Abstract
Stem cells maintain homeostasis in all regenerating tissues during the lifespan of an organism. Thus, age-related functional decline of such tissues is likely to be at least partially explained by molecular events occurring in the stem cell compartment. Some of these events involve epigenetic changes, which may dictate how an aging genome can lead to differential gene expression programs. Recent technological advances have made it now possible to assess the genome-wide distribution of an ever-increasing number of epigenetic marks. As a result, the hypothesis that there may be a causal role for an altered epigenome contributing to the functional decline of cells, tissues, and organs in aging organisms can now be explored. In this paper, we review recent developments in the field of epigenetic regulation of stem cells, and how this may contribute to aging.
Introduction
Aging is associated with a progressive decline in function of adult tissues and organs observed in all mammals. Adult stem cells have now been characterized in almost all mammalian tissues, including blood, skeletal muscle, intestine, skin, and brain. These tissue-specific stem cells possess self-renewal potential and the ability to generate mature cells: characteristics they need in order to maintain tissue homeostasis and regeneration of the tissue after stress or cell loss. Within many aged tissues, a loss of the regenerative capacity of adult stem cells has been documented. Therefore, impaired stem cell function, more than intrinsic changes in differentiated cells, has been considered as a driver of the aging process of multiple regenerating tissues, and as such may contribute to organismal aging. Such stem cell-intrinsic events could theoretically involve either genetic or epigenetic changes. Whereas the role of an accumulation of genetic lesions in stem cell functioning during aging has been recently reviewed elsewhere (Behrens et al. 2014), in the current manuscript we focus on the role of age-associated epigenetic changes. “Epigenetics” is a term used to classify heritable changes of gene expression that are not attributed to changes in the DNA sequence (Goldberg et al. 2007). Due to the fundamental role of epigenetics in the regulation of gene expression and the putative reversibility of such epigenetic marks, there is an increasing interest in the role of epigenetic processes as mediators of the aging process of stem cells. In this review, we discuss the biology of stem cell aging with a particular focus on the epigenetic contribution to the aging process. We briefly explain current methods to evaluate epigenetic marks in the context of biological aging and discuss to what extent these have revealed a common epigenetic pattern in stem cell aging.
Do aging stem cells contribute to the functional decline of organs?
As individuals age, there is a gradual loss of homeostasis of most tissues and, as a consequence, a decline in organ function. A large body of data suggests that in many tissues age-associated loss of homeostasis is caused by an age-related decline in the ability of stem cells to replace damaged cells, (reviewed in Rando 2006; Drummond-Barbosa 2008; Liu and Rando 2011). For example, skeletal muscle possesses remarkable regenerative ability upon injury, a process that is mediated by the resident muscle stem cells. However, muscle stem cells isolated from aged animals have a higher propensity to undergo fibrogenic differentiation (Brack et al. 2007). As a result, upon aging there is an increase in tissue fibrosis and the subsequent aged-related reduction in the mass of muscle tissue contributes to an impaired motor activity in the elderly. Similarly, aging in the nervous system leads to the loss of neuronal stem cells (NSCs) (Molofsky et al. 2006). NSCs in the adult brain give rise to new granule layer neurons that integrate into functional neuronal circuits (Song et al. 2002), supporting processes such as learning and memory formation (Clelland et al. 2009), which are often impaired as individuals age. Also in the skin, melanocyte stem cells that pigment new hair drop in number upon aging (Maslov et al. 2004), leading to the very common phenotype observed in the elderly, hair loss and graying (Nishimura et al. 2005). However, in mammals, not every organ is directly dependent on stem cell activity. Aging-related alterations in organs like eyes, inner ears, or bones are more difficult to attribute to impaired stem cell activity. Retinal stem cells can potentially account for age-related diseases like macular degeneration, but not for the changes in corneal curvature or in the condensation of the vitreous gel that cause alteration in refraction and decreased sight capacity in elderly. Similarly, ear sensory cells do not regenerate if lost (Groves 2010); therefore, aged-associated loss of hearing has so far not been associated to stem cell exhaustion.
Understanding the basic properties of the various types of tissue-specific stem cells and cataloguing the molecular changes that accumulate in these cells as they age is of great interest. In particular, insight into molecular changes that could potentially be reversible, such as epigenetic alterations, may open options to develop therapeutic approaches for age-related diseases based on interventions to delay or prevent stem cell aging.
Functional and molecular manifestations of stem cell aging
In the section above, we introduced the aged-associated decline of function at the level of tissues and organs. In the following sections, we discuss the main functional manifestation and molecular changes that occur in several regenerating tissues as stem cells age. In particular, we will focus on age-related changes that appear to occur commonly versus changes that are tissue specific.
Stem cell pool size
In almost all mammalian tissues that are capable to regenerate, the number of adult stem cells is affected by aging. However, the directionality of this change is variable. Stem cells in the hematopoietic tissue have been reported to increase in number (de Haan and Van Zant 1999), and this age-dependent expansion of HSCs is a transplantable, cell-intrinsic aspect of hematopoietic stem cells. On the contrary, skin and muscle stem cells display an age-dependent decrease in number (Nishimura et al. 2005; Renault et al. 2002). In brain, changes in the neuronal stem cell pool appear to be region-specific (Kuhn et al. 1996; Maslov et al. 2004; Hattiangady and Shetty 2008). Currently, we know very little about how stem cell pool size in these various tissues is actually regulated. However, despite the diverse directionality of the change in stem cell pool size, alterations in the numbers of stem cells during aging suggest that deregulation of self-renewal and cell fate programs, i.e., a loss of control of stem cell pool size, might be a common molecular event occurring during aging. It is worth considering, however, that measurements of stem cell compartment size is confounded by the current inability to purify stem cells to homogeneity, as well as the lack of adequate models to test the function of many stem cell types.
Impaired functionality and altered lineage commitment
The decline in stem cell functionality is a shared feature among the majority of adult stem cell compartments. Defects of aged HSCs in long-term reconstitution of the immune system have been demonstrated in competitive transplantation assays (Kamminga et al. 2005; Rossi et al. 2005). Neurogenesis potential of neuronal stem cells in the nervous system declines with age (Bondolfi et al. 2004; Enwere et al. 2004). Similarly, aged muscle satellite cells display impaired muscle regeneration after injury (Conboy et al. 2003; Carlson and Conboy 2007). The loss of functionality in a stem cell compartment directly translates into a decline in the function of progenitors cells and ultimately into an altered differentiation program. Thus, an aging-associated feature shared among many adult stem cell compartments is the aberrant lineage specification of stem cell progeny. Within the hematopoietic system, stem cells from both old humans and old mice show an increased propensity to differentiate along the myeloid, rather than the lymphoid lineage (Sudo et al. 2000; Rossi et al. 2005; Dykstra et al. 2007; Cho et al. 2008). In the brain, there is an increased production of astrocytes (astroglial lineage skewing) (Peinado et al. 1998). Aged muscles are characterized by the increased tendency of satellite cells to convert from myogenic to fibroblastic and adipogenic lineages (Taylor-Jones et al. 2002; Brack et al. 2007). The accumulation of abnormal progenies in all the tissues reported above critically contributes to the gradual deterioration of tissue structure and function associated with aging.
