Abstract
Impulsivity is a multi-faceted construct that, while characterized by a set of correlated dimensions, is centered around a core definition that involves acting suddenly in an unplanned manner without consideration for the consequences of such behavior. Several psychiatric disorders include impulsivity as a criterion, and thus it has been suggested that it may link a number of different behavioral disorders, including substance abuse. Native Americans experience some of the highest rates of substance abuse of all US ethnic groups. The described analyses used data from a low coverage whole genome sequence scan to conduct a genome-wide association study of an impulsivity phenotype in an American Indian community sample (n=658). Demographic and clinical information were obtained using a semi-structured interview. Impulsivity was assessed using a scale derived from the Maudsley personality inventory that combines both novelty-seeking and lack of planning items. The impulsivity score was tested for association with each variant adjusted for demographic variables, and corrected for ancestry and kinship, using EMMAX. Simulations were conducted to calculate empirical p-values. Genome-wide significant findings were observed for a variant 50 kb upstream from catenin cadherin-associated protein, alpha 2 (CTNNA2), a neuronal specific catenin, in the REG gene cluster. A meta-analysis of genome-wide association studies had previously identified common variants in CTNNA2 as being associated with excitement seeking. A second locus upstream of NEIL3 on chromosome 4 also achieved genome-wide significance. The association between sequence variants in these regions suggests their potential roles in the genetic regulation of this phenotype in this population.
Keywords: American Indian, association analyses, catenin cadherin-associated protein, chromosome 2, genetics, impulsivity, Native American, single nucleotide polymorphisms, substance dependence, whole genome sequencing
Introduction
Impulsivity is a multi-faceted construct that has been hypothesized to represent a core aspect of personality and psychopathology. Most commonly, impulsivity has been defined as a set of correlated dimensions that include a desire to engage in novel and/or thrill-seeking behavior, impulsive behavior associated with attempts to relieve negative emotion, as well as a heightened responsiveness to short-term rewards without consideration of long-term consequences (Whiteside & Lynam, 2001; Sharma et al., 2014). The latter aspect is of particular interest because it is most closely linked to the construct of disinhibition that has been posited as a core feature of the externalizing spectrum disorders that include alcohol and other substance use disorders, attention-deficit hyperactivity disorder (ADHD), conduct disorder, and antisocial personality disorder. As a further reflection of this, several psychiatric disorders defined in the Diagnostic and Statistical Manual of Psychiatric Disorder, 5th edition (DSM-5) (American Psychiatric Association, 2013) include impulsivity as a core feature, including the disorders mentioned above as well as borderline personality disorder (which shows relations with multiple aspects of impulsivity), pathological gambling, pyromania and kleptomania, paraphilias, and others. Thus, it has been suggested that specific facets of impulsivity may represent important endophenotypes for the molecular genetic study of these disorders (Kreek et al., 2005).
Twin studies have provided evidence to suggest that externalizing disorders may share a genetic vulnerability that includes impulsivity-related personality constructs, such as behavioral undercontrol and disinhibited personality (Krueger et al., 2002; Slutske, 2001; Young et al., 2000). Quantitative genetic studies have also demonstrated that about 30–50% of the variance in these impulsivity-related constructs is heritable (Bezdjian et al., 2011), yet the specific genetic variants that contribute to impulsivity-related traits remain largely unknown (Gizer et al., 2015). Candidate gene studies have identified a few variants that have been significantly associated with impulsivity, including variants located in or near genes related to dopaminergic and serotonergic function (Benko et al., 2010; Congdon et al., 2008). In an American Indian community, impulsivity was also significantly associated with the cannabinoid receptor gene CNR1 (Ehlers et al., 2007). However, the findings reported in this and in the other described studies did not achieve significance when correcting for multiple comparisons at the genome-wide level. In addition, of the few genome-wide association studies (GWAS) of trait impulsivity which have been published, only one reported a genome-wide significant result. That study conducted a multi-site GWAS analysis of the impulsivity subfacet related to “excitement-seeking,” and reported that a single variant located in the catenin cadherin-associated protein, alpha 2 (CTNNA2) gene achieved genome-wide significance (Terracciano et al., 2011). This finding demonstrates the potential of molecular genetic studies to identify genetic variants related to impulsivity-related traits.
In recent years, the development and refinement of next generation sequencing technologies has made their application to the study of complex traits more feasible, and importantly, this technology has the potential to further our understanding of the genetic architecture underlying these traits beyond what has been obtained from GWA studies using microarrays. For example, GWAS microarrays have been primarily designed to measure common genetic variants (i.e., minor allele frequencies [MAF] > 0.05), and thus, are not well-positioned to capture genetic variants with lower allele frequencies (Nelson et al., 2013). In contrast, sequencing technologies, which directly interrogate each variant, do not have this limitation. Further, GWAS microarrays have typically been designed to capture common variation in individuals of specific ancestral groups (e.g., European ancestry), and as a result, may yield reduced coverage when studying populations outside of these groups (e.g., American Indians).
The present report is part of a larger study exploring risk factors for substance dependence in a Native American Indian community sample (Ehlers et al., 2004a,b). The lifetime prevalence of substance dependence in this Indian population is high and evidence for heritability and linkage to specific chromosome locations and associations with candidate genes have been demonstrated (see Ehlers & Gizer, 2013). DNA obtained from this community sample has recently been sequenced using low coverage whole genome sequencing (LWGS) (see Bizon et al., 2014). Given the described evidence suggesting that facets of impulsivity may represent an important endophenotype for the study of a number of mental disorders, including alcohol and other substance use disorders, the aim of the present study was to conduct an association analysis of a specific impulsivity phenotype using the LWGS data.
Materials and Methods
Participants
American Indian participants were recruited from eight geographically contiguous reservations with a total population of about 3,000 individuals, Participants were recruited using a combination of a venue-based method for sampling hard-to-reach populations and a respondent-driven procedure which has been described elsewhere (Gilder et al., 2004). To be included in the study, participants had to report at least 1/16th Native American heritage, be between the ages of 18 and 85 years, and be mobile enough to be transported from his or her home to The Scripps Research Institute (TSRI). The protocol for the study was approved by the Institutional Review Board (IRB) of TSRI, and the Indian Health Council, a tribal review group overseeing health issues for the reservations where recruitment was undertaken. Written informed consent was obtained from each participant after the study was fully explained.
Measures
Impulsivity, as defined in the present study, was assessed using a scale drawn from the Maudsley personality inventory (MPI; Eysenck, 1959) that was described by Eysenck and colleagues in subsequent studies (e.g., Eaves & Eysenck, 1975). Notably, the MPI was later revised as the Eysenck Personality Inventory (Eysenck & Eysenck, 1964) and Eysenck Personality Questionnaire (Eysenck & Eysenck, 1975), which have been used widely as measures of personality. The following 7 items were positively keyed: Do you long for excitement? Are you usually carefree? Do you generally do and say things quickly without stopping to think? Would you do almost anything for a dare? Do you often do things on the spur of the moment? When people shout at you, do you shout back? Do you like doing things in which you have to act quickly? The following 2 items were negatively keyed: Do you stop and think things over before doing anything? Are you slow and unhurried in the way you move? The resulting score (0–9) was used in the genetic analyses.
