Abstract
Malaria is a worldwide disease caused by Plasmodium parasites. A sesquiterpene endoperoxide artemisinin isolated from Artemisia annua was discovered and has been accepted for its use in artemisinin-based combinatorial therapies, as the most effective current antimalarial treatment. However, the quantity of this compound produced from the A. annua plant is very low, and the availability of artemisinin is insufficient to treat all infected patients. In addition, the emergence of artemisinin-resistant Plasmodium has been reported recently. Several techniques have been applied to enhance artemisinin availability, and studies related to its mode of action and the mechanism of resistance of malaria-causing parasites are ongoing. In this review, we summarize the application of modern technologies to improve the production of artemisinin, including our ongoing research on artemisinin biosynthetic genes in other Artemisia species. The current understanding of the mode of action of artemisinin as well as the mechanism of resistance against this compound in Plasmodium parasites is also presented. Finally, the current situation of malaria infection and the future direction of antimalarial drug development are discussed.
Keywords: Artemisia annua, Artemisinin, Resistant parasites, Malaria, Metabolic engineering, Mode of action
Introduction
As a worldwide disease, malaria has been one of the main cause of illness and death in humans for over a century, especially in sub-Saharan Africa and Southeast Asia. More than 200 million cases of malaria are reported every year; in 2015, there were 214 million cases and 438,000 related deaths [1]. This disease is caused by five species of Plasmodium parasites: P. falciparum, P. vivax, P. malariae, P. ovale, and P. knowlesi. Among these, P. falciparum is the major cause of malaria infection in Africa, and P. vivax is the most widely distributed malaria-causing parasite globally [1]. Several antimalarial drugs have been developed since the seventeenth century. However, malaria-causing parasites have developed resistance to these conventional drugs, leading to treatment failure.
In response to the urgent need for new antimalarial drugs, Chinese scientists Professor Youyou Tu and her research group discovered artemisinin, the most effective antimalarial drug derived from Artemisia annua in 1971 [2]. Artemisinin is a sesquiterpene lactone with an endoperoxide bridge, which is necessary for antimalarial activity during multiple stages of parasite development [3–7]. Owing to its rapid action and high effectiveness against malaria, the combination of artemisinin derivatives and other antimalarial drugs, so-called artemisinin-based combination therapies (ACTs), has been recommended as the first-line treatments against malaria since 2006 [8]. ACTs have become the most powerful strategy to prevent malaria and related deaths. Professor Tu was then awarded the Nobel Prize in Physiology or Medicine in 2015 for the discovery of this effective antimalarial compound.
The demand for ACTs increases dramatically each year; yet, the production yield of artemisinin from A. annua is very low and varies widely from 0.01 to 2 % dry weight [9]. Alternative approaches, including plant breeding technologies, synthetic biology, and total and semi-syntheses of artemisinin, have been investigated to enhance the production and reduce the cost of this compound. In addition, the recent emergence of artemisinin-resistant Plasmodium parasites has also become a new challenge to scientists in the elucidation of the mechanism of resistance and identification of the new strategies for malaria treatment.
In this review, we summarize recent studies on the enhancement of artemisinin production and on artemisinin biosynthetic genes in other Artemisia species, conducted in our laboratory. In addition, the current understanding of the mode of action of artemisinin against malaria-causing parasites and, in turn, the mechanism of resistance of the parasites to this compound are also presented. Finally, the current situation of malaria infection and future directions, including ongoing studies on antimalarial drug development, are discussed.
Discovery of artemisinin
Before the discovery of artemisinin, powder derived from cinchona tree bark had been used to treat malaria since the seventeenth century. The active compound from this plant, quinine, was first isolated in 1820 and was used as the only effective antimalarial compound until the 1920s. The quinine derivative chloroquine was developed as a new effective antimalarial drug once quinine-resistant Plasmodium strains appeared. During that time, the insecticide DDT was widely used to control the spread of infected mosquitoes as well. However, in the 1960s, increasing of chloroquine-resistant Plasmodium strains and DDT-resistant mosquitoes became a critical sign of the failure of malaria prevention and treatment [10].
In response to the urgent need for new antimalarial drugs, the Chinese government launched a national project against malaria called Project 523 in 1967 [2]. The group, led by Professor Youyou Tu, investigated more than 2000 Chinese herbs used as traditional Chinese medicines to treat fever. Among these herbs, an extract from A. annua showed highly effective inhibition against growth of malaria-causing parasites. The active antimalarial components were then extracted from the leaves of mature plants in 1971 [2, 10–12]. After purification, the active antimalarial compound, named qinghaosu or artemisinin, was obtained as colorless needle-like crystals. Its stereochemistry and chemical and X-ray crystal structures were determined and reported several years later [2, 10, 11, 13]. Clinical trials involving either a non-toxic A. annua extract or pure artemisinin have been conducted since 1972 by several groups, and all patients in these trials quickly recovered from the disease [11, 12]. These results clearly indicated that artemisinin is an effective antimalarial compound with rapid action and low toxicity.
Despite showing effective antimalarial activity, the low solubility of artemisinin in both oil and water becomes a therapeutic limitation of this compound. To address this problem, many scientists have developed semi-synthetic drugs and synthesized artemisinin derivatives with higher solubility. Some of these artemisinin derivatives, which have been used until the present, include dihydroartemisinin, artemether, and artesunate [14]. In addition, the combination of artemisinin or its derivatives with other conventional antimalarial drugs greatly increased the parasite clearance rate in patients and was first recommended as a new strategy for malaria treatment in 1984 [15]. This strategy, known as ACT, has been recommended by the World Health Organization (WHO) as a first-line treatment for malaria to prevent recurrence and development of resistance in malaria-causing parasites, whereas the monotherapy is considered as an inappropriate treatment [2, 8, 13, 14, 16].