Molecular alterations underling stem cell aging
Several molecular changes, including perturbation of signaling pathways, mitochondrial dysfunction, metabolic and cell energetic deregulation, have been implicated in tissue-specific aging patterns and have been extensively reviewed elsewhere (Lopez-Otin et al. 2013). In general, cell-intrinsic changes can be broadly grouped into two classes: those that are genetic, and therefore typically irreversible, versus those that are epigenetic. The extent to which mostly irreversible genomic changes (including nuclear and mitochondrial DNA damage and telomere shortening) contribute to stem cell aging is an active area of investigation and has been extensively reviewed elsewhere (Adams et al. 2015). Mouse models involving DNA repair proteins have shown that an aging-like phenotype is accelerated in the presence of increased DNA damage (reviewed in Garinis et al. 2008). However, so far no experimental evidence has been provided which demonstrates that repressing the occurrence of DNA mutations leads to an extension of life span.
DNA mutations can also occur in genes coding for epigenetic modifiers, leading to an altered epigenetic regulation and possibly altered fate programs. Interestingly, common DNMT3A mutations have been recently described to arise early in the development of acute myeloid leukemia, a type of leukemia that preferentially occurs in the elderly. DNMT3a mutations appear to lead to a clonally expanded pool of pre-leukemic stem cells from which AML evolves (Shlush et al. 2014).
Whereas most genetic changes that occur in somatic stem cells are believed to be irreversible, molecular changes that affect epigenetic moieties are potentially reversible. What are the indications that such epigenetic changes are associated with stem cell aging?
Epigenetic control of stem cell regulation
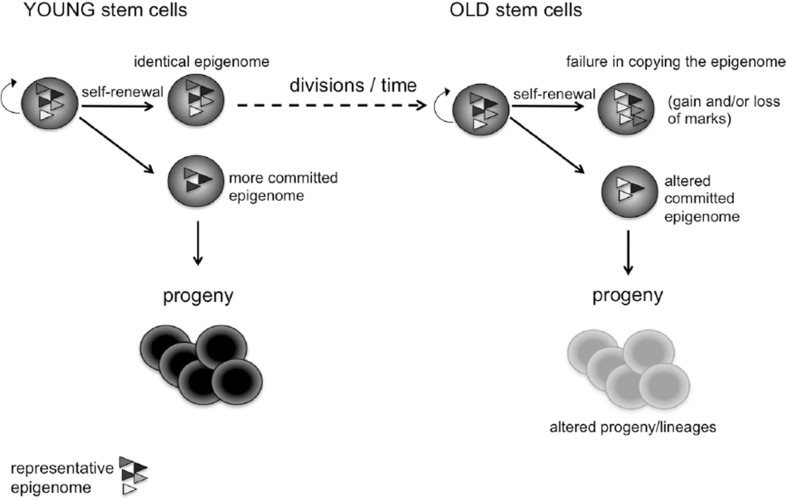
Epigenetics is the study of phenotypes or gene expression patterns that are heritable through cell divisions, but remain independent of DNA sequence (Berger et al. 2009; Goldberg et al. 2007; reviewed in Bonasio et al. 2010). Epigenetic regulation is required for the establishment and maintenance of biological states (Grewal and Klar 1996; Cavalli and Paro 1998; Martin and Zhang 2007). Thus, while in differentiated-end stage-cells epigenetic regulation is mainly used to regulate ongoing cellular processes, in stem cells the implications of the epigenome are wider. During a self-renewing division, a stem cell must not only replicate its genome flawlessly but also must copy all the relevant epigenetic markers, deposited by a large number of writers and erasers, to at least one of the two daughter cells, in the timeframe of a single cell division. Epigenetic regulation in adult stem cells must ensure the co-occurrence of self-renewal programs together with instructions for differentiation into cells with distinct potentials and, in some instances, into a large set of mature cells of different lineages with vast functional heterogeneity. Thus, epigenetic alterations that arise in stem cells can be amplified through self-renewal programs and propagated to the progeny upon differentiation. Infidelity in the epigenetic stem cell machinery in stem cells could result in detrimental losses or gains of a plethora of marks during aging (Fig. 1).
Fig. 1.
Epigenetic alterations in aging of stem cells. Upon multiple cell divisions that occur as a function of time, infidelity in exact replication of the epigenome to daughter cells might occur. Both detrimental losses and gains of epigenetic modifications can thus be amplified within the stem cell pool and propagated to the progeny upon differentiation
DNA methylation
Epigenetic regulation can occur at different levels. At the DNA level, epigenetic modification is shaped by the addition or removal of a methyl group or hydroxymethyl group at the 5th carbon atom of cytosine. DNA methyltransferase enzymes are responsible for both the establishment (DNMT3A and DNMT3B) and maintenance (DNMT1) of methylated nucleotides. Many studies have documented a role for these enzymes in balancing self-renewal and differentiation in multiple adult stem cell compartments (reviewed in Beerman and Rossi 2015). The aberrant stem cell functioning caused by dysregulation of DNA methylation leads to phenotypes frequently mirroring those observed with aging. Loss of Dnmt1 in HSCs leads to dysregulation of lineage output, with a skewing towards myelopoiesis (Broske et al. 2009), a phenotype largely associated with HSCs aging. Similarly, Dnmt3a ablation in HSCs predisposes mice to develop a spectrum of myeloid and lymphoid malignancies, and Dnmt3a-KO-derived myeloid malignancies and T cell acute lymphocytic leukemia/lymphoma show distinct methylation aberrations (Mayle et al. 2015). Interestingly, Dnmt3a has been found to be slightly but significantly downregulated with age (Sun et al. 2014).