The described items were initially taken from the Extraversion scale of the MPI, and are also contained within the Extraversion or Psychoticism dimensions in later versions of the questionnaire. As is evident, most of the items reflect a lack of planning, and thus are most closely related to the ‘lack of planning’ dimension of the widely used UPPS Impulsive Behavior Scale (Whiteside & Lynam, 2001). Nonetheless, some items also relate to excitement-seeking. As a result, the internal consistency of the scale is less than optimal (α=0.60; Eaves & Eysenck, 1975); however, as noted by the authors, investigations into the factor structure of this scale resulted in a highly correlated set of facets that did not provide a parsimonious solution. For this reason, the original scale was retained as the phenotype in the present study. Notably, this scale demonstrated a moderate correlation with the disinhibition dimension of the Zuckerman Sensation Seeking Scale (Zuckerman et al., 1978) in an independent sample (r = 0.311, p < 0.0001 providing an estimate of its relation to a commonly used measure of impulsivity.
Whole Genome Sequencing
Low coverage whole genome sequencing on all samples was performed using an Illumina HiSeq 2000 (Illumina, San Diego, CA). Resulting paired end (2x100) reads were aligned to GRCh37 with BWA version 0.5.8c. Sequence depth varied from sample to sample, with 80% of the samples having coverage between 3X and 12X. Variants were called using Thunder, which uses linkage disequilibrium in a manner analogous to genotype imputation. One effect of using imputation in variant calling is that a full genotype matrix is produced, so that the missing rate per sample or per variant is always zero.
As fully described in (Bizon et al., 2014), genotypes were validated by comparing these results to approximately 200,000 genotypes measured using a first generation Axiom Affymetrix Exome Chip (Affymetrix, Inc, Santa Clara, CA). Genotypes between low coverage whole genome sequencing and the genotyping chip have a 97.5% concordance rate. For variants with a minor allele frequency above 0.01 in the sample, the sequencing identifies over 97% of the variant sites detected with the genotyping chip. At lower allele frequencies, the sequencing detects fewer variant sites; at the lowest frequencies in the sample, corresponding to a single minor allele detected by the genotyping chip, sequencing detects 41% of the variant sites.
After the sequencing and variant calling was complete, the resulting genotypes were used to confirm sample identification by estimating IBD sharing proportions, which were generated using PREST-Plus (Sun et al., 2002), and comparing those estimates to the predicted kinship coefficients based on the self-reported pedigree structures provided by the study participants. Based on this process, 11 samples were excluded from the study because the observed genotype data could not be reconciled with the reported pedigree data, suggesting sample misidentification or contamination.
Statistical Analyses
Single-variant association analysis was performed using EMMAX, as implemented in the EPACTS package (Kang, 2014). EMMAX uses a linear mixed-model approach to control for both population substructure and nesting of individuals within families. Prior to association analysis, pair-wise kinship coefficients are calculated using measured genotype data. Tests of association are then conducted for each variant conditional on the calculated kinship matrix with the measured genotype and relevant covariates modeled as fixed effects and phenotype as the dependent variable. For the present report, covariates included age, age squared, and gender. Ancestry estimates were also included as covariates to further control for population stratification resulting from variants showing marked differences in allele frequencies across ancestral populations (Price et al., 2010). Ancestry covariates were calculated using the ancestry estimation ANC4 program (Libiger & Schork, 2012). ANC4 is a supervised clustering program that uses input from genotype data on 364,470 loci collected on reference individuals from global populations (European, African, Native American, and East Asian), included by permission from a recent Native American population history study (Reich et al., 2012). There were 697 individuals with sequence data suitable for ANC4 to calculate the percentage ancestry for each individual for these 4 ancestral groups. Because the ancestries sum to 1 for each individual, only the first three were used as independent covariates.
Permutation Generation and Empirical P-values
To determine empirical p-values for association results, while accounting for pedigree structure, we employed a permutation scheme inspired by gene-dropping. Briefly, to generate a single permutation, an initial allele frequency was chosen, and for each founder in the pedigree, genotypes were assigned based on this frequency. Subsequent generations were then assigned genotypes by randomly assigning one allele from each parent. Once genotypes were assigned for all successive generations, the number of minor alleles (MAC) assigned to samples with available phenotype and sequence data was determined. For a given measured variant, those permutations resulting in the same MAC were stored in a VCF file and test statistics were calculated for each simulated genotype using the described EPACTS pipeline. The permutations were performed in an iterative manner such that if the initial set of simulations yielded 3 or fewer values more extreme than the observed variant, additional simulations were performed. Because of computational costs, the number of permutations was limited to 109. The resulting test statistics thus provided a null distribution that was used to calculate an empirical p-value taking into account the pedigree structure of the data.
Results
Six hundred and fifty eight participants, which originated from 150 families, had both impulsivity phenotype data and genetic sequence data for the analyses. The demographic information on the sample is provided in table 1. A description of the sample has been reported previously (Ehlers et al., 2004a,b; Peng et al., 2014).
Table 1.
Sample Demographics.
| Samples | 658 |
| Families | 150 |
| Gender | Male: 284, Female: 374 |
| NA heritage (self-report) | 273 ≥ 50%, 385 < 50% |
| Age (mean±s.d.[min-max]) | 31.2±13.2 [18–82] |
| Income | 284 < $20k/yr, 321 ≥ $20k/yr |
| Education (mean±s.d. [min-max]) | 11.6±1.5 [3–17] yrs |
An examination of the empirical p-values generated from the gene-dropping simulations revealed some asymmetry in the distribution of the test statistic. To account for this asymmetry, the associated (i.e., alternate) alleles of all variants were modeled under two directional tests, the first modeling the associated allele as having a protective effect and the second modeling the associated allele as having a risk effect. When combined, the results of the two models allowed for more precise estimates of significance of the observed data at each end of the distribution of the test statistic. The associated lambda values calculated to estimate deviations of the observed p-values from the expected uniform distribution were 1.01 and 0.99 suggesting that the empirical p-values follow closely the expected distribution. To further ensure that the top association signals were not artifacts resulting from low-quality variant calls, Mendelian error rates were calculated for the 20 most highly associated variants. Across these variants, only a single error was observed, suggesting that the calls for the variants described below were of high quality.
