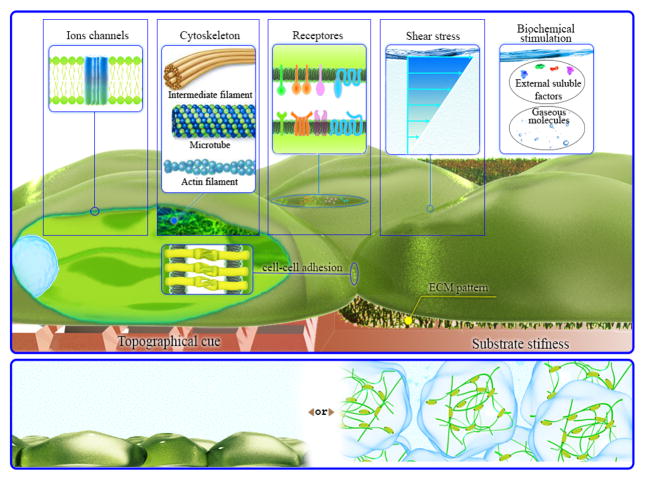
The combination of the particular advantages of microfluidics, and the range of possibilities provided by stem cell technologies, may provide solutions for the management of neurodegenerative diseases such as Alzheimer’s and Parkinson’s and other disorders or injuries of the central or peripheral nervous system. This approach has even gone so far as to propose the creation of devices that have become known as a “brain-on-a-chip” 22–25 . Figure 1 schematically illustrates mimicking of the native ECM via microfluidics with the potential to control the spatiotemporal interactions of stem cells with the ECM, with the provision of internal or external stimuli and potential cellular targets. Two main approaches of microfluidic-based cell/stem cells culture, gel free- or gel supported substrates, are also shown.
Figure 1.
Stem cells in a microfluidic device. The figure demonstrates the possible physic-chemical and biomolecular stimuli, which could be provided by microfluidics (top). Schematic illustration of different stem cell culturing approaches (supported via gel matrix or not) is also shown (bottom).
To explain the synergistic combination of microfluidics and stem cell research, we begin with the introduction of different types of stem cells, their sources and specific microenvironment, as well as the limitations of traditional stem cell culture techniques. Next microfluidics, and its physico-mechanical and biochemical properties are discussed with a particular focus on tissue engineering applications. We also review the recent applications of microfluidics in stem cell-based neural tissue engineering and neural stem cell culture.
2. Stem cells and tissue engineering
The absence of any effective therapy for spinal cord injury (SCI), prevalent neurodegenerative diseases, not to mention strokes and traumatic brain injuries has led to the possibility of using stem cell engineering as an innovative approach for the regeneration of damaged neural tissue. In this regard, finding appropriate sources of stem cells that are able to differentiate into different types of mature neuronal cells, including neurons, glial cells, astrocytes and oligodendrocytes, has become the first step towards stem cell-based neural tissue engineering 26.
2.1 Stem cells' sources for Neural Tissue engineering
With the discovery of multipotent and pluripotent stem cells (PSCs), new avenues for tissue engineering involving the formation of various soft and hard tissues have emerged 27–29. Among the different kinds of stem cells available, embryonic stem cells (ESCs) 30, neural stem cells (NSCs) 31, human induced pluripotent stem cells (hiPSCs) 32, mesenchymal stem cells (MSCs) 33 and adipose tissue-derived stem cells (ATSCs) 34 have all shown promising results for applications in neural tissue engineering. Intrinsic mechanisms such as the expression and activation of transcription factors, and extrinsic signals provided by the microenvironment (niche) such as growth factors, ECM-cell interactions, and cell-cell interactions have improved the ability to control the fate of stem cells 35, 36. On the other hand, essential elements of cell sources must be considered to develop the cell/tissue replacement and promote the outcome efficiency. First they must be allogeneic to reduce the unwanted immune-responses 37, further they should represent higher surviving rate to promote the clinical applications 38. Also the cell sources must be able to be prepared by standard methods to control the expression of undesired phenotype and risk of dyskinesia 39.
2.1.1 Pluripotent stem cells (PSCs)
PSCs were obtained from a mouse embryo for the first time in 1981, and at that time were called embryonic stem cells (ESCs) to distinguish them from stem cells derived from other sources such as teratocarcinomas 40. The discovery of the unique properties of these stem cells, their self-renewing ability, and their responsiveness to particular stimulations by undergoing differentiation to different specific cell types, paved the way for a revolution in regenerative medicine 41. For example Iwai et al 37, demonstrated that ESC obtained from allogeneic sources could be used to generate neurons and to from synaptic connections in a non-human primate. Kassmer et al. 42 reported new type of PSCs known as very small embryonic-like cells that have potential ability to generate all cell types including the neurons. Although the PSCs possess favorable properties but their expansion is difficult in culturing conditions and they lose their multipotency in vitro. Chromosomal abnormality takes place when they grow in culturing medium and need strict selection at the time of engrafting to prevent transforming of undifferentiated cells and teratoma formation. Moreover they required a feeder layer in culture dish to support their continues growth direction and suppress spontaneous differentiation 43.
2.1.2 Induced-pluripotent stem cells
Other major approach that is often discussed involves the treatment of somatic stem cells under certain specific conditions with manipulation of genetic factors that would cause them to revert to a type of immortal and PSCs, called induced-PSCs (iPSCs) 44. At the initiation of these iPSCs efforts, the efficiency of reprogramming was less than 1%, but in 2013 it was reported to have reached 100% by genetic regulation 45. Further studies found that the efficiency of differentiation of these cells into the desired tissue type was less effective than ESCs, although this effectiveness can be improved through optimized treatment methods 26. Other researchers have used iPSCs isolated from different sources such as reprogramming of adult fibroblasts that can be transformed into brain cells. This technique could provide the possibility to wider application of iPSCs in tissue engineering, because the possibility of triggering an immune response and subsequent rejection would be reduced 46, 47. New methods have been investigated to allow direct transformation of somatic cells into neural progenitor cells that are called “induced neural stem cells” (iNSCs) in order to reduce the possible tumorigenesis of iPSCs 48. Although the iPSCs have expanded potential therapeutic application, but their utilization is limited due to requirement of genetic manipulation and cell regulation as well as ethical consideration.
2.1.3 Neural stem cells (NSCs)
Neural stem cells are part multipotent stem cells and exist in all main subdivision of central nervous systems including non-neurogenetic regions. In 1992, Reynolds et al. established new methods for in vitro cell culture, and showed adult-derived brain cells were able to proliferate and generate both neurons and astrocytes 49. Primary adult neural progenitors known as a major candidates for brain regeneration. Although oligodentrocyte progenitors and astrocytes have the ability of self-renewing, they do not consider as neural stem cell, due to their disability of multipotency 50. Ependimal cells, epithelial cells of brain's ventricular system and spinal core's central canal, are in vitro potential cells to generate all types of neural cells 51. Cave et al. 52 in a review introduce adult subventricular zone cells as a suitable source for replacing the dopaminerging neural cells.
2.1.4 Other non-neural stem cells
Nowadays non-neural stem cells are the main source of cells to be differentiated into neurons and other neural cells. For example cells derived from bone marrow (a rich source of hematopoietic stem cells (HSCs)), are often used in different aspects of tissue engineering, as well as for the production of glial cells and neurons53. Other varieties of non-neural stem cells and non-PSCs have been reported that can serve as multipotent stem cells. The variety of their differentiation and proliferation abilities is less pronounced than those of PSCs, and they have been identified as multi-cloned stem cells. Moreover, under normal growth conditions they are only able to produce a limited number of cell types. However, under certain specialized conditions and using appropriate techniques, all of them are able to generate neural progenitor cells. Different types of MSCs which are present in various tissues such as skeletal stem cells, bone marrow stromal cells, hepatoblasts and chondroblasts are other example that have been studied 54. Furthermore it was represented that cells obtained from human liposuction fat have special ability to divide into other cell types. These MSCs were called adipose-drived stem cells (ASCs) 55. ASCs are abundant and can be harvested with minimal invasive procedure. Their transferring into the host is safe and effective and manipulation of them is facile by current manufacturing guideline 56. In addition placenta-derived multipotent stem cells (PDMCs) can be obtained from human placenta with the potential to differentiate into different cell lineages without any ethical problems 57. Some studies have reported derivation of neuroepithelial stem cells (NEPSCs) from ESCs with the potential to differentiate into neural tube and neural crest lineages under the appropriate biochemical conditions 58, 59.
2.2 Neural stem cell biology and microenvironment
Generally, the biology and fate of stem cells is under control of a range of internal or external factors, including the ECM, the interaction of stem cells with surrounding cells, various physical-chemical stimuli, and soluble growth factors 60. The combination of these factors as a whole, determines the regulatory environment for neuronal cells, which is called the “neurogenic niche” 61. The interactions of stem cells with their environment and with surrounding cells govern their function, proliferation and differentiation. The neighboring cells (including mature neurons, astrocytes, endothelial cells, microglia, etc) interact with the NSCs and govern their final fate. In fact, this network controls the behavior of NSCs in terms of differentiation, proliferation and migration 62.
The ECM proteins of the neural tissue play a dual role in nerve development; firstly they provide a suitable physical support for structural shape of the mature nerves especialy collagen, and secondly they regulate the NSCs responses to the growth factors especially tenascin 63, 64. The most important ECM proteins are summarized in Table 1.
Table 1.
The most important ECM proteins in the neural stem cell microenvironment
| ECM Protein | Location | Function | Ref |
|---|---|---|---|
| HSPG1 | Neuroepithelial cells | Pluripotency and neural differentiation | 65 |
| Collagen | Neocortex | Regulation of corticogenesis, promotion of neuronal differentiation | 66 |
| Laminins | Neuroepithelial cells | Promotes polarization and neuroepithelium formation | 67 |
| Heparin | Neocortex | Proliferation of NSC | 68 |
| CSPG2 | Neocortex and ganglionic eminence | Promotes self-renewal and neurogenesis | 69 |
| Vitronectin | Spinal cord | Promotes differentiation into oligodendrocytes | 32 |
| Reelin | Developing cortex | Controls differentiation, migration, and proliferation of NSC | 64 |
| Tenascin | Neocortex and RGC3 | Promotes epidermal growth factor (EGF) response | 63 |
HSPG: heparan sulfate proteoglycan,
CSPG: chondroitin sulfate proteoglycan,
RGC: radial glial cells. Data derived from ref 70.
In development of the embryonic central nervous system and neural tube, two main anatomical areas have been established; the ventricular zone (VZ) which is the origin of the all the different nervous cell types, and the sub-ventricular zone (SVZ) which consists of intermediate progenitors that ultimately produce neurons and glia 70. During the maturation process, neuro-epithelial cells are exposed to several extracellular factors and components and migrate from the VZ toward the basal parts (SVZ) where they are transformed into RGC 52. The NSCs niche is a modulated system in which several components interact with each other to development nerve tissue, and to preserve the neural progenitor pool under control of “Sonic Hedgehog” (Shh) proteins 71. Notch family proteins influence the gene expression pattern in the NSCs, and their mutation has been shown to deplete the neural pool 72 Moreover, morphogens such as Wnt and bone morphogenic proteins (BMPs) also participate in the maturation and distribution of NSCs in the SVZ 73. The major growth factors and hormones that participate in the embryonic NSCs microenvironment are summarized in Table 2.