The gold standard for direct analysis of DNA methylation is the sequencing-based quantification of cytosine methylation following bisulfite conversion, which allows identification and quantification of DNA methylation at single nucleotide resolution. A description of current techniques and methods used for analysis of DNA methylation has been recently reported (Plongthongkum et al. 2014). An ever-increasing effort has been made towards the application of assays aimed to quantify DNA methylation in a limited amount of input material (Table 1), such as highly purified populations of stem cells. Experiments that use low numbers of cells are predicted to reduce bias originating from cell-to-cell variability, even within a relatively homogeneous cell population. Early studies examined global DNA methylation patterns in aged tissues, and largely demonstrate age-associated hypo-methylation (Wilson and Jones 1983; Wilson et al. 1987; Heyn et al. 2012; Florath et al. 2014; Jung and Pfeifer 2015). Nonetheless, global DNA hypo-methylation does not exclude the possibility that individual loci become hypermethylated during aging. Indeed, along with a genome-wide analysis of the global DNA methylation landscape, several studies report have uncovered locus-specific DNA hyper-methylation associated with aging tissues (Maegawa et al. 2010; Rakyan et al. 2010; Zykovich et al. 2014). Interestingly, DNA methylation of a limited set of specific loci has been hypothesized to provide biomarkers for the aging process (Horvath 2013; Weidner et al. 2014). It is worth noticing that studies in neuronal, hematopoietic, and skin stem cells in which the DNA methylome was correlated with the transcriptome revealed that altered DNA methylation profiles not always directly correlated with altered transcription in stem cells (Guo et al. 2011; Bock et al. 2012; Beerman et al. 2013; Sun et al. 2014).
Table 1.
Analysis of the epigenome: recent methods for single cell or ultra-low-input assays
| Assay | Developer | Input material |
|---|---|---|
| Nano-ChIP-seq | Adli and Bernstein 2011 | 10,000 cells for H3k4me3 histone mark |
| Linear DNA amplification (LinDA) | Shankaranarayanan et al. 2011 | For transcription factors using 5000 cells and for the H3K4me3 histone modification using 10,000 cells |
| Ultra-low-input micrococcal nuclease-based native ChIP (ULI-NChIP) | Brind’Amour et al. 2015 | 10,000 for histone marks |
| ChIPmentation | Schmidl et al. 2015 | For several histone marks 10,000 cells per IP, and 100,000 cells for transcription factors |
| Single-cell ChIP-seq | Rotem et al. 2015 | One cell. ChIP-seq for H3k4me3 and H3k4me2 |
| Single-cell reduced-representation bisulfite sequencing scRRBS | Guo et al. 2015 | One cell. Non-targeted enrichment DNA methylation analysis |
| Low-input and single-cell whole-genome bisulfite sequencing (μWGBS, scWGBS) | Farlik et al. 2015 | High-throughput bisulfite sequencing assay for low-input and single-cell samples |
| Single-cell bisulfite sequencing scBS-seq | Smallwood et al. 2014 | One cell. Targeted enrichment DNA methylation analysis |
Histone variants, histone modifications, and higher order chromatin structure
Core histones proteins within a nucleosome can be substituted by several variants. Each variant histone contains specific physical properties, which differentially regulate expression of nearby DNA sequences. Very little is known about the contribution of different histone variants to the aging process. Beyond different histone variants, a next layer of epigenetic regulation occurs at histone tails that can be modified by an array of post-translational modifications, including acetylation, methylation, ubiquitylation, phosphorylation, sumoylation, ribosylation, and citrullination. These post-translational histone modifications affect transcription both directly, through changes in higher order chromatin structures, and indirectly, by recruitment of downstream effectors. Current techniques to analyze the modification of the histone tails and their implication in the aging process will be discussed into more detail in the next section. The combined effect of all these epigenetic mechanisms ultimately alters the higher order structure of the chromatin itself and the position of chromatin in the cell nucleus, allowing the recruitment or dismissal of transcription factors.
Substantial efforts have been made to understand the higher, three-dimensional genome organization within the nucleus. Chromosome conformation capture allows to analyze physical contacts between different regions of a chromosome and between the different chromosomes in cell populations (Dekker et al. 2002). In the past year, 4C, 5C, and Hi-C techniques have added new layers of information to the structure of chromosomes (reviewed in Cattoni et al. 2015). The development of single-cell approaches to reach single-cell, high-throughput, and high-resolution maps will be of great relevance as to what extent the 3-dimensional chromatin architecture in a nucleus of an aged (stem) cell is different from a young cell, and if so, whether or not this contributes to impaired stem cell functioning.
A third layer of epigenetic information is provided by nucleosome positioning and nucleosome occupancy. Recently, data have suggested that the variation in the total number of histone proteins may be altered during aging. The expression of core histones has been found to be reduced during replicative aging in yeast (Feser et al. 2010), as well as in aged muscle stem cells (Liu et al. 2013). Interestingly, increasing the cellular supply of histones increased the replicative lifespan of yeast by up to 50 % (Feser et al. 2010). The molecular mechanisms underlying this process, the genome-wide consequences of reduced nuclear histone content, and the mechanisms by which core histone expression levels are transferred to the daughter cells are still to be elucidated. Nonetheless, it is interesting to hypothesize that histone availability may have direct consequences on the epigenetic landscape of histone modifications and on transcription factor activity, as they compete with nucleosomes for binding to DNA.
Epigenetic regulation by histone modifications
We will now focus in more detail on the epigenetic regulation by the spatial–temporal chemical modifications at the histone tails, and how these change upon aging.
Before the advent of genome-wide techniques to study the epigenome, research had been mainly focused on investigating whether specific histone modifications globally decreased or increased upon aging. These studies typically used Western blotting or immunostaining in order to directly quantify the amount of specific histone modifications in a given cell population. However, with this approach no information can be retrieved concerning differentially affected loci. Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) is nowadays the standard technique for identifying genome-wide loci and biochemical modifications of chromatin-bound proteins and histone modifications. Promoters can feature multiple activating and repressive marks, although only few are typically assessed in a single experiment. Together with promoters, distal enhancers refine timing and extent of gene expression. Active or repressive enhancers are marked by specific chromatin moieties that can be identified in ChIP-seq analyses.
