Abstract
The 5′leader of the HIV-1 genomic RNA is a multifunctional region that folds into secondary/tertiary structures that regulate multiple processes during viral replication including translation initiation. In this work we examine the internal ribosome entry site (IRES) located in the 5′leader that drives translation initiation of the viral Gag protein under conditions that hinder cap-dependent translation initiation. We show that activity of the HIV-1 IRES relies on ribosomal protein S25 (eS25). Additionally, a mechanistic and mutational analysis revealed that the HIV-1 IRES is modular in nature and that once the 40S ribosomal subunit is recruited to the IRES, translation initiates without the need of ribosome scanning. These findings elucidate a mechanism of initiation by the HIV-1 IRES whereby a number of highly structured sites present within the HIV-1 5′leader leads to the recruitment of the 40S subunit directly at the site of initiation of protein synthesis.
Keywords: HIV-1, RNA, IRES, protein synthesis
INTRODUCTION
Translation initiation of most eukaryotic mRNAs occurs by a cap-dependent mechanism [1], whereby the 5′cap structure (m7GpppN) is recognized by eukaryotic translation initiation factors (eIFs): the cap-binding protein (eIF4E), the ATP-dependent RNA helicase (eIF4A), and the scaffolding protein (eIF4G), which form the eIF4F complex. The eIF4F complex bridges the 5′ and the 3′ ends of the mRNA by also binding to the poly(A) binding protein (PABP) bound to the poly(A) tail [1]. These factors recruit the 40S ribosomal subunit as a 43S pre-initiation complex composed of the 40S subunit, eIF2/GTP/Met-tRNAi (ternary complex), eIF1A, eIF1 and eIF3, which scans the 5′untranslated region (5′UTR) in a 5′ to 3′ direction until the initiation codon, commonly an AUG in the optimal context ((A/G)XXAUGG, where X is any nucleotide), is encountered [1]. Once the start codon is recognized the 60S ribosomal subunit joins to form the 80S ribosome and translation ensues [1]. An alternative cap-independent mechanism exists in which the 40S ribosomal subunit is recruited to an internal site within the 5′UTR of the mRNA without requiring a 5′cap. This alternative mechanism of recruitment, initially described for the uncapped mRNAs from the Picornaviridae family, is dependent on an RNA element termed the internal ribosomal entry site (IRES) [2, 3]. Functionally, the IRES was identified by inserting the poliovirus (PV) or the encephalomyocarditis virus (EMCV) 5′UTR into the intercistronic spacer of a bicistronic reporter encoding two proteins [2, 3]. Expression of the second cistron is independent of the first and only occurs when the inserted sequence can recruit the ribosome internally. The fact that the ribosome enters the RNA internally was demonstrated using a closed circular mRNA, such that only RNAs that contained an IRES and thus could recruit the ribosome internally were translated [4]. Subsequent studies demonstrated that this alternative mechanism of translation initiation was also used by the mRNAs of other viral families, including members of the Flaviviridae [5], Retroviridae [6], and Herpesviridae [7]. IRESs have also been found in the RNAs of insect [8, 9], and plant viruses [10–12], as well as insect and rodent retrotransposons [13–15]. The mechanism of internal initiation is not restricted to viruses; IRESs have been increasingly identified in cellular mRNAs [16].
The 5′UTR of the full length or genomic RNA of the human immunodeficiency virus type-1 (HIV-1), prototype member of the lentivirus genus of the Retroviridae and the etiologic agent of AIDS, contains multiple functional elements that are critical for virus replication [17]. Studies of the structure of the HIV-1 leader RNA revealed that most of these regulatory elements consist of discrete hairpin structures [15, 17–20]. The functional RNA elements include the trans-activating region (TAR), polyadenylation (PolyA) signal, primer binding site (PBS), dimerization initiation site (DIS), splice donor (SD), and packaging signal (Psi). These structural elements are involved in transcription, mRNA splicing, genomic RNA dimerization, genomic RNA encapsidation, reverse transcription, and viral mRNA translation [17]. Like a number of retroviral mRNAs [6, 21, 22], the HIV-1 5′UTR can drive internal initiation of translation, enabling protein synthesis under conditions that hinder cap-dependent translation initiation [23–29]. For example, the HIV-1 IRES facilitates protein synthesis during oxidative and osmotic stress [25, 30], G2/M cell cycle arrest [23, 26], or when eIF4G and the poly(A) binding protein (PABP), two proteins that are critical for cap-dependent translation initiation, are cleaved by the HIV-1 protease [31–34]. In the context of a viral infection, a report shows that at early stages of HIV-1 infection viral protein synthesis is driven by a cap-dependent mechanism, however at later time points, viral protein synthesis switches to an IRES-mediated translation initiation mode [27]. This would explain how the expression of the structural protein Gag is maintained during HIV-1 replication in poliovirus co-infected cells [24]. Together these observations suggest that the HIV-1 IRES drives the synthesis of the viral structural protein Gag when cap-dependent translation is hindered [24, 27]. To date, a number of studies have focused on characterizing the activity of the HIV-1 IRES [23–30]. However, very little is still known about the mechanism used by the HIV-1 IRES to recruit the translational machinery of the host and how cellular proteins impact HIV-1 IRES activity [23, 30, 35, 36]. In addition, little is known about how the different RNA structural elements present within the HIV-1 5′UTR participate, if at all, on internal initiation [23, 25, 28, 37]. In this study we provide new insights into the mechanism used by the HIV-1 IRES in order to promote internal initiation. We show that the HIV-1 IRES is modular in nature, similar to what has been observed for many cellular IRESs, and that the PBS, DIS, SD, and Psi hairpins are required for HIV-1 IRES activity. Additionally, we show that the HIV-1 IRES recruits the ribosome directly to the start codon not requiring scanning of the 40S subunit for initiation. Furthermore, we establish that the activity of the HIV-1 IRES is dependent upon the ribosomal protein S25 (RPS25, eS25), which has been shown to be required for other viral and cellular IRESs, but does not significantly affect cap-dependent translation [38, 39].
RESULTS
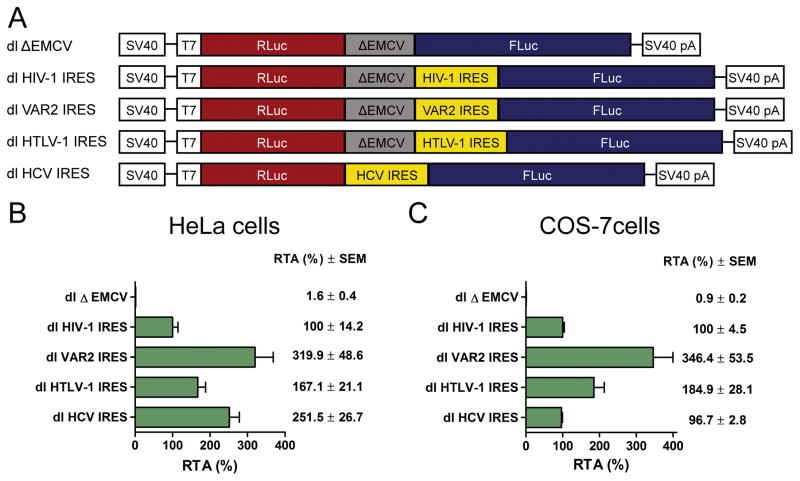
The HIV-1 5′UTR mediates translation initiation in HeLa and COS-7 cells when in the context of a bicistronic vector
The 5′UTR of the HIV-1 full-length mRNA harbors an IRES [24–26, 28, 29]. HIV-1 IRES activity is high in T-cell lines, when compared to other cell types [28]. However, in our hands, most commercially available transfection reagents were inefficient in the transfecting cells that grow in suspension. Thus, for this study we chose to use HeLa and COS-7 cells, two easily transfectable cell lines that yielded highly reproducible results. Several studies, conducted using different in vitro translation systems [26, 29, 35, 40] or in different cellular backgrounds [23–26, 28, 29], support the presence of a functional IRES within the HIV-1 full length mRNA. However, other studies directly question the existence of an IRES within the HIV-1 full length mRNA [41, 42]. Due to this controversy, we first sought to establish if the 5′UTR of the HIV-1 full length RNA exhibits IRES activity when evaluated in the context of HeLa and COS-7 cell lines. For this, we used a dual luciferase (dl) reporter containing an upstream Renilla luciferase (RLuc) and a downstream firefly luciferase (FLuc) coding region that harbors either the 5′UTR of the full-length mRNA of the HIV-1 infectious recombinant proviral clone NL4.3 (pNL4.3) named dl HIV-1 IRES [26], or the 5′UTR from a clinically sourced RNA isolate VAR2, the dl VAR2 IRES [29] (Fig. 1A). Both plasmids were transfected into HeLa or COS-7 cells as previously described [21–24, 26, 29, 30, 40, 43]. The amount of FLuc activity was taken as a measurement of the IRES activity, while the RLuc open reading frame was translated by a cap-dependent mechanism and served as an internal control for transfection efficiency. The dl ΔEMCV vector (Fig. 1A), containing the defective EMCV IRES inserted upstream of the FLuc reporter, was used as negative control for IRES activity [26]. The positive controls for IRES activity were the dl HCV IRES and dl HTLV-1 IRES [21, 44] reporters, which harbored the hepatitis C virus (HCV, genotype 1b) and human lymphotrophic virus type 1 (HTLV-1) IRESs, respectively (Fig. 1A). For each reporter the IRES activity (FLuc) was normalized to cap-dependent translation (RLuc) and the results are expressed as relative translation activity (RTA) to the dl HIV-1 IRES, which was set to 100% (Figs. 1B and 1C). Consistent with previous reports [24, 26, 27, 29], the 5′UTR of pNL4.3 was capable of driving translation of the second cistron in the context of a bicistronic reporter (Figs.1B and 1C). In agreement with a previous report, the IRES activity from the pNL4.3 5′UTR was lower than that of the HTLV-1 or HCV IRESs (Figs.1B and 1C) [21]. Nonetheless, in both cell lines translation driven by the pNL4.3 5′UTR was 2 logs higher than the background (Figs. 1B and 1C; compare dl HIV-1 IRES to dl ΔEMCV). In agreement with our previous findings [29], the IRES activity of a natural variant named VAR2 exhibited higher IRES activity than that of the pNL4.3, HCV and HTLV-1 IRESs in both HeLa and COS-7 cells (Figs. 1B and 1C). These results suggest that the 5′UTR of the HIV-1 mRNA, pNL4.3 and VAR2 can initiate translation internally in both HeLa and COS-7 cells.
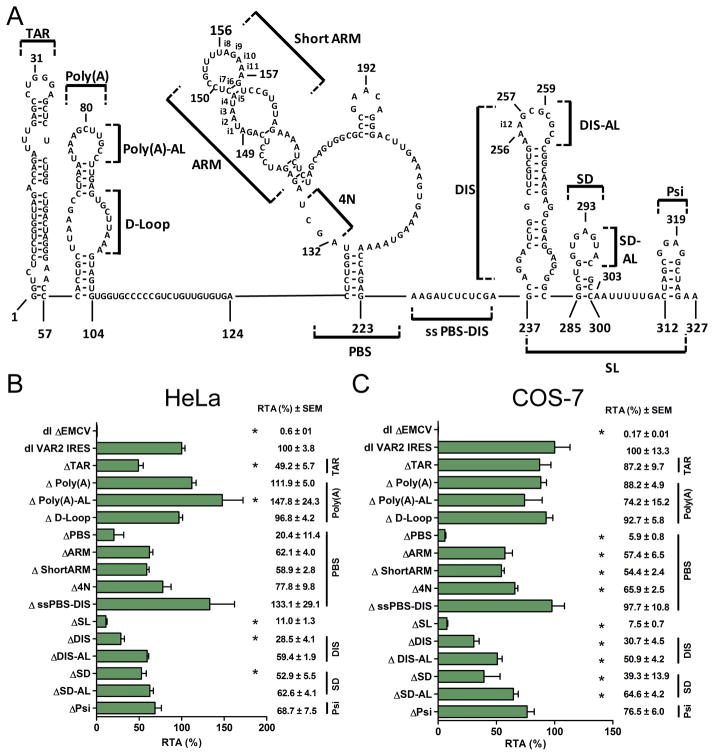
Figure 1. The HIV-1 IRES is functional in HeLa and COS-7 cells.