Table 2.
Major growth factors and hormones in the embryonic NSCs niche
| Growth factor or Hormone | Location | Function | Ref |
|---|---|---|---|
| Growth hormone | Neocortex | Stimulates the differentiation and proliferation of cells | 76 |
| Cystatin-C | Neocortex | Promotes astrogenesis and suppresses oligodendrogenesis | 77 |
| Insulin Growth Factors | Neocortex | Promotes self-renewal of NSCs | 78 |
| TGFβ-1 | Neocortex | Promotes astrocytosis | 75 |
| Ciliary neurotrophic factor | VZ | Enhances proliferation and survival of NSCs | 79 |
| Leukemia inhibitory factor | VZ | Enhances proliferation and survival of NSCs | 80 |
| Platelet derived growth factor (PDGF)-A & B | SVZ | Promotes oligodendrocytsis and astrocytosis | 71 |
| Bone morphogenetic proteins | SVZ | Promotes SVZ response to EGF | 73 |
| VEGF | RGC | Angiogenic and mitogenic factor | 81 |
| FGF | RGC | Promotes self-renewal and multipotency | 82 |
| Erythropoietin | Ganglionic eminences | Enhances neural progenitor cell production | 78 |
| BDNF1 | Forebrain | Promotes NSC proliferation | 83 |
| EGF | Striatal primordial | Promotes NSC proliferation | 68 |
| Glial cell line derived neurotrophic factor | Ventral mesencephalon | Neurotrophic factor for NSC | 82 |
| Ghrelin | Spinal cord | Promotes NSC proliferation | 84 |
brain-derived neurotrophic factor. Data derived from Ref 70.
Vascularization of the embryonic central nervous system takes place simultaneously, along with the process of neural expansion. Vascularization is governed by non-neural cells like pericyte cells of the brain, and products secreted by RGCs and endothelial cells, such as vascular endothelial growth factor (VEGF), fibroblast growth factor-2 (FGF2) and Notch ligands 74. This blood vessel network has an important role in the maintenance of NSCs and their proliferation and differentiation, and can further enhance neural growth and protection by promoting the migration of astroglial cells and helps the distribution of several soluble regulatory factors. The endothelial cells of the vessels also generate transforming growth factor β-2 (TGFβ-2) that encourages neurogenesis 75.
Knowledge of the regulatory mechanisms of neural progenitor cells is the major key to design suitable in vitro conditions which is required for generation of mature nerves. Mimicking the behavior of ECM with biocompatible scaffolds and conserving the neural extracellular components are necessary for efficient cell culture. In this manner, microfluidic systems could allow each single cell to interact with signaling molecules and adopt its own natural behavior. Thus detailed studies of the neural microenvironment and the impact of ECM are important for maximizing cell viability and controlling neural cell differentiation.
2.3 Traditional cell/stem cell culture techniques; limitations and solutions
Traditional 2D culture methods were the first strategy used for culturing stem cells and screening for the importance of different components. In this method cells are attached to a plastic dish containing medium and usually grow in a flat monolayer. The 2D cell culture systems provide the basic information of cell growth requirement, but are unsuitable to mimic the entire in vivo microenvironment especially since they interact with an unnatural (plastic) surface. The production yield of these systems is low and cell growth take place in an unnatural and uncontrolled manner in response to external factors. Furthermore, the viability of cells in these systems is low, and they require further improvement to increase cell-cell interactions and normalize signaling 9, 85. On the other hand, 3D cell culture systems have been designed to provide the ability to better sustain cell cultures in which cells are naturally attached to each other and normal gap junction are produced between them that enable the individual cells to communicate with each other by transfer of electrical currents, ions and small molecules. The use of biocompatible extracellular scaffolds allows the cells to move and migrate in a 3D culture system and exert different growth factors and each differentiated cell can act as it would in vivo 86. Since 2D static cultures do not resemble in vivo conditions and have poor scalability, different culture methods such as cell aggregates, nano-carriers, microencapsulation and microfluidic based approaches have been investigated. The cell aggregation approach (cell spheroids) has advantages including easy handling, with an ability for a high yield of cell production, can better mimic the native microenvironment due better to cell-cell interactions, and can allow efficient differentiation. However it has disadvantages including being difficult to control the aggregation size, the aggregates require dissociation, the long-term outcome of the culture is uncertain, and physical forces might damage the cells if the aggregate becomes too big 87. In the nanocarrier strategy, the production yield is high and due to the high surface to volume (S/V) ratio typical of nanostructures, there is no limitation for diffusion of nutrients. This system can be set up for large scale production with easy handling and reduced consumption of materials (growth factors and medium). However, this method requires cell bead separation technique and clump controlling. The cost of materials in this system can be high, cells might be lysed owing to mechanical forces, also the monitoring of the culture is not easy 88. 3D cell-cell and cell-matrix interactions are well conserved in the microencapsulation strategy. In this system a polymeric biomaterial is engineered to enhance cell culture performance, which is the main benefit of this method, but cell harvesting is complicated for microencapsulated cultures 89.
3. Microfluidics and tissue engineering
Microfluidic technology refers to manipulation of fluids at microliter to picoliter levels in specific environments, structures and devices 90, 91. Microfluidics is a multidisciplinary field involving different scientific disciplines such as biotechnology, engineering, and physics 91. The scale of these devices is considered to be a favorable environment for cells and tissues. In other words, by applying the tools of microfluidics, researchers can control the microenvironment of cells under nearly optimum conditions 9, 92. Microfluidic devices and methods help biologists to culture, maintain and analyze the cell behavior in a controlled microenvironment. For example a series of microchannels was fabricated in order to obtain a standard culture medium to maintain neuronal differentiation of C17.2 NSCs and therefore to prevent unfavorable changes in the cell phenotype 93. The combination of microfluidic systems with stem cell technology can provide appropriate conditions for stem cell culture compared to the use of other cell types, and other traditional culture approaches. For example in neuroregeneration, these systems allow the generation of uniform populations of neuronal cells and glial cells 20. Combining diffusion and laminar flow, better controlled signaling, and the ability to co-culture cells in a 3D arrangement are the most important advantages of microfluidic cell culture 94. Different applications of these characteristics in neural regeneration include the culture of single ESCs 95, study of ESC differentiation and monitoring of their migration 96, and gradient-mediated NSC chemotaxis 97.
3.1 Microfluidics and the physico-chemical and physico-mechanical properties of stem cells
The microenvironment of stem cells arises from different physiological, physico-chemical and physico-mechanical cues such as cell-ECM interactions, growth factor stimulation, shear stress, and the rigidity and topography of the microenvironment (Fig 1) 9, 18, 98. The effects of these factors may be detectable in improved cell-to-cell and cell-to-matrix interactions, cellular signaling and interactions with the microenvironment 98. Stem cell behavior such as proliferation and differentiation are governed by these factors and interactions 98. For example the stiffness of ECM affects the differentiation pathway of MSCs; soft matrix increases neural differentiation while tough or rigid matrix increases myogenic and osteogenic differentiation 98. Microfluidics can provide precise control over the stem cell/cell numbers and growth conditions, and enables researchers to arrange or design the cells in spatially controlled positions, and to track cell responses to different internal/external mechanical, chemical, and optical stimuli. Moreover, microfluidics techniques allows single cells to be studied in a high-throughput manner in microenvironments closely mimicking biologically -relevant conditions by creating gradients of mechanical forces and different chemical agents 99, 100. These advantages can be categorized into four groups consisting of (i) biophysico-mechanical, (ii) biomaterial composition, (iii) biochemical properties and (iv) fabrication characteristics. We briefly explain these properties and their applications in tissue engineering and stem cell culture. Extensive reviews of microfluidics based stem cell culture and implementation of different stimuli can be found elsewhere 9, 18, 98.
3.1.1 Biophysico-mechanical properties
In biophysico-mechanical properties, microfluidics can allow control over different physical and mechanical factors including spatial confinement, biomolecular tensions, shear stress, substrate rigidity and topography. Each individual stimulus has impacts, and the combination of different factors can be determinative on the phenotypes and fate of the stem cells. Microfluidics can provide a high level of confinement as found in vivo and a large S/V ratio 18, that are not possible using conventional stem cell culture methods. As a result, a controlled 3D spatial environment and appropriate oxygen tension and pH/temperature gradients together with diffusion-based nutrient delivery and gas exchange are possible. For example an investigation into early embryogenesis and differentiation using a microfluidic based temperature gradient has been reported 101. Enhanced spreading and migration of stem cells was reported in response to the combination of properties of the microfluidic surface including roughness and stiffness 102.
On the other hand, control over the fluidic flow that affects dynamic cell culture conditions, availability of shear stress as naturally found in different organs, availability of different medium compositions 18 are other advantages of microfluidics in stem cell research. Microfluidics, based on the application of fluid dynamic methods, can be divided into two main categories; continuous flow microfluidics and droplet-based microfluidics 91, 103. Continuous flow microfluidics follows the principles of continuum mechanics in microenvironments. Droplet-based microfluidics is based on the use of immiscible phases 103. Diverse methods have been applied to make the droplets including flow-focusing, T-junction and electro-wetting, resulting in droplets with nanoliter to picoliter volume range 91, 103. Loading hydrogel structures with biomolecules via droplet-microfluidic methods has been applied in stem cell/cell cultures. For example the feasibility of a co-culture system was investigated via alteration of the flow rate of two cell streams in a hydrogel microbead generating system 104. Furthermore, droplet-based microfluidics has been used to encapsulate stem cells in synthetic hydrogels, providing in vitro 3D cell culture systems with enhanced maintenance of cell viability 103. For example this approach was used to encapsulate human MSCs (hMSCs) in 4-armed polyethylene glycol maleimide hydrogels (PEG-4MAL), and showed improved cytocompatibility without any loss of hMSCs after 7 days continuous culture 105. In addition to continuous microfluidics that has numerous applications in stem cell based neuroregeneration, the potential advantages of droplet based microfluidics could also be a promising strategy for future investigations of neural tissue engineering in a more controlled manner.
3.1.2 Biomaterial properties
The biomaterial properties of microfluidics-based tissue engineering devices consist of different factors; various biomaterial components related to ECM and biological materials have been used for microfabrication. ECM of cells and stem cells is a composite of different kinds of biological molecules such as proteins, proteoglycans and soluble factors (Tables 1 and 2) 98. The interaction of cells with their microenvironment and substrate can produce a cellular mechanical force which can be sensed by myosin motors and integrin molecules, and affects cellular behavior via focal adhesion complexes and the actin cytoskeleton 98. Different natural or synthetic materials have been used for fabrication of these microdevices, and they can be subsequently modified to mimic the ECM. Using microfluidics, well-defined geometric patterning of different molecules and combinations 13 can be achieved, something that is not possible using individual culture dishes.