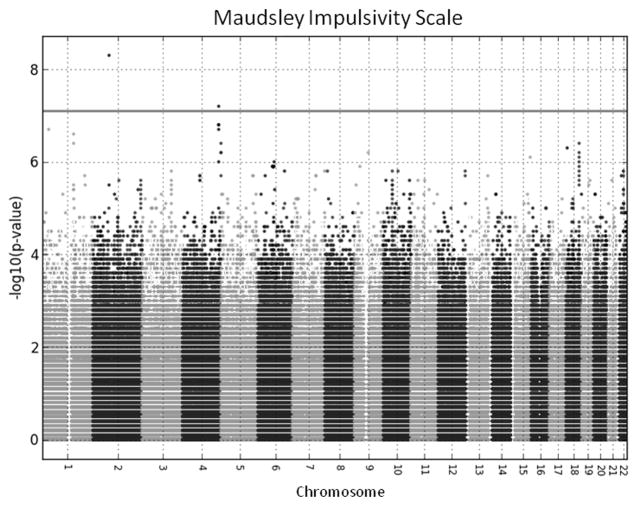
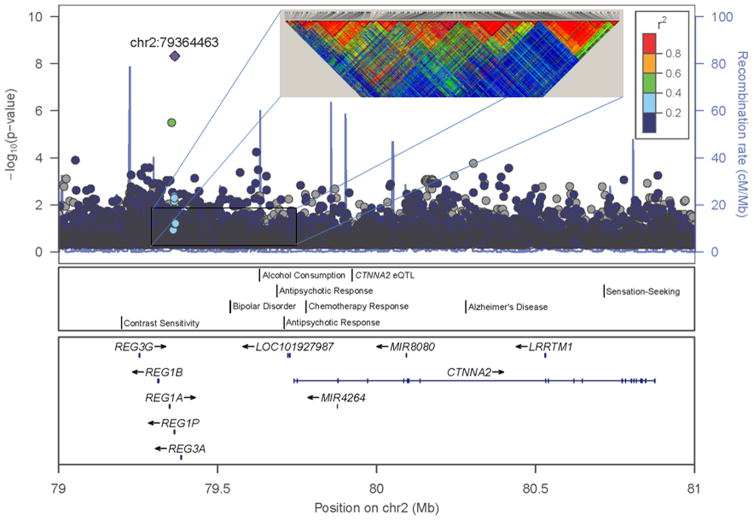
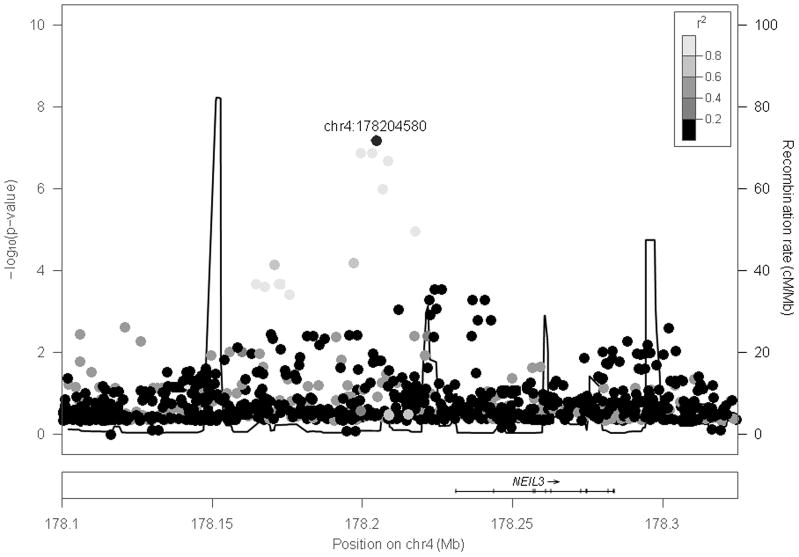
After these corrections, the described single variant association tests for the impulsivity phenotype revealed two genome-wide significant findings. A Manhattan plot for the two tailed tests is shown in Figure 1, and the top 20 results are displayed in Table 2. The most significant result emerged for a variant on chromosome 2 (rs879022, bp 79364463, p = 4.72e-09) located in a cluster of genes that encode for a family of proteins primarily excreted by the pancreas that are associated with islet regeneration (REG proteins encoded by REG1A, REG1B, REG3A, REG3G), but are also expressed in the CNS and related to neurodevelopment in the fetal brain (de la Monte et al., 1990) and neuronal sprouting and synaptogenesis in the adult brain (Acquatella-Tran Van Ba et al., 2012). The associated variant was located in the pseudogene, REG1P, that was likely produced by a duplication event related to one of the primary genes in the cluster (Figure 2). Of note, this gene cluster is located approximately 400 kb upstream of the CTNNA2 gene, a cell adhesion gene involved in cell differentiation in the nervous system and synaptic plasticity. Variants located in theREG gene cluster and CTNNA2 as well as the intergenic region between them have been associated with numerous psychiatric phenotypes characterized by impulsivity as described below. A second genome-wide significant locus was observed on chromosome 4 (rs1588052, bp 178204580, p = 6.62e-08) approximately 25 kb upstream of the nei endonuclease VIII-like 3 (E. coli), NEIL3, gene (Figure 3). It is located within a transcription factor binding site, and thus may be involved in NEIL3 expression.
Figure 1. GWAS of impulsivity results.
Manhatten plot of the -log10 p-values for the tests of association between single variants and the Maudsley Impulsivity Scale. Variants are ordered according to chromosomal position, and the p-values are alternately shaded light and dark gray to differentiate between variants on adjacent chromosomes.
Table 2.
Top 20 results of the single variant association analyses of the impulsivity phenotype.
| Chrom. | Base Position | rsNumber | MAF | β | p-value | Nearest Gene |
|---|---|---|---|---|---|---|
| 2 | 79364463 | rs879022 | 0.0308 | 1.624 | 4.72E-09 | REG1P, REG1A, REG1B, REG3A, CTNNA2 |
| 4 | 178204580 | rs1588052 | 0.0395 | −1.43 | 6.62E-08 | NEIL3 |
| 4 | 178199530 | rs1976178 | 0.0386 | −1.391 | 1.43E-07 | NEIL3 |
| 4 | 178206051 | rs6552225 | 0.0386 | −1.391 | 1.43E-07 | NEIL3 |
| 1 | 31128165 | -- | 0.0115 | 2.136 | 1.98E-07 | MATN1 |
| 4 | 178208476 | rs2046825 | 0.037 | −1.396 | 2.10E-07 | NEIL3 |
| 1 | 153893023 | rs10494303 | 0.4815 | 0.4911 | 2.67E-07 | GATAD2B, DENND4B |
| 1 | 153856498 | rs10908512 | 0.4764 | −0.4847 | 3.97E-07 | GATAD2B, DENND4B |
| 4 | 189795983 | rs151153084 | 0.1952 | −0.5948 | 4.03E-07 | LOC401164 |
| 18 | 64508801 | rs11151309 | 0.3297 | 0.5115 | 4.44E-07 | CDH19 |
| 18 | 5645946 | rs141252122 | 0.0061 | 2.875 | 4.82E-07 | EPB41L3 |
| 18 | 64506756 | rs4891571 | 0.3292 | 0.4991 | 5.92E-07 | CDH19 |
| 18 | 64508650 | rs12607996 | 0.3299 | 0.5013 | 5.92E-07 | CDH19 |
| 18 | 64513094 | rs2098950 | 0.3292 | 0.4991 | 5.92E-07 | CDH19 |
| 18 | 64514336 | rs7237644 | 0.3292 | 0.4991 | 5.92E-07 | CDH19 |
| 18 | 64520540 | rs12455928 | 0.3292 | 0.4991 | 5.92E-07 | CDH19 |
| 18 | 64530136 | rs11151313 | 0.3312 | 0.5007 | 5.92E-07 | CDH19 |
| 9 | 66529193 | -- | 0.0053 | 3.167 | 6.58E-07 | LOC403323 |
| 4 | 189796065 | rs72499936 | 0.1943 | −0.5955 | 6.68E-07 | LOC401164 |
| 4 | 189797261 | rs1375272 | 0.1943 | −0.5955 | 6.68E-07 | LOC401164 |
Figure 2. Evidence for association between variants in the CTNNA2 region and impulsivity.