(A) Schematic representation of the dual luciferase bicistronic DNA reporters used. The bicistronic reporters are expressed from the SV40 promoter and receive a poly(A) tail by the SV40 poly(A) signal. The dl ΔEMCV [26], dl HIV-1 IRES [26], dl VAR2 IRES [29], dl HTLV-1 IRES [21], or the dl HCV IRES [44] (200 ng) vectors were co-transfected into HeLa (B) or COS-7 (C) cells with the pcDNA3.1 lacZ plasmid (50 ng) that expresses β-Galactosidase from a cap-dependent transcript [21]. Co-transfection of β-Galactosidase serves as an internal control for transfection efficiency. The FLuc/RLuc ratio was used as an index of IRES activity and results are expressed as RTA (%), relative to the dl HIV-1 IRES [26], which was set to 100%. Values are the mean (+/− SEM) for three independent experiments, each performed in duplicate.
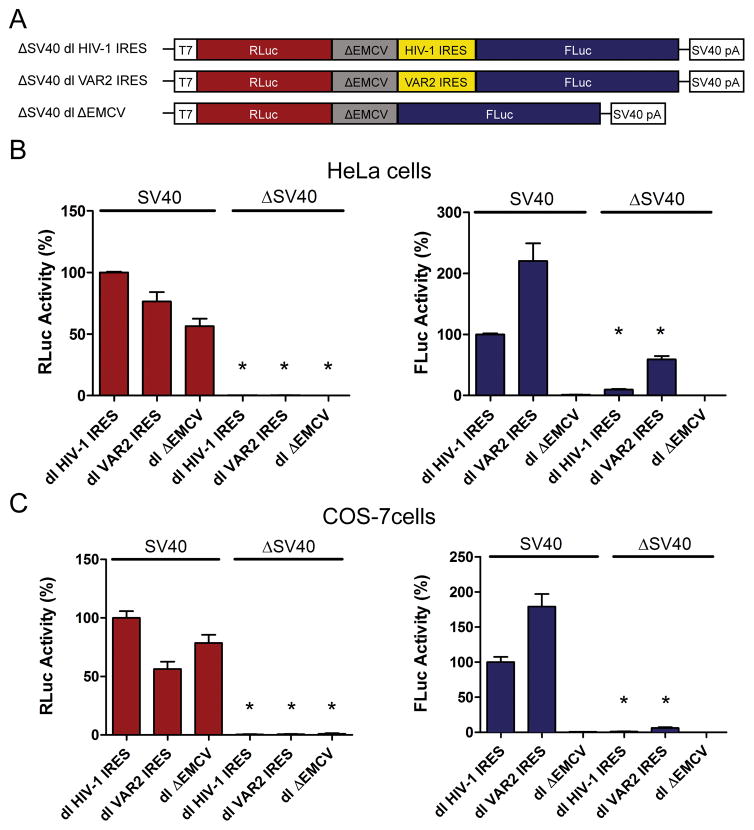
Next we sought to evaluate whether FLuc expression from the dl HIV-1 IRES and dl VAR2 IRES plasmids could be attributed to cryptic promoter activity. For this, the SV40 promoter was deleted (ΔSV40) from the plasmid. This strategy allows us to determine if any FLuc activity is generated in the absence of transcription of the full-length bicistronic mRNA. HeLa and COS-7 cells were transfected with the dl HIV-1 IRES, dl VAR2 IRES, or dl ΔEMCV reporters that either have or lack the eukaryotic SV40 promoter (Fig. 2A). In the absence of the SV40 promoter both the RLuc and FLuc activities from the reporters were significantly diminished in HeLa (Fig. 2B) and COS-7 (Fig. 2C) cells. In the absence of the SV40 promoter FLuc activity of the dl HIV-1 IRES plasmid was reduced in more than 90% in HeLa cells (Fig. 2B) and almost 99% in COS-7 cells (Fig. 2C), while the FLuc activity of the dl VAR2 IRES plasmid was reduced in more than 73% in HeLa cells (Fig. 2B) and about 97% in COS-7 cells (Fig. 2C). The detected residual FLuc activity is most likely associated with an mRNA encoding a functional FLuc protein generated by a cryptic promoter. Interestingly, there is some difference in the level of cryptic promoter activity between the two cell lines with the HeLa cells having slightly more cryptic promoter activity than the COS-7 cells. Nonetheless, the findings indicate that the majority of the FLuc activity detected in the assay is associated with the bicistronic mRNA.
Figure 2. Evaluating the presence of a cryptic promoter in the bicistronic vectors.
(A) Schematic representation of the bicistronic constructs. The SV40 promoter from dl ΔEMCV, dl HIV-1 IRES or dl VAR2 IRES was removed to generate the promoterless (ΔSV40) vectors. SV40pA represents the SV40 polyadenylation signal. HeLa (B) or COS-7 (C) cells were transfected with DNA (200 ng) corresponding to the vectors depicted in (A) together with the pcDNA3.1 lacZ (50 ng) plasmid. RLuc (left panel) and FLuc (right panel) activities were determined and normalized to the β-Galactosidase activity. The [RLuc/β-Galactosidase] and [FLuc/β-Galactosidase] activities obtained for the dl HIV-1 IRES vector was defined as 100%. Values shown are the mean (+/− SEM) for three independent experiments, each performed in triplicate. Statistical analysis was performed by a one way ANOVA test followed by a Bonferroni’s multiple test comparison between the SV40 and the promoterless version of each dl vector (*P<0.05).
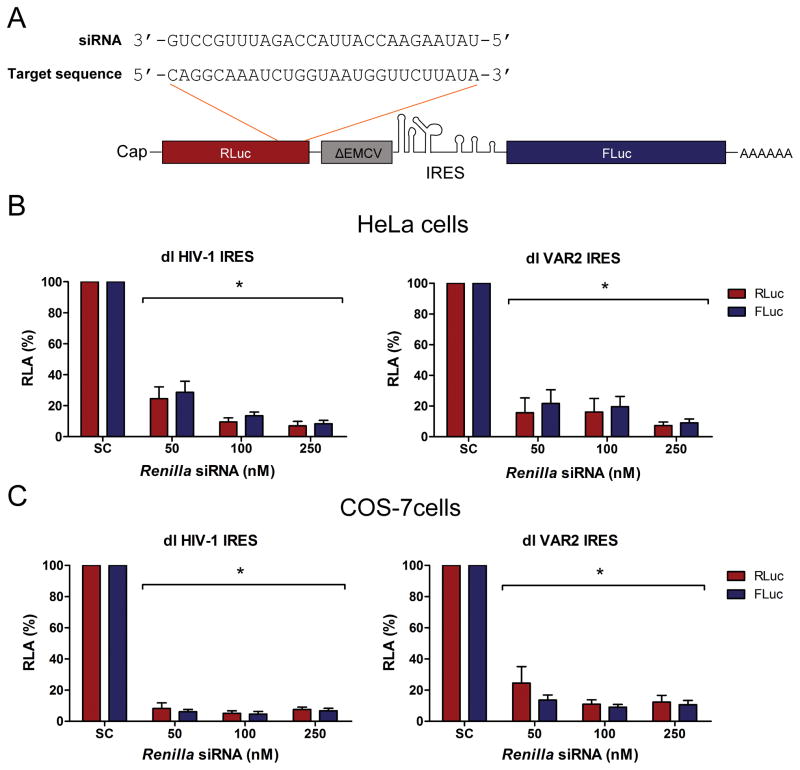
The possibility remains that an alternative splicing event could generate a monocistronic cap-dependent mRNA that encodes for a functional FLuc protein. Therefore, we targeted the Renilla ORF with a short interfering RNA (siRNA) such that if there were only full-length bicistronic transcripts generated then RLuc and FLuc activities would decrease proportionally, however, if spliced transcripts or a monocistronic mRNA from a cryptic promoter encoding a functional FLuc were generated, then FLuc activities would remain high while RLuc activities decreased. The Renilla siRNA or a scrambled control (SC) siRNA were co-transfected with the reporters and RLuc and FLuc activities were determined. In the presence of the siRNA targeting Renilla both RLuc and FLuc activities were reduced equivalently in HeLa (Fig. 3A) and COS-7 (Fig. 3B) cells. This observation strongly suggests that both RLuc and FLuc are encoded on the same bicistronic transcript. Thus, FLuc expression is dependent upon the IRES activity of the HIV-1 5′UTR. Taken together, the findings presented in figures 1, 2 and 3 confirm that the HIV-1 5′UTR isolated from the pNL4.3 infectious clone and the clinical isolate, VAR2, exhibit IRES activity in both HeLa and COS-7 cells. In addition, these observations validate the use of these cell lines to study HIV-1 IRES activity.
Figure 3. A monocistronic mRNA encoding a functional FLuc is not generated in HeLa and COS-7 cells.
A) Schematic representation of the RLuc RNA region targeted by the siRNA. HeLa (B) or COS-7 (C) cells were transfected with dl HIV-1 IRES or dl VAR 2 plasmid together with a scrambled control RNA (−) or a siRNA-RLuc that targets the RLuc RNA. RLuc (white bars) and FLuc (black bars) activities were measured 12 h post transfection and expressed relative to the scrambled control siRNA set to 100% (RLA: relative luciferase activity). Values shown are the mean (+/− SEM) for at least three independent experiments. Statistical analysis was performed by a one way ANOVA test followed by a Bonferroni’s multiple test comparison (square bracket indicates data that are significantly different from SC (*P<0.05)
HIV-1 IRES activity is dependent on ribosomal protein S25
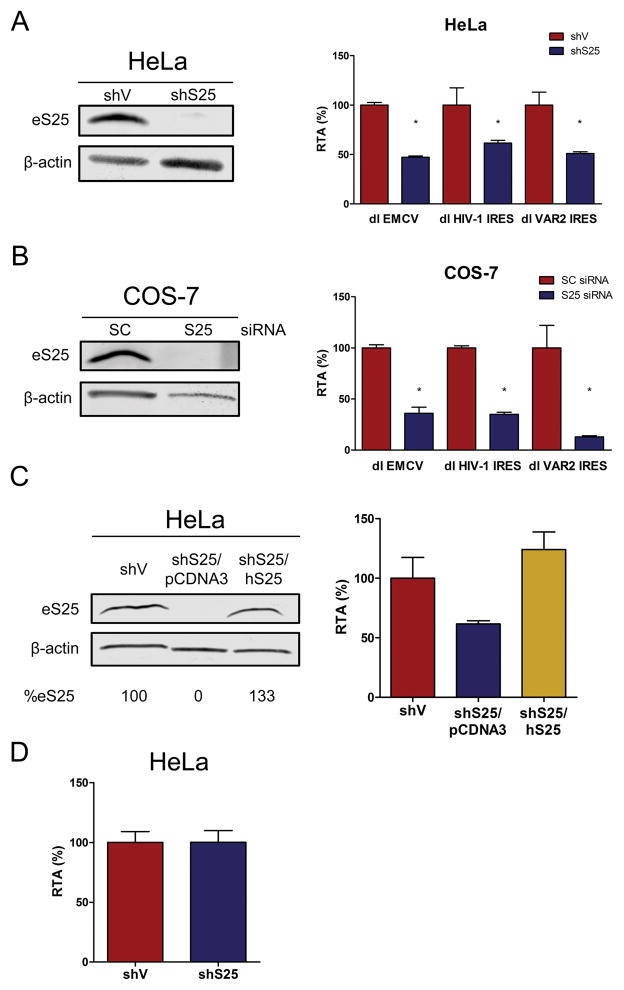
Little is known about the mechanism used by the HIV-1 IRES to initiate translation. The mechanism of ribosome recruitment by various viral IRESs differs greatly [45, 46]. Despite structural and functional differences between various viral and cellular IRESs, previous studies [38, 39], suggests that IRESs share a common mechanism that is dependent on eS25 for cap-independent initiation. Based on these findings we next asked whether translation initiation driven by the HIV-1 IRES requires eS25. Knowing that the HIV-1 IRES is functional in HeLa cells (Figs. 1–3) we transfected the dl HIV-1 IRES or the dl VAR2 IRES plasmids into clonal cell line derived from the previously published stable cell lines [38] that constitutively expresses an shRNA against eS25 (HeLashS25) or a control empty vector (HeLashV). Knockdown of eS25 (HeLashS25) was confirmed by western analysis (Fig. 4A, left panel). As an additional control HeLashS25 were transfected with the dl EMCV IRES plasmid harboring the EMCV IRES, which is known to be dependent on eS25 for full IRES activity [38]. Transfection of a positive control dl EMCV IRES plasmid resulted in a 53% decrease in IRES activity when eS25 was knocked down (Fig. 4A, right panel; see dl EMCV IRES). Translation driven by the pNL4.3 and VAR2 IRESs were also significantly reduced to 61% and 51%, respectively, when eS25 was targeted (Fig. 4A, right panel). These findings indicate that the IRESs from two different HIV-1 isolates, pNL4.3 and VAR2, are dependent on eS25 for their activity.