Polydimethylsiloxane (PDMS) is the most widely investigated material for fabrication of microfluidic devices for tissue regeneration including the neural tissue engineering field. PDMS is a silicon elastomer material considered to possess high gas permeability, low cost, simple fabrication, and the possibility of mass production 73. Furthermore, PDMS can readily undergo molding, has low levels of auto-fluorescence and good transparency which facilitates cell imaging; these properties make it the first choice of bioengineers 106, 107. In regard to neuroregeneration, numerous studies have taken advantage of these benefits to investigate the use of PDMS-based microdevices in stem cell biology, and manipulation of their fate 108–110. However, despite these advantages, there is some difficulty in the handling and fabrication of high aspect-ratio channels, the material has low stiffness, a hydrophobic nature, and can undergo unexpected evaporation due to its porous structure, as well as possessing biocompatibility and sterilization issues, that collectively comprise its challenges 9, 92, 107. In recent years, new alternative materials have been introduced such as different polymers and some combined strategies. For example in the neural tissue-engineering field, hybrid PDMS-glass platforms have been used for transdifferentiation of human adipose tissue-derived stem cells (hATSCs) 111. Microfluidic devices based on naturally-occurring polymers have been fabricated by using fibrin, agarose and collagen to mimic the in vivo condition 73. For example the application of a gelatin methacrylate (GelMA) polymer was reported as a photo-crosslinkable physical barrier in a PDMS based microdevice for NSC culture 112. Another study used a gelatin-based hydrogel microdevice for ESC culture and neural differentiation 113. The hydrophobic nature of PDMS and its porous structure may be problematic for tissue engineering and biological studies, since hydrophobic molecules are absorbed well onto the surface of this material 92, 107. This issue can be resolved by surface modification of the PDMS with methods such as chemical vapor deposition (CVD), plasma treatment as well as application of surfactants, polymers and nanomaterials, and PDMS-patterning approaches such as chemical and topographical patterning (Reviewed by ref 114). Furthermore, joining hydrophilic groups to the surface of microfluidic devices seems a promising approach to functionalize them. In some cases, especially with cell culture applications, charged molecules such as poly-D-lysine, or ECM proteins, including laminin, fibronectin and collagen, can be applied and coated onto the surface of PDMS 99. Collagen type 1 115, poly-L-lysine (PLL) 116 and fibronectin 111 coatings have all been used to overcome the shortcomings of PDMS, and other polymeric material-based microdevices. For example Pluronic F-127 treatment was used to prevent absorption of fluorescent molecules into the PDMS matrix, and lessening noise and background during fluorescence imaging 110.
3.1.3 Biochemical properties
Many biochemical molecules have been used in microdevices to more closely imitate the physiological properties of the native tissue of interest. Stem cells are in contact with different soluble signaling cues in their in vivo microenvironment (Fig 1) (Table 2) such as extracellular calcium ions, various growth factors, nutrients and oxygen. Automated culture systems, soluble gradients and temporal exposure regimens 98 are the most investigated conventional and microfluidics strategies to meet this purpose. Microfluidics can be advantageous for controlling the chemical composition of the microenvironment and to produce spatiotemporal control of soluble gradients 98. In this regard, microfluidics uses various mechanical devices such as pneumatic valves, osmotic pumps or diffusion-based gradient generation to study the signaling pathways, growth factor /nutrient gradients, long-term culture, and stem cell differentiation 117–119. Generation of continuous and stable gradients in a precise manner is achievable by application of microfluidic tools and devices 18. In addition, automated temporal control of the delivery of soluble factors can be achieved using microfluidic systems. For example a continuous growth factor gradient with a mixture of PDGF, EGF and FGF2 was created using a gradient-generating microfluidic device produced from PDMS by rapid prototyping and soft lithography 117. Controlled autocrine and paracrine signaling and growth factor-dependent NSC differentiation into astrocytes was reported using this device. Another study used a microdevice in which a gradient was generated by two laminar flow streams and an osmotic pump to recapitulate the biological effects of different biomolecules (including cytokine gradients (FGF8, agonist Shh and antagonist (BMP4) 120. Long-term culture and Shh concentration-dependent generation of a complex neural network was achieved using human ESC-derived neurons guided to differentiate and proliferate. Stem cells behaviours in native tissue are in contact with multi stimuli with different origin. Generally appropriate migration or differentiation isn’t a single factor dependent mechanism. Study the effect of multi biochemical factors or in combination with effects of some other cues such as shear stress can represent new insights about their effects on stem cells proliferation and differentiation.
3.1.4 Fabrication characteristics
Soft lithography is the most often exploited method for fabrication of microfluidic devices in the neural tissue-engineering and biological research. This method involves replica molding, embossing, and printing 91, 121. The process uses a master (mold) with a topographic pattern, and then the liquid polymer is poured over the master. In this way, the pattern on the master is replicated 122. Soft lithography-based fabrication methods have been used for production of different simple and complex microchips which have been employed in stem cell-mediated neural tissue engineering 57, 108, 115. In this regard two additional topics should be noticed; large scale assays and integration of sensors. Microfluidic large scale integration can be obtained using valves or droplet-based microfluidics for parallelization of the assays, and design of a single platform to accomplish a series of successive steps 18. This strategy can be used to study the effect of various biochemical factors on stem cell behavior and fate as well as controlled gene/drug delivery systems or for nanotoxicological assays. Despite the advantages of these high-throughput systems over the conventional costly and time-consuming methods, their integration into traditional set-ups especially concerning read-out equipment needs more effort. Rotating culture systems and stirred culture systems are two methods using fluidic flow to maximize the yields in stem cell research. Rotatory culture systems have low shear stress and a more homogeneous environment along with efficient gas transfer ability, but the aggregate size must be controlled. In stirred systems, the culture environment is controlled efficiently and nutrients and gases can easily transfer through the medium 88.
On the other hand, precise, in situ, real-time and non-invasive monitoring of the cellular microenvironment and cell activity 18, 99 can be achieved using bio-sensors incorporated into the microdevice. Microelectromechanical system technology can be a key part of the design and fabrication of such devices. These approaches can be used in neural tissue engineering applications 18 such as for monitoring oxygen tension, physical parameters such as temperature, cellular stress levels, cell/tissue metabolism parameters, electrochemical and chemical activity, and the differentiation status of stem cells.
4. Stem cells in microfluidic-based neural tissue engineering
Various types and sources of different stem cells have been employed in stem cell-based neural tissue engineering using microfluidic systems. In addition to the afore-mentioned advantages, some other properties are important for application of microfluidics in stem cell based neuroregeneration. ESCs and NSCs have been the two principle categories of stem cells used for this purpose, and different shapes and designs of microdevices have been employed to achieve the best results. In this regard, the geometry of the microfluidic device is very important. Separated and confined structures are the best options for neuroregeneration, especially for neural outgrowth and for the study of the impact of different stimuli 18. Using microtechnology, fabrication of devices with different dimensions and controlled geometry, using the soft lithography technique, and rapid prototyping for various applications can be possible. For example compartmentalized microdevices have been used for guided neuron growth and cell-cell interactions. In addition, co-culture of different neural cells like glial cells, astrocytes or Schwann cells is possible, allowing modeling of diseases such as neurodegeneration (and neuroregeneration) can be achieved using various microdevices. Simple microfluidic chips, compartmentalized microdevices, hydrogel-based microfluidic devices, microfluidic bioreactors, and microfluidic arrays have all been reported in recent years for studying stem cell culture, migration and differentiation.
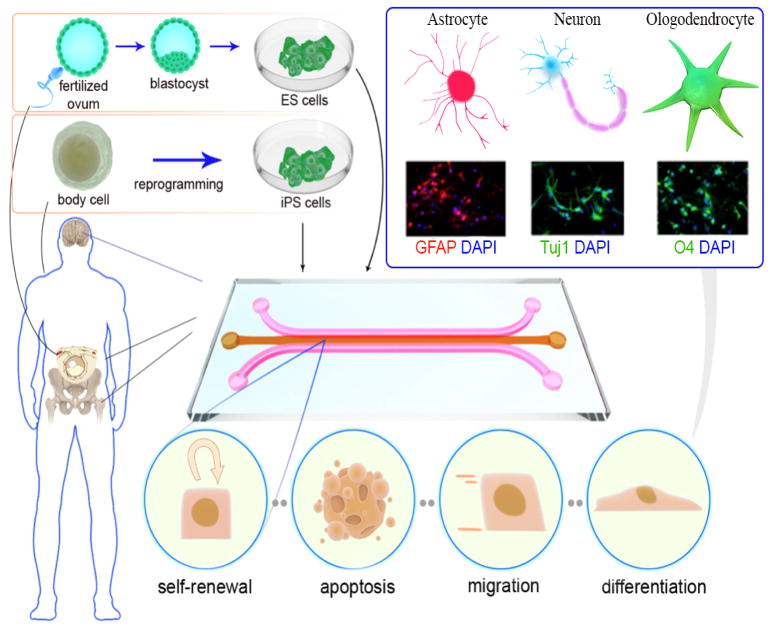
Likewise, the combination of microfluidics with other approaches has been investigated in order to monitor the differentiation of the stem cells, and for understanding their biology. Figure 2 shows the overall process of stem cell derivation and isolation, as well as microfluidic stem cell culture and their possible behavior. Different microfluidic-based neural tissue engineering applications are listed in Table 3. More details are given in the following sections focusing on reports of the use of stem cells for neural regeneration.
Figure 2.
Illustration of the different stem cell sources, microfluidic stem cell culture, and different stem cells behaviors in a microchip, including self-renewal, apoptosis, migration and differentiation. Direct differentiation of stem cells to various neural phenotypes is also shown. (Experimental figures reprinted from Ref. 109. Copyright 2015 with permission from Elsevier).
Table 3.
Different stem cells and application of microdevices for neural tissue engineering.
| Stem cell | Microdevice | Highlights | Ref |
|---|---|---|---|
| ESCs | Compartmental culture system with grooved microchannels |
|
96 |
| Large-scale, concave-microwell culture plate containing arrayed cylindrical well structures | 123 | ||
| Dual-micropillar microfluidic platform |
|
95 | |
| Compartmentalized microfluidic device with open wells |
|
124 | |
| Microfluidic biochip with four separate porous membrane, micro-chambers and biosensors |
|
125 | |
| Gelatin hydrogel microfluidic chip placed at the bottom of a standard multi-well plate |
|
113 | |
| ESC-Ns2 | Triple-chamber compartmentalized micro-grooved microfluidic device with holes in the main channel |
|
116 |
| NSCs | Microfluidic gradient generator using microcapillaries as connectors between chambers |
|
97 |
| 3D ECM containing microfluidic array |
|
108 | |
| Low oxygen tension mimicking microfluidic array |
|
109 | |
| Collagen coated 3D co-culture microdevice |
|
115 | |
| Microfluidic dual-well cell-culture device |
|
126 | |
| 3D microenvironments containing microchannels |
|
8 | |
| On-chip LEPD3 |
|
110 | |
| Micro-engineered gradient generator consisting of mirrored serpentine channels connected by rectangular and triangular gradient chambers |
|
127 | |
| Photo-crosslinkable GelMA hydrogel 3D microfluidic Device |
|
112 | |
| Collagen gel supported microfluidic device |
|
128 | |
| hATSCs | Gel-free 3D microfluidic culture containing microchannel and reservoir layers |
|
111 |
| NEPSCs | Phase-guided 3D microfluidic cell culture bioreactors |
|
58 |
| PDMCs | Microfluidic chip with physico–chemical stimulation |
|
57 |
EBs: Embryoid Bodies;
ESC-Ns: ESC-derived neurons;
LEPD: Localized electroporation device
4.1 Embryonic stem cells
One of the best sources for the isolation of stem cells is the inner cell mass of the blastocyst that contains pluripotent “embryonic SCs” (ESCs) 44. The other major approach that is often discussed, involves the treatment of somatic stem cells under certain specific conditions that allow manipulation of gene expression to produce a pluripotent cell type called “induced PSCs” (iPSCs) 44. The term “pluripotency” refers to the ability of these cells to eventually produce all the different cell types that comprise an entire organism, and of course, neural cells can also be obtained from differentiation of PSCs (ESCs or iPSCs). One of the first lineages that can be obtained from differentiation of PSCs can often be neural cells. In other words, in a simple culture medium ESCs can spontaneously differentiate to form neural cells 129. Several studies have investigated the wide range of application of PSCs for repairing diseases of the nervous system, including genetic disorders, neurodegenerative diseases, and mitigating the symptoms of damage to the brain and spinal cord 29, 129–132. Microfluidics can provide desirable conditions to facilitate culture and neural differentiation pathway of ESCs. Microfluidic-based cell and stem cell culture systems are generally categorized into two classes, gel-supported- and gel-free approaches. Figure 1 illustrates these two main microfluidic-based stem cell culture methods. Each of these microfluidic cell culture systems has specific advantages and disadvantages. The gel-free approaches may be ineffective for long-term cell/stem cell culture because of limited space, evaporation of the medium, effects of shear stress, and limitations in passive and diffusive mass transport 73. The gel-supported approach provides good cell/biomolecule encapsulation properties and more closely resembles the in vivo environment to overcome the limitations of traditional culture techniques. However this method can suffer problems with cell viability due to limitation in nutrient and oxygen transport, or due to lack of sufficient ECM in some circumstances 73. Different gel-free techniques 111 and gel-supported techniques using collagen 112, 115 have been reported for neural differentiation of stem cells.