Currently, there is great interest in profiling enhancer-associated chromatin features in order to produce genome-wide maps of regulatory elements of clusters of genes. An outstanding contribution to this field has recently been provided by the Encyclopedia of DNA Elements (ENCODE), a public research project launched by the US National Human Genome Research Institute (NHGRI) in September 2003 (Feinglod Science 2004). As a follow-up of the Human Genome Project, ENCODE is ultimately dedicated to identify all functional elements in the human genome. In the Roadmap Epigenomics Project, epigenomic information has now been gathered for 127 cell types from most human tissues (Farh et al. 2015; Gjoneska et al. 2015; Leung et al. 2015; Polak et al. 2015; Roadmap Epigenomics et al. 2015; Ziller et al. 2015). This has resulted in the identification of hundreds of thousands of enhancer-like regions in the mammalian genome that regulate gene expression at long range. Although at present it is not yet feasible to determine how many features are required for a minimal representation of the epigenome, the consortium has focused so far on DNA methylation, six histone modifications (H3K4me1, H3K4me3, H3K9me3, H3K9ac, H3K27me3, and H3K36me3), and chromatin accessibility. The emerging and may be not too surprising picture is that combinations of multiple modifications predict gene activity in ways that a single type of modification does not.
Although these data provide a massive amount of epigenetic information in a large set of human cell types, it is important to notice that these experiments, and indeed most studies, are still based on ChIP-seq protocols that require a large numbers of cells (~10e6) as starting material. This limitation precludes the analysis of rare primary stem cell populations. Therefore, in recent past, attempts have been made to optimize new protocols suitable for low-input ChIP-seq. Some of the recent methods for single cell or ultra-low-input assay for the study of histone modifications and DNA methylation are summarized in Table 1. Challenges that need to be overcome relate to the fact that PCR amplification from limited ChIP input material more easily leads to amplification artifacts and duplicate reads, and to a reduced complexity of the immunoprecipitated material recovered. In addition, different bioinformatic approaches are used to analyze and present data, which ultimately results in substantial discordance of conclusions between seemingly similar studies generated from different laboratories.
Do histone modifications alter during stem cell aging?
As mentioned in the previous sections, the direct analysis of histone modification in rare adult stem cell populations during aging has been challenging for a long time, as most of chromatin IP protocols require a high number of cells as a starting material. Consequently, as of to date only two recent studies have addressed whether an altered epigenetic regulation of histone modifications in mammalian stem cells is associated with aging. However, although this review focuses on epigenetic profiling in stem cell aging in mammals, it is important to consider that global loss or gain of histone modifications during aging have been reported in other model organisms (reviewed in Benayoun et al. 2015).
Recently, Sun et al. profiled two chromatin marks associated with active transcription, H3K4me3 and H3K36me3, and the repressive mark H3k27me3 in young and old HSCs using low-input ChIP-seq (Sun et al. 2014). Their analysis revealed that old HSCs exhibited a slight increase in the genome-wide number of H3K4me3 peaks, although no differences in expression levels of the various H3K4 methyltransferases were detected. Also, some H3K4me3 domains became broader upon aging. Interestingly, broad H3K4me3 domains have been reported to mark genes that are important for cell identity (Benayoun et al. 2014). Therefore, spreading of H3K4me3 during HSCs aging could account for the increase self-renewal and loss of lineage differentiation observed in aged HSCs. In contrast to what has been observed in HSCs, examination of the same H3K4me3 mark in muscle satellite cells showed few or no differences between cells isolated from young or aged mice (Liu et al. 2013).
The repressive Polycomb-mediated H3k27me3 mark, aged HSCs showed a similar number of peaks in aged compared to young HSCs, but these peaks appeared to become broader and more intense upon aging (Sun et al. 2014). This same effect was also observed in old muscle stem cells (Liu et al. 2013), both at the TSS and in intergenic regions. It is interesting to note that while the increase in H3k27me3 in activated versus quiescent muscle stem cells in young mice correlated with an increase in the expression of the H3K27 methyltransferase Ezh2, the accumulation of H3k27me3 in aging did not correlated with changes in the expression of neither Ezh2 nor with the H3K27 demethylase Jmjd3. Also, over 30 % of the genes that acquired H3K27me3 at their TSSs with age were not expressed in either young or old muscle stem cells, suggesting that the gain of the repressive H3K27me3 mark can underlie other mechanisms different from transcription suppression, and might be driven by processes that do not include canonical methyltransferases or demethylases (for example, decreased histone turnover and preferential accumulation of some histone marks).
As pointed out before, it is important to note that at present it is not trivial to cross-compare ChIP-Seq datasets generated from different groups, due to the huge variety of algorithms and custom-made pipelines. While commonly used peak calling algorithms identify whether a locus is being covered or not by a specific histone marks, it has become relevant also to use pipelines capable of identifying the extent to which individual loci are covered by different histone marks. Due to these issues, and due to the fact that at present only two studies have reported ChIP-seq profiles in aged stem cells (Sun et al. 2014; Liu et al. 2013), it is premature to conclude whether or not a common epigenetic stem cell signature exists in aged stem cells from different tissues.
Mouse models to unravel the mechanism of epigenetic regulation
Next to biochemical assays to directly study chromatin modifications, epigenetic regulation in stem cells can be investigated using animal models in which enzymes responsible for the deposition or removal of histone marks (chromatin modifiers) are disrupted. This review will now focus on some of the most relevant findings generated from these studies.
The H3K27 methylation mark is added by the Polycomb repressive Complex 2 (PRC2), a complex that includes EED, SUZ12, and the catalytically active subunits EZH2/1. Alterations in expression enzymes involved in the transfer or removal of the methyl group on the lysine 27 of histone H3 have been shown to modulate organismal longevity in models other than mouse (reviewed in Benayoun et al. 2015; Jin et al. 2011; Maures et al. 2011; Ni et al. 2012; Siebold et al. 2010). In mammals, no studies have been reported that show that lifespan can be extended by perturbation of expression of epigenetic enzymes, but the effect of such proteins on (lifespan of) adult stem cells is well established. Most of our knowledge on the role of the PcG complexes in stem cells is based on studies in HSCs. For example, the PRC1 subunit Bmi-1 was shown to play an essential role in the generation of self-renewing adult HSCs (Park et al. 2003). Similarly, the Cbx7-PcG subunit was shown to regulate the balance between self-renewal and differentiation (Klauke et al. 2013). Overexpression of Ezh2 in HSCs prevents stem cell exhaustion during serial transplantation (Kamminga et al. 2006). A similar role of Ezh2 has been recently described in muscle satellite cell self-renewal (Woodhouse et al. 2013). Conversely, loss of Ezh1 induces significant depletion of adult HSCs (Hidalgo et al. 2012).