Biosynthesis of artemisinin and expression pattern of artemisinin biosynthetic genes in A. annua
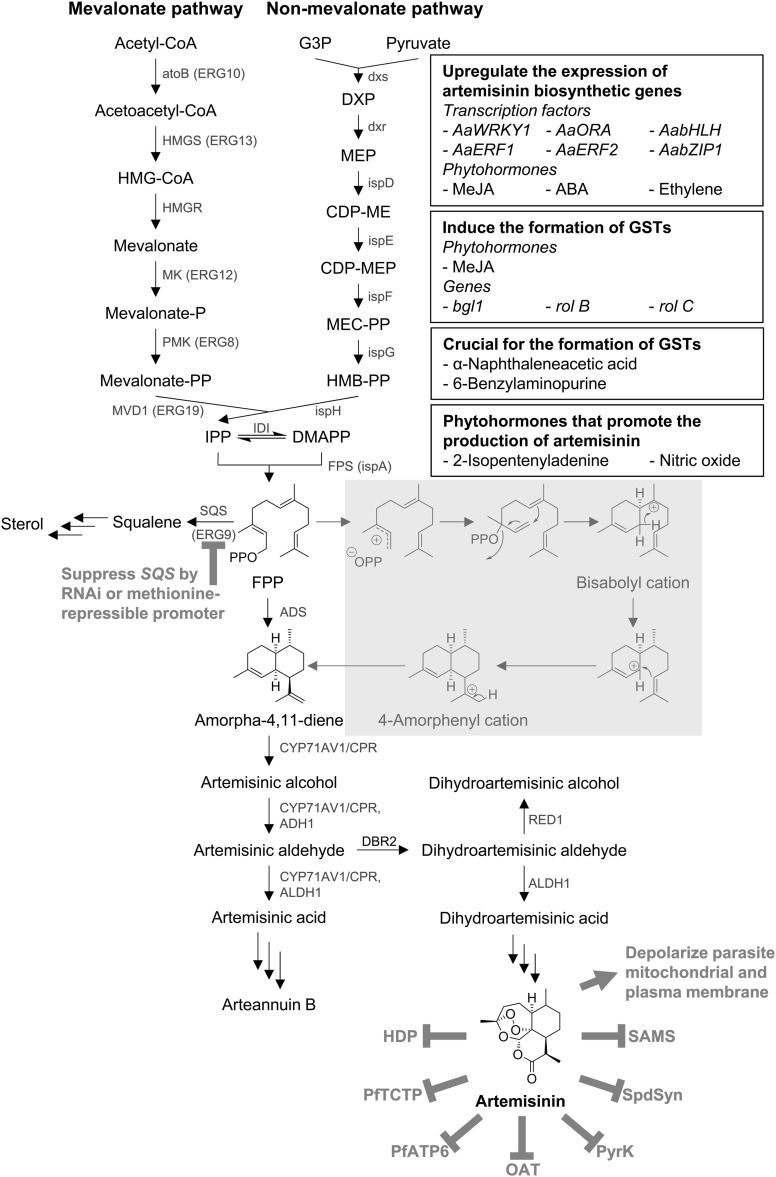
A precursor of artemisinin, farnesyl pyrophosphate (FPP, C15), is synthesized from two C-5 isoprenoid units derived from the cytosolic mevalonate (MVA) pathway and one isoprenoid unit derived from the non-mevalonate (MEP or DXP) pathway [17, 18]. FPP is cyclized to amorpha-4,11-diene by amorpha-4,11-diene synthase (ADS) [19–21] via the generation of bisabolyl and 4-amorphenyl cation intermediates [22, 23] (Fig. 1). The following step is the oxidation of amorpha-4,11-diene to artemisinic alcohol by amorpha-4,11-diene 12-monooxygenase (CYP71AV1) [24]. This enzyme also catalyzes the oxidation of artemisinic alcohol to artemisinic aldehyde and artemisinic acid. In addition, alcohol dehydrogenase 1 (ADH1) and aldehyde dehydrogenase 1 (ALDH1) also show specific oxidation activity on artemisinic alcohol into artemisinic aldehyde and on artemisinic aldehyde into artemisinic acid, respectively [25, 26]. Artemisinic acid was thought to be the last precursor of artemisinin. However, it has been revealed that this compound is converted non-enzymatically into arteannuin B and related compounds, rather than artemisinin [27]. The next step of artemisinin biosynthesis is the reduction of artemisinic aldehyde into dihydroartemisinic aldehyde by artemisinic aldehyde Δ11(13) reductase (DBR2) [28]. Then, ALDH1 oxidizes dihydroartemisinic aldehyde into dihydroartemisinic acid, which is converted non-enzymatically into artemisinin [26, 29], as shown in Fig. 1. Rydén et al. [30] discovered dihydroartemisinic aldehyde reductase 1 (RED1), which reduces dihydroartemisinic aldehyde into dihydroartemisinic alcohol. Although the role of RED1 in artemisinin biosynthesis is still unclear, it has been suggested that the silencing of RED1 might increase the production of artemisinin in A. annua.
Fig. 1.
Summary of artemisinin biosynthesis, transgenic approaches to enhance artemisinin production, and artemisinin mode of action. The enzymes responsible for each reaction are indicated next to the arrows. Suppression of competing pathways and artemisinin activity and its targets are shown in bold. Transgenic approaches regulating artemisinin production are shown in black boxes. Cyclization mechanism of FPP to generate amorpha-4,11-diene is highlighted in gray. Full names of intermediates and enzymes involved in the pathway are as follows: HMG-CoA 3-hydroxy-3-methylglutaryl-coenzyme A, G3P glycerol-3-phosphate, DXP 1-deoxy-d-xylulose 5-phosphate, MEP 2C-methyl-d-erythritol 4-phosphate, CDP-ME 4-diphosphocytidyl-2C-methyl d-erythritol, CDP-MEP CDP-ME 2-phosphate, MEC-PP 2C-methyl-d-erythritol 2,4-cyclodiphosphate, HMB-PP (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate, IPP isopentenyl pyrophosphate, DMAPP dimethylallyl pyrophosphate, atoB (ERG10) acetoacetyl-CoA thiolase, HMGS (ERG13) HMG-CoA synthase, HMGR HMG-CoA reductase, MK (EGR12) mevalonate kinase, PMK (ERG8) phosphomevalonate kinase, MVD1 (ERG19) mevalonate pyrophosphate decarboxylase, dxs DXP synthase, dxr DXP reductase, ispD CDP-ME synthase, ispE CDP-ME kinase, ispF MEC-PP synthase, ispG HMB-PP synthase, ispH HMB-PP reductase, IDI IPP isomerase, FPS (ispA) farnesyl pyrophosphate (FPP) synthase, SQS (ERG9) squalene synthase, ADS amorpha-4,11-diene synthase, CYP71AV1 amorpha-4,11-diene 12-monooxygenase, CPR cytochrome P450 reductase, ADH1 alcohol dehydrogenase 1, ALDH1 aldehyde dehydrogenase 1, DBR2 artemisinic aldehyde Δ11(13) reductase, RED1 dihydroartemisinic aldehyde reductase 1 (color figure online)
Artemisinin is produced mainly in glandular secretory trichomes (GSTs) and its accumulation level declines as plants mature. Olofsson et al. [31] showed that GSTs of A. annua are found in all aerial tissues of plants, but not in roots or hairy roots. The density of GSTs is highest in flower buds and young leaves and decreases as leaves age.
The expression pattern of genes involved in the artemisinin biosynthetic pathway has been investigated extensively for over a decade. The expression of genes in the upstream pathway shows no correlation with the density of GSTs or the accumulation levels of artemisinin intermediates [32]. In contrast, the expression of genes in the downstream pathway is consistent with the density of GSTs in each tissue. The expression of ADS is highest in GSTs, high in flower buds and young leaves, low in stems, negligible in old leaves and hairy roots, and not detected in roots [31, 33–37]. CYP71AV1, DBR2, and ALDH1 showed similar expression patterns: highest in GSTs and very low in stems and roots. In hairy roots, the expression levels of CYP71AV1 and DBR2 are relatively low, but the expression of ALDH1 is negligible [24, 26, 28, 31]. The expression levels of CYP71AV1 and DBR2 in leaves and flowers show similar patterns, as they are high in leaf primordia and flower buds but decrease as leaves and flowers develop [38–40]. The expression pattern of ALDH1 in leaves at different stages is similar to those of CYP71AV1 and DBR2 [31]. Although there is no report on the expression level of ALDH1 during different stages of flowering, this gene shows higher expression in flowers than in leaves [26, 31]. The expression of RED1 is relatively low in flower buds, young leaves, and stems. In contrast, the expression of this gene is much higher in old leaves and roots than in young leaves [30, 31]. Interestingly, the expression of RED1 is approximately 50-fold higher in hairy roots compared with old leaves. Nevertheless, the function of RED1 in hairy roots has not been established [31].
The expression levels of ADS and ALDH1, as well as their enzymatic activities in high-artemisinin-producing and low-artemisinin-producing A. annua cultivars, show no differences. Even though the expression levels of CYP71AV1 in these two cultivars are similar, CYP71AV1 in a high-artemisinin-producing cultivar shows lower enzyme activity, which is suitable for the change in metabolic flux to dihydro-analogues and artemisinin production [41]. In contrast, the activity of DBR2 in both cultivars shows no significant difference, but the gene encoding this enzyme shows considerably higher expression levels in high-artemisinin-producing cultivars than in low-artemisinin-producing cultivars [42].