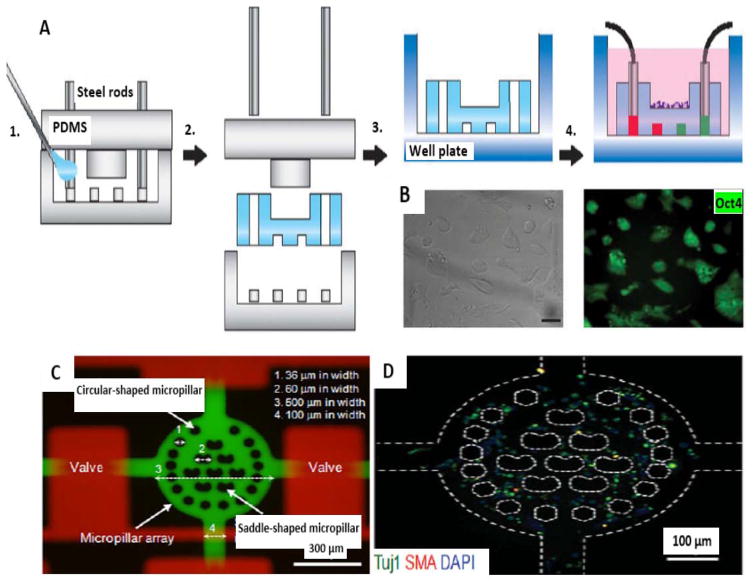
In this regard, to overcome the challenges of gel-free culture and to investigate the effect of biomolecular gradients, a study developed a hydrogel microfluidic device that allowed precise delivery of biomolecules to mouse ESCs that were cultured through a combination of macroscale and microscale approaches 113. The chip consisted of a macroscale culture substrate, containing embedded microchannels for spatiotemporal control of biomolecule delivery (Fig. 3A). Neural differentiation of ESCs was induced by a gradient of the morphogen, retinoic acid. Furthermore, the strongest neuronal differentiation and a greater occurrence of EBs were reported using this gradient. Moreover, ESC pluripotency was confirmed by Oct4 expression (GFP) and efficient colony formation and colony morphology (Fig. 3B). The size limitation of microfluidic platforms (and therefore the low scalability of these systems) may prevent their widespread clinical application. However advanced designs such as previous example would be helpful to eliminate the restriction of microfluidic-based tools for the construction of real-sized grafts and tissues.
Figure 3.
Fabrication of microchips and ESC culture. A) Schematic illustration of fabrication of the hydrogel microfluidic chip. 1. Mold fabrication and injection of hydrogel monomers. 2. Gelation, and mold removal. 3. Transfer to a well plate. 4. Cell seeding and system activation for cell-based assays. B) ESC culture on the surface of the chip (reproduced from ref. 113. Copyright 2015 with permission from nature publishing group). (C) Dual-micropillar-based microfluidic platform containing microvalves. (D) Neural-like cells in microchip with stained Tuj1 (antineuronal classIII, β_tubulin), SMA (anti-α-smooth muscle actin) and DAPI (blue-fluorescent DNA stain) (reproduced from ref. 95. Copyright 2015 with permission from John Wiley & Sons.).
A popular method in PSC biology is called direct differentiation, in which differentiation to specific cell type is obtained by application of a tailored array of external signals in a controlled environment 29. In this regard, a study developed a dual-micropillar-based microfluidic device in order to direct differentiation of single ESCs to a neural phenotype 95. In this study, eight inner saddle-shaped micropillars and 16 circular-shaped outer pillars were used to allow reduction of cell docking (via a higher hydrodynamic resistance) and control the shear stress, respectively. Furthermore, after 6 days of culture the ESCs showed 72% differentiation into neural-like cells (Fig. 3C and D).
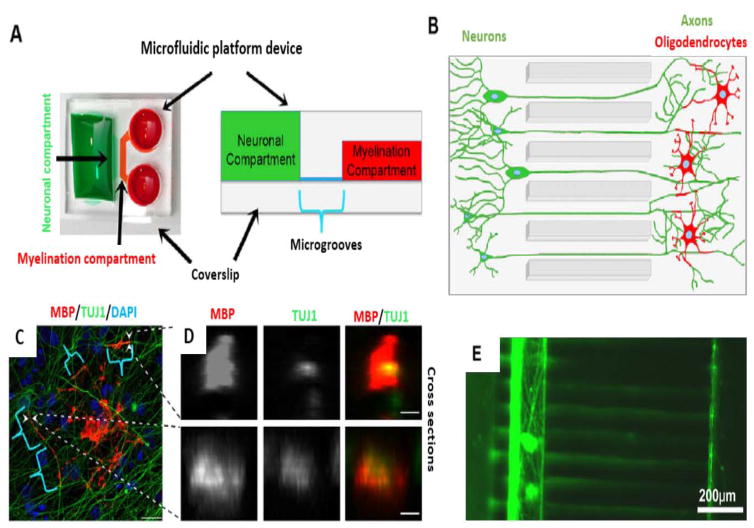
Oligodendrocytes are one of the important cell types in the structure of the brain, that produce myelin which acts as an insulator for axons 29. The actual mechanism of myelination (myelin sheath) formation around the axons by oligodendrocytes is a less investigated field. An investigation developed a combination of microfluidic technology with stem cell biology to cause differentiation of ESCs into myelinating oligodendrocytes, and to allow assessment of myelin formation 124 (Fig 4. A–D). Quantification of myelin formation was obtained through an automated method. Results of this study are relevant, not only for long-term live imaging, but also for the search for new treatments for demyelinating diseases like multiple sclerosis (MS).
Figure 4.
Microfluidic based myelin formation and axonal isolation. A, B) Schematic of microfluidic devices for myelination. C) Microfluidic myelinating oligodendrocytes (MBP), neurites (TUJ1) and cell nuclei (DAPI). Brackets mark indicates myelin, tubes of oligodendrocyte extensions wrapping neuritis. D) Arrowheads marked tubes showed by optical transverse sections (Scale bars: 20 μm in C and 3 μm in D) (Reproduced from Ref. 124. Copyright 2015 with permission from The Company of Biologists Ltd). E) Three-chamber microfluidic devices for Map5 stained axonal isolation of ESC_Ns (Reproduced from Ref. 116. Copyright 2015 with permission from Springer).
In recent years, numerous studies have reported the application of compartmentalized microfluidic platforms for neurobiological research 133–135. The ability to observe the different parts of neurons (axons and cell bodies) in compartmentalized microfluidic devices can be useful for studying neurodegenerative diseases. For example Hwa Sung Shin et al., used embryonic stem cell (ESC)-derived neurons (ESC_Ns) (derived from mouse ESCs) and showed that the axons traversed the microchannels, and were finally isolated from the somatic cell bodies to allow the study of axonal biology 116 (Fig. 4E).
4.2 Neural stem cells
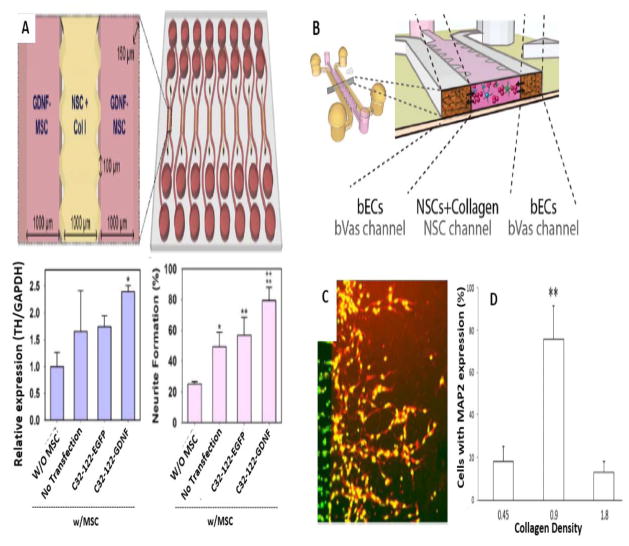
Adult neural progenitor cells (NPCs), in the presence of particular factors such as EGF and FGF can differentiate into different types of neuronal cells 49. Although this type of “neural-engineered adult stem cells” (NESCs) cells can indeed be generated, owing to restricted resources, and a narrow range of cells they can differentiate into, they are not considered to be good candidates for human clinical trials 136. The ability of these differentiation pathways to be guided by the use of appropriate factors, illustrates the benefit of the employment of microfluidic approaches. The ability to mimic the stem cell niche (which plays an important role in NSCs self-renewal and differentiation) can be a useful for NSC culture. A study reported a PDMS-based microfluidic device that could reconstitute the NSC-vascular niche, containing a 3D brain vasculature (bVas) and an ECM microenvironment that allowed NSCs to adopt physiologically relevant phenotypes. Through the combined effects of ECM components, chemical gradients, and signaling molecules, neuronal differentiation was suppressed, but NSC differentiation into astrocytes and oligodendrocytes was promoted 115. In addition, monitoring of the self-renewal and differentiation of NSCs was assessed using quantitative reverse-transcription polymerase chain reaction (qRT-PCR). A soft lithography process was used to fabricate the microdevice with a central channel for collagen type I hydrogel that was seeded with NSCs, and two adjacent surrounding channels contained the bVas (Fig. 5B).
Figure 5.
Investigation of NSCs biology via microfluidics. A) Microfluidic platform for in vivo paracrine signaling mimicking for enhanced NSC differentiation. Top: Schematic illustration of Microfluidic array for NSCs and GDNF-MSCs coculturing. Bottom (left): qRTPCR experiments with upregulated expression of the TH marker in hNSCs. Bottom (right): Neurite formation level of GDNF-hMSCs in comparison with control groups (Reproduced from Ref. 108. Copyright 2015 with permission from Elsevier). B) Schematic illustration of microdevice for interaction of cultured NSCs with bVas (Reprinted from Ref. 115. Copyright 2015 with permission from John Wiley & Sons). C and D) Neural stem/progenitor cells migration and differentiation in the collagen matrix. C) Fluorescent labeling (red) of MAP2 protein expression in appropriate collagen density (0.9 mg/ml). D) Neural stem/progenitor cells percentage with MAP2 expression within different collagen matrices (Reproduced from Ref. 128. Copyright 2015 with permission from the Royal Society of Chemistry).