Figure 4. Full HIV-1 IRES activity requires eS25.
(A) HeLashV and HeLashS25 cells were transfected with dl HIV-1 IRES or dl VAR 2 IRES plasmids together with a β-galactosidase control plasmid. (A) After 48 h total proteins were extracted and eS25 and β-actin were detected by western analysis (left panel). IRES activity was measured, normalized to the transfection control β-galactosidase and expressed as RTA (%) relative to the activity measured in the HeLashV cells transfected with pcDNA3 (right panel). (B) COS-7 cells were transfected with a scrambled RNA or a siRNA designed to target eS25 mRNA together with the dl HIV-1 IRES or dl VAR 2 IRES plasmids as indicated in Material and Methods. Knockdown of eS25 was confirmed by western (left panel). IRES activity was measured, normalized to the transfection control β-galactosidase and expressed as RTA (%) relative to the activity measured in the COS-7 cells transfected with the scrambled RNA (right panel). (C) HeLashV and HeLashS25 cells were transfected with dl HIV-1 IRES plasmid together with a β-galactosidase control plasmid and where indicated a plasmid expressing human eS25 harboring nucleotide changes that render it resistant to the shRNA, named hS25 or an empty vector (pcDNA3). After 48 h IRES activity was measured, normalized to the transfection control β-galactosidase and expressed as RTA (%) relative to the activity measured in the HeLashV cells transfected with pcDNA3 (right panel). eS25 and β-actin were also detected by Western analysis (left panel). (D) HeLashV and HeLashS25 cells were transfected with a β-galactosidase reporter that is translated by a cap-dependent mechanism driven by the CMV promoter. β-galactosidase activity was measured and expressed as RTA (%) and expressed relative to activity in the HeLashV cells standard error is shown (n=57). Statistical analysis was performed by a one way ANOVA test followed by a Bonferroni’s multiple test comparison (*P<0.05).
To exclude the possibility that the reduction in HIV-1 IRES activity that was observed when eS25 is knocked down is exclusively associated to HeLa cells, we confirmed these findings in a different cell line, COS-7. For this a siRNA designed to specifically target eS25 mRNA (eS25-siRNA) or a scrambled siRNA control (SC siRNA) was transiently co-transfected into COS-7 cells along with the bicistronic reporter plasmids. Western analysis revealed a complete knockdown in the levels of endogenous eS25 following eS25-siRNA (S25) transfection in to the COS-7 cells (Fig. 4B, left panel). The activity of the EMCV IRES was also reduced, to 36% (64 % decrease), in COS-7 cells when eS25 was knocked down (Fig. 4B, right panel). Consistent with what was observed in HeLashS25 cells (Fig. 4A), knockdown of eS25 in COS-7 cells significantly reduced pNL4.3 (65%) and VAR2 (87%) IRES activity (Fig. 4B, right panel). Together, the results presented in figure 4, show that like other IRESs [38], optimal HIV-1 IRES activity is dependent on eS25.
To determine whether the observed reduction in IRES activity in the HeLashS25 cells was indeed associated with the knockdown of eS25 and not due to some off target effects, we rescued eS25 protein levels by expressing a shRNA resistant human eS25 from a plasmid. As before, the reporter we used contained the 5′UTR of the pNL4.3 infectious clone [26]. The eS25 rescue plasmid was co-transfected into the HeLashS25 cells together with the dl HIV-1 IRES vector. Western analysis revealed a complete rescue of eS25 (133%) in the presence of the hS25 plasmid (Fig 4C, left panel). Consistent with our previous results, the activity of the pNL4.3 IRES was significantly reduced (61 %) in HeLashS25 cells compared to the IRES activity in HeLashV cells (Fig. 4C, right panel). Importantly, transfection of hS25 plasmid in HeLashS25 cells fully rescued the HIV-1 IRES activity (Fig. 4C, right panel). Thus, the reduction in HIV-1 IRES activity observed in HeLashS25 cells is indeed due to the lower eS25 levels. Previous reports show that eS25 is required for IRES-mediated translation, but not for cap-dependent initiation [38, 39]. As an additional control we evaluated the impact of eS25 knockdown on cap-dependent initiation. In agreement with previous reports eS25 knockdown did not impact on cap-dependent initiation (Fig. 4D), these findings further support our conclusion that the HIV-1 5′UTR harbors an IRES which is active in HeLa cells.
Defining the RNA structures that participate in HIV-1 IRES-mediated translation initiation
A recent study suggests that the HIV-1 IRES is modular in nature [37]. Intrigued by these observations, we sought to evaluate if indeed this was the case. Several studies do support the modular nature of some IRESs [47–50]. In these cases, each module is composed of discrete RNA secondary/tertiary structures that contribute to the overall process of recruiting the translational machinery and initiating translation [47–50]. As depicted in figure 5A [29], the HIV-1 5′UTR has well-defined RNA structural elements, all of which are required for different stages during the viral life cycle [15, 17–20]. These structures correspond to the TAR, poly(A) stem-loop, PBS, DIS, SD and Psi. To evaluate the role of each of these structural elements on HIV-1 IRES activity we used the VAR2 5′UTR [29] because it exhibits an increased (3- to 4-fold) IRES activity as compared to pNL4.3 when evaluated in HeLa cells (Fig. 1 and [29]), yet, RNA structural domains observed in the context of the pNL4.3 5′UTR are largely conserved in VAR2 [29].
Figure 5. RNA Structural domains within the 5′untranslated region of the HIV-1 full-length mRNA influence IRES activity.
(A) Schematic representation of VAR 2 5′UTR adapted from [29] with the main domains of the 5′UTR region indicated. The nucleotide numeration is according to the infectious clone pNL4.3 (AF 324493). The previously described nucleotide insertions (i), with respect to pNL4.3 are indicated [29]. The deletion mutants that were assayed are indicated. VAR2 5′UTR mutants were cloned in the intercistronic region of the dl vector. Bicistronic reporters carrying the mutant or wild-type VAR2 IRESs [29], or the ΔEMCV sequence (negative control) were transfected into HeLa (B) or COS-7 (C) cells. After 24 h the FLuc/RLuc ratio was used as an index of IRES activity and the activity of the mutants are expressed as relative translational activity (RTA (%)) with respect to the dl VAR2 IRES set to 100% (+/− SEM). Values are the mean ± SEM (error bars) of at least three independent experiments. Statistical analysis was performed by a one way ANOVA test followed by a Dunnett’s multiple test comparison (*P<0.05).
Mutant VAR2 HIV-1 IRESs containing deletions of the various structural elements were introduced into the bicistronic reporter harboring the VAR2 HIV-1 IRES [29]. A diagram depicting these regions is shown in figure 5A. The bicistronic reporter plasmids, named according to the deleted region (Fig. 5B), were transfected in HeLa or COS-7 cells and luciferase activities were measured after 24 h. Changes in IRES activities as a result of the mutation were largely consistent between HeLa and COS-7 cells (compare Figs. 5B and 5C). One exception was the ΔTAR, which had a decreased IRES activity, by about 51%, in HeLa cells (Fig. 5A), but only a marginal impact on IRES activity in COS-7 cells (13%) (Fig. 5B). These differences cannot be readily explained but may have to do with differences in expression of cellular proteins between the two cell types which is consistent with the observation that VAR2 HIV-1 IRES activity varies strongly from one cell line to another [28]. Deletion of the poly(A) loop did not dramatically alter IRES activity (Fig. 5). Next, we analyzed the contribution of specific structural domains of the poly(A) hairpin on VAR2 HIV-1 IRES activity. For this, eight nucleotides (U91-A98) that form the internal bulge (D-loop), or nucleotides in the apical loop (AL) of the poly(A) stem-loop were independently deleted (Fig. 5A). Even though the poly(A) stem-loop is not part of the minimal HIV-1 IRES [26, 28], previous studies have suggested that the D-loop and AL become less accessible to RNA modifications in the presence of cell extracts suggesting that cellular proteins may bind to these elements, influencing IRES activity [23]. Furthermore, a previous report [25], describes that in T-cell lines the poly(A) hairpin is a negative regulator of HIV-1 IRES activity. Notably, in HeLa cells the IRES activity increased (by 48%) when the AL was removed from the VAR2 HIV-1 IRES (Fig. 5B). However, in the COS-7 cells, removal of the AL decreased IRES activity by 26% (Fig. 5C). Deletion of the D-loop of the poly(A) hairpin had no effect on VAR2 HIV-1 IRES activity (Fig. 5B and 5C). Again, differences obtained COS-7 and HeLa cells could be due to differences in the expression levels of cellular proteins in the different cell types.
In full agreement with previous studies that mapped the minimal HIV-1 IRES to the region comprising the PBS to the Gag protein initiation codon [26, 28], the PBS proved to be an important determinant of VAR2 HIV-1 IRES activity in both HeLa and COS-7 cell lines (Figs. 5B and 5C). Deletion of the PBS was highly deleterious in the context of COS-7 cells (94 % reduction) when compared to HeLa cells (80 % reduction). Even though we cannot readily explain this difference it is interesting to note that about 26 % of FLuc activity in HeLa cells is due to a cryptic promoter activity (compare dl VAR2 IRES and ΔSV40 dlVAR2 IRES in Fig. 2). To determine whether specific subdomains of the PBS were required for IRES activity smaller deletion mutants were evaluated. Selection of the targets was based on the RNA protection pattern exhibited by the HIV-1 5′leader obtained by SHAPE conducted in the presence of cell extracts [23, 29], and also on a study that suggested that the PBS-ARM harbors an IRES inhibitory element [25]. Deletion of the PBS-ARM or the PBS-Short-ARM resulted in similar reductions in IRES activity in both HeLa and COS-7 cells (ranging from 38–46% down). Furthermore, deletion of the PBS-4N region reduced IRES activity by 21% (HeLa) and 34% (COS-7) (Figs. 5B and 5C). We next evaluated the influence of the single-stranded linker region that exists between the PBS and the DIS loop (Δss-PBS-DIS in Fig. 5B and 5C). SHAPE assays revealed that the accessibility of 1-methyl-7-nitroisatoic anhydride (1M7) to the linker region varies in the presence of G2/M cell extracts, suggesting that this region is modified under conditions that favor HIV-1 IRES activity [23]. However, the deletion of the ssPBS-DIS region had no impact on IRES-mediated translation in COS-7 cells (Fig. 5C) or it increased IRES activity, but not significantly, by 33% in HeLa cells. The SL region (Fig. 5A), which comprises the DIS, SD and Psi hairpin loops, is modified under conditions that favor HIV-1 IRES activity [23]. SHAPE analysis revealed that the accessibility of 1M7 to the apical loops of the DIS, SD, and Psi varies in the presence of different cell extracts [23]. Consistent with a possible role in HIV-1 IRES-mediated translation initiation, the deletion of the SL region reduces HIV-1 IRES activity to 11% in HeLa cells and to 7% in COS-7 cells (Figs. 5B and 5C). These data indicate that the PBS or the SL structures alone do not exhibit significant IRES activity.