Mode of action of artemisinin
Before artemisinin can exert its action, the endoperoxide bridge has to be activated to generate the free radical species. Two activation pathways of artemisinin have been suggested, namely the mitochondrial and heme-mediated degradation pathways [43]. Mitochondria-activated artemisinin is involved in lipid peroxidation inducing cytotoxicity via the generation of reactive oxygen species (ROS) and depolarization of the parasite mitochondrial and plasma membranes [43–47]. In the heme-mediated pathway, two activation models (i.e., a reductive scission model and an open peroxide model) have been proposed, both of which lead to the generation of an active carbon-centered radical [48]. Even though the non-heme Fe2+ ion was suggested to bind and activate artemisinin [7], recent studies showed that heme plays a predominant role in artemisinin activation rather than the Fe2+ ion [5]. In Plasmodium spp., heme is produced via endogenous heme biosynthesis at the early ring stage and via hemoglobin digestion at the trophozoite stage. However, the level of heme biosynthesized endogenously in the parasites is much lower than its production via hemoglobin digestion, suggesting that hemoglobin-derived heme plays a major role in artemisinin activation [5, 49]. Recently, Xie et al. [50] reported that falcipains FP2a and FP3 (two main cysteine protease hemoglobinases) are also involved in the potential activation of artemisinin at an early ring stage.
After hemoglobin digestion, the heme detoxification protein (HDP) can trigger the conversion of free heme to hemozoin, which is essential for parasite survival [4, 51]. However, the formation of the artemisinin-free heme complex shows an inhibitory effect on this conversion [51]. A translationally controlled tumor protein (PfTCTP) was also reported as a potential target of artemisinin, as it could form a covalent bond with this protein, resulting in protein malfunction [52, 53]. Eckstein-Ludwig et al. [54] showed that artemisinin specifically mediated the inhibition of PfATP6, an orthologous sarco/endoplasmic reticulum Ca2+-ATPase (SERCA), outside the food vacuole. Recently, five enzymes involved in the key metabolic pathways of the parasite were also reported as potential targets of artemisinin, namely ornithine aminotransferase (OAT), pyruvate kinase (PyrK), l-lactate dehydrogenase (LDH), spermidine synthase (SpdSyn), and S-adenosylmethionine synthetase (SAMS). All of them are covalently modified by the interaction with artemisinin, resulting in the irreversible malfunction of enzyme activities [5].
Enhancement of artemisinin production
The demand for artemisinin increases every year. Even though total synthesis of artemisinin from commercially available chemicals or semi-synthesis from its intermediates have been reported, all of those methods are costly and require several synthesis steps [55, 56]. In this review, we summarize recent studies regarding four approaches to enhance the production of artemisinin: (1) plant breeding technologies, (2) overexpression of genes involved in the artemisinin biosynthetic pathway, (3) direct or indirect upregulation of artemisinin biosynthesis, and (4) heterologous production.
Plant breeding technologies
Conventional plant breeding techniques to select high-artemisinin-producing cultivars have been used for decades. These techniques include cultivation of A. annua and collection of cultivars with the desired properties. At present, a robust hybrid A. annua is generated from the combination of high-artemisinin-producing and vigorous cultivars to increase the production yield of artemisinin to more than 2 % dry weight [57–59]. Recently, an alternative approach to increase the production of artemisinin from the cultivation of high-artemisinic acid or dihydroartemisinic acid-producing cultivars was proposed, since a method for the semi-synthesis of artemisinin from these two precursors has been developed [56, 60].
Scientists at the University of York used advanced breeding techniques to evaluate the distribution of traits that contribute to artemisinin yield [61]. From the screening of 23,000 strains, they succeeded in identifying genes and molecular markers for fast-track breeding, enabling the construction of a detailed genetic map of A. annua with nine linkage groups. The established quantitative trait loci (QTL) map is also applicable for rapid identification of A. annua parental lines with useful traits for plant breeding. Two hybrids, called Hybrid 1209r Shennong and Hybrid 8001r Zenith, were developed with high artemisinin productivity of up to 36.3 and 54.5 kg/ha, respectively. The diallel cross approach to determine the combining ability of the robust parental lines for the production of artemisinin high-yielding A. annua hybrids was also developed by the same group and showed consistent results with the QTL-based molecular breeding approach [62].
Hairy root culture is another method to enhance the production of secondary (specialized) metabolites, owing to its rapid growth capabilities [63]. Transformation protocols to obtain hairy roots containing artemisinin from this plant have been reported [64, 65]. In our laboratory, we also attempted to establish the conditions for A. annua hairy root cultivation. However, we still could not detect even trace amounts of artemisinin or its intermediates from the extract of hairy root cultures by GC–MS (unpublished data). Artemisinin biosynthetic genes are highly expressed in trichomes but almost negligible in root tissue [31, 33–40], suggesting that the production of this compound by hairy root cultures could be somewhat difficult. Therefore, the most suitable conditions for hairy root cultures to enhance production of artemisinin must be investigated. In addition, the identification of artemisinin production from root extracts requires extreme care, and NMR spectroscopic and mass spectrometric analyses are required.
Overexpression of genes involved in artemisinin biosynthetic pathway
Metabolic engineering of A. annua by overexpressing genes involved in artemisinin biosynthesis has been given more attention during the last 20 years. To obtain successful transformants, several parameters for Agrobacterium tumefaciens-mediated transformation, such as the concentration of antibiotics, method and duration of co-cultivation, and phytohormones supplied for plant regeneration, have been optimized [66–71]. Among various explants available for transformation, stem internodes and young inflorescence seem to be the most appropriate [70–72]. Phytohormones α-naphthaleneacetic acid and 6-benzylaminopurine are crucial for GST development in young leaves, and root generation also affects GST size [73]. Recently, Kiani et al. [72] developed miniprep methods using A. tumefaciens- and Agrobacterium rhizogenes-mediated transformation. This method exhibits higher transformation rates with faster development of transformants within 3–4 weeks compared with other methods.
The overexpression of several genes involved in artemisinin biosynthesis in A. annua has been evaluated. Overexpression of farnesyl pyrophosphate synthase (FPS) increased artemisinin production up to 2- to 3.6-fold higher than that in the control [74, 75]. Overexpressing CYP71AV1 and its redox partner cytochrome P450 reductase (CPR) in artemisinin biosynthesis could increase artemisinin content in planta by 38 % [76]. Xiang et al. [77] generated dxr- and CYP71AV1/CPR-overexpressing A. annua and found that both transformants increased the production of artemisinin. The overexpression of DBR2 increased the production of artemisinin as well as its precursor dihydroartemisinic acid, up to twofold, compared with non-transgenic plants. It also increased production of artemisinic acid up to 5.48- to 9.06-fold and arteannuin B up to twofold [78]. The reason why overexpression of DBR2 enhanced biosynthesis of artemisinic acid and arteannuin B has not been revealed. However, Yuan et al. [78] hypothesized that excess dihydroartemisinic acid might be converted into artemisinic acid in planta.
Overexpression of multiple genes involved in artemisinin biosynthesis could greatly increase the production of artemisinin in planta. Chen et al. [79] showed that the co-overexpression of FPS, CYP71AV1, and CPR increased artemisinin levels in A. annua up to 3.6-fold. The co-overexpression of HMGR and FPS increased production of artemisinin up to 1.8-fold higher than that in the control [80]. Alam et al. [81, 82] co-overexpressed HMGR and ADS in A. annua and found greatly increased artemisinin levels, up to 7.65-fold, in this transgenic line.