Biological paracrine signals produced by the surrounding cells may affect the differentiation of stem cells. To enhance the differentiation of human NSCs (hNSCs) to functional neuronal cells, a study investigated the effect of signaling on the hNSC differentiation in a 3D microfluidic array 108. ECM hydrogel was used for hNSC culture, and genetic engineering of human mesenchymal stem cells (hMSCs) was used to increase expression of glial cell-derived neurotrophic factor (GDNF) (performed by anionic polymeric nanoparticles). Paracrine signaling took place between hMSCs and hNSCs using the microfluidic design with a soft lithography-based central channel for hNSC culture, and lateral channels for GDNF-expressing hMSCs. Enhanced differentiation of hNSCs to neuronal cells was achieved with functional electrophysiological features and glial differentiation was suppressed (Fig 5. A). The in vivo effectiveness of this technique was confirmed by sterotactic transplantation into the brain of mice with neonatal hypoxic-ischemic brain injury. A hyaluronic acid (HA) hydrogel was used to transplant GDNF-hMSC cells (and control hMSC cells) into the striatum. The mice that were transplanted with GDNF-hMSCs showed better neurological performance on the Rotarod, grip strength, and passive avoidance tasks.
The generation of a biomolecular gradient in a controlled manner can be an effective method in neural regeneration. Conventional pieces of apparatus for studying chemotaxis in vitro cell culture, suffer from limitations such as uncontrolled concentration gradients, and limitations on direction and speed of cell migration. A study investigated a microfluidic-based gradient of CXCL12 (aka stromal cell-derived factor 1 α) for investigation of the effect of BDNF pretreatment on NSC chemotaxis 97. Enhancement of NSC chemotaxis with better directionality of cell migration was obtained; however this chemotaxis required CXCR4-CXCL12 receptor-activation.
A recent study reported culture of encapsulated NPCs in a collagen microfluidic device 128. Improved NPC migration and differentiation was shown at an optimal nerve growth factor concentration. Synchronized and organized migration of cells, and better neuronal differentiation with good cell-cell connections was obtained in the appropriate conditions (Fig 5. C,D).
4.3 Other types of stem cells
Differentiation of other types of stem cells with the goal of neural regeneration and neural tissue engineering has also been investigated. HSCs derived from bone marrow (that can generate all types of different blood cells) 137, and different types of MSCs found in other tissues such as skeletal, bone marrow are some examples of this subject. Adipose derived SCs (ADSCs) are multipotent MSCs that exist in subcutaneous fat deposits and are easily isolated from material obtained using lipoplasty (fat removal) 55.
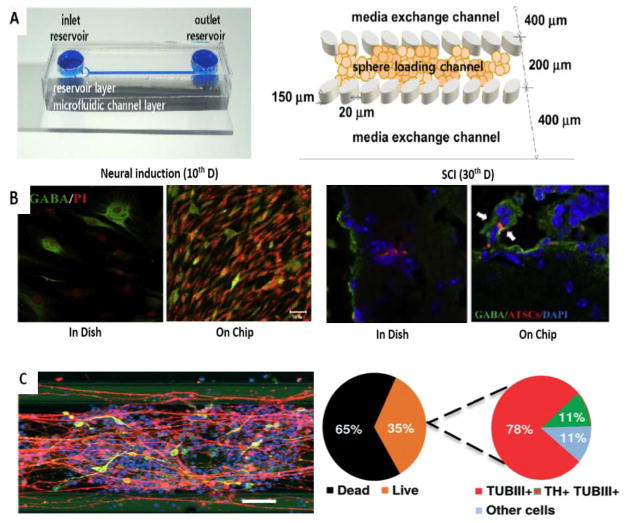
Similar to other stem cell types, employment of specific stimuli including different intracellular or extracellular factors that affect growth and differentiation delivered in microfluidic-based devices are the most investigated in neurogenesis. For example Jeein Choi et al., reported self-renewal and trans-differentiation of multipotent hADSCs to neurons which was induced by activating the Wnt5A/β-catenin signaling pathway in a static cell culture using a 3D microfluidic device 111. A gel-free microfluidic cell culture system with low oxygen partial pressure was developed that caused increased hADSCs growth (in comparison with 2D culture) and a significant increase in neuronal-like cell structures. HIF1α (Hypoxia-inducible factor 1-alpha) expression was induced by low oxygen levels, which caused increased expression of Wnt5A/β-catenin and Oct4. HIF1a binding to regulatory sites of Oct4 and β-catenin genes induced hADSCs neural differentiation (particularly into motor neurons useful for repairing of SCI) and gamma-aminobutyric acid (GABA) secreting neurons (Fig 6. A–B).
Figure 6.
Application of microfluidics for other types of stem cells. A–B) Neuronal differentiation in a gel-free microfluidic chip. A) Microchip with two inlets (exchanging medium and loading neurospheres) and one outlet (left) and schematic concept of immobilization of the neurospheres in the sphere-loading channel with elliptical pillars (right). C) Efficient neurogenesis of hATSC neurospheres into GABA secreting neurons (60% of DAPI) in comparison to dish culture (30% of DAPI) (left). High rate of neurogenesis to give motor neurons (NF160+) in chip culture of hATSC neurospheres compared to dish culture in the lesions of mouse injured spinal cord tissue (10–15% transdifferentiation of engrafted cells in comparison with less than 5%, respectively) (right) (Reproduced from Ref. 111. Copyright 2015 with permission from Elsevier). C) Immunostaining of differentiated neurons with nuclei, TUBβIII and TH stains in a microfluidic bioreactor for differentiation of human NEPSCs into neurons (left). Scale bar: 100μm. Live–dead cell correlation and efficiency of differentiation (right). (Reproduced from Ref. 58. Copyright 2015 with permission from the Royal Society of Chemistry).
One of the most investigated examples of neurodegenerative diseases is Parkinson’s disease, that affects about 2% percent of humans after 65 years old 138. Progressive loss of dopaminergic neurons (DNs) is the hallmark of Parkinson's disease. In this regard a study investigated the application of 3D microfluidic cell culture technology for differentiation of NEPSCs derived from human iPSC to DNs (Fig 6. D,E) 58. After 30 days DNs were produced that were electrophysiologically active, and exhibited long neurites typical of mature neurons forming an interconnected network in this biocompatible phase-guided microfluidic cell culture bioreactor. These innovative studies were proposed to be useful for Parkinson's disease investigation and drug discovery.
PDMCs represent a new source of stem cells that are relatively simple to obtain, and have good ability to differentiate into neurons. A recent study reported a microfluidic chip that used physical methods to modulate the differentiation PDMCs 57. Enhanced neuronal cell differentiation was achieved with physical stimulation using shear stress (produced by varying the injection flow rate) in comparison to chemical stimulation with 1-methyl-3-isobutylxanthine (IBMX).
Creation of even more sophisticated parts of the neural system has been investigated using micro/nanofabrication techniques and microfluidic culture of neural cells with goal of creating a “brain-on-a-chip”. For example the creation of a blood-brain barrier (BBB) using endothelial and astrocyte co-culture and silicon nitride membrane has been reported 139. A two-layer membrane-based microdevice working through biochemical and biomechanical stimulation 140, and an integrated device consisting of four patterned PDMS sub-layers, electrode layers, and the sandwiched polycarbonate membrane 141 have also been reported in recent years. Another study used plastic, disposable and optically clear synthetic materials to model the microvasculature of the BBB (SyM-BBB microdevice) and create a functional BBB model 142. Similarly, creation of neural circuit models using different microdevices has been investigated. For example creation of a lower motor neuron–neuromuscular junction circuit using microgrooves 143, a 3D neural circuit using a microdevice consisting of ECM components and micropillar arrays 144, and acompartmentalized microsystem 145 have all been reported. In regard to application of stem cells and microfabrication devices for this purpose, one example reported a microfabricated compartmentalized chamber with proteins micropatterned on the surfaces for investigating the effects of fibroblast growth factor receptor (FGFR) signaling on guidance and outgrowth of ESCs-derived axons, and creation of functional neural circuits 146.
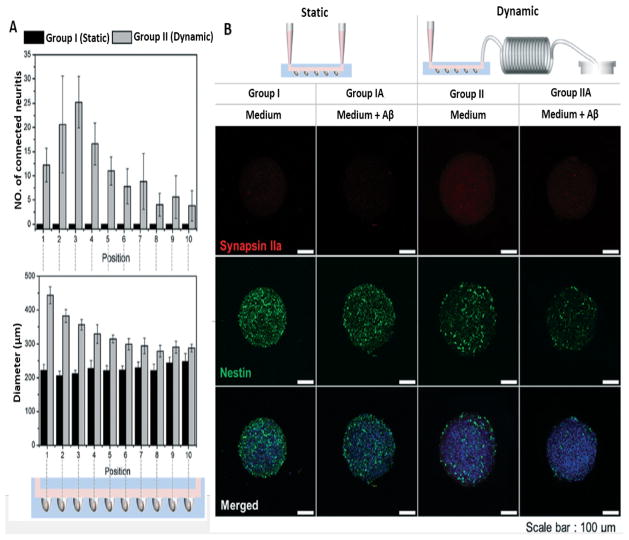
As one of the next steps toward a brain-on-a-chip, Park et al. combined an osmotic micropump-based system with concave microwell arrays, and modulated the constant flow of fluid to evaluate the effect of 0.15 μL min 1 flow (that mimics the in vivo situation) on 3D neurospheroids 147. Neural progenitor/stem cells were cultured for 10 days to form neurospheroids allocated into different dynamic and static groups (neurospheroids cultured with and without flow, respectively). They followed up the neurospheroid size and the formation of neural networks in dynamic and static flow conditions, generally observing a larger neurospheroid size in the dynamic group. They reported greater neurite extension in the dynamic group compared to the static group resulting in formation of a more robust neural network (Figure 7. A). Quantitative analysis and optical imaging confirmed this result. Furthermore, neural progenitor/stem cell marker staining showed enhanced differentiation into neurons. In this model, they exposed the neurospheroids to amyloid-β treatment and compared their viability with those that were not treated with amyloid-β. Amyloid-β staining with thioflavin showed a greater amount of amyloid-β and a lower percentage of viable cells in the dynamic group (Figure 7. B).
Figure 7.
Neural network formation and neurotoxic effects of amyloid-β. A) Average number of neurites extending (upper graph) and average size of neurospheroids (lower graph) in different microdevice sections of two main group. B) Destruction of neural networks shown by immunofluorescence images staining synapsin IIa (synaptic marker) and nestin (neural progenitor/stem cell marker) in neurospheroids in different groups (Scale bar:100 μm). (Reproduced from Ref. 147. Copyright 2015 with permission from the Royal Society of Chemistry).