Next we sought to evaluate the contribution of each stem-loop within the SL region to HIV-1 IRES activity. Deletion of the DIS stem-loop reduced HIV-1 IRES activity by 71% (HeLa) and 69% (COS-7), while deletion of the DIS apical loop (DIS-AL) reduced translational activity by 41% (HeLa) and 49% (COS-7). Deletion of the SD stem-loop reduced IRES activity by 47% (HeLa) and 61% (COS-7), while elimination of its AL caused a 37 to 35% reduction of HIV-1 IRES activity (Figs. 5B and 5C). Deletion of the Psi-stem loop reduced IRES activity by 31% (HeLa) and 23 % (COS-7) (Fig. 5B). Together results suggest that the PBS and the SL structures significantly contribute to the overall IRES activity. Furthermore, they also indicate the presence of both the PBS and the SL structures is required in order to conform a functional IRES.
Translation initiation driven by the HIV-1 5′UTR is insensitive to edeine
IRESs are known to recruit the 40S subunit either directly at the start codon or upstream of it followed by scanning in the 5′ to 3′ direction to reach the start codon [51]. Since the RNA structures required for HIV-1 IRES spans approximately 100 nts upstream of the AUG of the Gag initiation codon, it was of interest to determine whether the 40S ribosomal subunit is recruited directly at the Gag initiation codon or upstream of it. For this, in vitro translation assays using rabbit reticulocyte lysate (RRL) were performed in the presence of edeine, which is a strong basic, linear oligopeptide antibiotic. At high concentrations edeine inhibits elongation, however, at low concentrations edeine interferes with initiation codon (AUG) recognition by the scanning 40S-ternary complexes [52–55]. To establish conditions that inhibit 40S recognition of the start codon during scanning but not elongation, RRL was programmed with in vitro synthesized, capped dl PV IRES (harboring the poliovirus IRES) or dl HCV IRES bicistronic mRNAs, which contain IRESs that are known to either require 40S scanning or not, respectively. In vitro translation was conducted in the absence or in the presence of increasing concentrations of edeine as previously described [21, 56]. Results show that cap-dependent translation (RLuc activity) in the absence of edeine were similar for both bicistronic mRNAs indicating the RRL was programmed with equivalent amounts of RNA (Figs. 6A and 6B, left panels). As predicted, for both bicistronic mRNAs cap-dependent translation (RLuc) was inhibited in the presence of 0.25 μM and 0.5 μM of edeine (Figs. 6A and 6B; left panel). Expression of the downstream cistron mediated by the PV IRES was also inhibited (Fig. 6A; right panel), consistent with the requirement for 40S scanning in order for start codon recognition to occur [21, 56]. In contrast, translation initiation mediated by the HCV IRES remained unaffected at 0.25 μM edeine (Fig. 6B; right panel). These results are consistent with the ability of the HCV IRES to directly recruit the 40S ribosomal subunit to the initiation codon without scanning [56, 57]. Inhibition of HCV IRES activity at 0.5 μM of edeine has been previously described [21], and likely reflects inhibition of elongation, which is observed at high concentrations of the drug [56].
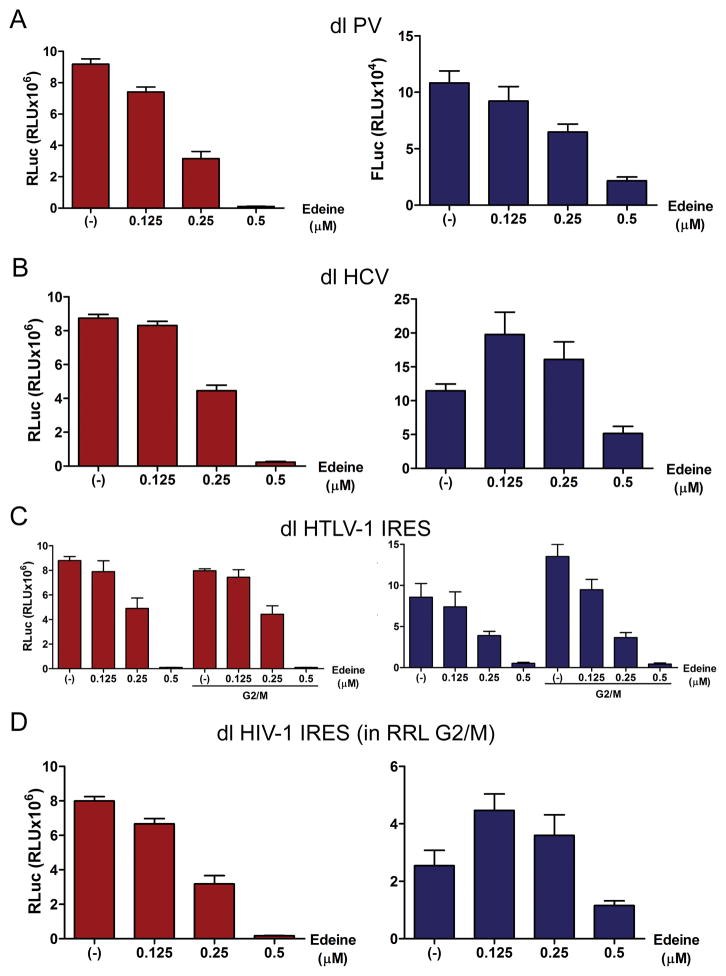
Figure 6. Translation initiation from the HIV-1 5′UTR is insensitive to edeine in RRL/G2M.
The in vitro transcribed capped dl PV IRES (A) or dl HCV-1 IRES (B) RNAs were in vitro translated in RRL in the absence (−) or the presence of increasing concentrations of edeine (0.125, 0.25, or 0.5 μM). RLuc (white bars, left panel) and FLuc (black bars, right panels) activities were measured. The RLuc and FLuc activity for each bicistronic reporter are shown as relative light units (RLU). Values shown are the mean (+/− SEM) for four independent experiments, each conducted in duplicate. (C)The in vitro transcribed capped dl HTLV-1 IRES RNA was in vitro translated in RRL or in RRL supplemented with HeLa extracts (S10) generated from G2/M arrested cells (RRL/G2M) in the absence (−) or the presence of increasing concentrations of edeine (0.125, 0.25, or 0.5 μM). RLuc (white bars, left panels) and FLuc (black bars, right panels) activities were measured. The RLuc and FLuc activity for each bicistronic reporter are shown as relative light units (RLU). Values shown are the mean (+/− SEM) for four independent experiments, each conducted in duplicate. (D) The in vitro transcribed capped dl HIV-1 IRES was in vitro translated in RRL/G2M in the absence (−) or the presence of increasing concentrations of edeine (0.125, 0.25, or 0.5 μM). RLuc (white bars, left panels) and FLuc (black bars, right panels) activities were measured. The RLuc and FLuc activity for each bicistronic reporter are shown as relative light units (RLU). Values shown are the mean (+/− SEM) for four independent experiments, each conducted in duplicate.
The HIV-1 IRES is functional in RRL only when it is supplemented with S10 cell extracts generated from G2/M arrested HeLa cells (here referred to as RRL/G2M) [23]. So we next sought to establish if the presence of cell extracts impacts on how an IRES recruits the initiation complex. We used the dl HTLV-1 IRES as a control RNA, since the HTLV-1 IRES is functional in both RRL and in RRL/G2M [21]. In vitro synthesized, capped dl HTLV-1 RNA was used to program RRL or RRL/G2M in the presence or absence of edeine. These results recapitulate previous findings [21], and confirm that translation of both RLuc and FLuc reporters of the dl HTLV-1 IRES plasmid require 40S scanning since they are both inhibited by 0.25 μM edeine (Fig. 6C). As expected [21], HTLV-1 IRES-mediated translation initiation (Fig. 6C, right panel), but not cap-dependent translation (Fig. 6C, left panel), was increased in RRL/G2M. Importantly, the addition of G2/M HeLa extracts to the RRL did not alter the susceptibility of the HTLV-1 IRES to edeine (Fig. 6C). This result suggest that for the HTLV-1 IRES the presence of cell extracts does not impact on the mechanism used by the IRES to recruit the initiation complex. Similar assays were then performed using in vitro synthesized capped dl HIV-1 IRES RNA. Translation assays were conducted in RRL/G2M in the absence or presence of increasing amounts of edeine. In agreement with our previous observations (Figs. 6A, 6B, 6C and [21]), the presence of edeine negatively impacted on cap-dependent (RLuc) translation (Fig. 6D, left panel). However, in sharp contrast to what was observed for the PV and HTLV-1 IRESs but similar to the HCV IRES (Figs. 6A, 6B and 6C), translation driven by the HIV-1 IRES was not inhibited at a concentration of 0.25 μM of the drug (Fig. 6D, right panel). This observation suggests that the HIV-1 IRES recruits the 40S subunit to the start codon and does not require a scanning mechanism for initiation.
Translation initiation driven by the HIV-1 5′UTR-gag region is insensitive to edeine
An important caveat of the above experiments is that in the dl HIV-1 IRES bicistronic mRNA the HIV-1 IRES is directly followed by the FLuc ORF without any Gag coding region sequences following the start codon. Several reports have suggested that the context of the Gag start codon is important for modulating HIV-1 mRNA translation [20, 26, 28, 40, 58]. To determine whether Gag coding sequences following the initiation codon affected the sensitivity of the HIV-1 IRES to edeine (Fig. 6D), reporters with 18 or 660 nucleotides of the Gag ORF fused 5′ to the FLuc reporter to generate plasmids dl HIV-1 1-353 and dl HIV-1 1-996, respectively. The 18 nucleotides of Gag ORF encode the first amino acids of the matrix (MA) protein, while the 660 nucleotides of Gag ORF encode the full MA protein and part (88 first amino acids) of the capsid (CA) protein. These regions, MA and part of CA, were included in this study since they have been suggested to participate in HIV-1 RNA packaging and are expected to inhibit translation initiation [20, 26, 28, 40, 58]. The capped dl HIV-11-353 and dl HIV-1 1-996 in vitro transcribed RNAs were programmed into RRL/G2M. The RLuc activities were similar for both bicistronic mRNAs indicating that RRL and RRL/G2M were programmed with equivalent amounts of RNA (Fig. 7A, left panels). As shown previously, addition of G2/M HeLa extracts did not impact cap-dependent translation (RLuc activity) (Fig. 6C, left and [29]). The activity of the HIV-1 IRES is reduced when the translation assay is conducted in RRL (Fig. 7A, right panel, compare dl ΔEMCV and dl HIV-1 IRES RNAs), as previously reported [23, 26]. As expected, the addition of G2/M extracts increased IRES activity driven by the HIV-1 5′UTR (Fig. 7A, right panel) [23, 26]. Results show that both the HIV-1 derived 1-353 and 1-996 regions are capable of driving IRES activity in RRL/G2M (Fig. 7A, right panel).