Whereas the Polycomb group proteins are predominantly involved in histone methylation and lead to gene repression, histone acetylation, which is associated with transcriptional activation, is controlled by the opposing activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs). Although direct manipulation of HAT, and HDACs in stem cell with regard to age has not been reported yet, both HATs and HDACs have been implicated in modulating lifespan in mammals. Recent studies in transgenic mice overexpressing the histone deacetylase Sirtuin-1 in the brain showed significant life span extension, supporting a role for histone acetylation in health and lifespan (Satoh et al. 2013). Also, a role of the deacetylase Sirt6 in regulating mammalian longevity has been demonstrated. Mice carrying a constitutive deletion of Sirt6 show reduced lifespan and premature aging, while the overexpression of Sirt6 significantly increased lifespan in male mice (Mostoslavsky et al. 2006; Kanfi et al. 2012). The major mechanism of action of the HDACs is the deacetylation of the histone tails and the consequent transcriptional repression. However, it has also been shown that HDACs may exert their pro-longevity role by promoting increased genomic stability (Oberdoerffer et al. 2008; Wang et al. 2008; Toiber et al. 2013; Van Meter et al. 2014). A link between HATs and aging has also been provided in a progeroid mouse model (HGPS) where the decreased association of HATs with the nuclear periphery showed decreased H4K16ac and decreased lifespan (Krishnan et al. 2011). Interestingly, low H4K16ac levels have been correlated with aging in mouse hematopoietic stem cells (Florian et al. 2012). To what extent these lifespan changes result from stem cell-specific effects remains to be determined. It would therefore be of great interest to further investigate the role of HATs and HDACs during the aging of the multiple types of adult stem cells, especially as a potential therapeutic intervention aimed to selectively target these enzymes could contribute to prevention of aging-related stem cell impairment, and possibly organismal aging.
The genetics of epigenetics
It is clear that epigenetic changes occur in cells as they age, and it appears plausible that such epigenetic changes contribute to the aging process. However, it is also interesting to notice that the rate of aging is quite distinct between genetically distinct individuals, or between different strains of mice (Yuan et al. 2009). Thus, a fascinating question in the field is whether it is possible that some of the differences in epigenetic marks that arise during stem cell aging are in fact controlled by genetic polymorphisms. Such polymorphisms could, for example, reside in loci encoding for the many epigenetic writers and erasers (thus affecting enzymatic activity) or in non-coding regulatory loci. Studies in humans have contributed to the identification of quantitative trait loci (QTLs) or polymorphisms that affect histone modifications or RNA polymerase II occupancy (McVicker et al. 2013; Grubert et al. 2015). As part of the Roadmap Epigenomics Consortium, the integrative analysis of 111 reference human epigenomes and the impact of DNA sequence and genetic variation on epigenomic state was investigated. From this analysis, it appeared that indeed histone modifications and DNA methylation can be predicted by the underlying DNA sequence using DNA motifs analysis in ES cells. Moreover, allelic bias in both transcript levels and epigenomic marks was found for each epigenome analyzed (Roadmap Epigenomics et al. 2015).
While studies from the ENCODE consortium were carried out in human cells, other mammalian models can also be used. For example, the use of genetically diverse inbred mouse strains with different lifespans allows quantitative trait locus analyses to determine the genomic locations of loci that are associated with the aging process. Early studies tested a subset of the BXD (C57BL/6J × DBA/2J) recombinant inbred panel of mice for lifespan (de Haan et al. 1998), and demonstrated that intra-strain variability of lifespan appeared to be genetically controlled. This suggested the presence of loci that induce individual variability among members of an inbred strain of mouse. Such loci would be predicted to encode for epigenetic modifiers, where polymorphisms would result in robust or disorganized epigenetic memory. More recently, other QTLs associated with aging in mice have been mapped (Klebanov et al. 2001; Miller et al. 2002). Therefore, it would be highly interesting to search for and identify the molecular nature of genetic loci that control the extent of epigenetic regulation in stem cells as they age.
Perspectives
Aging is a complex process from which no living organism escapes. Identifying the molecular mechanisms that contribute to, if not cause, aging is a major challenge in aging research. The possibility that epigenetic alterations in aged stem cells contribute to aging of specific tissues and thus to organismal aging has received an ever-increasing attention, partly due to the appealing potential reversibility of such epigenetic modifications. A growing body of evidence, discussed in this review, has shown that epigenetic alterations accumulate as stem cells age, although most of these observations are often species or tissue specific. Substantial effort is required in order to identify whether a common epigenetic signature in aged stem cells exists. This is gradually becoming feasible due to technological advancements to screen multiple histone marks genome-wide. Critical in such analysis will be the use of highly purified and homogeneous cell populations, to rule out bias originating from cellular heterogeneity. In most tissues, purified stem cell populations are only available in limited quantities and therefore they have been so far excluded from genome-wide analyses that require high-input protocols. However, this will soon change as technological advancements will allow highly sensitive analyses using a low number of (stem) cells. It will be very interesting to address how the epigenome of a stem cell changed over its lifespan that is during aging. It will also be very important to standardize pipelines for the data analysis of genome-wide epigenetic data. Multiple complementary efforts have been undertaken by individual laboratories in understanding how the organization of the epigenome varies across rare cell types and different states, included aging. However, due to the early stage of low-input ChIP-seq assays and to the fact that often custom-made pipelines were used for the analysis of these datasets, the results are still in their infancy and are very difficult to be cross-compared between different laboratories. At the moment, it is not obvious that common epigenetic stem cell aging patterns can indeed be identified or rather whether aging at the epigenetic level is a completely random and unpredictable process. Either way, if epigenetic alterations can be causally linked to stem cell aging, targeting of such alterations in aged stem cells might be feasible in the near future. This would open the possibility to reverse, at least some, aging-associated deleterious epigenetic modifications.
Acknowledgments
The authors would like to acknowledge Dr. Leonid Bystrykh for inspiring discussions for the writing of the manuscript. This work was supported by FP7-PEOPLE-2011-ITN Project HEM-ID.