Suppressing the expression of genes involved in the pathways competing with artemisinin biosynthesis is another approach to enhance artemisinin content in planta. Zhang et al. [83] used RNAi techniques to suppress the expression of SQS, the first committed gene in sterol biosynthesis. The suppression of this gene enhanced the production of artemisinin up to 3.14-fold.
Direct or indirect upregulation of artemisinin biosynthesis
The effect of several stresses on production of artemisinin in A. annua has been analyzed since the 1990s. These stresses usually lead to the generation of ROS (required for the last non-enzymatic step in artemisinin biosynthesis) or upregulate the expression of artemisinin biosynthetic genes [84–87]. Details of the stresses placed on artemisinin production have been summarized previously [88, 89], and the appropriate cultivation conditions of A. annua were suggested [9].
Some transcription factors upregulated the expression of artemisinin biosynthetic genes and promoted production of artemisinin in A. annua. The WRKY1 transcription factor is thought to bind to the W-box cis-acting elements of promoters to promote gene expression. It is also involved in the regulation of plant defense responses and developmental and physiological processes. Ma et al. [33] showed that the transcript levels of HMGR, ADS, CYP71AV1, and DBR2 were induced in transient AaWRKY1-overexpressing leaves. Furthermore, the specific overexpression of this transcription factor in GSTs increased transcript levels of CYP71AV1 up to 33-fold, compared with the wild type [90]. AaORA, one of the APETALA2/ethylene response factor (AP2/ERF) transcription factor involved in plant responses to biotic and abiotic stresses, showed a similar expression pattern to those of ADS, CYP71AV1, and DBR2. The overexpression of this transcription factor led to the upregulation of the expression levels of ADS, CYP71AV1, and DBR2in planta and promoted artemisinin production [91]. Yu et al. [92] also reported the enhancement of artemisinin production via overexpression of two transcription factors from the same family, AaERF1 and AaERF2, which bind to the promoter regions of ADS and CYP71AV1. Another transcription factor that positively regulates the biosynthesis of artemisinin is a basic helix-loop-helix (bHLH) transcription factor, involved in metabolic regulation of various hormones, developmental processes, and regulation of light signaling, iron and phosphate homeostasis, and various abiotic stresses [93]. Recently, Zhang et al. [94] reported that a basic leucine zipper transcription factor (AabZIP1) binds to the ABA-responsive elements (ABRE) of ADS and CYP71AV1 promoters and upregulates the expression of ADS, CYP71AV1, DBR2, and ALDH1.
Several phytohormones upregulating artemisinin biosynthesis have been reported. Treatment with salicylic acid upregulates the expression of HMGR and ADS, as well as induces ROS generation, driving the conversion of dihydroartemisinic acid into artemisinin [95]. Methyl jasmonate (MeJA) promotes the formation of GSTs and enhances the expression of several genes involved in the artemisinin biosynthetic pathway and related transcription factors (ORA and ERF1), leading to the enhancement of artemisinin production [96–98]. This phytohormone also regulates trichome-specific fatty acyl-CoA reductase 1 (TFAR1), ABCG transporter unigenes (AaABCG6 and AaABCG7), and allene oxide cyclase (AaAOC) [96, 99, 100]. TFAR1 is involved in the formation of cuticular wax during GST expansion in A. annua. AaABCG6 and AaABCG7 are ATP-binding cassette transporter G, involved in the development of trichome cuticle and may share a common regulatory system with ADS and CYP71AV1. AaAOC is involved in JA biosynthesis. The expression of this gene may be upregulated by treatment with not only MeJA but also ABA and ethylene [100]. The overexpression of the ABA receptor, AaPYL9, also improves the sensitivity of ABA and promotes artemisinin biosynthesis after ABA treatment [99, 101].
The enhancement of artemisinin production can be achieved by increased GST density. Singh et al. [102] reported that the expression of bgl1, encoding β-glucosidase from Trichoderma reesei, in A. annua improved the density of GSTs in flowers up to 66 % and increased the production of artemisinin up to five-fold compared with the control. The expression of rolB and rolC of A. rhizogenes also increases GST density and upregulates the expression of ADS, CYP71AV1, ALDH1, and TFAR1. Artemisinin content is then increased 2- to 9-fold and 4-fold in rolB- and rolC-expressing plants, respectively [103].
Co-cultivation of an endophytic fungus Piriformospora indica and a nitrogen-fixing bacterium Azotobacter chroococcum with A. annua increases artemisinin content up to 70 % [104]. This dual symbiosis also shows a positive effect on plant height, dry weight, and leaf yield. Another example of using symbiosis to increase the production of artemisinin was reported using Glomus mosseae and Bacillus subtilis [105]. Although clear evidence for the effect of this symbiosis on the enhancement of artemisinin production is still unknown, Arora et al. [104] suggested that it might be due to improved growth and nutrient status of the plant.
Heterologous production
Metabolic engineering of several platforms, such as Nicotiana benthamiana or chloroplasts, has been conducted. Although ADS and CYP71AV1 were introduced into N. benthamiana, the production of artemisinic acid 12-β-diglucoside, instead of artemisinic acid, was detected at 39.5 mg/kg fresh weight (FW) [106]. The production yield of artemisinic acid in tobacco chloroplasts was also very low (0.1 mg/g FW) [107].
The production of plant natural compounds in microorganisms is an alternative approach with several advantages. The metabolic pathways in microorganisms could be modified to produce various types of natural compounds, including isoprenoids, alkaloids, and phenylpropanoids. Microorganisms can grow rapidly, allowing shorter production time compared with the biosynthesis of desired natural compounds in plants. Scaling up production to industrial scale is also possible [108].
The production of artemisinin precursors in microorganisms was first reported in 2003. Martin et al. [109] expressed entire genes encoding the MVA pathway from yeast Saccharomyces cerevisiae in Escherichia coli to increase the intracellular concentration of FPP. To prevent the rapid loss of highly volatile amorpha-4,11-diene during culturing, the culture media was overlaid with dodecane to trap amorpha-4,11-diene, referred to as a two-phase partitioning bioreactor. As a result, they recovered the volatilized amorpha-4,11-diene, improving production titers from 24 mg/L to approximately 500 mg/L in a fed-batch bioreactor [110].
The coexpression of MevT operon with extra copies of HMGR reduced the accumulation of toxic HMG-CoA and increased production of mevalonate by threefold [111]. The replacement of lac by lacUV5 promoter with a codon-optimized MevT and an additional copy of MK also led to the increase in artemisinin production [112]. Tsuruta et al. [113] succeeded in enhancing amorpha-4,11-diene production in E. coli up to 27.4 g/L by replacing yeast HMGS and HMGR with the equivalent enzymes from gram-positive bacteria Staphylococcus aureus.
Engineering of the MEP pathway and membrane efflux transporters to improve the production of amorpha-4,11-diene in E. coli has been reported as well [114–117]. However, there are many issues regarding the expression of membrane-bound cytochrome P450s in this bacterium posing a limitation on the production of the subsequent oxidized compounds. To overcome these problems, Chang et al. [118] engineered the N-terminal transmembrane domain of the codon-optimized CYP71AV1 and coexpressed it with CPR from A. annua. As a result, production of artemisinic acid (105 mg/L) in this E. coli strain was obtained. Two years later, the same group replaced CYP71AV1 by engineered P450 from gram-positive bacteria Bacillus megaterium (P450BM3) and could produce artemisinic-11S,12-epoxide at higher than 250 mg/L successfully [119]. From this finding, a novel semi-biosynthetic route for the production of artemisinin stemming from the cleavage of this epoxide followed by several oxidation steps was proposed.