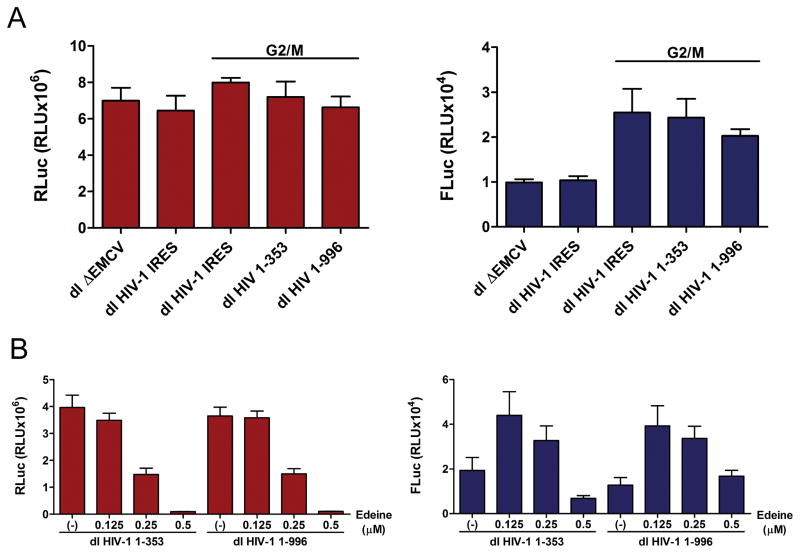
Figure 7. Translation initiation from the HIV-1 5′UTR-Gag construct is insensitive to edeine in RRL/G2M.
(A) The capped dl ΔEMCV, dl HIV-1 IRES, dl HIV-1 1-353 and dl HIV-1 1-996 RNAs were in vitro translated in RRL or RRL/G2M as indicated. The RLuc (left; white bars) and FLuc (right; black bars) luciferase activities (relative light units; RLU) were measured. (B) The in vitro transcribed capped dl HIV-1 1-353 and dl HIV-1 1-996 RNAs were in vitro translated in RRL/G2M in the absence (−) or the presence of increasing concentrations of edeine (0.125, 0.25, or 0.5 μM). RLuc (white bars, left panels) and FLuc (black bars, right panels) activities were measured. The RLuc and FLuc activity for each bicistronic reporter are shown as relative light units (RLU). Values shown are the mean (+/− SEM) for four independent experiments, each conducted in duplicate.
Next, the in vitro synthesized, capped dl HIV-1 1-353 and dl HIV-1 1-996 RNAs were translated in RRL/G2M in the absence or presence of increasing concentrations of edeine. Results show that as before cap-dependent (RLuc) expression was inhibited in the presence of 0.25 μM and 0.5 μM of edeine (Fig. 7B, left panel). In full agreement with the results shown in figure 6D, FLuc translation from the both plasmids remained largely unaffected at 0.25 μM edeine (Fig. 7B, right panel). Together, these results strongly suggest that the HIV-1 IRES recruits the 40S ribosomal subunit directly to the translation initiation codon and that IRES recruitment strategy is not affected by sequence context downstream of the initiation codon.
DISCUSSION
The capped and polyadenylated full-length genomic RNA of HIV-1 encodes for the Gag and Gag-Pol polyproteins [59]. Translation initiation of this mRNA can occur by a canonical cap-dependent [60, 61], a non-canonical cap-dependent eIF4E-independent [62, 63], or through an IRES-dependent mechanism [23, 25, 26, 28, 29, 37, 64]. Interestingly, results suggest that Gag but not Gag-Pol synthesis can be driven following a cap-independent translation initiation mechanism [24]. This potential redundancy of cap-, non canonical-cap, and IRES-dependent mechanisms that drive the synthesis of Gag protein strongly suggests that regulating the synthesis of this viral protein is essential for HIV-1 replication. This possibility is strongly supported by reports showing that in Gag drives most of the steps associated to the late stages of the HIV-1 replication cycle [65]. For example, Gag selects the HIV-1 genomic RNA from the cytoplasm for encapsidation [66–68], directs virion assembly and is sufficient for the organization, budding and release of virus-like particles from cells [65]. After particle budding, Gag is processed within the viral particle giving rise to several structural proteins each of which is required for essential steps during HIV-1 infection [69]. Based on these multiple functions of the Gag polyprotein it is conceivable that the virus has evolved several different strategies to ensure translation of the genomic RNA in order to guarantee the adequate expression of the Gag protein throughout the viral replication cycle. Noteworthy, this potential redundancy of cap- and IRES-dependent mechanisms that drive the synthesis of the HIV-1 Gag protein seems conserved in other retroviruses [6, 21, 22, 24, 26, 27, 43, 60].
Due to the apparent relevance in driving HIV-1 Gag protein synthesis when cap-dependent translation initiation is hindered [23–27, 30], in this study we focused on understanding the mechanism of translation driven by the IRES that is located within the 5′UTR of the HIV-1 full length mRNA (HIV IRES), [25, 26, 28]. To date, the molecular basis of HIV-1 IRES mediated translation initiation remains poorly understood (reviewed in [70, 71]). The HIV-1 IRES is active in HeLa and COS-7 cells (Figs, 1, 2, 3, and 4), although, relative to other viral IRESs its activity is low in HeLa cells (Fig. 1B and [21, 28]). Nevertheless, the HIV-1 IRES activity is 100-fold higher than background and thus HeLa cells can be used to evaluate and characterize the molecular mechanisms driving HIV-1 IRES mediated translation initiation. We also demonstrate that the HIV-1 IRES is active in COS-7 cell lines (Fig. 1 to 4). These results together with previous reports [24, 25, 28], suggest that HIV-1 IRES is active in different cell lines, although its activity varies between different cell types. Although the HIV IRES is more active in T-cell lines [28], they are inefficiently transfected in comparison to what we obtained when using HeLa or COS-7 cells. Although the magnitude of the HIV-1 IRES activity varies when comparing T-cells and HeLa cell lines, the data and conclusions obtained when directly comparing both cell lines are consistent (compare data from [26, 28]), suggesting that the mechanism of HIV-1 IRES mediated translation is the same regardless of the cell line.
Using HeLa and COS-7 cells we established that translation driven by the HIV-1 5′UTR is dependent on eS25 (Fig. 4). Interestingly, to date all evaluated IRES from either cellular or viral origin have demonstrated to be dependent on eS25 for their activity [21, 38, 39]. The stable clonal HeLashS25 cell line, which constitutively expresses an shRNA against eS25 [38] or COS-7 cells that have eS25 knocked down by transfection of an siRNA exhibit a decrease in HIV-1 IRES activity (Fig. 4). The overall impact of eS25 reduction in the HeLashS25 cell line has been well characterized [38]. In fact, knocked down of eS25 does not impact on HeLashS25 cell growth, cell morphology, nor does it have a significant effect on the rate of global translation [38]. What is most relevant to this study, knockdown of eS25 does not alter the rate of cellular cap-dependent translation initiation [38, 39]. Given that depletion of eS25 has no impact on cap-dependent translation initiation (Fig. 4D, and [38, 39]), reporting a reduction in translation initiation driven by the HIV-1 5′UTR alone provides novel evidence in support of the existence of an IRES within this region of the HIV-1 full length mRNA and further validates HIV-1 IRES activity in HeLa and COS-7 cells (Fig. 4). Next, we focused on understanding the molecular mechanism driving HIV-1 IRES initiation. Previous reports have shown that single nucleotide modifications have no apparent impact on HIV-1 IRES function [23, 28, 29]. Additionally, several reports propose that the HIV-1 5′UTR is a structurally plastic RNA capable of adopting several different conformations, consistent with the diverse roles it plays during viral infection [20, 58, 72]. These observations lead us to evaluate the contribution of the well defined RNA structures present in the HIV-1 mRNA 5′UTR on IRES activity. By conducting 5′ and 3′ deletions of the HIV-1 5′UTR Brasey et al. (2003) reported that the poly(A) loop (nucleotide 58–104) was not part of the minimal IRES (nucleotide 104–336), but identified the PBS structure, the SD stem-loop, and the Psi stem-loop as crucial for HIV-1 IRES activity [26]. By also using 5′ and 3′ truncations and mutational analysis Plank et al. (2013) further confirmed these results [28]. Both studies show that when alone the PBS or the SL RNA structures lack IRES activity [26, 28]. Using yet a different approach (Fig. 5), here we identified the structural elements required for optimal IRES activity. These findings fully support the notion of a “core IRES structure” that is defined by the PBS, DIS, and SD RNA structures (Fig. 5) [26, 37].
In a recent study, Plank et al. (2014) established that translation initiation driven by the HIV-1 IRES is eIF4A-dependent. Since eIF4A is an RNA helicase, they suggested that initiation may involve a “land and scan” mechanism of internal initiation, whereby the 40S subunit is recruited upstream of the start codon and scans in a 5′ to 3′ direction to the start codon. However, requirement of eIF4A is not by itself an indication of ribosomal scanning. For example, the EMCV, foot-and-mouth disease virus and Theiler’s murine encephalomyelitis viral IRESs recruit the ribosome directly to the AUG start codon and initiate translation without scanning, yet they require eIF4A for their function [46, 73]. In the case of EMCV, eIF4A induces conformational changes in the 3′border of the IRES to facilitate direct attachment of 43S complexes to the initiation codon [73]. Since, the core IRES elements for HIV are located directly upstream of the Gag AUG initiation codon, this positioning suggested that the IRES may recruit the ribosome directly to the start codon. Therefore, it became apparent that a direct analysis of the mechanism of initiation was required. Using edeine, a drug that inhibits the initiation step of protein synthesis by blocking the recognition of the initiation codon by scanning ribosomes [52, 55, 56, 74] revealed that the 40S ribosomal subunit is directly loaded on the RNA at or close to the Gag initiation codon without scanning (Figs. 6 and 7), in contrast to what was previously suggested [37]. A scanning mechanism of initiation was observed independent of the context of the AUG initiation codon since similar data was obtained when the HIV-1 5′UTR was directly followed by the FLuc reporter coding region or by sequences from the Gag ORF (Figs. 6 and 7). This suggests that the Gag ORF does not affect the mechanism of initiation by the HIV-1 IRES in RRL/G2M.
Together, the findings presented herein give new insights into the molecular basis of HIV-1 IRES function by showing that the HIV-1 IRES is modular in nature with the core RNA elements that participate in IRES activity being the PBS, DIS and SD RNA structures. Importantly, we show for the first time that the HIV-1 IRES activity is dependent on the ribosomal protein eS25, which has been shown to be important for a number of cellular and viral IRESs [21, 38, 39], regardless of their mechanism of ribosome recruitment and positioning on the initiation codon or their dependency on initiation factors. Importantly, eS25 is not essential for cap-dependent translation, which further validates the reporter assays used as reflecting IRES activity, since reporter activity that resulted from artifacts of the reporter such as cryptic promoters, splicing or readthrough would not be affected by eS25 knockdown since these transcripts would be translated cap-dependently. Interestingly, the HIV-1 IRES activity was reduced about 50% without eS25, which is similar to the decrease observed for many cellular IRESs and the picornaviral IRESs [38]. The molecular basis that explains the requirement of eS25 for optimal IRES-mediated translation initiation remains unknown, however, some models have been put forward in previous studies [38, 39]. Further experiments, not included in this report, are needed to fully explain the role of eS25 in HIV-1 IRES-mediated translation initiation.
Following ribosome recruitment, the HIV-1 IRES does not require scanning suggesting that the 40S ribosomal subunit is directly recruited to the initiation codon. Together these findings are consistent with a model in which the viral Gag protein is translated late in infection, when cap-dependent translation is down regulated [24, 27], using a modular IRES, consisting of multiple high affinity binding elements to recruit the ribosome internally directly at the start codon.