References
- Adams PD, Jasper H, Rudolph KL. Aging-induced stem cell mutations as drivers for disease and cancer. Cell Stem Cell. 2015;16(6):601–612. doi: 10.1016/j.stem.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adli M, Bernstein BE. Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nat Protoc. 2011;6(10):1656–1668. doi: 10.1038/nprot.2011.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman I, Rossi DJ. Epigenetic control of stem cell potential during homeostasis, aging, and disease. Cell Stem Cell. 2015;16(6):613–625. doi: 10.1016/j.stem.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman I, Bock C, Garrison BS, Smith ZD, Gu H, Meissner A, Rossi DJ. Proliferation-dependent alterations of the DNA methylation landscape underlie hematopoietic stem cell aging. Cell Stem Cell. 2013;12(4):413–425. doi: 10.1016/j.stem.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Behrens A, van Deursen JM, Rudolph KL, Schumacher B. Impact of genomic damage and ageing on stem cell function. Nat Cell Biol. 2014;16(3):201–207. doi: 10.1038/ncb2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED, Devarajan K, Daugherty AC, Kundaje AB, Mancini E, Hitz BC, Gupta R, Rando TA, Baker JC, Snyder MP, Cherry JM, Brunet A. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014;158(3):673–688. doi: 10.1016/j.cell.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Brunet A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat Rev Mol Cell Biol. 2015;16(10):593–610. doi: 10.1038/nrm4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23(7):781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Beerman I, Lien WH, Smith ZD, Gu H, Boyle P, Gnirke A, Fuchs E, Rossi DJ, Meissner A. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell. 2012;47(4):633–647. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330(6004):612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondolfi L, Ermini F, Long JM, Ingram DK, Jucker M. Impact of age and caloric restriction on neurogenesis in the dentate gyrus of C57BL/6 mice. Neurobiol Aging. 2004;25(3):333–340. doi: 10.1016/S0197-4580(03)00083-6. [DOI] [PubMed] [Google Scholar]
- Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, Rando TA. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317(5839):807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- Brind’Amour J, Liu S, Hudson M, Chen C, Karimi MM, Lorincz MC. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat Commun. 2015;6:6033. doi: 10.1038/ncomms7033. [DOI] [PubMed] [Google Scholar]
- Broske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, Nerlov C, Leutz A, Andrade-Navarro MA, Jacobsen SE, Rosenbauer F. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41(11):1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- Carlson ME, Conboy IM. Loss of stem cell regenerative capacity within aged niches. Aging Cell. 2007;6(3):371–382. doi: 10.1111/j.1474-9726.2007.00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoni DI, Valeri A, Le Gall A, Nollmann M. A matter of scale: how emerging technologies are redefining our view of chromosome architecture. Trends Genet. 2015;31(8):454–464. doi: 10.1016/j.tig.2015.05.011. [DOI] [PubMed] [Google Scholar]
- Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93(4):505–518. doi: 10.1016/S0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Cho RH, Sieburg HB, Muller-Sieburg CE. A new mechanism for the aging of hematopoietic stem cells: aging changes the clonal composition of the stem cell compartment but not individual stem cells. Blood. 2008;111(12):5553–5561. doi: 10.1182/blood-2007-11-123547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clelland CD, Choi M, Romberg C, Clemenson GD, Jr, Fragniere A, Tyers P, Jessberger S, Saksida LM, Barker RA, Gage FH, Bussey TJ. A functional role for adult hippocampal neurogenesis in spatial pattern separation. Science. 2009;325(5937):210–213. doi: 10.1126/science.1173215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Smythe GM, Rando TA. Notch-mediated restoration of regenerative potential to aged muscle. Science. 2003;302(5650):1575–1577. doi: 10.1126/science.1087573. [DOI] [PubMed] [Google Scholar]
- de Haan G, Van Zant G. Dynamic changes in mouse hematopoietic stem cell numbers during aging. Blood. 1999;93(10):3294–3301. [PubMed] [Google Scholar]
- de Haan G, Gelman R, Watson A, Yunis E, Van Zant G. A putative gene causes variability in lifespan among genotypically identical mice. Nat Genet. 1998;19(2):114–116. doi: 10.1038/465. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295(5558):1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Drummond-Barbosa D. Stem cells, their niches and the systemic environment: an aging network. Genetics. 2008;180(4):1787–1797. doi: 10.1534/genetics.108.098244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykstra B, Kent D, Bowie M, McCaffrey L, Hamilton M, Lyons K, Lee SJ, Brinkman R, Eaves C. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1(2):218–229. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Enwere E, Shingo T, Gregg C, Fujikawa H, Ohta S, Weiss S. Aging results in reduced epidermal growth factor receptor signaling, diminished olfactory neurogenesis, and deficits in fine olfactory discrimination. J Neurosci. 2004;24(38):8354–8365. doi: 10.1523/JNEUROSCI.2751-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJ, Shishkin AA, Hatan M, Carrasco-Alfonso MJ, Mayer D, Luckey CJ, Patsopoulos NA, De Jager PL, Kuchroo VK, Epstein CB, Daly MJ, Hafler DA, Bernstein BE. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518(7539):337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farlik M, Sheffield NC, Nuzzo A, Datlinger P, Schonegger A, Klughammer J, Bock C. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 2015;10(8):1386–1397. doi: 10.1016/j.celrep.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. Elevated histone expression promotes life span extension. Mol Cell. 2010;39(5):724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florath I, Butterbach K, Muller H, Bewerunge-Hudler M, Brenner H. Cross-sectional and longitudinal changes in DNA methylation with age: an epigenome-wide analysis revealing over 60 novel age-associated CpG sites. Hum Mol Genet. 2014;23(5):1186–1201. doi: 10.1093/hmg/ddt531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florian MC, Dorr K, Niebel A, Daria D, Schrezenmeier H, Rojewski M, Filippi MD, Hasenberg A, Gunzer M, Scharffetter-Kochanek K, Zheng Y, Geiger H. Cdc42 activity regulates hematopoietic stem cell aging and rejuvenation. Cell Stem Cell. 2012;10(5):520–530. doi: 10.1016/j.stem.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garinis GA, van der Horst GT, Vijg J, Hoeijmakers JH. DNA damage and ageing: new-age ideas for an age-old problem. Nat Cell Biol. 2008;10(11):1241–1247. doi: 10.1038/ncb1108-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjoneska E, Pfenning AR, Mathys H, Quon G, Kundaje A, Tsai LH, Kellis M. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature. 2015;518(7539):365–369. doi: 10.1038/nature14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Klar AJ. Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell. 1996;86(1):95–101. doi: 10.1016/S0092-8674(00)80080-X. [DOI] [PubMed] [Google Scholar]