Yeast is another attractive host for the production of artemisinin precursors as it produces FPP for sterol biosynthesis via the MVA pathway. Since the MVA pathway in S. cerevisiae has been characterized, ADS was introduced into this yeast, and an amorpha-4,11-diene-producing yeast strain was generated successfully [120]. While there are many issues concerning the expression of cytochrome P450s in E. coli, the expression of this gene in yeast is much more feasible. Therefore, CYP71AV1 and CPR were coexpressed, and all genes involved in the MVA pathway were upregulated either directly or indirectly. The competing pathway (sterol biosynthetic pathway) was also downregulated using a methionine-repressible promoter to improve the production of artemisinic acid in the yeast expression system. As a result, this transgenic yeast strain produced artemisinic acid at up to 100 mg/L [121, 122]. Several factors were further optimized for the production of artemisinic acid in an industrial fermenter. For example, the carbon source for growing yeast in a fermenter was switched from glucose to galactose, and the oxygen transfer rate was controlled. With this development, called the galactose fed-batch process controlled by the DO-stat algorithm, the artemisinic acid titer increased to 2.5 g/L [123].
Despite conferring a higher production yield of artemisinic acid, the use of galactose is costly and not applicable, especially in developing countries. Thus, lower-cost chemicals are needed as carbon sources. Yeast with GAL1, GAL7, GAL10, and GAL80 deletions was generated to exclude the use of galactose, and ethanol was alternatively used as a carbon source. Two additional copies of truncated HMGR (tHMG1) were integrated into this yeast strain. As a result, the production of amorpha-4,11-diene was increased up to more than 40 g/L [124]. Further development was performed by the introduction of artemisinin biosynthetic genes, CYP71AV1, CPR, ADH1, and ALDH1, to oxidize amorpha-4,11-diene into artemisinic acid. Cytochrome b5 (CYB5) was also introduced into this strain as it can accelerate cytochrome P450 reactions [125]. High-level production of artemisinic acid, at 25 g/L, was thereby achieved. The semi-synthesis of artemisinin from artemisinic acid was also optimized, and the overall yield after purification increased to 40–45 % [126, 127]. A potent coupled chromatography–crystallization method to purify artemisinin was then developed, and the recovery yield of this antimalarial compound from the reaction mixture increased to 61.5 %, with 99 % purity [128]. All of the transgenes and modifications to several heterologous hosts mentioned here are summarized in Table 1.
Table 1.
Heterologous production of artemisinin intermediates
| Host | No. | Transgenes or modifications | Product | Yield | References |
|---|---|---|---|---|---|
| N. benthamiana | 1 | P 35S-tHMGR-FPS-ADS, P 35S-CYP71AV1 | Artemisinic acid 12-β-diglucoside | 39.5 mg/kg FW | [106] |
| Tobacco chloroplasts | 2 | P rrn16S-atoB-HMGS-HMGR-MK-PMK-MVD1, P psbA-E. coli IDI-FPS-ADS-CYP71AV1-AaCPR | Artemisinic acid | 0.1 mg/g FW | [107] |
| E. coli | 3 | P lac-MevT a, P lac-MBIS b, P trc-ADS | Amorpha-4,11-diene | 24 mg/L | [109] |
| 4 | Same as 3 but overlaid with dodecane | Amorpha-4,11-diene | 500 mg/L | [110] | |
| 5 | P BAD-MevT, P BAD-tHMGR1 | Mevalonate | Threefold from CTc | [111] | |
| 6 | P lacUV5-MevT (codon opt.)-MBIS, P trc-ERG12 (codon opt.)-ADS | Amorpha-4,11-diene | 293 mg/L | [112] | |
| 7 | P lacUV5-MevT (codon opt.) with HMGS and HMGR from S. aureus, P lac-MBIS, P lac-ADS | Amorpha-4,11-diene | 27.4 g/L | [113] | |
| 8 | P BAD-dxs-IDI-ispDF, ADS with Δpts and optimized medium | Amorpha-4,11-diene | 182 mg/L | [114] | |
| 9 | P TM2-galP-glk, P T7-dxs-IDI-ispA-ADS | Amorpha-4,11-diene | 201.2 mg/L | [115] | |
| 10 | AcrB, TolC (x2), ADS (codon opt.) | Amorpha-4,11-diene | 404.83 mg/L | [116] | |
| 11 | P BAD-dxs-IDI-ispDF, P araBAD-ADS, P TM1-macAB-TolC | Amorpha-4,11-diene | ~30 mg/L/OD | [117] | |
| 12 | Same as 3 with CYP71AV1 (codon opt., engineered N-terminal transmembrane)-AaCPR | Artemisinic acid | 105 mg/L | [118] | |
| 13 | Same as 12 but replaced CYP71AV1 with P450 BM3 | Artemisinic-11S,12-epoxide | 250 mg/L | [119] | |
| S. cerevisiae | 14 | P GAL1-ADS | Amorpha-4,11-diene | 600 μg/L | [120] |
| 15 | P GAL1-tHMGR P GAL1-upc2-1 erg9::P MET3-ERG9 P GAL1-tHMGR P GAL1-ERG20, P GAL1-ADS P GAL10-CYP71AV1 P GAL1-AaCPR | Artemisinic acid | 100 mg/L | [121, 122] | |
| 16 | Same as 15 with optimized culture condition | Artemisinic acid | 2.5 g/L | [123] | |
| 17 | gal80Δ::nat r MAT a erg9Δ∷kan r P MET3-ERG9, leu2-3,112∷HIS P GAL1-MVD1 P GAL10-ERG8 his3Δ1∷HIS P GAL1-ERG12 P GAL10-ERG10ade1Δ∷P GAL1-tHMG1 P GAL10-IDI1 ADE1 ura3-52∷P GAL1-tHMG1 P GAL10-ERG13 URA3trp1-289∷P GAL1-tHMG1 P GAL10-ERG20 TRP1 [pAM322] | Amorpha-4,11-diene | 41 g/L | [124] | |
| 18 | Same as 17. but replaced gal80Δ::nat r with gal1Δ gal7Δ gal10Δ::hphA | Amorpha-4,11-diene | 37 g/L | [124] | |
| 19 | gal1Δ,gal10Δ,gal7Δ::P GAL3-CPR1natA, erg9Δ::dsdAP CTR3- ERG9, leu2-3,112::kanAP GAL7-AaCYB5P GAL1-ERG19P GAL10- ERG8, ade1Δ::P GAL1-tHMG1P GAL10-IDI1_ADE1, his3Δ1::hphAP GAL7-AaALDH1P GAL1-ERG12P GAL10-ERG10, ura3-52:: P GAL1-tHMG1P GAL10-ERG13hisG, trp1-289:: P GAL1- tHMG1P GAL10-ERG20TRP1, ndt80Δ::P TDH1-HEM1HIS3PPGK1-CTT1, gal80Δ::URA3P GAL7-AaADH1, [pAM552: 2μ-LEU2d P GAL1-ADS P GAL10-CYP71AV1] | Artemisinic acid | 25 g/L | [126, 127] |
a MevT operon consists of atoB-HMGS-tHMGR
b MBIS operon consists ofERG12-ERG8-MVD1-IDI-ispA
cProduction yield as compared to control (CT)
Artemisinin biosynthetic genes in non-artemisinin-producing Artemisia species
Some studies reported that artemisinin is produced in other Artemisia species [129–134]. However, we attempted to isolate artemisinin from other Artemisia species but failed to detect any trace amounts of artemisinin or its intermediates (unpublished data). Thus, we analyzed the expression of genes highly homologous to artemisinin biosynthetic genes in these species. Firstly, we selected A. afra and A. absinthium as they are widely cultivated in Africa and exhibit anti-plasmodial activity [135–138]. Putative ADS orthologs were not expressed in either A. afra or A. absinthium [139]. However, we detected the expression of putative CYP71AV1 orthologs in both species. Functional analysis revealed that these orthologous enzymes show similar catalytic activities to their correspondent in A. annua on the oxidation of amorpha-4,11-diene into artemisinic acid [139]. We also detected the expression of DBR2 ortholog in A. absinthium, and the encoded enzyme showed comparable activity to that of A. annua DBR2 [140]. In addition, we showed that this plant can convert the fed artemisinin intermediates into the following products along the biosynthetic pathway of artemisinin [140]. Our findings suggest that ADS might be a limiting factor for the production of artemisinin in planta, and A. absinthium could be an alternative host for artemisinin production. The introduction of ADS into A. absinthium might lead to the generation of artemisinin-producing A. absinthium, which could be used as an alternative approach to produce artemisinin in other Artemisia species. To prove this hypothesis, this research is now ongoing in our laboratory.