MATERIALS AND METHODS
Plasmid construction
The dl ΔEMCV, dl HIV-1 IRES, dl HIV-1 1-996, dl VAR 2 IRES, dl HTLV-1 IRES, dl HCV IRES, dl PV IRES, ΔSV40 dl ΔEMCV, ΔSV40 dl HIV-1 IRES, ΔSV40 dl VAR2 IRES plasmids have been previously described [9, 21, 26, 29, 75]. The dl ΔEMCV was kindly provided by Dr. P. Sarnow (Stanford University, California, USA) and dl HIV-1 IRES, dl HIV-1 1-996 plasmids were kindly provided by N. Sonenberg (McGill University, Montreal, Canada). To construct the dl HIV-1 1-353 plasmid the 5′UTR followed by the first 18 nucleotides of Gag coding region was amplified from the pNL4.3 plasmid (Genbank accession number AF 324493) by PCR using primers 5′-TTTGAAAAACACGAATTCGGTCTCTCTG-3′ and 5′-CCATGGGCGACGCTCTCGCACCCATC-3′. The resulting amplicon was digested with EcoRI (#ER0271, Thermo Fisher Scientific Inc), and NcoI (#ER0571, Thermo Fisher Scientific Inc) enzymes (restriction sites added by PCR) and inserted into the intercistronic region of dl HIV-1 IRES plasmid [26], previously digested with the same restriction enzymes. The new bicistronic vectors harboring the 5′UTR of natural variants were constructed according to [29]. The generation of the deletions mutants of VAR 2 were performed using the primers described in Table 1, using the strategy of overlap extension PCR (OE-PCR) [76]. In this assay EMCVF (5′-TGGATCCCCCCTCTCCCTCCCCCTAACG-3′)andTOEFluc120(5′-GTCCACCTCGATATGTGCATC-3′) were used as external primers. A final amplification was performed using primers HIV1-F (5′-CACGAATTCGGTCTCTCTG-3′) and HIV1-R (5′-CCATGGTCTCTCTCCTTC-3′). The generated amplicon was digested with EcoRI and NcoI (both restriction sites added by PCR) and inserted into the intercistronic region of dl HIV-1 IRES plasmid [26], previously digested with the same enzymes (Thermo Fisher Scientific Inc.). All plasmids were confirmed by sequencing (Macrogen Corp, Rockville, MD, USA).
Table 1.
List of primers used to generate the deletion mutants in the VAR 2 5′UTR background
| Primer Nucleotide Sequence | Deletion | Domain |
|---|---|---|
| F: 5′-GAATTCCACTGCTTAAGCC-3′ | ΔTAR | TAR |
| F: 5′-GGAACCTGTGCCCGTCTGTTG-3′ | ΔPoly(A) | Poly(A) |
| R: 5′-CACAGGTTCCCTAGTCAGCC-3′ | ||
| F: 5′-GCCTCAATTGAGTGCTTAAAG-3′ | ΔPoly(A)-AL | |
| R: 5′-CTCAATTGAGGCTTAAGCAGTGGGTTC-3′ | ||
| F: 5′-GAGGTAGTGTGTGCCCGTC-3′ | ΔD-Loop | |
| R: 5′-CACTACCTCAAGGCAAGCTTTATTG-3′ | ||
| F: 5′-GTGTGAAAGATCTCTCGACGCAG-3′ | ΔPBS | PBS |
| R: 5′-GATCTTTCACACAACAGACGGG-3′ | ||
| F: 5′-GTAGCTAGCAGTGGCGCCCG-3′ | ΔARM | |
| R: 5′-CTGCTAGCTACCAGAGTCAC-3′ | ||
| F: 5′-CCCTCGAAAATCTCTAGCAGTGGC-3′ | ΔShortARM | |
| R: 5′-GATTTTCGAGGGATCTCTAG-3′ | ||
| F: 5′-GACTCTGGAGAGATCCCTCAG-3′ | Δ4N | |
| R: 5′-CTCTCCAGAGTCACACAACAGAC-3′ | ||
| F: 5′-CCAGATCTCTCGACGCAGGACTC-3′ | ΔssPBS-DIS | |
| R: 5′-CGAGAGATCTGGTTTTACTTTC-3′ | ||
| F: 5′-GACCATGGAAGACGCCAAAAACATAAAG-3′ | ΔSL | DIS/SD/Psi |
| R: 5′-TCTTCCATGGTCATCTTCTCTGG-3′ | ||
| F: 5′-CTCGACGCGGCTGGTGAGTACGC-3′ | ΔDIS | DIS |
| R: 5′-CAGCCGCGTCGAGAGATCTTCTC-3′ | ||
| F: 5′-GCTGCGGCAAGAGGCGAGGAG-3′ | ΔDIS-AL | |
| R: 5′-CTTGCCGCAGCAAGCCGAGTCC-3′ | ||
| F: 5′-GAGCGGAATTTTTGACTAGCGGAG-3′ | ΔSD | SD |
| R: 5′-CAAAAATTCCGCTCCTCGCCTCTTGC-3′ | ||
| F: 5′-GCGGCTGCCAATTTTTGACTAGCGGAG-3′ | ΔSD-AL | |
| R: 5′-CAAAAATTGGCAGCCGCCGCTCCTCGC-3′ | ||
| F: 5′-CAATTTTTGAGGAGAGAGACCATGGAAGAC-3′ | ΔPsi | Psi |
| R: 5′-CTCTCCTCAAAAATTGGCGTACTCACC-3′ |
F: Forward primer; R: Reverse primer.
In vitro transcription
The dual luciferase plasmids were digested with XbaI (#ER0681, Thermo Fisher Scientific Inc.) and used as template to generate bicistronic RNAs. Capped RNAs were synthesized using the mMESSAGE mMACHINE® kit (#AM1344 Ambion, Thermo Fisher Scientific Inc.) according to the manufacturer’s protocol. The template DNA was degraded with DNase I, and the RNA was precipitated with 2.5 M LiCl. The RNA was resuspended in nuclease-free water (Integrated DNA Technologies, IDT, Coralville, Iowa, USA). RNA concentrations were determined spectrophotometrically (NanoDrop Technology, Wilmington, Delaware, USA) and RNA integrity was assessed by electrophoresis on denaturing formaldehyde (6.66%) agarose gel in 1X MOPS (40mM MOPS, pH 7.0, 10mM Sodium Acetate, 1mM EDTA) buffer.
In vitro translation
In vitro translations were carried out in nuclease treated rabbit reticulocyte lysate (RRL #L496, Promega Corporation, Madison, USA), at 35 % (v/v), supplemented with 20 mM amino acids (Promega), 0.8 U/μl Ribolock RNase inhibitor (#EO0381, Thermo Fisher Scientific Inc.) and 1 ng/μl of capped RNA. When HeLa cell extracts (S10) prepared as described in [35] were used, bicistronic RNA was pre-incubated for 10 min with the cell extracts prior to the addition of the RRL translation mix. For in vitro translation reactions conducted in the presence of edeine, a kind gift from Dr. I. Brierley (Division of Virology, Department of Pathology, University of Cambridge, United Kingdom), RRL was incubated at 30 °C for 10 min in presence (ranging from 0.125–0.5 μM) or absence of edeine, prior to addition of bicistronic RNAs used for in vitro translations assay as previously described [21]. The optimal salt concentration used to in vitro translate the dl HCV IRES (100 mM KCl and 0.75 mM MgCl2), dl PV IRES (80 mM KOAc and 0.25 mM MgOAc), dl HTLV-1 IRES (60 mM KOAc and 0.25 mM MgOAc), and dl HIV IRES (120 mM KOAc and 0.5 mM MgOAc) mRNAs were previously described [21, 23, 26, 75, Barria, 2009 #20, 77]. All translation reactions were incubated at 30 °C for 90 min. Renilla luciferase (RLuc) and Firefly luciferase (FLuc) activities were measured using the DLR™ Assay System (Promega) according to manufacturer’s instructions on a Sirius Single Tube Luminometer Lumat 9507 (Berthold Detection Systems GmbH, Pforzheim, Germany).
Cell culture
HeLa cells (ATCC, CCL-2TM), COS-7 cells (ATCC® CRL-1651), and HeLaShV and HeLashS25 cell clones from transduced cell lines [38] were cultured in Dulbecco’s modified Eagle’s medium (DMEM; #SH30022, HyClone, GE Healthcare Life Sciences, Logan, Utah, USA), 1 % penicilin-streptomicin (1000 U/mL) (#SV30010, Hyclone, GE Healthcare Life Sciences), 2.5 μg/ml amphotericin B (Fungizone, #SV30078.01, Hyclone, GE Healthcare Life Sciences), and 10 % fetal bovine serum (#SH30910 Hyclone, GE Healthcare Life Sciences) at 37 °C in a 5 % CO2 atmosphere.
DNA transfection
Cells were seeded at 1 ×105 cells/well in 12-well culture plates or at 5 ×104 cells per well in a 24 well plate on the day prior to transfection. For the bicistronic reporter assays DNA transfections were performed on cells that were 60 to 80 % confluent using polyethyleneimine (PEI, GIBCO, ThermoFisher Scientific Inc, Waltham, Massachusetts, USA) or Lipofectamine 2000 (#11668019, Invitrogen, ThermoFisher Scientific Inc) according to manufacturer’s protocols. Unless indicated in the text, cells were transfected with 200 ng bicistronic DNA plasmids together with 50 ng of pcDNA 3.1-LacZ plasmid, encoding the β-galactosidase enzyme, used as a control for the transfection efficiency. For transfection into the HeLashV and HeLashS25 cell lines, 400 ng of bicistronic DNA, 400 ng of pcDNA 3.1-LacZ plasmid, and 400 ng of either pSRT376 (human eS25 rescue plasmid) [38] or the empty vector pcDNA3.1 (Invitrogen, ThermoFisher Scientific Inc) were co-transfected. 24 hours post-transfection, the cells were washed once with phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 · 7H2O, 1.4 mM KH2PO4, pH 7.4) and lysed with 100 μL lysis buffer (100 mM potassium phosphate pH 7.8, 0.2 % Triton X-100, Applied Biosystems, ThermoFisher Scientific Inc). For the eS25 rescue experiments luciferase assays were performed 48 h post transfection to allow sufficient time for eS25 synthesis and incorporation into ribosomes [38]. The IRES activity (Firefly luciferase) was measured as above and normalized to the cap-dependent translation of β-galactosidase and expressed as a percentage of the activity of HeLashV cells with the empty vector. All assays were performed three independent times in triplicate. The activity of β-galactosidase was measured from the same lysates using the Beta-Glo™ Assay System according to manufacturer’s protocols (#E4720, Promega Corporation) or Galacto-light Plus kit (#T1007, Applied Biosystems, ThermoFisher Scientific Inc) according to the manufacturer’s protocols.
siRNA transfection
HeLa or COS-7 cells were seeded at 5.5 ×104 cells per well in a 24-well culture plate. Transfections were performed at 70–80 % confluency using the Lipofectamine 2000 system (#11668019, Invitrogen, ThermoFisher Scientific Inc) according to the manufacturer’s protocol. 100 or 200 ng of dl HIV-1 or dl VAR2 plasmid was co-transfected with different final concentrations (50, 100 or 250 nM) of an siRNA that targets the RLuc open reading frame (Renilla siRNA, 5′UAUAAGAACCAUUACCAGAUUUGCCUG-3′; IDT, Coralville, Iowa, USA) or with a Negative Control siRNA (# 4390844, Silencer Select Negative N°1 Ambion, ThermoFisher Scientific Inc) at the same concentrations. The RLuc and FLuc activities were measured after 12h as described above. siRNAs 5′-GGACUUAUCAAACUGGUUUtt-3′ and 5′-AAACCAGUUUGAUAAG-UCCtt-3′ (siRNA identification number 142220; Ambion; the TTDNA overhang is in lowercase) were used to knockdown eS25 in COS-7 cells. The Dicector DS scrambled negative-control duplex was used as a negative control (Integrated DNA Technologies, IDT, Coralville, Iowa, USA). First, 200nM siRNA was transfected into 6.5 × 105 COS-7 cells using the Neon® Transfection System (Life Technologies, ThermoFisher Scientific Inc). 24 hours post-transfection cells were plated into a 24-well plate at a concentration of 2.5 × 104 cells/well. 24h later, a second knockdown with 200 nM siRNA was performed in conjunction with a co-transfection using 0.5 μg DNA per well consisting of the dual luciferase reporter dl HIV-1 IRES or dl VAR2 IRES and the pcDNA 3.1-LacZ using Lipofectamine 2000 (Invitrogen, ThermoFisher Scientific Inc), according to the manufacturer’s recommendations. 48 h after the second transfection, cells were lysed in 1X PBS and luciferase (Promega Corporation) and β-galactosidase assays (Applied Biosystems, ThermoFisher Scientific Inc) were performed according to the manufacturer’s protocol.