- Groves AK. The challenge of hair cell regeneration. Exp Biol Med (Maywood) 2010;235(4):434–446. doi: 10.1258/ebm.2009.009281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubert F, Zaugg JB, Kasowski M, Ursu O, Spacek DV, Martin AR, Greenside P, Srivas R, Phanstiel DH, Pekowska A, Heidari N, Euskirchen G, Huber W, Pritchard JK, Bustamante CD, Steinmetz LM, Kundaje A, Snyder M. Genetic control of chromatin states in humans involves local and distal chromosomal interactions. Cell. 2015;162(5):1051–1065. doi: 10.1016/j.cell.2015.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, Zhang K, Ming GL, Gao Y, Song H. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14(10):1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Zhu P, Guo F, Li X, Wu X, Fan X, Wen L, Tang F. Profiling DNA methylome landscapes of mammalian cells with single-cell reduced-representation bisulfite sequencing. Nat Protoc. 2015;10(5):645–659. doi: 10.1038/nprot.2015.039. [DOI] [PubMed] [Google Scholar]
- Hattiangady B, Shetty AK. Aging does not alter the number or phenotype of putative stem/progenitor cells in the neurogenic region of the hippocampus. Neurobiol Aging. 2008;29(1):129–147. doi: 10.1016/j.neurobiolaging.2006.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, Diez J, Sanchez-Mut JV, Setien F, Carmona FJ, Puca AA, Sayols S, Pujana MA, Serra-Musach J, Iglesias-Platas I, Formiga F, Fernandez AF, Fraga MF, Heath SC, Valencia A, Gut IG, Wang J, Esteller M. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci USA. 2012;109(26):10522–10527. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo I, Herrera-Merchan A, Ligos JM, Carramolino L, Nunez J, Martinez F, Dominguez O, Torres M, Gonzalez S. Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell. 2012;11(5):649–662. doi: 10.1016/j.stem.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Li J, Green CD, Yu X, Tang X, Han D, Xian B, Wang D, Huang X, Cao X, Yan Z, Hou L, Liu J, Shukeir N, Khaitovich P, Chen CD, Zhang H, Jenuwein T, Han JD. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 2011;14(2):161–172. doi: 10.1016/j.cmet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Jung M, Pfeifer GP. Aging and DNA methylation. BMC Biol. 2015;13:7. doi: 10.1186/s12915-015-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga LM, van Os R, Ausema A, Noach EJ, Weersing E, Dontje B, Vellenga E, de Haan G. Impaired hematopoietic stem cell functioning after serial transplantation and during normal aging. Stem Cells. 2005;23(1):82–92. doi: 10.1634/stemcells.2004-0066. [DOI] [PubMed] [Google Scholar]
- Kamminga LM, Bystrykh LV, de Boer A, Houwer S, Douma J, Weersing E, Dontje B, de Haan G. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107(5):2170–2179. doi: 10.1182/blood-2005-09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483(7388):218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- Klauke K, Radulovic V, Broekhuis M, Weersing E, Zwart E, Olthof S, Ritsema M, Bruggeman S, Wu X, Helin K, Bystrykh L, de Haan G. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat Cell Biol. 2013;15(4):353–362. doi: 10.1038/ncb2701. [DOI] [PubMed] [Google Scholar]
- Klebanov S, Astle CM, Roderick TH, Flurkey K, Archer JR, Chen J, Harrison DE. Maximum life spans in mice are extended by wild strain alleles. Exp Biol Med (Maywood) 2001;226(9):854–859. doi: 10.1177/153537020122600908. [DOI] [PubMed] [Google Scholar]
- Krishnan V, Chow MZ, Wang Z, Zhang L, Liu B, Liu X, Zhou Z. Histone H4 lysine 16 hypoacetylation is associated with defective DNA repair and premature senescence in Zmpste24-deficient mice. Proc Natl Acad Sci USA. 2011;108(30):12325–12330. doi: 10.1073/pnas.1102789108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn HG, Dickinson-Anson H, Gage FH. Neurogenesis in the dentate gyrus of the adult rat: age-related decrease of neuronal progenitor proliferation. J Neurosci. 1996;16(6):2027–2033. doi: 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D, Jung I, Rajagopal N, Schmitt A, Selvaraj S, Lee AY, Yen CA, Lin S, Lin Y, Qiu Y, Xie W, Yue F, Hariharan M, Ray P, Kuan S, Edsall L, Yang H, Chi NC, Zhang MQ, Ecker JR, Ren B. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature. 2015;518(7539):350–354. doi: 10.1038/nature14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Rando TA. Manifestations and mechanisms of stem cell aging. J Cell Biol. 2011;193(2):257–266. doi: 10.1083/jcb.201010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cheung TH, Charville GW, Hurgo BM, Leavitt T, Shih J, Brunet A, Rando TA. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013;4(1):189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegawa S, Hinkal G, Kim HS, Shen L, Zhang L, Zhang J, Zhang N, Liang S, Donehower LA, Issa JP. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20(3):332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Curr Opin Cell Biol. 2007;19(3):266–272. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Maslov AY, Barone TA, Plunkett RJ, Pruitt SC. Neural stem cell detection, characterization, and age-related changes in the subventricular zone of mice. J Neurosci. 2004;24(7):1726–1733. doi: 10.1523/JNEUROSCI.4608-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maures TJ, Greer EL, Hauswirth AG, Brunet A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell. 2011;10(6):980–990. doi: 10.1111/j.1474-9726.2011.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayle A, Yang L, Rodriguez B, Zhou T, Chang E, Curry CV, Challen GA, Li W, Wheeler D, Rebel VI, Goodell MA. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2015;125(4):629–638. doi: 10.1182/blood-2014-08-594648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G, van de Geijn B, Degner JF, Cain CE, Banovich NE, Raj A, Lewellen N, Myrthil M, Gilad Y, Pritchard JK. Identification of genetic variants that affect histone modifications in human cells. Science. 2013;342(6159):747–749. doi: 10.1126/science.1242429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RA, Chrisp C, Jackson AU, Galecki AT, Burke DT. Coordinated genetic control of neoplastic and nonneoplastic diseases in mice. J Gerontol A Biol Sci Med Sci. 2002;57(1):B3–B8. doi: 10.1093/gerona/57.1.B3. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443(7110):448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124(2):315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Ni Z, Ebata A, Alipanahiramandi E, Lee SS. Two SET domain containing genes link epigenetic changes and aging in Caenorhabditis elegans. Aging Cell. 2012;11(2):315–325. doi: 10.1111/j.1474-9726.2011.00785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura EK, Granter SR, Fisher DE. Mechanisms of hair graying: incomplete melanocyte stem cell maintenance in the niche. Science. 2005;307(5710):720–724. doi: 10.1126/science.1099593. [DOI] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135(5):907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423(6937):302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Peinado MA, Quesada A, Pedrosa JA, Torres MI, Martinez M, Esteban FJ, Del Moral ML, Hernandez R, Rodrigo J, Peinado JM. Quantitative and ultrastructural changes in glia and pericytes in the parietal cortex of the aging rat. Microsc Res Tech. 1998;43(1):34–42. doi: 10.1002/(SICI)1097-0029(19981001)43:1<34::AID-JEMT6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Plongthongkum N, Diep DH, Zhang K. Advances in the profiling of DNA modifications: cytosine methylation and beyond. Nat Rev Genet. 2014;15(10):647–661. doi: 10.1038/nrg3772. [DOI] [PubMed] [Google Scholar]