Next challenge: artemisinin-resistant Plasmodium parasites
Artemisinin is the most effective antimalarial drug and has been used as an ACT to treat malaria for over a decade. However, the emergence of artemisinin-resistant Plasmodium parasites in Southeast Asia, prolonging the parasite clearance rate in patients, has been reported recently and has become a critical issue [141–144]. No correlation between resistance and other previously proposed candidate targets of artemisinin (PfATP6 and PfTCTP) was detected [145]. However, it has been suggested that the resistance occurs predominantly during the early ring stage of parasite development as a result of the multiple forms of mutations in the PF3D7_1343700 kelch propeller domain (K13-propeller) on chromosome 13 [146–155]. K13-propeller mutations lead to the increase of phosphatidylinositol-3-kinase (PfPI3K), which is required for the mediation of cell signaling and survival [156, 157], and prolong parasite development at the ring stage when the activation level of artemisinin is rather low [5, 7, 158]. The B subfamily of ABC transporters, known as multidrug resistance proteins (MDR), also promotes artemisinin resistance. In artemether–lumefantrine post-treatment infections, alleles of Pfmdr1 tended to have 86N, 184F, and 1246D, rather than the common YYY haplotype, and increased the number of treatment failures [159]. The deletion of Pfmdr5 induced greater sensitivity to artemisinin treatment, suggesting that this gene might contribute to artemisinin resistance as well [160].
Current situation of malaria infection and ongoing studies on antimalarial drug development
Since ACTs have become the major treatment for malaria and strict preventive measures against parasite-infected mosquitoes have been implemented, the malaria-related mortality rate and case incidence have decreased gradually during the past 10 years [1]. Although artemisinin-resistant Plasmodium parasites have emerged and show a significant delay in clearance rate, the response of dihydroartemisinin against either wild-type parasites or mutants exhibits similar Km values suggesting that dihydroartemisinin does not lose its activity against the mutants [161]. Extending the treatment courses could be an effective strategy to clear resistant parasite infection. However, the parasites can still develop complete resistance against artemisinin-based treatment at any point in the future. In addition, the proportion of malaria-infected patients is concentrated in countries with low national income levels. Among these, more than 68 million infected children do not receive any ACTs [1]. Therefore, large amounts of low-cost artemisinin for ACTs, by either increasing the cultivation of high-artemisinin-producing A. annua plants or developing cheaper synthetic biological processes in the long term, are required to prevent any further development of parasites and meet the demand of ACTs worldwide. Moreover, novel effective antimalarial treatments must be developed continually. Recently, low-cost plant-based artemisinin combination therapy (pACT) has driven attention on the production of no semi-synthetic artemisinin in planta as this treatment showed higher antimalarial activity, and the synergistic effect of artemisinin and the plant matrix overcame resistance to artemisinin [162–169]. Several scientists have also focused on the investigation of novel potential drug targets [170–180] and on the synthesis of novel antimalarial compounds including artemisinin hybrids [181–186]. Still, further studies on these avenues are required.
Conclusion
Several approaches to enhance the production of artemisinin have been investigated for over a decade. As a result, the availability of artemisinin for ACTs is increasing, and the number of malaria-related deaths is decreasing gradually. Although artemisinin is still effective against malaria-causing parasites, the emergence of artemisinin-resistant strains has posed a new challenge to scientists worldwide. Therefore, elucidating the mode of action of artemisinin and the mechanism of resistance against this compound in Plasmodium parasites is important for further development of antimalarial drugs. We hope that the current understanding of artemisinin as summarized in this review will provide clues for further investigation and development of antimalarial treatments to overcome artemisinin resistance in Plasmodium parasites in the future.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research (23780104) and a research grant from the Shorai Foundation for Science and Technology to H.S.; Frontier Research Base for Global Young Researcher, Osaka University from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) to E.O.F.; and the Monbukagakusho Scholarship to P.M.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.World Health Organization . World malaria report 2015. Geneva: World Health Organization; 2015. [Google Scholar]
- 2.Tu Y. The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat Med. 2011;17:1217–1220. doi: 10.1038/nm.2471. [DOI] [PubMed] [Google Scholar]
- 3.de Ridder S, van der Kooy F, Verpoorte R. Artemisia annua as a self-reliant treatment for malaria in developing countries. J Ethnopharmacol. 2008;120:302–314. doi: 10.1016/j.jep.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 4.van Agtmael MA, Eggelte TA, van Boxtel CJ. Artemisinin drugs in the treatment of malaria: from medicinal herb to registered medication. Trends Pharmacol Sci. 1999;20:199–205. doi: 10.1016/S0165-6147(99)01302-4. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Zhang CJ, Chia WN, Loh CCY, Li Z, Lee YM, He Y, Yuan LX, Lim TK, Liu M, Liew CX, Lee YQ, Zhang J, Lu N, Lim CT, Hua ZC, Liu B, Shen HM, Tan KSW, Lin Q. Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum. Nat Commun. 2015;6:10111. doi: 10.1038/ncomms10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willcox M. Artemisia species: from traditional medicines to modern antimalarials—and back again. J Altern Complement Med. 2009;15:101–109. doi: 10.1089/acm.2008.0327. [DOI] [PubMed] [Google Scholar]
- 7.Klonis N, Creek DJ, Tilley L. Iron and heme metabolism in Plasmodium falciparum and the mechanism of action of artemisinins. Curr Opin Microbiol. 2013;16:722–727. doi: 10.1016/j.mib.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization . Guidelines for the treatment of malaria. 1. Geneva: World Health Organization; 2006. [Google Scholar]
- 9.Ferreira JFS, Laughlin JC, Delabays N, de Magalhães PM. Cultivation and genetics of Artemisia annua L. for increased production of the antimalarial artemisinin. Plant Genet Resour. 2005;3:206–229. doi: 10.1079/PGR200585. [DOI] [Google Scholar]