Western analysis
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1 % Nonidet P-40, 0.5 % Sodium Deoxycholate, 1mM EDTA, 0.1 % SDS). 10 μg of total protein was separated by 16 % Tricine SDS-PAGE and transferred onto an Immobilon-FL polyvinylidene difluoridemembrane (#IPFL00010, EMD Millipore Co, Milford, MA), using the Genie wet transfer system (Idea Scientific Company, Minneapolis, MN). Membranes were probed with either the mouse monoclonal eS25 (generated in house at the UAB Analytical Imaging and Immunoreagent Core) or the β-actin (#200-301-E69S, Rockland Immunochemical Inc, Limerick, PA) primary antibodies and the fluorochrome-conjugated secondary anti-mouse antibody (Li-Cor, Lincoln, NE). Westerns were visualized and quantitated using Odyssey scanner and software (Li-Cor, Lincoln, NE).
Acknowledgments
We are grateful to Drs. N. Sonenberg (McGill University, Montreal, Canada), and P. Sarnow (Stanford University, California, USA) for kindly providing the plasmids used in this study. We thank Dr. I. Brierley (University of Cambridge, Cambridge, UK) for sharing the drug edeine. We thank the UAB Analytical Imaging and Immunoreagent Core for generation of the monoclonal eS25 antibody. This work was supported by grant FONDECYT 1130270 from the Fondo Nacional de Ciencia y Tecnología del Gobierno de Chile, P09/016-F of Iniciativa Científica Milenio del Ministerio de Economía, Fomento y Turismo, and the Poyecto Anillo ACT1408 to MLL; the National Institutes of Health (grants R01GM084547 and 3R01GM084547-01A1S1 to SRT) and a UAB Cancer Center HIV-Associated Malignancy pilot research grant (UAB Comprehensive Cancer Center core support grant P30 CA13148) to SRT. The monoclonal eS25 antibody was generated as part of Grant RDCC P30 AR48311 to SRT. MV conducted this work as a Postdoctoral fellow of the Instituto Milenio funded by grant P09/016-F de la Iniciativa Científica Milenio del Ministerio de Economía, Fomento y Turismo. While developing this study FC, NC, CJC, were Ph.D. fellows from the Comisión Nacional de Ciencia y Tecnología de Chile (CONICYT). EO was supported by UCH0604 MECESUP-USACH doctoral fellowships. AL was initially supported by a VRAI fellowship granted by the Pontificia Universidad Católica de Chile followed by a CONICYT fellowship.
Abbreviations
- IRES
internal ribosomal entry site
- 5′UTR
5′ untranslated region
- HIV-1
human immunodeficiency virus type 1
- AIDS
acquired immunodeficiency syndrome
- HTLV-1
Human T Cell Lymphotropic Virus Type 1
- PV
poliovirus
- EMCV
encephalomyocarditis virus
- HCV
hepatitis C virus
- SV40
simian virus 40
- RRL
rabbit reticulocyte lysate
- G2/M
G2/M cell cycle stage arrested cells extract
- eIFs
eukaryotic translation initiation factors
- TAR
trans-activating region
- Poly(A)
polyadenylation domain
- PBS
primer binding site domain
- DIS
dimerization domain
- SD
splicing donor domain
- Psi
encapsidation domain
- SL
stem-loop region
- “dl”
bicistronic reporter vector
- RLuc
Renilla luciferase
- FLuc
Firefly luciferase
- RTA
relative translation activity
- RLA
relative luciferase activity
- PEI
polyethyleneimine
- UV
ultraviolet
- ORF
open reading frame
- eS25
ribosomal protein S25
- shRNA
small hairpin RNA
- siRNA
short interfering RNA
- SC
nonrelated scrambled siRNA
Footnotes
AUTHOR CONTRIBUTION STATEMENT
MLL, FC, MV, SRT planned experiments. FC, MV, AL, constructed plasmids used in the study. NC conducted and analyzed the RLuc siRNA experiments. BAW, MIH and SRT, conducted and analyzed all experiments regarding eS25 and confirmed the RLuc siRNA experiments. EO conducted and analyzed the experiments using edeine. CJC evaluated and analyzed data regarding the cryptic promoter activity and confirmed the edeine experiments. FC, MV, KP, conducted and repeated experiments in the different cell lines. MLL integrated all the data and wrote the first draft of the manuscript. SRT edited the manuscript. All authors corrected the manuscript.
References
- 1.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. nrm2838 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 4.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 5.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balvay L, Lopez Lastra M, Sargueil B, Darlix JL, Ohlmann T. Translational control of retroviruses. Nat Rev Microbiol. 2007;5:128–140. doi: 10.1038/nrmicro1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths A, Coen DM. An unusual internal ribosome entry site in the herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci U S A. 2005;102:9667–9672. doi: 10.1073/pnas.0504132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasaki J, Nakashima N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J Virol. 1999;73:1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson JE, Powell MJ, Hoover SE, Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol Cell Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong SM, Koh DC, Liu D. Identification of plant virus IRES. Methods Mol Biol. 2008;451:125–133. doi: 10.1007/978-1-59745-102-4_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Roberts R, Rakotondrafara AM. The role of the 5′ untranslated regions of Potyviridae in translation. Virus Res. 2015;206:74–81. doi: 10.1016/j.virusres.2015.02.005. S0168-1702(15)00058-1 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Gallie DR. Cap-independent translation conferred by the 5′ leader of tobacco etch virus is eukaryotic initiation factor 4G dependent. J Virol. 2001;75:12141–12152. doi: 10.1128/JVI.75.24.12141-12152.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez-Lastra M, Ulrici S, Gabus C, Darlix JL. Identification of an internal ribosome entry segment in the 5′ region of the mouse VL30 retrotransposon and its use in the development of retroviral vectors. J Virol. 1999;73:8393–8402. doi: 10.1128/jvi.73.10.8393-8402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meignin C, Bailly JL, Arnaud F, Dastugue B, Vaury C. The 5 ′ untranslated region and gag product of Idefix, a long terminal repeat-retrotransposon from Drosophila melanogaster, act together to initiate a switch between translated and untranslated states of the genomic mRNA. Molecular and Cellular Biology. 2003;23:8246–8254. doi: 10.1128/Mcb.23.22.8246-8254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ronfort C, De Breyne S, Sandrin V, Darlix JL, Ohlmann T. Characterization of two distinct RNA domains that regulate translation of the Drosophila gypsy retroelement. RNA. 2004;10:504–515. doi: 10.1261/rna.5185604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229–240. doi: 10.4161/cc.10.2.14472. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res Mol Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 18.Paillart JC, Dettenhofer M, Yu XF, Ehresmann C, Ehresmann B, Marquet R. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J Biol Chem. 2004;279:48397–48403. doi: 10.1074/jbc.M408294200. M408294200 [pii] [DOI] [PubMed] [Google Scholar]
- 19.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. 07-PLBI-RA-2987 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, et al. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science. 2011;334:242–245. doi: 10.1126/science.1210460. 334/6053/242 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olivares E, Landry DM, Caceres CJ, Pino K, Rossi F, Navarrete C, Huidobro-Toro JP, Thompson SR, Lopez-Lastra M. The 5′ untranslated region of the human T-cell lymphotropic virus type 1 mRNA enables cap-independent translation initiation. J Virol. 2014;88:5936–5955. doi: 10.1128/JVI.00279-14. JVI.00279-14 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vallejos M, Ramdohr P, Valiente-Echeverria F, Tapia K, Rodriguez FE, Lowy F, Huidobro-Toro JP, Dangerfield JA, Lopez-Lastra M. The 5′-untranslated region of the mouse mammary tumor virus mRNA exhibits cap-independent translation initiation. Nucleic Acids Res. 2010;38:618–632. doi: 10.1093/nar/gkp890. gkp890 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vallejos M, Deforges J, Plank TD, Letelier A, Ramdohr P, Abraham CG, Valiente-Echeverria F, Kieft JS, Sargueil B, Lopez-Lastra M. Activity of the human immunodeficiency virus type 1 cell cycle-dependent internal ribosomal entry site is modulated by IRES trans-acting factors. Nucleic Acids Res. 2011;39:6186–6200. doi: 10.1093/nar/gkr189. gkr189 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monette A, Valiente-Echeverria F, Rivero M, Cohen EA, Lopez-Lastra M, Mouland AJ. Dual mechanisms of translation initiation of the full-length HIV-1 mRNA contribute to gag synthesis. PLoS One. 2013;8:e68108. doi: 10.1371/journal.pone.0068108. PONE-D-13-12498 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gendron K, Ferbeyre G, Heveker N, Brakier-Gingras L. The activity of the HIV-1 IRES is stimulated by oxidative stress and controlled by a negative regulatory element. Nucleic Acids Res. 2011;39:902–912. doi: 10.1093/nar/gkq885. gkq885 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brasey A, Lopez-Lastra M, Ohlmann T, Beerens N, Berkhout B, Darlix JL, Sonenberg N. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J Virol. 2003;77:3939–3949. doi: 10.1128/JVI.77.7.3939-3949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amorim R, Costa SM, Cavaleiro NP, da Silva EE, da Costa LJ. HIV-1 transcripts use IRES-initiation under conditions where Cap-dependent translation is restricted by poliovirus 2A protease. PLoS One. 2014;9:e88619. doi: 10.1371/journal.pone.0088619. PONE-D-13-10338 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plank TD, Whitehurst JT, Kieft JS. Cell type specificity and structural determinants of IRES activity from the 5′ leaders of different HIV-1 transcripts. Nucleic Acids Res. 2013;41:6698–6714. doi: 10.1093/nar/gkt358. gkt358 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vallejos M, Carvajal F, Pino K, Navarrete C, Ferres M, Huidobro-Toro JP, Sargueil B, Lopez-Lastra M. Functional and Structural Analysis of the Internal Ribosome Entry Site Present in the mRNA of Natural Variants of the HIV-1. PLoS One. 2012;7:e35031. doi: 10.1371/journal.pone.0035031. PONE-D-11-19755 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monette A, Ajamian L, Lopez-Lastra M, Mouland AJ. Human immunodeficiency virus type 1 (HIV-1) induces the cytoplasmic retention of heterogeneous nuclear ribonucleoprotein A1 by disrupting nuclear import: implications for HIV-1 gene expression. J Biol Chem. 2009;284:31350–31362. doi: 10.1074/jbc.M109.048736. M109.048736 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alvarez E, Castello A, Menendez-Arias L, Carrasco L. HIV protease cleaves poly(A)-binding protein. Biochem J. 2006;396:219–226. doi: 10.1042/BJ20060108. BJ20060108 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castello A, Franco D, Moral-Lopez P, Berlanga JJ, Alvarez E, Wimmer E, Carrasco L. HIV-1 protease inhibits Cap- and poly(A)-dependent translation upon eIF4GI and PABP cleavage. PLoS One. 2009;4:e7997. doi: 10.1371/journal.pone.0007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohlmann T, Prevot D, Decimo D, Roux F, Garin J, Morley SJ, Darlix JL. In vitro cleavage of eIF4GI but not eIF4GII by HIV-1 protease and its effects on translation in the rabbit reticulocyte lysate system. J Mol Biol. 2002;318:9–20. doi: 10.1016/S0022-2836(02)00070-0. S0022-2836(02)00070-0 [pii] [DOI] [PubMed] [Google Scholar]