Regional plot surrounding the CTNNA2 gene of the -log10 p-values for the tests of association between single variants and the Maudsley Impulsivity Scale. The -log10 p-values of the single variant analysis are plotted according to the physical location in base pairs on chromosome 2. Coloring of the data points from blue to red indicate increasing linkage disequilibrium (R2) with the top result in the region (at base pair 79364463). The inlaid linkage disequilibrium plot shows the pairwise d′ values between variants in the region with coloring from blue to red indicating increasing d′ values. The blue line indicates amount of recombination in the region expressed as the ratio of centiMorgans to megabases as assessed in the Ceph samples of the HapMap dataset. The panels below depict results from other genomewide association studies, and the position and structure (exons/introns) of genomic elements in the region mapped according to their physical location.
Figure 3. Evidence for association between variants in the NEIL3 region and impulsivity.
Regional plot surrounding the NEIL3 gene of the -log10 p-values for the tests of association between single variants and the Maudsley Impulsivity Scale. The -log10 p-values of the single variant analysis are plotted according to the physical location in base pairs on chromosome 4. Coloring of the data points from black to light gray indicate increasing linkage disequilibrium (R2) with the top result in the region (at base pair 178204580). The black line indicates amount of recombination in the region expressed as the ratio of centiMorgans to megabases as assessed in the Ceph samples of the HapMap dataset. The panel below depicts the position and structure (exons/introns) of genomic elements in the region mapped according to their physical location.
To examine the relation of rs879022 to other GWAS signals reported in this REG-CTNNA2 region, data from the 1000 Genomes project were accessed to evaluate linkage disequilibrium across this region. Given that the previous GWAS studies were conducted in predominantly European ancestry samples, data from the 174 individuals of European ancestry contained in the 1000 Genomes dataset were used to calculate d′ values for variant pairs with a MAF > 0.05 in the sub region from 79350kb to 79750kb. The d′ values are displayed in the inlaid heatmap at the top of Figure 2 with blue to red coloring indicating increasing LD values. The figure demonstrates that substantial LD is observed across broad areas of the region. LD as measured by R2 between rs879022 and the nine variants in the region that are included in the GWAS catalog are relatively low, though d′ values are higher for some of these variants, indicating that the minor allele occurs primarily on a single haplotype with these variants (Table 3). Finally, the National Center for Biotechnology Information (NCBI) eQTL browser was queried to determine whether any variants in the region represent an eQTL for CTNNA2 or any of the REG protein genes. A single variant in CTNNA2 (rs7597912) was reported to be correlated with CTNNA2 expression in liver tissue (Schadt et al., 2008). Similar to those variants identified in the GWAS catalog, rs879022 showed little evidence of LD with this variant as measured by R2, but did show a modest d′ value (Table 3).
Table 3.
Linkage disequilibrium statistic between rs879022 on Chromosome 2 and surrounding variants identified in the NCBI GWAS catalog and eQTL browser.
| Position (bp) | Marker ID | Phenotype | Gene | R2 | d′ |
|---|---|---|---|---|---|
| 79198167 | rs11683503 | Contrast sensitivity1 | Intergenic | 0.034 | 0.2 |
| 79539987 | rs13409348 | Bipolar disorder2 | Intergenic | 0.022 | 0.292 |
| 79632346 | rs2100290 | Alcohol consumption3 | Intergenic | 0.001 | 0.131 |
| 79687251 | rs399885 | Response to antipsychotic treatment4 | Intergenic | 0 | 0.044 |
| 79709353 | rs7570469 | Response to antipsychotic treatment4 | Intergenic | 0.009 | 0.516 |
| 79778112 | rs7597912 | CTNNA2 eQTL6 | CTNNA2 | 0.004 | 0.465 |
| 79922801 | rs11695685 | Protein quantitative trait loci5 | CTNNA2 | 0.033 | 1 |
| 80281172 | rs6738962 | Alzheimer’s disease (cognitive decline) 7 | CTNNA2 | 0.003 | 0.068 |
| 80715149 | rs7600563 | Excitement-Seeking8 | CTNNA2 | 0.004 | 0.335 |
Discussion
Significant evidence suggesting a heritable component underlying aspects of impulsivity and other substance dependence related traits has been previously reported in this American Indian population (Ehlers & Gizer, 2013; Ehlers et al., 2007). As described, the phenotype explored in the present population includes aspects of impulsivity that are related to both a preference for novelty and thrill seeking as well as the tendency to act on short-term desires without considering potential long-term consequences resulting from such behavior. Thus, in relation to the Five Factor Model (FFM) of personality (Costa & McCrae, 1985), it includes aspects of extraversion as well as conscientiousness (Sharma et al., 2014). Although the heritability estimate for the impulsivity phenotype derived from the Maudsley personality inventory, estimated in the present study population in a previous study as the variance explained by degree of familial relatedness, was found to be modest (h2 = 0.20) (Ehlers et al., 2007), this estimate is similar in magnitude to a previous report estimating the heritability (h2=0.36) of a similarly derived scale in a primarily European ancestry twin sample (Eaves & Eysenck, 1975). To investigate the genetic contributions to this phenotype, the aim of the present study was to conduct the first GWAS of this impulsivity phenotype, derived from the Maudsley Personality inventory, using whole genome sequence data in a Native American Indian community sample.
The present study identified a variant on chromosome 2 in the REG1P pseudogene that is contained within a cluster of 4 regenerating family protein (REG) genes. The associated variant has been previously reported to alter the structure of a NAGNAG motif (El Sharawy et al., 2009), and thus could be involved in alternative splicing, though given that it lies in a pseudogene, the relevance of this is unclear. REG proteins, also referred to as lithostathines, were first identified in studies isolating the protein family as playing a central role in pancreatic β-cell regeneration (Terazono et al., 1988). More recent studies have demonstrated that REG proteins are also expressed in the central nervous system where they are involved in inflammatory responses (Duplan et al., 2001). Further, the REG-1α protein, encoded by REG1A, has been shown to be elevated in the cerebrospinal fluid (CSF) of Alzheimer’s patients and present in the senile plaques and neurofibrillary tangles of post-mortem brain tissue of Alzheimer’s patients (de la Monte et al., 1990). Finally, studies of a primate model of Alzheimer’s disease have also observed increased expression of REG1P (Marchal et al., 2012). This gene family has not, however, been implicated in other psychiatric disorders.
An alternative possibility is that the associated variant plays a regulatory role in the expression of a nearby gene. Notably, the REG gene cluster and the associated variant implicated in the present report are located 400 kb from the catenin, cadherin-associated protein, alpha 2 gene (CTNNA2) gene on chromosome 2 gene. CTNNA2 is a large gene that is conserved across species, and microarray expression data indicate that it is expressed primarily in central nervous system and also in the testis. CTNNA2 encodes for a cell-adhesion protein (alpha N-catenin) which has been shown to regulate synaptic plasticity, and is involved in the binding of cadherins and the actin cytoskeleton and as such is important for maintaining the stability of dendritic spines (Abe et al., 2004). There is a homologue gene in mice (Catna2) that when deleted causes hippocampal and cerebellar lamination defects, axon migration deficits, and other changes in brain morphogenesis (Park et al., 2002; Uemura & Takeichi, 2006). Mice with this deletion also show impaired responding in fear conditioning, enhanced acoustic startle responses, and cerebellar ataxias, a phenotype that was shown to be rescued through expression of Catna2 transgene (Park et al., 2002). These studies suggest a plausible role for CTNNA2 in the regulation of personality features.