5. Conclusions and future perspective
The complex structure and intricate function of the nervous system, together with the intrinsic limitation on its ability to regenerate, cause considerable difficulties for the field of neural tissue engineering. The ability to culture and differentiate stem cells in a controlled in vitro environment also presents some problems, considering the vast range of stimuli that can critically determine the fate of the stem cells. Providing an appropriate and adequate range of different stimuli, including specific ECM proteins, adequate shear stress, sufficient substrate stiffness and recoil force, appropriate oxygen tension, as well as desirable nano-features and micro-topography can all be important for the close mimicking of the in vivo microenvironment or stem cell niche.
Microfluidic technology has the ability to overcome the limitations of traditional 2D and 3D cell and stem cell culture techniques, producing improved results that more closely resemble those naturally obtained in vivo. Microfluidic devices have some excellent advantages that are important for stem cell-based tissue engineering, especially in the area of neural regeneration. Good control over the spatiotemporal factors that determine the extracellular microenvironment of stem cells, particularly their interactions with other cells and with the ECM, precise control over the biochemical gradients, and providing the most appropriate physical microenvironment are the most important properties of the microdevices that are used to replicate the stem cell niche. Various microfluidic systems have been designed and fabricated for the purpose of neural tissue engineering. Enhanced neural migration and differentiation, a better ability to monitor these processes, as well as understanding of stem cells and their behavior in different microenvironments have been obtained through application of microfluidic based devices to stem cell culture and neural tissue engineering.
Due to the complexity of stem cell biology, deeper investigations into the biological and physicochemical cues exerted by the native microenvironment that affect stem cell biology are required, and microfluidic devices can play a role with their ability to replicate the natural microenvironment. For example improved compartmentalized microfluidic devices can allow examination of different parts of neural cells, and co-culture of several different cell types.
Some potential applications of microfluidics in neural tissue engineering are the utilization of natural or synthetic materials as a biocompatible scaffold to guide neural system development. Microfluidics can be a promising approach for screening of these different biomaterials. The potential advantages of microfluidics are its use of minimum quantities of material, testing combinations of different biomaterials, and investigating their biological interactions and comparative efficacy. On the other hand, long-term cultures and assays are a challenge in microfluidics as well in neural tissue engineering. Precise quantification of measurements in most biological investigations including tissue engineering methods need both long-term and high through-put assays. Application of microfluidics especially via high through-put devices and parallelization of multiple assays in neural tissue engineering can be advantageous for long-term stem cell culture, characterizing their proliferative and differentiation capacity and modeling of neural diseases (for example Alzheimer’s, malignancies and abnormalities of the BBB) and for drug/biomaterial screening. These studies can also be useful for investigation of the impacts of drugs, nanoparticles, growth factors and cytokines on differentiation and proliferation of stem cells. In these regard, nanotechnology approaches are promising candidate for biology, medicine, tissue engineering, as well as for neuroscience and neural tissue engineering 5, 148–150. Various nanostructures have been reported with potential applications in these fields 150–153. Employment of nanoparticles in tissue regeneration and neural tissue engineering can be used as a potential research field, for example stem cell tracking by nanoparticles in a microdevice or studying their toxicity on neural cells.
Future studies should also consider some less-investigated subjects in microfluidic based neural tissue engineering. For example there has been much attention on the application of MSCs in neural tissue engineering because the ethical issues involving ESCs and cells derived from ESCs such as NSCs. Hence more microfluidics-based neural tissue engineering studies using ADSCs, placenta and bone narrow derived stem cells would be a potential future application. Also there has been an extensive discussion on the impact of inflammation on tissue regeneration, and whether it is overall considered desirable or undesirable. One interesting study could be the investigation of the impacts of immune cells and inflammatory cells on the neural regeneration process using stem cells in a microdevice. On the other hand, utilization of computational simulations for better imitation of the native microenvironment, and improved control of microdevices is another potential application. For example simulation of native shear stress of the neural niche and its ECM components, and integration of these for tissue engineering approaches would be useful to demonstrate the synergistic combination of microfluidics, tissue engineering and in silico studies.
Through the multidisciplinary overlap of biology and engineering combined with emerging new trends such as microfluidics, stem cells and nanotechnology, the fabrication of an artificial human organ (even going as far as a brain) is beginning to be considered possible; this has been imagined as a “brain-on-a-chip” 147 . Organs on a chip will provide much better mimicking of real human physiology, and will be useful for tissue engineering, disease modeling and drug screening; however much more well-designed studies are still required in these fields.
Supplementary Material
Acknowledgments
Michael R Hamblin was supported by US NIH grant number R01AI050875
Contributor Information
Mahdi Karimi, Email: m_karimy2006@yahoo.com.
Sajad Bahrami, Email: sajadbahrami2021@yahoo.com.
Hamed Mirshekari, Email: hamedmirshekari83@gmail.com.
Seyed Masoud Moosavi Basri, Email: moosavi@europe.com.
Amirala Bakhshian Nik, Email: amir.bakhshian@gmail.com.
Amir R. Aref, Email: Amir_aref@hms.harvard.edu.
Mohsen Akbari, Email: makbari@uvic.ca.
Michael R. Hamblin, Email: hamblin@helix.mgh.harvard.edu.
References
- 1.Saracino GA, Cigognini D, Silva D, Caprini A, Gelain F. Chemical Society Reviews. 2013;42:225–262. doi: 10.1039/c2cs35065c. [DOI] [PubMed] [Google Scholar]
- 2.Mack GS. Nature biotechnology. 2011;29:95–97. doi: 10.1038/nbt0211-95. [DOI] [PubMed] [Google Scholar]
- 3.Place ES, Evans ND, Stevens MM. Nature materials. 2009;8:457–470. doi: 10.1038/nmat2441. [DOI] [PubMed] [Google Scholar]
- 4.Zlokovic BV. Nature medicine. 2010;16:1370–1371. doi: 10.1038/nm1210-1370. [DOI] [PubMed] [Google Scholar]
- 5.Dvir T, Timko BP, Kohane DS, Langer R. Nature nanotechnology. 2011;6:13–22. doi: 10.1038/nnano.2010.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Béduer A, Vieu C, Arnauduc F, Sol J-C, Loubinoux I, Vaysse L. Biomaterials. 2012;33:504–514. doi: 10.1016/j.biomaterials.2011.09.073. [DOI] [PubMed] [Google Scholar]
- 7.Li Y-C, Tsai L-K, Wang J-H, Young T-H. Biomaterials. 2014;35:1192–1204. doi: 10.1016/j.biomaterials.2013.10.066. [DOI] [PubMed] [Google Scholar]
- 8.Han S, Yang K, Shin Y, Lee JS, Kamm RD, Chung S, Cho S-W. Lab on a Chip. 2012;12:2305–2308. doi: 10.1039/c2lc21285d. [DOI] [PubMed] [Google Scholar]
- 9.Young EW, Beebe DJ. Chemical Society Reviews. 2010;39:1048–1036. doi: 10.1039/b909900j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamayol A, Najafabadi AH, Aliakbarian B, Arab-Tehrany E, Akbari M, Annabi N, Juncker D, Khademhosseini A. Advanced healthcare materials. 2015;4:2146–2153. doi: 10.1002/adhm.201500492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akbari M, Tamayol A, Annabi N, Juncker D, Khademhosseini A. 2014 [Google Scholar]
- 12.Akbari M, Tamayol A, Laforte V, Annabi N, Najafabadi AH, Khademhosseini A, Juncker D. Advanced functional materials. 2014;24:4060–4067. doi: 10.1002/adfm.201303655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sackmann EK, Fulton AL, Beebe DJ. Nature. 2014;507:181–189. doi: 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- 14.Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Analytical chemistry. 2009;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- 15.Weibel DB, DiLuzio WR, Whitesides GM. Nature Reviews Microbiology. 2007;5:209–218. doi: 10.1038/nrmicro1616. [DOI] [PubMed] [Google Scholar]
- 16.Shin Y, Han S, Jeon JS, Yamamoto K, Zervantonakis IK, Sudo R, Kamm RD, Chung S. Nature protocols. 2012;7:1247–1259. doi: 10.1038/nprot.2012.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghorbanian S, Qasaimeh MA, Akbari M, Tamayol A, Juncker D. Biomedical microdevices. 2014;16:387–395. doi: 10.1007/s10544-014-9842-8. [DOI] [PubMed] [Google Scholar]
- 18.Harink B, Le Gac S, Truckenmüller R, van Blitterswijk C, Habibovic P. Lab on a chip. 2013;13:3512–3528. doi: 10.1039/c3lc50293g. [DOI] [PubMed] [Google Scholar]
- 19.Park JW, Vahidi B, Taylor AM, Rhee SW, Jeon NL. Nature protocols. 2006;1:2128–2136. doi: 10.1038/nprot.2006.316. [DOI] [PubMed] [Google Scholar]
- 20.Park JW, Kim HJ, Kang MW, Jeon NL. Lab on a Chip. 2013;13:5.521–09. doi: 10.1039/c2lc41081h. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y, Williams JC, Johnson SM. Lab on a Chip. 2012;12:2103–2117. doi: 10.1039/c2lc21142d. [DOI] [PubMed] [Google Scholar]
- 22.Pamies D, Hartung T, Hogberg HT. Exp Biol Med (Maywood) 2014;239:1096–1107. doi: 10.1177/1535370214537738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J, Lee BK, Jeong GS, Hyun JK, Lee CJ, Lee S-H. Lab on a Chip. 2015;15:141–150. doi: 10.1039/c4lc00962b. [DOI] [PubMed] [Google Scholar]
- 24.Tang Y, Bernaudin M. Methods Mol Biol. 2007;399:153–166. doi: 10.1007/978-1-59745-504-6_11. [DOI] [PubMed] [Google Scholar]