- 10.Chang Z. The discovery of qinghaosu (artemisinin) as an effective anti-malaria drug: a unique China story. Sci China Life Sci. 2015;59:1–8. doi: 10.1007/s11427-015-4988-z. [DOI] [PubMed] [Google Scholar]
- 11.Miller LH, Su X. Artemisinin: discovery from the Chinese herbal garden. Cell. 2011;146:855–858. doi: 10.1016/j.cell.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su XZ, Miller LH. The discovery of artemisinin and the Nobel Prize in Physiology or Medicine. Sci China Life Sci. 2015;58:1175–1179. doi: 10.1007/s11427-015-4948-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao F. Discovery of artemisinin (qinghaosu) Molecules. 2009;14:5362–5366. doi: 10.3390/molecules14125362. [DOI] [Google Scholar]
- 14.World Health Organization . Guidelines for the treatment of malaria. 3. Geneva: World Health Organization; 2015. [PubMed] [Google Scholar]
- 15.Li G, Arnold K, Guo X, Jian H, Fu L. Randomised comparative study of mefloquine, qinghaosu, and pyrimethamine–sulfadoxine in patients with falciparum malaria. Lancet. 1984;2:1360–1361. doi: 10.1016/S0140-6736(84)92057-9. [DOI] [PubMed] [Google Scholar]
- 16.Brown GD. The biosynthesis of artemisinin (qinghaosu) and the phytochemistry of Artemisia annua L. (qinghao) Molecules. 2010;15:7603–7698. doi: 10.3390/molecules15117603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Towler MJ, Weathers PJ. Evidence of artemisinin production from IPP stemming from both the mevalonate and the nonmevalonate pathways. Plant Cell Rep. 2007;26:2129–2136. doi: 10.1007/s00299-007-0420-x. [DOI] [PubMed] [Google Scholar]
- 18.Schramek N, Wang H, Römisch-Margl W, Keil B, Radykewicz T, Winzenhörlein B, Beerhues L, Bacher A, Rohdich F, Gershenzon J, Liu B, Eisenreich W. Artemisinin biosynthesis in growing plants of Artemisia annua. A 13CO2 study. Phytochemistry. 2010;71:179–187. doi: 10.1016/j.phytochem.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 19.Bouwmeester HJ, Wallaart TE, Janssen MHA, van Loo B, Jansen BJM, Posthumus MA, Schmidt CO, De Kraker JW, König WA, Franssen MCR. Amorpha-4,11-diene synthase catalyses the first probable step in artemisinin biosynthesis. Phytochemistry. 1999;52:843–854. doi: 10.1016/S0031-9422(99)00206-X. [DOI] [PubMed] [Google Scholar]
- 20.Chang YJ, Song SH, Park SH, Kim SU. Amorpha-4,11-diene synthase of Artemisia annua: cDNA isolation and bacterial expression of a terpene synthase involved in artemisinin biosynthesis. Arch Biochem Biophys. 2000;383:178–184. doi: 10.1006/abbi.2000.2061. [DOI] [PubMed] [Google Scholar]
- 21.Mercke P, Bengtsson M, Bouwmeester HJ, Posthumus MA, Brodelius PE. Molecular cloning, expression and characterization of amorpha-4,11-diene synthase, a key enzyme of artemisinin biosynthesis in Artemisia annua L. Arch Biochem Biophys. 2000;381:173–180. doi: 10.1006/abbi.2000.1962. [DOI] [PubMed] [Google Scholar]
- 22.Kim SH, Heo K, Chang YJ, Park SH, Rhee SK, Kim SU. Cyclization mechanism of amorpha-4,11-diene synthase, a key enzyme in artemisinin biosynthesis. J Nat Prod. 2006;69:758–762. doi: 10.1021/np050356u. [DOI] [PubMed] [Google Scholar]
- 23.Picaud S, Mercke P, He X, Sterner O, Brodelius M, Cane DE, Brodelius PE. Amorpha-4,11-diene synthase: mechanism and stereochemistry of the enzymatic cyclization of farnesyl diphosphate. Arch Biochem Biophys. 2006;448:150–155. doi: 10.1016/j.abb.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 24.Teoh KH, Polichuk DR, Reed DW, Nowak G, Covello PS. Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett. 2006;580:1411–1416. doi: 10.1016/j.febslet.2006.01.065. [DOI] [PubMed] [Google Scholar]
- 25.Polichuk D, Teoh KH, Zhang Y, Ellens KW, Reed DW, Covello PS (2010) Nucleotide sequence encoding an alcohol dehydrogenase from Artemisia annua and uses thereof. Patent No. WO2010/012074
- 26.Teoh KH, Polichuk DR, Reed DW, Covello PS. Molecular cloning of an aldehyde dehydrogenase implicated in artemisinin biosynthesis in Artemisia annua. Botany. 2009;87:635–642. doi: 10.1139/B09-032. [DOI] [Google Scholar]
- 27.Brown GD, Sy LK. In vivo transformations of artemisinic acid in Artemisia annua plants. Tetrahedron. 2007;63:9548–9566. doi: 10.1016/j.tet.2007.06.062. [DOI] [Google Scholar]
- 28.Zhang Y, Teoh KH, Reed DW, Maes L, Goossens A, Olson DJH, Ross ARS, Covello PS. The molecular cloning of artemisinic aldehyde Δ11(13) reductase and its role in glandular trichome-dependent biosynthesis of artemisinin in Artemisia annua. J Biol Chem. 2008;283:21501–21508. doi: 10.1074/jbc.M803090200. [DOI] [PubMed] [Google Scholar]
- 29.Brown GD, Sy LK. In vivo transformations of dihydroartemisinic acid in Artemisia annua plants. Tetrahedron. 2004;60:1139–1159. doi: 10.1016/j.tet.2003.11.070. [DOI] [Google Scholar]
- 30.Rydén AM, Ruyter-Spira C, Quax WJ, Osada H, Muranaka T, Kayser O, Bouwmeester H. The molecular cloning of dihydroartemisinic aldehyde reductase and its implication in artemisinin biosynthesis in Artemisia annua. Planta Med. 2010;76:1778–1783. doi: 10.1055/s-0030-1249930. [DOI] [PubMed] [Google Scholar]
- 31.Olofsson L, Engström A, Lundgren A, Brodelius PE. Relative expression of genes of terpene metabolism in different tissues of Artemisia annua L. BMC Plant Biol. 2011;11:45. doi: 10.1186/1471-2229-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arsenault PR, Vail D, Wobbe KK, Erickson K, Weathers PJ. Reproductive development modulates gene expression and metabolite levels with possible feedback inhibition of artemisinin in Artemisia annua. Plant Physiol. 2010;154:958–968. doi: 10.1104/pp.110.162552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma D, Pu G, Lei C, Ma L, Wang H, Guo Y, Chen J, Du Z, Wang H, Li G, Ye H, Liu B. Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4,11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 2009;50:2146–2161. doi: 10.1093/pcp/pcp149. [DOI] [PubMed] [Google Scholar]
- 34.Li ZQ, Liu Y, Liu BY, Wang H, Ye HC, Li GF. Cloning, E. coli expression and molecular analysis of amorpha-4,11-diene synthase from a high-yield strain of Artemisia annua L. J Integr Plant Biol. 2006;48:1486–1492. doi: 10.1111/j.1744-7909.2006.00381.x. [DOI] [Google Scholar]
- 35.Pu GB, Ma DM, Wang H, Ye HC, Liu BY. Expression and localization of amorpha-4,11-diene synthase in Artemisia annua L. Plant Mol Biol Rep. 2013;31:32–37. doi: 10.1007/s11105-012-0472-0. [DOI] [Google Scholar]