This is notable given that a previously published meta-analysis of the excitement-seeking scale derived from the NEO, reported a genome-wide significant result for a variant in CTNNA2 (Terracciano et al., 2011). Combining data in a meta-analysis of six European ancestry samples (n=7860), the authors found a genome-wide significant association with an intronic SNP of CTNNA2 (rs7600563; P=2X10−8). Excitement seeking, which is related to the preference for novelty and thrill seeking and is a primary component of the multifaceted impulsivity construct, is assessed, in part, by the Maudsley impulsivity scale (see Eysenck & Eysenck, 1967; Whiteside & Lynam, 2001). Thus, the present report provides further evidence suggesting that variants in this region may contribute to the development of impulsivity-related traits.
As described earlier, however, it is important to note that the Maudsley impulsivity scale differs from pure excitement-seeking scales. The former includes facets of both extraversion and disinhibition or a lack of conscientiousness, whereas the latter focuses solely on facets of extraversion. This is notable for two reasons. First, the differences in how impulsivity was operationalized across the present study and that of Terracciano et al. (2011) could account for the differences in associated variants across studies. Second, it could also explain why results from the present study did not overlap with previous large-scale meta-analyses of personality traits derived from the Five Factor Model (de Moor et al., 2012). Despite these differences, the use of the Maudsley impulsivity scale to examine genetic influences on externalizing psychopathology is warranted. For example, an early study examining the relation of the FFM personality traits to psychopathology noted that a profile characterized by high extraversion and low conscientiousness identified a non-depressed substance abuse dimension (Trull & Sher, 1994), a pattern that has been generalized to externalizing psychopathology more broadly defined (DeYoung et al., 2008).
This is of direct relevance to the present study in that several previous GWA studies of externalizing disorders have reported suggestive associations with variants within CTNNA2 as well as in the upstream region near the REG gene cluster and the associated variant identified in the present report. For example, an intronic variant, rs13395022, in CTNNA2 was reported among the top hits in a GWAS of ADHD (Lesch et al., 2008), and an upstream variant, rs2100290, located 40 kb from the associated variant reported in this study was among the top hits in a GWAS of an alcohol consumption phenotype (McGue et al., 2013). Further, a genome-wide linkage analysis and an independent GWAS reported evidence suggesting a relation between genetic variants in this region and conduct disorder (Dick et al., 2011; Kendler et al., 2006). Finally, several studies have also reported associations between variants in this region and other psychiatric phenotypes. For example, significant associations have been reported between variants in the region and bipolar disorder (Scott et al., 2009), contrast sensitivity, which has been suggested as a putative endophenotype for autism and schizophrenia (Goodbourn et al., 2014), Alzheimer’s disease (Sherva et al., 2014), and response to antipsychotic medication (Adkins et al., 2011). Thus, the association between rs879022 and the impulsivity phenotype used in the present report is supported by the previous literature, an important consideration given the relatively small sample size of the current study.
Notably, the associated variants, with the exception of the variant associated with Alzheimer’s Disease, have all been observed upstream of CTNNA2, suggesting the associations may reflect relations with variants influencing the REG protein family genes or regulatory regions of CTNNA2. Nonetheless, an examination of the annotation data regarding the variants near CTNNA2 associated with the Maudsley impulsivity phenotype in the present report were not located in known regulatory elements or ncRNA coding regions, and were not in regions displaying epigenetic marks making it difficult to draw specific conclusions of how the associated variants might be related to CTNNA2 expression. Further, the variants that achieved genome-wide significance in previous studies show little evidence of LD with the variant associated with the Maudsley impulsivity phenotype in the present report, in either European reference populations or in the present study population. Thus, it may be that multiple variants in the region influencing either the REG genes or CTNNA2 are relevant to psychiatric phenotypes. As a result, it seems likely the observed association between rs879022 and the Maudsley impulsivity phenotype is independent of previous associations reported in this region.
There are several reasons why this might be. First, if either of these genes are relevant to externalizing psychopathology, then multiple variants within the gene and surrounding regulatory elements could be relevant with nuances in each dataset (e.g., ancestry differences) contributing to which variants exhibit evidence of association. Second, differences in LD patterns in the region across studies, which are likely given the present study was conducted in a highly admixed population of Native American and European ancestry individuals, could account for differences in the associated variants if the associated variants are not causal but rather represent tag SNPs in LD with the causal variant. Third, on a related note, the present study used next-generation sequencing technologies to derive genotypes rather than tagging variants on a microarray, and thus, this more exhaustive interrogation of the region could have identified a functional variant that was not well tagged by previous studies.
It is important to note, however, that while each of these explanations are plausible, the lack of replication to a single variant across the cited studies highlights the need for further exploration of the region using standardized, transdiagnostic phenotypes to elucidate which, if any, specific facet of impulsivity or a related construct is influenced by variants in this region. At present, the use of different psychiatric and personality-related phenotypes, including that used in the present study, makes it difficult to draw strong conclusions regarding the relevance of genetic variation in this region to the etiology of impulsivity-related phenotypes and also makes it difficult to draw strong conclusions regarding which genes may be involved in the etiology of these traits. Thus, future studies using a set of standardized, transdiagnostic phenotypes are sorely needed to clarify which genes are relevant to the etiology of impulsivity-related psychiatric disorders and the multi-faceted constructs that undergird them.
In addition to the chromosome 2 result, a second locus upstream of NEIL3 may also have relevance to impulsivity and substance use phenotypes. The protein encoded by NEIL3 belongs to a class of glycosylases that initiate DNA base excision repair resulting from reactive oxygen species by creating a DNA strand break via a lyase reaction (Liu et al., 2010). Notably, neurodevelopment studies have shown NEIL3 to play an important role in neurogenesis in the fetal brain (Hilderstrand et al., 2009), as well as a continued role in neurogenesis in the hippocampus of the adult brain (Regnell et al., 2012). Previous studies have reported significant genome-wide associations with both indices of body fat and heart rate variability in the Framingham Heart Study (Newton-Cheh et al., 2007). Additionally, a recent study conducted in the COGA sample reported suggestive associations between a SNP located in this region to a composite substance dependence phenotype (alcohol, cannabis, cocaine and opioid/heroin) as well as a continuous measure of substance dependence derived from a factor analysis of symptom level dependence data related to the same 4 substances (Wetherill et al., 2014).
In conclusion, these data represent the first whole genome sequence analysis of an impulsive behavior phenotype to report an association with a variant upstream from CTNNA2 near the REG gene cluster. The results of this study should, however, be interpreted in the context of several limitations. First, the findings may not generalize to other Native American communities or represent all Native Americans within this population. Second, comparisons of association findings to non-Indian populations may be limited by differences in a host of potential genetic and environmental variables. Third, because this population has significant admixture, estimates of allele frequencies may produce biased results although this was accounted for in the analyses. Finally, given the small sample size, the reported findings require replication. A power analysis suggested that the present study was sufficiently powered (i.e., 0.80) to detect variants that could explain ~5% of the variation in a studied trait (Feng et al., 2011). GWAS of psychiatric traits suggest that variants of such large effect are unlikely to be found, and thus, the reported results likely reflect an overestimate of the true magnitudes of the reported associations. Despite these limitations, this report represents an important step in an ongoing investigation to understand the genetic determinants associated with the development of substance use disorders in this high risk and understudied ethnic group.