- 25.Wheeler BC. Conf Proc IEEE Eng Med Biol Soc. 2008;2008:1604–1606. doi: 10.1109/IEMBS.2008.4649479. [DOI] [PubMed] [Google Scholar]
- 26.Montgomery A, Wong A, Gabers N, Willerth SM. Biomaterials Science. 2015;3:401–413. doi: 10.1039/c4bm00299g. [DOI] [PubMed] [Google Scholar]
- 27.Bianco P, Robey PG. Nature. 2001;414:118–121. doi: 10.1038/35102181. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz SD, Hubschman J-P, Heilwell G, Franco-Cardenas V, Pan CK, Ostrick RM, Mickunas E, Gay R, Klimanskaya I, Lanza R. The Lancet. 2012;379:713–7.20. doi: 10.1016/S0140-6736(12)60028-2. [DOI] [PubMed] [Google Scholar]
- 29.Tabar V, Studer L. Nature Reviews Genetics. 2014;15:82–92. doi: 10.1038/nrg3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinbeck JA, Choi SJ, Mrejeru A, Ganat Y, Deisseroth K, Sulzer D, Mosharov EV, Studer L. Nature biotechnology. 2015 doi: 10.1038/nbt.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park KI, Teng YD, Snyder EY. Nature biotechnology. 2002;20:1111–1117. doi: 10.1038/nbt751. [DOI] [PubMed] [Google Scholar]
- 32.Robinson M, Yau S-y, Sun L, Gabers N, Bibault E, Christie BR, Willerth SM. 2015 doi: 10.4137/BMI.S20064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prabhakaran MP, Venugopal JR, Ramakrishna S. Biomaterials. 2009;30:4996–5003. doi: 10.1016/j.biomaterials.2009.05.057. [DOI] [PubMed] [Google Scholar]
- 34.Georgiou M, Golding JP, Loughlin AJ, Kingham PJ, Phillips JB. Biomaterials. 2015;37:242–251. doi: 10.1016/j.biomaterials.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 35.Watt FM, Huck WT. Nature Reviews Molecular Cell Biology. 2013;14:467–473. doi: 10.1038/nrm3620. [DOI] [PubMed] [Google Scholar]
- 36.Discher DE, Mooney DJ, Zandstra PW. Science. 2009;324:1677–1673. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwai H, Shimada H, Nishimura S, Kobayashi Y, Itakura G, Hori K, Hikishima K, Ebise H, Negishi N, Shibata S. Stem cells translational medicine. 2015:sctm. 2014–0215. doi: 10.5966/sctm.2014-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kefalopoulou Z, Politis M, Piccini P, Mencacci N, Bhatia K, Jahanshahi M, Widner H, Rehncrona S, Brundin P, Björklund A. JAMA neurology. 2014;71:83–87. doi: 10.1001/jamaneurol.2013.4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Politis M, Wu K, Loane C, Quinn NP, Brooks DJ, Oertel WH, Björklund A, Lindvall O, Piccini P. Science translational medicine. 2012;4:128ra141–128ra141. doi: 10.1126/scitranslmed.3003391. [DOI] [PubMed] [Google Scholar]
- 40.Martin GR. Proceedings of the National Academy of Sciences. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomson J. Science. 1998;282:1827–1827. [Google Scholar]
- 42.Kassmer SH, Krause DS. Molecular reproduction and development. 2013;8:690–677. doi: 10.1002/mrd.22168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choumerianou DM, Dimitriou H, Kalmanti M. Tissue Engineering Part B: Reviews. 2008;14:53–60. doi: 10.1089/teb.2007.0216. [DOI] [PubMed] [Google Scholar]
- 44.Robinton DA, Daley GQ. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M. Nature. 2013;502:65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- 46.Oki K, Tatarishvili J, Wood J, Koch P, Wattananit S, Mine Y, Monni E, Tornero D, Ahlenius H, Ladewig J. Stem Cells. 2012;30:1120–1133. doi: 10.1002/stem.1104. [DOI] [PubMed] [Google Scholar]
- 47.Tornero D, Wattananit S, Madsen MG, Koch P, Wood J, Tatarishvili J, Mine Y, Ge R, Monni E, Devaraju K. Brain. 2013:awt278. doi: 10.1093/brain/awt278. [DOI] [PubMed] [Google Scholar]
- 48.Yamashita T, Abe K. Cell transplantation. 2014;23:435–439. doi: 10.3727/096368914X678274. [DOI] [PubMed] [Google Scholar]
- 49.Reynolds BA, Weiss S. science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 50.Stenudd M, Sabelström H, Frisén J. JAMA neurology. 2015;72:235–237. doi: 10.1001/jamaneurol.2014.2927. [DOI] [PubMed] [Google Scholar]
- 51.Göritz C, Dias DO, Tomilin N, Barbacid M, Shupliakov O, Frisén J. Science. 2011;333:238–242. doi: 10.1126/science.1203165. [DOI] [PubMed] [Google Scholar]
- 52.Cave JW, Wang M, Baker H. Frontiers in neuroscience. 2014;8 doi: 10.3389/fnins.2014.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang A, Tang Z, Park I-H, Zhu Y, Patel S, Daley GQ, Li S. Biomaterials. 2011;32:5023–5032. doi: 10.1016/j.biomaterials.2011.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hilfiker A, Kasper C, Hass R, Haverich A. Langenbeck's Archives of Surgery. 2011;396:489–497. doi: 10.1007/s00423-011-0762-2. [DOI] [PubMed] [Google Scholar]
- 55.Higuchi A, Wang C-T, Ling Q-D, Lee HH-c, Kumar SS, Chang Y, Alarfaj AA, Munusamy MA, Hsu S-T, Wu G-J. Scientific reports. 2015;5 doi: 10.1038/srep10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mizuno H. Journal of Nippon Medical School. 2009;76:56–66. doi: 10.1272/jnms.76.56. [DOI] [PubMed] [Google Scholar]
- 57.Cheng Y-C, Tsao C-W, Chiang M-Z, Chung C-A, Chien C-C, Hu W-W, Ruaan R-C, Li C. Microfluidics and Nanofluidics. 2015;18:587–598. [Google Scholar]
- 58.Moreno EL, Hachi S, Hemmer K, Trietsch SJ, Baumuratov AS, Hankemeier T, Vulto P, Schwamborn JC, Fleming RM. Lab on a Chip. 2015;15:2419–2428. doi: 10.1039/c5lc00180c. [DOI] [PubMed] [Google Scholar]
- 59.Reinhardt P, Glatza M, Hemmer K, Tsytsyura Y, Thiel CS, Höing S, Moritz S, Parga JA, Wagner L, Bruder JM. PloS one. 2013;8:e59252. doi: 10.1371/journal.pone.0059252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Azarin SM, Palecek SP. Biochemical engineering journal. 2010;48:378–384. doi: 10.1016/j.bej.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morrison SJ, Scadden DT. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lo EH. Nature medicine. 2008;14:497–500. doi: 10.1038/nm1735. [DOI] [PubMed] [Google Scholar]
- 63.Garcion E, Halilagic A, Faissner A. Development. 2004;131:3423–3432. doi: 10.1242/dev.01202. [DOI] [PubMed] [Google Scholar]
- 64.Aimone JB, Li Y, Lee SW, Clemenson GD, Deng W, Gage FH. Physiological reviews. 2014;94:991–1026. doi: 10.1152/physrev.00004.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moon JJ, Matsumoto M, Patel S, Lee L, Guan JL, Li S. Journal of cellular physiology. 2005;203:166–176. doi: 10.1002/jcp.20220. [DOI] [PubMed] [Google Scholar]
- 66.Park JS, Chu JS, Tsou AD, Diop R, Tang Z, Wang A, Li S. Biomaterials. 2011;32:3921–3930. doi: 10.1016/j.biomaterials.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woodford C, Zandstra PW. Current opinion in biotechnology. 2012;23:810–819. doi: 10.1016/j.copbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akita K, von Holst A, Furukawa Y, Mikami T, Sugahara K, Faissner A. Stem Cells. 2008;26:798–809. doi: 10.1634/stemcells.2007-0448. [DOI] [PubMed] [Google Scholar]
- 69.Ramasamy S, Yu F, Hong Yu Y, Srivats H, Stewart Dawe G, Ahmed S. Stem Cells. 2014;32:1636–1648. doi: 10.1002/stem.1645. [DOI] [PubMed] [Google Scholar]
- 70.Regalado-Santiago C, Juárez-Aguilar E, Olivares-Hernández JD, Tamariz E. Stem Cells International. 2015 doi: 10.1155/2016/1513285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu G, Shen J, Ishii Y, Fukuchi M, Dang T, Zheng Y, Hamashima T, Fujimori T, Tsuda M, Funa K. Neuroscience. 2013;238:195–208. doi: 10.1016/j.neuroscience.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 72.Kong JH, Yang L, Dessaud E, Chuang K, Moore DM, Rohatgi R, Briscoe J, Novitch BG. Developmental cell. 2015;33:373–387. doi: 10.1016/j.devcel.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bond AM, Bhalala OG, Kessler JA. Developmental neurobiology. 2012;72:1068–1084. doi: 10.1002/dneu.22022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bjornsson CS, Apostolopoulou M, Tian Y, Temple S. Developmental cell. 2015;32:435–446. doi: 10.1016/j.devcel.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martínez-Canabal A. International Journal of Neuroscience. 2015;125:9–1. doi: 10.3109/00207454.2014.903947. [DOI] [PubMed] [Google Scholar]
- 76.Pathipati P, Gorba T, Scheepens A, Goffin V, Sun Y, Fraser M. Neuroscience. 2011;190:409–427. doi: 10.1016/j.neuroscience.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 77.D'Adamio L. The American journal of pathology. 2010;177:2163–2165. doi: 10.2353/ajpath.2010.100829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Russo V, Gluckman P, Feldman E, Werther G. Endocrine reviews. 2005;26:916–943. doi: 10.1210/er.2004-0024. [DOI] [PubMed] [Google Scholar]
- 79.Pasquin S, Sharma M, Gauchat J-F. Cytokine & growth factor reviews. 2015;26:507–515. doi: 10.1016/j.cytogfr.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 80.Nicola NA, Babon JJ. Cytokine & growth factor reviews. 2015;26:533–544. doi: 10.1016/j.cytogfr.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Santos E, Hernández RM, Pedraz JL, Orive G. Trends in biotechnology. 2012;30:331–341. doi: 10.1016/j.tibtech.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 82.Fan C-G, Zhang Q-j, Zhou J-r. Stem Cell Reviews and Reports. 2011;7:195–207. doi: 10.1007/s12015-010-9168-8. [DOI] [PubMed] [Google Scholar]
- 83.Chen BY, Wang X, Wang ZY, Wang YZ, Chen LW, Luo ZJ. Journal of neuroscience ,research. 2013;91:30–41. doi: 10.1002/jnr.23138. [DOI] [PubMed] [Google Scholar]
- 84.Watanabe J, Matsumoto M, Kageyama H, Murai N, Sasaki S, Hirako S, Wada N, Arata S, Shioda S. Peptides. 2015;69:40–46. doi: 10.1016/j.peptides.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 85.Mohamet L, Lea ML, Ward CM. PLoS One. 2010;5:e12921. doi: 10.1371/journal.pone.0012921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lei Y, Schaffer DV. Proceedings of the National Academy of Sciences. 2013;110:E5039–E5048. doi: 10.1073/pnas.1309408110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baraniak PR, McDevitt TC. Cell and tissue research. 2012;347:701–711. doi: 10.1007/s00441-011-1215-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Serra M, Brito C, Correia C, Alves PM. Trends in biotechnology. 2012;30:35.359. doi: 10.1016/j.tibtech.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 89.Agarwal P, Zhao S, Bielecki P, Rao W, Choi JK, Zhao Y, Yu J, Zhang W, He X. Lab on a Chip. 2013;13:4525–4533. doi: 10.1039/c3lc50678a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Whitesides GM. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 91.Jiang L, Korivi NS. In: Nanolithography. Feldman M, editor. Woodhead Publishing; 2014. pp. 424–443. [Google Scholar]