- Polak P, Karlic R, Koren A, Thurman R, Sandstrom R, Lawrence MS, Reynolds A, Rynes E, Vlahovicek K, Stamatoyannopoulos JA, Sunyaev SR. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518(7539):360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Maslau S, Andrew T, Yang TP, Beyan H, Whittaker P, McCann OT, Finer S, Valdes AM, Leslie RD, Deloukas P, Spector TD. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010;20(4):434–439. doi: 10.1101/gr.103101.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando TA. Stem cells, ageing and the quest for immortality. Nature. 2006;441(7097):1080–1086. doi: 10.1038/nature04958. [DOI] [PubMed] [Google Scholar]
- Renault V, Thornell LE, Eriksson PO, Butler-Browne G, Mouly V. Regenerative potential of human skeletal muscle during aging. Aging Cell. 2002;1(2):132–139. doi: 10.1046/j.1474-9728.2002.00017.x. [DOI] [PubMed] [Google Scholar]
- Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi DJ, Bryder D, Zahn JM, Ahlenius H, Sonu R, Wagers AJ, Weissman IL. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci USA. 2005;102(26):9194–9199. doi: 10.1073/pnas.0503280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol. 2015;33(11):1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED, Yamada KA, Imai S. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013;18(3):416–430. doi: 10.1016/j.cmet.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl C, Rendeiro AF, Sheffield NC, Bock C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat Methods. 2015;12(10):963–965. doi: 10.1038/nmeth.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaranarayanan P, Mendoza-Parra MA, Walia M, Wang L, Li N, Trindade LM, Gronemeyer H. Single-tube linear DNA amplification (LinDA) for robust ChIP-seq. Nat Methods. 2011;8(7):565–567. doi: 10.1038/nmeth.1626. [DOI] [PubMed] [Google Scholar]
- Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW, McLeod JL, Doedens M, Medeiros JJ, Marke R, Kim HJ, Lee K, McPherson JD, Hudson TJ, Consortium HP-LGP, Brown AM, Yousif F, Trinh QM, Stein LD, Minden MD, Wang JC, Dick JE. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebold AP, Banerjee R, Tie F, Kiss DL, Moskowitz J, Harte PJ. Polycomb Repressive Complex 2 and Trithorax modulate Drosophila longevity and stress resistance. Proc Natl Acad Sci USA. 2010;107(1):169–174. doi: 10.1073/pnas.0907739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, Peat J, Andrews SR, Stegle O, Reik W, Kelsey G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods. 2014;11(8):817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HJ, Stevens CF, Gage FH. Neural stem cells from adult hippocampus develop essential properties of functional CNS neurons. Nat Neurosci. 2002;5(5):438–445. doi: 10.1038/nn844. [DOI] [PubMed] [Google Scholar]
- Sudo K, Ema H, Morita Y, Nakauchi H. Age-associated characteristics of murine hematopoietic stem cells. J Exp Med. 2000;192(9):1273–1280. doi: 10.1084/jem.192.9.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Luo M, Jeong M, Rodriguez B, Xia Z, Hannah R, Wang H, Le T, Faull KF, Chen R, Gu H, Bock C, Meissner A, Gottgens B, Darlington GJ, Li W, Goodell MA. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014;14(5):673–688. doi: 10.1016/j.stem.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Jones JM, McGehee RE, Rando TA, Lecka-Czernik B, Lipschitz DA, Peterson CA. Activation of an adipogenic program in adult myoblasts with age. Mech Ageing Dev. 2002;123(6):649–661. doi: 10.1016/S0047-6374(01)00411-0. [DOI] [PubMed] [Google Scholar]
- Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor B, Giacosa S, D’Urso A, Naar AM, Kingston R, Rippe K, Mostoslavsky R. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell. 2013;51(4):454–468. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, Gorbunova V. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun. 2014;5:5011. doi: 10.1038/ncomms6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, Jia R, Zheng ZM, Appella E, Wang XW, Ried T, Deng CX. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14(4):312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner CI, Lin Q, Koch CM, Eisele L, Beier F, Ziegler P, Bauerschlag DO, Jockel KH, Erbel R, Muhleisen TW, Zenke M, Brummendorf TH, Wagner W. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014;15(2):R24. doi: 10.1186/gb-2014-15-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220(4601):1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- Wilson VL, Smith RA, Ma S, Cutler RG. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem. 1987;262(21):9948–9951. [PubMed] [Google Scholar]
- Woodhouse S, Pugazhendhi D, Brien P, Pell JM. Ezh2 maintains a key phase of muscle satellite cell expansion but does not regulate terminal differentiation. J Cell Sci. 2013;126(Pt 2):565–579. doi: 10.1242/jcs.114843. [DOI] [PubMed] [Google Scholar]
- Yuan R, Tsaih SW, Petkova SB, Marin de Evsikova C, Xing S, Marion MA, Bogue MA, Mills KD, Peters LL, Bult CJ, Rosen CJ, Sundberg JP, Harrison DE, Churchill GA, Paigen B. Aging in inbred strains of mice: study design and interim report on median lifespans and circulating IGF1 levels. Aging Cell. 2009;8(3):277–287. doi: 10.1111/j.1474-9726.2009.00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Edri R, Yaffe Y, Donaghey J, Pop R, Mallard W, Issner R, Gifford CA, Goren A, Xing J, Gu H, Cacchiarelli D, Tsankov AM, Epstein C, Rinn JL, Mikkelsen TS, Kohlbacher O, Gnirke A, Bernstein BE, Elkabetz Y, Meissner A. Dissecting neural differentiation regulatory networks through epigenetic footprinting. Nature. 2015;518(7539):355–359. doi: 10.1038/nature13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zykovich A, Hubbard A, Flynn JM, Tarnopolsky M, Fraga MF, Kerksick C, Ogborn D, MacNeil L, Mooney SD, Melov S. Genome-wide DNA methylation changes with age in disease-free human skeletal muscle. Aging Cell. 2014;13(2):360–366. doi: 10.1111/acel.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]