Acknowledgments
This work was supported by the National Institutes of Health (NIH), 5R37 AA010201 to CLE; R01 DA030976 to CLE, IRG, WS, NJS, KCW; U19 AG023122, R01 MH094483, R01 HL089655; R01 AG035020, R01 MH093500 to NJS]. Dr. Schork and his lab are also supported in part by grants from Human Longevity, Inc., Johnson and Johnson, the Tanner Foundation, and the Stand-Up-to-Cancer organization. NIAAA, NIDA, NIA, NIMH, NHLBI, Human Longevity, Inc, Johnson and Johnson, The Tanner Foundation and Stand-Up-to-Cancer had no further role in study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the article for publication. Dr. Ehlers received compensation from RAPTOR Pharmaceuticals and Neurocrine Biosciences for consulting and received the James H. Tharp Award from the Research Society on Alcoholism. Dr. Slutske has received compensation from the Massachusetts Gaming Commission and the National Center for Responsible Gaming for consulting. Dr. Schork is also a founder and stock holder in Cypher Genomics and paid consultant for the following companies: Human Longevity, Inc., MD Revolution, and Click Therapeutics. The authors state that they don’t have any conflict of interest related to material presented in the manuscript.
The authors wish to acknowledge the technical support of Linda Corey, David Gilder, Philip Lau, Evie Phillips, Shirley Sanchez and Gina Stouffer.
References
- Abe K, Chisaka O, Van RF, Takeichi M. Stability of dendritic spines and synaptic contacts is controlled by alpha N-catenin. Nature Neuroscience. 2004;7:357–363. doi: 10.1038/nn1212. [DOI] [PubMed] [Google Scholar]
- Acquatella-Tran Van Ba I, Marchal S, Francois F, Silhol M, Lleres C, Michel B, Benyamin Y, Verdier JM, Trousse F, Marcilhac A. Regenerating islet-derived 1alpha (Reg-1alpha) protein is new neuronal secreted factor that stimulates neurite outgrowth via exostosin Tumor-like 3 (EXTL3) receptor. Journal of Biological Chemistry. 2012;287:4726–4739. doi: 10.1074/jbc.M111.260349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkins DE, Aberg K, McClay JL, Bukszar J, Zhao Z, Jia P, Stroup TS, Perkins D, McEvoy JP, Lieberman JA, Sullivan PF, van den Oord EJ. Genomewide pharmacogenomic study of metabolic side effects to antipsychotic drugs. Molecular Psychiatry. 2011;16:321–332. doi: 10.1038/mp.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders (DSM-5) 5. American Psychiatric Publishing; Washington, DC; London, England: 2013. [Google Scholar]
- Benko A, Lazary J, Molnar E, Gonda X, Tothfalusi L, Pap D, Mirnics Z, Kurimay T, Chase D, Juhasz G, Anderson IM, Deakin JF, Bagdy G. Significant association between the C(-1019)G functional polymorphism of the HTR1A gene and impulsivity. American Journal of Medical Genetics Part B Neuropsychiatric Genetics. 2010;153B:592–599. doi: 10.1002/ajmg.b.31025. [DOI] [PubMed] [Google Scholar]
- Bezdjian S, Baker LA, Tuvblad C. Genetic and environmental influences on impulsivity: a meta-analysis of twin, family and adoption studies. Clinical Psychology Review. 2011;31:1209–1223. doi: 10.1016/j.cpr.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizon C, Spiegel M, Chasse SA, Gizer IR, Li Y, Malc EP, Mieczkowski PA, Sailsbery JK, Wang X, Ehlers CL, Wilhelmsen KC. Variant calling in low-coverage whole genome sequencing of a Native American population sample. BMC Genomics. 2014;15:85. doi: 10.1186/1471-2164-15-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congdon E, Lesch KP, Canli T. Analysis of DRD4 and DAT polymorphisms and behavioral inhibition in healthy adults: implications for impulsivity. American Journal of Medical Genetics Part B Neuropsychiatric Genetics. 2008;147B:27–32. doi: 10.1002/ajmg.b.30557. [DOI] [PubMed] [Google Scholar]
- Costa PT, McCrae RR. The NEO Personality Inventory Manual. Psychological Assessment Resources; Odessa, FL: 1985. [Google Scholar]
- de la Monte SM, Ozturk M, Wands JR. Enhanced expression of an exocrine pancreatic protein in Alzheimer’s disease and the developing human brain. Journal of Clinical Investigation. 1990;86:1004–1013. doi: 10.1172/JCI114762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor MH, Costa PT, Terracciano A, Krueger RF, De Geus EJ, Toshiko T, Penninx BW, Esko T, Madden PA, Derringer J, Amin N, Willemsen G, Hottenga JJ, Distel MA, Uda M, Sanna S, Spinhoven P, Hartman CA, Sullivan P, Realo A, Allik J, Heath AC, Pergadia ML, Agrawal A, Lin P, Grucza R, Nutile T, Ciullo M, Rujescu D, Giegling I, Konte B, Widen E, Cousminer DL, Eriksson JG, Palotie A, Peltonen L, Luciano M, Tenesa A, Davies G, Lopez LM, Hansell NK, Medland SE, Ferrucci L, Schlessinger D, Montgomery GW, Wright MJ, Aulchenko YS, Janssens AC, Oostra BA, Metspalu A, Abecasis GR, Deary IJ, Raikkonen K, Bierut LJ, Martin NG, van Duijn CM, Boomsma DI. Meta-analysis of genome-wide association studies for personality. Molecular Psychiatry. 2012;17:337–349. doi: 10.1038/mp.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung CG, Peterson JB, Seguin JR, Tremblay RE. Externalizing behavior and the higher order factors of the Big Five. Journal of Abnormal Psychology. 2008;117:947–953. doi: 10.1037/a0013742. [DOI] [PubMed] [Google Scholar]
- Dick DM, Aliev F, Krueger RF, Edwards A, Agrawal A, Lynskey M, Lin P, Schuckit M, Hesselbrock V, Nurnberger J, Jr, Almasy L, Porjesz B, Edenberg HJ, Bucholz K, Kramer J, Kuperman S, Bierut L. Genome-wide association study of conduct disorder symptomatology. Molecular Psychiatry. 2011;16:800–808. doi: 10.1038/mp.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplan L, Michel B, Boucraut J, Barthellemy S, Desplat-Jego S, Marin V, Gambarelli D, Bernard D, Berthezene P, Alescio-Lautier B, Verdier JM. Lithostathine and pancreatitis-associated protein are involved in the very early stages of Alzheimer’s disease. Neurobiology of Aging. 2001;22:79–88. doi: 10.1016/s0197-4580(00)00182-2. [DOI] [PubMed] [Google Scholar]