- 92.Wang Z, Chen Z, Liu Z, Shi P, Dong K, Ju E, Ren J, Qu X. Biomaterials. 2014;35:9678–9688. doi: 10.1016/j.biomaterials.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 93.Wang B, Jedlicka S, Cheng X. 2014 doi: 10.1371/journal.pone.0109815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.van Duinen V, Trietsch SJ, Joore J, Vulto P, Hankemeier T. Current opinion in biotechnology. 2015;35:118–126. doi: 10.1016/j.copbio.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 95.Lee JM, Kim Je, Borana J, Chung BH, Chung BG. Electrophoresis. 2013;34:1931–1938. doi: 10.1002/elps.201200578. [DOI] [PubMed] [Google Scholar]
- 96.Lee N, Park JW, Kim HJ, Yeon JH, Kwon J, Ko JJ, Oh S-H, Kim HS, Kim A, Han BS. Molecules and cells. 2014;37:497. doi: 10.14348/molcells.2014.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu H, Heilshorn SC. Small. 2013;9:585–595. doi: 10.1002/smll.201202208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gupta K, Kim D-H, Ellison D, Smith C, Kundu A, Tuan J, Suh K-Y, Levchenko A. Lab on a Chip. 2010;10:2019–2031. doi: 10.1039/c004689b. [DOI] [PubMed] [Google Scholar]
- 99.Halldorsson S, Lucumi E, Gómez-Sjöberg R, Fleming RM. Biosensors and Bioelectronics. 2015;63:218–231. doi: 10.1016/j.bios.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 100.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 101.Lucchetta EM, Lee JH, Fu LA, Patel NH, Ismagilov RF. Nature. 2005;434:1134–1138. doi: 10.1038/nature03509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Menon NV, Chuah YJ, Phey S, Zhang Y, Wu Y, Chan V, Kang Y. ACS applied materials & interfaces. 2015;7:17095–17103. doi: 10.1021/acsami.5b03753. [DOI] [PubMed] [Google Scholar]
- 103.Allazetta S, Lutolf MP. Current opinion in biotechnology. 2015;35:86–93. doi: 10.1016/j.copbio.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 104.Tumarkin E, Tzadu L, Csaszar E, Seo M, Zhang H, Lee A, Peerani R, Purpura K, Zandstra PW, Kumacheva E. Integrative Biology. 2011;3:653–662. doi: 10.1039/c1ib00002k. [DOI] [PubMed] [Google Scholar]
- 105.Headen DM, Aubry G, Lu H, García AJ. Advanced Materials. 2014;26:3003–3008. doi: 10.1002/adma.201304880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Iliescu C, Taylor H, Avram M, Miao J, Franssila S. Biomicrofluidics. 2012;6:016505. doi: 10.1063/1.3689939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mehling M, Tay S. Current opinion in Biotechnology. 2014;25:95–102. doi: 10.1016/j.copbio.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 108.Yang K, Park H-J, Han S, Lee J, Ko E, Kim J, Lee JS, Yu JH, Song KY, Cheong E. Biomaterials. 2015;63:177–188. doi: 10.1016/j.biomaterials.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 109.Yang K, Han S, Shin Y, Ko E, Kim J, Park KI, Chung S, Cho S-W. Biomaterials. 2013;34:6607–6614. doi: 10.1016/j.biomaterials.2013.05.067. [DOI] [PubMed] [Google Scholar]
- 110.Kang W, Giraldo-Vela JP, Nathamgari SSP, McGuire T, McNaughton RL, Kessler JA, Espinosa HD. Lab on a Chip. 2014 doi: 10.1039/c4lc00721b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Choi J, Kim S, Jung J, Lim Y, Kang K, Park S, Kang S. Biomaterials. 2011;32:7013–7022. doi: 10.1016/j.biomaterials.2011.05.090. [DOI] [PubMed] [Google Scholar]
- 112.Lee Y, Lee JM, Bae PK, Chung IY, Chung BH, Chung BG. Electrophoresis. 2015;36:994–1001. doi: 10.1002/elps.201400465. [DOI] [PubMed] [Google Scholar]
- 113.Cosson S, Lutolf M. Scientific reports. 2014;4 doi: 10.1038/srep04462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou J, Khodakov DA, Ellis AV, Voelcker NH. Electrophoresis. 2012;33:89–104. doi: 10.1002/elps.201100482. [DOI] [PubMed] [Google Scholar]
- 115.Shin Y, Yang K, Han S, Park HJ, Seok Heo Y, Cho SW, Chung S. Advanced healthcare materials. 2014;3:1457–1464. doi: 10.1002/adhm.201300569. [DOI] [PubMed] [Google Scholar]
- 116.Shin HS, Kim HJ, Min SK, Kim SH, Lee BM, Jeon NL. Biotechnology letters. 2010;32:1063–1070. doi: 10.1007/s10529-010-0280-2. [DOI] [PubMed] [Google Scholar]
- 117.Chung BG, Flanagan LA, Rhee SW, Schwartz PH, Lee AP, Monuki ES, Jeon NL. Lab on a Chip. 2005;5:401–406. doi: 10.1039/b417651k. [DOI] [PubMed] [Google Scholar]
- 118.Melin J, Quake SR. Annu Rev Biophys Biomol Struct. 2007;36:213–231. doi: 10.1146/annurev.biophys.36.040306.132646. [DOI] [PubMed] [Google Scholar]
- 119.Abhyankar VV, Lokuta MA, Huttenlocher A, Beebe DJ. Lab on a Chip. 2006;6:389–393. doi: 10.1039/b514133h. [DOI] [PubMed] [Google Scholar]
- 120.Park JY, Kim SK, Woo DH, Lee EJ, Kim JH, Lee SH. Stem cells. 2009;27:2646–2654. doi: 10.1002/stem.202. [DOI] [PubMed] [Google Scholar]
- 121.Anderson JR, Chiu DT, Wu H, Schueller O, Whitesides GM. Electrophoresis. 2000;21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 122.McDonald JC, Whitesides GM. Accounts of chemical research. 2002;35:491–499. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- 123.Jeong GS, Song JH, Kang AR, Jun Y, Kim JH, Chang JY, Lee SH. Advanced healthcare materials. 2013;2:119–125. doi: 10.1002/adhm.201200070. [DOI] [PubMed] [Google Scholar]
- 124.Kerman BE, Kim HJ, Padmanabhan K, Mei A, Georges S, Joens MS, Fitzpatrick JA, Jappelli R, Chandross KJ, August P. Development. 2015 doi: 10.1242/dev.116517. dev. 116517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sandoz A, Charvet I, Stoppini L. Procedia Engineering. 2013;59:46–50. [Google Scholar]
- 126.Lin C-H, Hsiao Y-H, Chang H-C, Yeh C-F, He C-K, Salm EM, Chen C, Chiu M, Hsu C-H. Lab on a Chip. 2015 doi: 10.1039/c5lc00541h. [DOI] [PubMed] [Google Scholar]
- 127.Keenan T, Grinager J, Procak A, Svendsen C. Integrative Biology. 2012;4:1522–1531. doi: 10.1039/c2ib20074k. [DOI] [PubMed] [Google Scholar]
- 128.Shamloo A, Heibatollahi M, Mofrad MR. Integrative Biology. 2015;7:335–344. doi: 10.1039/c4ib00144c. [DOI] [PubMed] [Google Scholar]
- 129.Eiraku M, Sasai Y. Nature protocols. 2012;7:69–79. doi: 10.1038/nprot.2011.429. [DOI] [PubMed] [Google Scholar]
- 130.Fox IJ, Daley GQ, Goldman SA, Huard J, Kamp TJ, Trucco M. Science. 2014;345:1247391. doi: 10.1126/science.1247391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Goldman SA, Nedergaard M, Windrem MS. Science. 2012;338:491–495. doi: 10.1126/science.1218071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shi P, Kirwan Y, Livesey FJ. Nature protocols. 2012;7:1836–1846. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- 133.Taylor A, Jeon NL. Critical Reviews™ in Biomedical Engineering. 2011;39 doi: 10.1615/critrevbiomedeng.v39.i3.20. [DOI] [PubMed] [Google Scholar]
- 134.Park J, Koito H, Li J, Han A. Biomedical microdevices. 2009;11:1145–1153. doi: 10.1007/s10544-009-9331-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yamada A, Vignes M, Bureau C, Mamane A, Venzac B, Descroix S, Viovy J-L, Villard C, Peyrin J-M, Malaquin L. Lab on a Chip. 2016 doi: 10.1039/c6lc00414h. [DOI] [PubMed] [Google Scholar]
- 136.Shim JW, Park CH, Bae YC, Bae JY, Chung S, Chang MY, Koh HC, Lee HS, Hwang SJ, Lee KH. Stem Cells. 2007;25:1252–1262. doi: 10.1634/stemcells.2006-0274. [DOI] [PubMed] [Google Scholar]
- 137.Omatsu Y, Sugiyama T, Kohara H, Kondoh G, Fujii N, Kohno K, Nagasawa T. Immunity. 2010;33:387–399. doi: 10.1016/j.immuni.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 138.Delcroix GJ-R, Schiller PC, Benoit J-P, Montero-Menei CN. Biomaterials. 2010;31:2120–2105. doi: 10.1016/j.biomaterials.2009.11.084. [DOI] [PubMed] [Google Scholar]
- 139.Ma SH, Lepak LA, Hussain RJ, Shain W, Shuler ML. Lab on a chip. 2005;5:74–85. doi: 10.1039/b405713a. [DOI] [PubMed] [Google Scholar]
- 140.Griep L, Wolbers F, De Wagenaar B, Ter Braak P, Weksler B, Romero IA, Couraud P, Vermes I, Van Der Meer A, Van den Berg A. Biomedical microdevices. 2013;15:145–150. doi: 10.1007/s10544-012-9699-7. [DOI] [PubMed] [Google Scholar]
- 141.Booth R, Kim H. Lab on a chip. 2012;12:1784–1792. doi: 10.1039/c2lc40094d. [DOI] [PubMed] [Google Scholar]
- 142.Prabhakarpandian B, Shen M-C, Nichols JB, Mills IR, Sidoryk-Wegrzynowicz M, Aschner M, Pant K. Lab on a Chip. 2013;13:1093–1101. doi: 10.1039/c2lc41208j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Southam KA, King AE, Blizzard CA, McCormack GH, Dickson TC. Journal of neuroscience methods. 2013;218:164–169. doi: 10.1016/j.jneumeth.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 144.Bang S, Na S, Jang JM, Kim J, Jeon NL. Advanced healthcare materials. 2015 doi: 10.1002/adhm.201500397. [DOI] [PubMed] [Google Scholar]
- 145.Berdichevsky Y, Staley KJ, Yarmush ML. Lab on a Chip. 2010;10:999–1004. doi: 10.1039/b922365g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shi P, Nedelec S, Wichterle H, Kam LC. Lab on a chip. 2010;10:1005–1010. doi: 10.1039/b922143c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Park J, Lee BK, Jeong GS, Hyun JK, Lee CJ, Lee SH. Lab Chip. 2015;15:141–15.0. doi: 10.1039/c4lc00962b. [DOI] [PubMed] [Google Scholar]
- 148.Karimi M, Avci P, Mobasseri R, Hamblin MR, Naderi-Manesh H. Journal of nanoparticle research. 2013;15:1–14. doi: 10.1007/s11051-013-1651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Karimi M, Avci P, Ahi M, Gazori T, Hamblin MR, Naderi-Manesh H. Journal of Nanopharmaceutics and Drug Delivery. 2013;1:278–266. doi: 10.1166/jnd.2013.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Karimi M, Ghasemi A, Zangabad PS, Rahighi R, Basri MSM, Mirshekari H, Amiri M, Pishabad ZS, Aslani A, Bozorgomid M. Chemical Society Reviews. 2016 doi: 10.1039/c5cs00798d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Karimi M, Mirshekari H, Basri SMM, Bahrami S, Moghoofei M, Hamblin MR. Advanced Drug Delivery Reviews. 2016 doi: 10.1016/j.addr.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Karimi M, Bahrami S, Ravari SB, Zangabad PS, Mirshekari H, Bozorgomid M, Shahreza S, Sori M, Hamblin MR. Expert Opinion on Drug Delivery. 2016 doi: 10.1080/17425247.2016.1193149. null-null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shi J, Votruba AR, Farokhzad OC, Langer R. Nano letters. 2010;10:3223–3230. doi: 10.1021/nl102184c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.