- 34.Perales C, Carrasco L, Ventoso I. Cleavage of eIF4G by HIV-1 protease: effects on translation. FEBS Lett. 2003;533:89–94. doi: 10.1016/s0014-5793(02)03764-x. [pii] [DOI] [PubMed] [Google Scholar]
- 35.Rivas-Aravena A, Ramdohr P, Vallejos M, Valiente-Echeverria F, Dormoy-Raclet V, Rodriguez F, Pino K, Holzmann C, Huidobro-Toro JP, Gallouzi IE, et al. The Elav-like protein HuR exerts translational control of viral internal ribosome entry sites. Virology. 2009;392:178–185. doi: 10.1016/j.virol.2009.06.050. S0042-6822(09)00412-7 [pii] [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Henao-Mejia J, Liu H, Zhao Y, He JJ. Translational regulation of HIV-1 replication by HIV-1 Rev cellular cofactors Sam68, eIF5A, hRIP, and DDX3. J Neuroimmune Pharmacol. 2011;6:308–321. doi: 10.1007/s11481-011-9265-8. [DOI] [PubMed] [Google Scholar]
- 37.Plank TD, Whitehurst JT, Cenic R, Pelletier J, Kieft JS. Internal translation initiation from HIV-1 transcripts is conferred by a common RNA structure. Translation. 2014:e27694. doi: 10.4161/trla.27694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hertz MI, Landry DM, Willis AE, Luo G, Thompson SR. Ribosomal protein S25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol Cell Biol. 2013;33:1016–1026. doi: 10.1128/MCB.00879-12. MCB.00879-12 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landry DM, Hertz MI, Thompson SR. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009;23:2753–2764. doi: 10.1101/gad.1832209. 23/23/2753 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valiente-Echeverria F, Vallejos M, Monette A, Pino K, Letelier A, Huidobro-Toro JP, Mouland AJ, Lopez-Lastra M. A cis-acting element present within the Gag open reading frame negatively impacts on the activity of the HIV-1 IRES. PLoS One. 2013;8:e56962. doi: 10.1371/journal.pone.0056962. PONE-D-12-32529 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miele G, Mouland A, Harrison GP, Cohen E, Lever AM. The human immunodeficiency virus type 1 5′ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J Virol. 1996;70:944–951. doi: 10.1128/jvi.70.2.944-951.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smirnova VV, Terenin IM, Khutornenko AA, Andreev DE, Dmitriev SE, Shatsky IN. Does HIV-1 mRNA 5′-untranslated region bear an internal ribosome entry site? Biochimie. 2016;121:228–237. doi: 10.1016/j.biochi.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 43.Ohlmann T, Lopez-Lastra M, Darlix JL. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J Biol Chem. 2000;275:11899–11906. doi: 10.1074/jbc.275.16.11899. [DOI] [PubMed] [Google Scholar]
- 44.Barria MI, Gonzalez A, Vera-Otarola J, Leon U, Vollrath V, Marsac D, Monasterio O, Perez-Acle T, Soza A, Lopez-Lastra M. Analysis of natural variants of the hepatitis C virus internal ribosome entry site reveals that primary sequence plays a key role in cap-independent translation. Nucleic Acids Res. 2009;37:957–971. doi: 10.1093/nar/gkn1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fitzgerald KD, Semler BL. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim Biophys Acta. 2009;1789:518–528. doi: 10.1016/j.bbagrm.2009.07.004. S1874-9399(09)00081-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kieft JS. Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci. 2008;33:274–283. doi: 10.1016/j.tibs.2008.04.007. S0968-0004(08)00091-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chappell SA, Edelman GM, Mauro VP. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc Natl Acad Sci U S A. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chappell SA, LeQuesne JP, Paulin FE, deSchoolmeester ML, Stoneley M, Soutar RL, Ralston SH, Helfrich MH, Willis AE. A mutation in the c-myc-IRES leads to enhanced internal ribosome entry in multiple myeloma: a novel mechanism of oncogene de-regulation. Oncogene. 2000;19:4437–4440. doi: 10.1038/sj.onc.1203791. [DOI] [PubMed] [Google Scholar]
- 49.Owens GC, Chappell SA, Mauro VP, Edelman GM. Identification of two short internal ribosome entry sites selected from libraries of random oligonucleotides. Proc Natl Acad Sci U S A. 2001;98:1471–1476. doi: 10.1073/pnas.98.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jang GM, Leong LE, Hoang LT, Wang PH, Gutman GA, Semler BL. Structurally distinct elements mediate internal ribosome entry within the 5′-noncoding region of a voltage-gated potassium channel mRNA. J Biol Chem. 2004;279:47419–47430. doi: 10.1074/jbc.M405885200. [DOI] [PubMed] [Google Scholar]
- 51.Filbin ME, Kieft JS. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol. 2009;19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dinos G, Wilson DN, Teraoka Y, Szaflarski W, Fucini P, Kalpaxis D, Nierhaus KH. Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin: the universally conserved residues G693 and C795 regulate P-site RNA binding. Mol Cell. 2004;13:113–124. doi: 10.1016/s1097-2765(04)00002-4. [pii] [DOI] [PubMed] [Google Scholar]
- 53.Obrig T, Irvin J, Culp W, Hardesty B. Inhibition of peptide initiation on reticulocyte ribosomes by edeine. Eur J Biochem. 1971;21:31–41. doi: 10.1111/j.1432-1033.1971.tb01436.x. [DOI] [PubMed] [Google Scholar]
- 54.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hunter AR, Jackson RJ, Hunt T. The role of complexes between the 40-S ribosomal subunit and Met-tRNA-Met-f in the initiation of protein synthesis in the wheat-germ system. Eur J Biochem. 1977;75:159–170. doi: 10.1111/j.1432-1033.1977.tb11513.x. [DOI] [PubMed] [Google Scholar]
- 56.Lancaster AM, Jan E, Sarnow P. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA. 2006;12:894–902. doi: 10.1261/rna.2342306. rna.2342306 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keane SC, Heng X, Lu K, Kharytonchyk S, Ramakrishnan V, Carter G, Barton S, Hosic A, Florwick A, Santos J, et al. RNA structure. Structure of the HIV-1 RNA packaging signal. Science. 2015;348:917–921. doi: 10.1126/science.aaa9266. 348/6237/917 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cochrane AW, McNally MT, Mouland AJ. The retrovirus RNA trafficking granule: from birth to maturity. Retrovirology. 2006;3:18. doi: 10.1186/1742-4690-3-18. 1742-4690-3-18 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ricci EP, Soto Rifo R, Herbreteau CH, Decimo D, Ohlmann T. Lentiviral RNAs can use different mechanisms for translation initiation. Biochem Soc Trans. 2008;36:690–693. doi: 10.1042/BST0360690. BST0360690 [pii] [DOI] [PubMed] [Google Scholar]
- 61.Berkhout B, Arts K, Abbink TE. Ribosomal scanning on the 5′-untranslated region of the human immunodeficiency virus RNA genome. Nucleic Acids Res. 2011;39:5232–5244. doi: 10.1093/nar/gkr113. gkr113 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharma A, Yilmaz A, Marsh K, Cochrane A, Boris-Lawrie K. Thriving under stress: selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathog. 2012;8:e1002612. doi: 10.1371/journal.ppat.1002612. PPATHOGENS-D-11-01322 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soto-Rifo R, Rubilar PS, Limousin T, de Breyne S, Decimo D, Ohlmann T. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 2012;31:3745–3756. doi: 10.1038/emboj.2012.220. emboj2012220 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buck CB, Shen X, Egan MA, Pierson TC, Walker CM, Siliciano RF. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J Virol. 2001;75:181–191. doi: 10.1128/JVI.75.1.181-191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gottlinger HG. The HIV-1 assembly machine. AIDS. 2001;15(Suppl 5):S13–20. doi: 10.1097/00002030-200100005-00003. [DOI] [PubMed] [Google Scholar]
- 66.Anderson EC, Lever AM. Human immunodeficiency virus type 1 Gag polyprotein modulates its own translation. J Virol. 2006;80:10478–10486. doi: 10.1128/JVI.02596-05. 80/21/10478 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Levesque K, Halvorsen M, Abrahamyan L, Chatel-Chaix L, Poupon V, Gordon H, DesGroseillers L, Gatignol A, Mouland AJ. Trafficking of HIV-1 RNA is mediated by heterogeneous nuclear ribonucleoprotein A2 expression and impacts on viral assembly. Traffic. 2006;7:1177–1193. doi: 10.1111/j.1600-0854.2006.00461.x. [DOI] [PubMed] [Google Scholar]
- 68.Poole E, Strappe P, Mok HP, Hicks R, Lever AM. HIV-1 Gag-RNA interaction occurs at a perinuclear/centrosomal site; analysis by confocal microscopy and FRET. Traffic. 2005;6:741–755. doi: 10.1111/j.1600-0854.2005.00312.x. TRA312 [pii] [DOI] [PubMed] [Google Scholar]
- 69.Scarlata S, Carter C. Role of HIV-1 Gag domains in viral assembly. Biochim Biophys Acta. 2003;1614:62–72. doi: 10.1016/s0005-2736(03)00163-9. [pii] [DOI] [PubMed] [Google Scholar]
- 70.Ohlmann T, Mengardi C, Lopez-Lastra M. Translation initiation of the HIV-1 mRNA. Translation. 2014;2:e960242. doi: 10.4161/2169074X.2014.960242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guerrero S, Batisse J, Libre C, Bernacchi S, Marquet R, Paillart JC. HIV-1 replication and the cellular eukaryotic translation apparatus. Viruses. 2015;7:199–218. doi: 10.3390/v7010199. v7010199 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huthoff H, Berkhout B. Multiple secondary structure rearrangements during HIV-1 RNA dimerization. Biochemistry. 2002;41:10439–10445. doi: 10.1021/bi025993n. [pii] [DOI] [PubMed] [Google Scholar]
- 73.Kolupaeva VG, Lomakin IB, Pestova TV, Hellen CU. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol Cell Biol. 2003;23:687–698. doi: 10.1128/MCB.23.2.687-698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kozak M, Shatkin AJ. Migration of 40 S ribosomal subunits on messenger RNA in the presence of edeine. The Journal of biological chemistry. 1978;253:6568–6577. [PubMed] [Google Scholar]
- 75.Poulin F, Gingras AC, Olsen H, Chevalier S, Sonenberg N. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J Biol Chem. 1998;273:14002–14007. doi: 10.1074/jbc.273.22.14002. [DOI] [PubMed] [Google Scholar]
- 76.Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. nprot.2007.132 [pii] [DOI] [PubMed] [Google Scholar]
- 77.Dorner AJ, Semler BL, Jackson RJ, Hanecak R, Duprey E, Wimmer E. In vitro translation of poliovirus RNA: utilization of internal initiation sites in reticulocyte lysate. J Virol. 1984;50:507–514. doi: 10.1128/jvi.50.2.507-514.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]