- Eaves L, Eysenck H. The nature of extraversion: a genetical analysis. Journal of Personality and Social Psychology. 1975;32:102–112. doi: 10.1037/h0076862. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wall TL, Betancourt M, Gilder DA. The clinical course of alcoholism in 243 Mission Indians. American Journal of Psychiatry. 2004a;161:1204–1210. doi: 10.1176/appi.ajp.161.7.1204. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Wall TL, Phillips E, Feiler H, Wilhelmsen KC. Genomic screen for loci associated with alcohol dependence in Mission Indians. American Journal of Human Genetics. 2004b;129B:110–115. doi: 10.1002/ajmg.b.30057. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wall TL, Corey L, Lau P, Gilder DA, Wilhelmsen K. Heritability of illicit drug use and transition to dependence in Southwest California Indians. Psychiatric Genetics. 2007;17:171–176. doi: 10.1097/01.ypg.0000242201.56342.1a. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gizer IR. Evidence for a genetic component for substance dependence in Native Americans. American Journal of Psychiatry. 2013;170:154–164. doi: 10.1176/appi.ajp.2012.12010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Sharawy A, Hundrieser B, Brosch M, Wittig M, Huse K, Platzer M, Becker A, Simon M, Rosenstiel P, Schreiber S, Krawczak M, Hampe J. Systematic evaluation of the effect of common SNPs on pre-mRNA splicing. Human Mutation. 2009;30:625–632. doi: 10.1002/humu.20906. [DOI] [PubMed] [Google Scholar]
- Eysenck HJ. Manual of the Maudsley personality inventory. University of London Press; London: 1959. [Google Scholar]
- Eysenck HJ, Eysenck SBG. Manual of the Eysenck personality inventory. Univ. of London Press; London: 1964. [Google Scholar]
- Eysenck HJ, Eysenck SB. On the unitary nature of extraversion. Acta Psychology (Amst) 1967;26:383–390. doi: 10.1016/0001-6918(67)90034-0. [DOI] [PubMed] [Google Scholar]
- Eysenck HJ, Eysenck SBG. Manual of the Eysenck personality questionnaire (junior and adult) London: Hodder and Stoughton; 1975. [Google Scholar]
- Feng S, Wang S, Chen CC, Lan L. GWAPower: a statistical power calculation software for genome-wide association studies with quantitative traits. BMC Genetics. 2011;12:12. doi: 10.1186/1471-2156-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilder DA, Wall TL, Ehlers CL. Comorbidity of select anxiety and affective disorders with alcohol dependence in Southwest California Indians. Alcoholism: Clinical and Experimental Research. 2004;28:1805–1813. doi: 10.1097/01.alc.0000148116.27875.b0. [DOI] [PubMed] [Google Scholar]
- Gizer IR, Otto JM, Ellingson J. Molecular genetics of the externalizing spectrum. In: Beauchaine T, Hinshaw S, editors. Oxford Handbook on Externalizing Spectrum Disorders. New York: Oxford University Press; 2015. pp. 149–169. [Google Scholar]
- Goodbourn PT, Bosten JM, Bargary G, Hogg RE, Lawrance-Owen AJ, Mollon JD. Variants in the 1q21 risk region are associated with a visual endophenotype of autism and schizophrenia. Genes, Brain and Behavior. 2014;13:144–151. doi: 10.1111/gbb.12096. [DOI] [PubMed] [Google Scholar]
- Hildrestrand GA, Neurauter CG, Diep DB, Castellanos CG, Krauss S, Bjoras M, Luna L. Expression patterns of Neil3 during embryonic brain development and neoplasia. BMC Neuroscience. 2009;10:45. doi: 10.1186/1471-2202-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM. Efficient and Parallelizable Association Container Toolbox (EPACT) University of Michigan Center for Statistical Genetics; 2014. [Accessed 6.11.15]. Available at: http://genome.sph.umich.edu/wiki/EPACTS. [Google Scholar]
- Kendler KS, Kuo PH, Todd WB, Kalsi G, Neale MC, Sullivan PF, Walsh D, Patterson DG, Riley B, Prescott CA. A joint genomewide linkage analysis of symptoms of alcohol dependence and conduct disorder. Alcoholism: Clinical and Experimental Research. 2006;30:1972–1977. doi: 10.1111/j.1530-0277.2006.00243.x. [DOI] [PubMed] [Google Scholar]
- Kreek MJ, Nielsen DA, Butelman ER, LaForge KS. Genetic influences on impulsivity, risk taking, stress responsivity and vulnerability to drug abuse and addiction. Nature Neuroscience. 2005;8:1450–1457. doi: 10.1038/nn1583. [DOI] [PubMed] [Google Scholar]
- Krueger RF, Hicks BM, Patrick CJ, Carlson SR, Iacono WG, McGue M. Etiologic connections among substance dependence, antisocial behavior, and personality: modeling the externalizing spectrum. Journal of Abnormal Psychology. 2002;111:411–424. [PubMed] [Google Scholar]
- Lesch KP, Timmesfeld N, Renner TJ, Halperin R, Roser C, Nguyen TT, Craig DW, Romanos J, Heine M, Meyer J, Freitag C, Warnke A, Romanos M, Schafer H, Walitza S, Reif A, Stephan DA, Jacob C. Molecular genetics of adult ADHD: converging evidence from genome-wide association and extended pedigree linkage studies. Journal of Neural Transmission. 2008;115:1573–1585. doi: 10.1007/s00702-008-0119-3. [DOI] [PubMed] [Google Scholar]
- Libiger O, Schork NJ. A method for inferring an individual’s genetic ancestry and degree of admixture associated with six major continental populations. Frontiers in Genetics. 2012;3:322. doi: 10.3389/fgene.2012.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proceedings of the National Academy of Sciences US A. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal S, Givalois L, Verdier JM, Mestre-Frances N. Distribution of lithostathine in the mouse lemur brain with aging and Alzheimer’s-like pathology. Neurobiology of Aging. 2012;33:431–25. doi: 10.1016/j.neurobiolaging.2011.01.002. [DOI] [PubMed] [Google Scholar]
- McGue M, Zhang Y, Miller MB, Basu S, Vrieze S, Hicks B, Malone S, Oetting WS, Iacono WG. A genome-wide association study of behavioral disinhibition. Behavior Genetics. 2013;43:363–373. doi: 10.1007/s10519-013-9606-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer D, Perry JRB, Hernandez D, Corsi A-M, Stevens K, Rafferty I, Lauretani F, Murray A, Gibbs JR, Paolisso G, Rafiq S, Simon-Sanchez J, Lango H, Scholz S, Weedon MN, Arepalli S, Rice N, Washecka N, Hurst A, Britton A, Henley W, Leemput Jvd, Li R, Newman AB, Tranah G, Harris T, Panicker V, Dayan C, Bennett A, McCarthy MI, Ruokonen A, Jarvelin M-R, Guralnik J, Bandinelli S, Frayling TM, Singleton A, Ferrucci L. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genetics. 2008;4(5):e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SC, Doheny KF, Pugh EW, Romm JM, Ling H, Laurie CA, Browning SR, Weir BS, Laurie CC. Imputation-based genomic coverage assessments of current human genotyping arrays. G3 Genes, Genomes, Genetics (Bethesda) 2013;3:1795–1807. doi: 10.1534/g3.113.007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton-Cheh C, Guo CY, Wang TJ, O’donnell CJ, Levy D, Larson MG. Genome-wide association study of electrocardiographic and heart rate variability traits: the Framingham Heart Study. BMC Medical Genetics. 2007;8(Suppl 1):S7. doi: 10.1186/1471-2350-8-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Falls W, Finger JH, Longo-Guess CM, Ackerman SL. Deletion in Catna2, encoding alpha N-catenin, causes cerebellar and hippocampal lamination defects and impaired startle modulation. Nature Genetics. 2002;31:279–284. doi: 10.1038/ng908. [DOI] [PubMed] [Google Scholar]
- Peng Q, Gizer IR, Libiger O, Bizon C, Wilhelmsen KC, Schork NJ, Ehlers CL. Association and ancestry analysis of sequence variants in ADH and ALDH using alcohol-related phenotypes in a Native American community sample. American Journal of Medical Genetics Part B Neuropsychiatric Genetics. 2014;165B:673–683. doi: 10.1002/ajmg.b.32272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nature Reviews. Genetics. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnell CE, Hildrestrand GA, Sejersted Y, Medin T, Moldestad O, Rolseth V, Krokeide SZ, Suganthan R, Luna L, Bjoras M, Bergersen LH. Hippocampal adult neurogenesis is maintained by Neil3-dependent repair of oxidative DNA lesions in neural progenitor cells. Cell Reports. 2012;2:503–510. doi: 10.1016/j.celrep.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Reich D, Patterson N, Campbell D, Tandon A, Mazieres S, Ray N, Parra MV, Rojas W, Duque C, Mesa N, Garcia LF, Triana O, Blair S, Maestre A, Dib JC, Bravi CM, Bailliet G, Corach D, Hunemeier T, Bortolini MC, Salzano FM, Petzl-Erler ML, Acuna-Alonzo V, Aguilar-Salinas C, Canizales-Quinteros S, Tusie-Luna T, Riba L, Rodriguez-Cruz M, Lopez-Alarcon M, Coral-Vazquez R, Canto-Cetina T, Silva-Zolezzi I, Fernandez-Lopez JC, Contreras AV, Jimenez-Sanchez G, Gomez-Vazquez MJ, Molina J, Carracedo A, Salas A, Gallo C, Poletti G, Witonsky DB, Alkorta-Aranburu G, Sukernik RI, Osipova L, Fedorova SA, Vasquez R, Villena M, Moreau C, Barrantes R, Pauls D, Excoffier L, Bedoya G, Rothhammer F, Dugoujon JM, Larrouy G, Klitz W, Labuda D, Kidd J, Kidd K, Di RA, Freimer NB, Price AL, Ruiz-Linares A. Reconstructing Native American population history. Nature. 2012;488:370–374. doi: 10.1038/nature11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, Zhu J, Millstein J, Sieberts S, Lamb J, Guhathakurta D, Derry J, Storey JD, Avila-Campillo I, Kruger MJ, Johnson JM, Rohl CA, van NA, Mehrabian M, Drake TA, Lusis AJ, Smith RC, Guengerich FP, Strom SC, Schuetz E, Rushmore TH, Ulrich R. Mapping the genetic architecture of gene expression in human liver. PLoS Biology. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LJ, Muglia P, Kong XQ, Guan W, Flickinger M, Upmanyu R, Tozzi F, Li JZ, Burmeister M, Absher D, Thompson RC, Francks C, Meng F, Antoniades A, Southwick AM, Schatzberg AF, Bunney WE, Barchas JD, Jones EG, Day R, Matthews K, McGuffin P, Strauss JS, Kennedy JL, Middleton L, Roses AD, Watson SJ, Vincent JB, Myers RM, Farmer AE, Akil H, Burns DK, Boehnke M. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proceedings of the National Academy of Sciences USA. 2009;106:7501–7506. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma L, Markon KE, Clark LA. Toward a theory of distinct types of “impulsive” behaviors: A meta-analysis of self-report and behavioral measures. Psychological Bulletin. 2014;140:374–408. doi: 10.1037/a0034418. [DOI] [PubMed] [Google Scholar]
- Sherva R, Tripodis Y, Bennett DA, Chibnik LB, Crane PK, de Jager PL, Farrer LA, Saykin AJ, Shulman JM, Naj A, Green RC. Genome-wide association study of the rate of cognitive decline in Alzheimer’s disease. Alzheimer’s & Dementia. 2014;10:45–52. doi: 10.1016/j.jalz.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutske WS. The genetics of antisocial behavior. Current Psychiatry Reports. 2001;3:158–162. doi: 10.1007/s11920-001-0014-1. [DOI] [PubMed] [Google Scholar]
- Sun L, Wilder K, McPeek MS. Enhanced pedigree error detection. Human Heredity. 2002;54:99–110. doi: 10.1159/000067666. [DOI] [PubMed] [Google Scholar]
- Terazono K, Yamamoto H, Takasawa S, Shiga K, Yonemura Y, Tochino Y, Okamoto H. A novel gene activated in regenerating islets. Journal of Biological Chemistry. 1988;263:2111–2114. [PubMed] [Google Scholar]
- Terracciano A, Esko T, Sutin AR, de Moor MH, Meirelles O, Zhu G, Tanaka T, Giegling I, Nutile T, Realo A, Allik J, Hansell NK, Wright MJ, Montgomery GW, Willemsen G, Hottenga JJ, Friedl M, Ruggiero D, Sorice R, Sanna S, Cannas A, Raikkonen K, Widen E, Palotie A, Eriksson JG, Cucca F, Krueger RF, Lahti J, Luciano M, Smoller JW, van Duijn CM, Abecasis GR, Boomsma DI, Ciullo M, Costa PT, Jr, Ferrucci L, Martin NG, Metspalu A, Rujescu D, Schlessinger D, Uda M. Meta-analysis of genome-wide association studies identifies common variants in CTNNA2 associated with excitement-seeking. Translational Psychiatry. 2011;1:e49. doi: 10.1038/tp.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trull TJ, Sher KJ. Relationship between the five-factor model of personality and Axis I disorders in a nonclinical sample. Journal of Abnormal Psychology. 1994;103:350–360. doi: 10.1037//0021-843x.103.2.350. [DOI] [PubMed] [Google Scholar]
- Uemura M, Takeichi M. Alpha N-catenin deficiency causes defects in axon migration and nuclear organization in restricted regions of the mouse brain. Developmental Dynamics. 2006;235:2559–2566. doi: 10.1002/dvdy.20841. [DOI] [PubMed] [Google Scholar]
- Wetherill L, Kapoor M, Agrawal A, Bucholz K, Koller D, Bertelsen SE, Le N, Wang JC, Almasy L, Hesselbrock V, Kramer J, Nurnberger JI, Jr, Schuckit M, Tischfield JA, Xuei X, Porjesz B, Edenberg HJ, Goate AM, Foroud T. Family-based association analysis of alcohol dependence criteria and severity. Alcoholism: Clinical and Experimental Research. 2014;38:354–366. doi: 10.1111/acer.12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteside SP, Lynam DR. The five factor model and impulsivity: using a structural model of personality to understand impulsivity. Personality and Individual Differences. 2001;30:669–689. [Google Scholar]
- Young SE, Stallings MC, Corley RP, Krauter KS, Hewitt JK. Genetic and environmental influences on behavioral disinhibition. American Journal of Medical Genetics. 2000;96:684–695. [PubMed] [Google Scholar]
- Zuckerman M, Eysenck S, Eysenck HJ. Sensation seeking in England and America: cross-cultural, age, and sex comparisons. J Consult Clin Psychol. 1978;46:139–149. doi: 10.1037//0022-006x.46.1.139. [DOI] [PubMed] [Google Scholar]