- 36.Kim SH, Chang YJ, Kim SU. Tissue specificity and development pattern of amorpha-4,11-diene synthase (ADS) proved by ADS promoter-driven GUS expression in the heterologous plant, Arabidopsis thaliana. Planta Med. 2008;74:188–193. doi: 10.1055/s-2008-1034276. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Olofsson L, Lundgren A, Brodelius PE. Trichome-specific expression of amorpha-4,11-diene synthase, a key enzyme of artemisinin biosynthesis in Artemisia annua L., as reported by a promoter-GUS fusion. Am J Plant Sci. 2011;2:619–628. doi: 10.4236/ajps.2011.24073. [DOI] [Google Scholar]
- 38.Wang Y, Yang K, Jing F, Li M, Deng T, Huang R, Wang B, Wang G, Sun X, Tang KX. Cloning and characterization of trichome-specific promoter of cpr71av1 gene involved in artemisinin biosynthesis in Artemisia annua L. Mol Biol. 2011;45:751–758. doi: 10.1134/S0026893311040145. [DOI] [PubMed] [Google Scholar]
- 39.Wang H, Han J, Kanagarajan S, Lundgren A, Brodelius PE. Trichome-specific expression of the amorpha-4,11-diene 12-hydroxylase (cyp71av1) gene, encoding a key enzyme of artemisinin biosynthesis in Artemisia annua, as reported by a promoter-GUS fusion. Plant Mol Biol. 2013;81:119–138. doi: 10.1007/s11103-012-9986-y. [DOI] [PubMed] [Google Scholar]
- 40.Jiang W, Lu X, Qiu B, Zhang F, Shen Q, Lv Z, Fu X, Yan T, Gao E, Zhu M, Chen L, Zhang L, Wang G, Sun X, Tang K. Molecular cloning and characterization of a trichome-specific promoter of artemisinic aldehyde Δ11(13) reductase (DBR2) in Artemisia annua. Plant Mol Biol Rep. 2014;32:82–91. doi: 10.1007/s11105-013-0603-2. [DOI] [Google Scholar]
- 41.Ting HM, Wang B, Rydén AM, Woittiez L, van Herpen T, Verstappen FWA, Ruyter-Spira C, Beekwilder J, Bouwmeester HJ, van der Krol A. The metabolite chemotype of Nicotiana benthamiana transiently expressing artemisinin biosynthetic pathway genes is a function of CYP71AV1 type and relative gene dosage. New Phytol. 2013;199:352–366. doi: 10.1111/nph.12274. [DOI] [PubMed] [Google Scholar]
- 42.Yang K, Monafared RS, Wang H, Lundgren A, Brodelius PE. The activity of the artemisinic aldehyde Δ11(13) reductase promoter is important for artemisinin yield in different chemotypes of Artemisia annua L. Plant Mol Biol. 2015;88:325–340. doi: 10.1007/s11103-015-0284-3. [DOI] [PubMed] [Google Scholar]
- 43.Sun C, Li J, CaoY Long G, Zhou B. Two distinct and competitive pathways confer the cellcidal actions of artemisinins. Microb Cell. 2015;2:14–25. doi: 10.15698/mic2015.01.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, Huang L, Li J, Fan Q, Long Y, Li Y, Zhou B. Artemisinin directly targets malarial mitochondria through its specific mitochondrial activation. PLoS One. 2010;5:e9582. doi: 10.1371/journal.pone.0009582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mercer AE, Copple IM, Maggs JL, O’Neill PM, Park BK. The role of heme and the mitochondrion in the chemical and molecular mechanisms of mammalian cell death induced by the artemisinin antimalarials. J Biol Chem. 2011;286:987–996. doi: 10.1074/jbc.M110.144188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hartwig CL, Rosenthal AS, Angelo JD, Griffin CE, Posner GH, Cooper RA. Accumulation of artemisinin trioxane derivatives within neutral lipids of Plasmodium falciparum malaria parasites is endoperoxide-dependent. Biochem Pharmacol. 2009;77:322–336. doi: 10.1016/j.bcp.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Antoine T, Fisher N, Amewu R, O’Neill PM, Ward SA, Biagini GA. Rapid kill of malaria parasites by artemisinin and semi-synthetic endoperoxides involves ROS-dependent depolarization of the membrane potential. J Antimicrob Chemother. 2014;69:1005–1016. doi: 10.1093/jac/dkt486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Neill PM, Barton VE, Ward SA. The molecular mechanism of action of artemisinin—the debate continues. Molecules. 2010;15:1705–1721. doi: 10.3390/molecules15031705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klonis N, Crespo-Ortiz MP, Bottova I, Abu-Bakar N, Kenny S, Rosenthal PJ, Tilley L. Artemisinin activity against Plasmodium falciparum required hemoglobin uptake and digestion. Proc Natl Acad Sci USA. 2011;108:11405–11410. doi: 10.1073/pnas.1104063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie SC, Dogovski C, Hanssen E, Chiu F, Yang T, Crespo MP, Stafford C, Batinovic S, Teguh S, Charman S, Klonis N, Tilley L. Haemoglobin degradation underpins the sensitivity of early ring stage Plasmodium falciparum to artemisinins. J Cell Sci. 2016;129:406–416. doi: 10.1242/jcs.178830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chugh M, Sundararaman V, Kumar S, Reddy VS, Siddiqui WA, Stuart KD, Malhotra P. Protein complex directs hemoglobin-to-hemozoin formation in Plasmodium falciparum. Proc Natl Acad Sci USA. 2013;110:5392–5397. doi: 10.1073/pnas.1218412110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhisutthibhan J, Pan XQ, Hossler PA, Walker DJ, Yowell CA, Carlton J, Dame JB, Meshnick SR. The Plasmodium falciparum translationally controlled tumor protein homolog and its reaction with the antimalarial drug artemisinin. J Biol Chem. 1998;273:16192–16198. doi: 10.1074/jbc.273.26.16192. [DOI] [PubMed] [Google Scholar]
- 53.Eichhorn T, Winter D, Büchele B, Dirdjaja N, Frank M, Lehmann WD, Mertens R, Krauth-Siegel RL, Simmet T, Granzin J, Efferth T. Molecular interaction of artemisinin with translationally controlled tumor protein (TCTP) of Plasmodium falciparum. Biochem Pharmacol. 2013;85:38–45. doi: 10.1016/j.bcp.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 54.Eckstein-Ludwig U, Webb RJ, van Goethem IDA, East JM, Lee AG, Kimura M, O’Neill PM, Bray PG, Ward SA, Krishna S. Artemisinin target the SERCA of Plasmodium falciparum. Nature. 2003;424:957–961. doi: 10.1038/nature01813. [DOI] [PubMed] [Google Scholar]
- 55.Covello PS. Making artemisinin. Phytochemistry. 2008;69:2881–2885. doi: 10.1016/j.phytochem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 56.Wang Z, Yang L, Yang X, Zhang X. Advances in the chemical synthesis of artemisinin. Synth Commun. 2014;44:1987–2003. doi: 10.1080/00397911.2014.884225. [DOI] [Google Scholar]
- 57.Jain DC, Mathur AK, Gupta MM, Singh AK, Verma RK, Gupta AP, Kumar S. Isolation of high artemisinin-yielding clones of Artemisia annua. Phytochemistry. 1996;43:993–1001. doi: 10.1016/S0031-9422(96)00369-X. [DOI] [Google Scholar]
- 58.Delabays N, Simonnet X, Gaudin M. The genetics of artemisinin content in Artemisia annua L. and the breeding of high yielding cultivars. Curr Med Chem. 2001;8:1795–1801. doi: 10.2174/0929867013371635. [DOI] [PubMed] [Google Scholar]
- 59.Cockram J, Hill C, Burns C, Arro RRJ, Woolley JG, Flockart I, Robinson T, Atkinson CJ, Davies MJ, Dungey N, Greenland AJ, Smith LLMJ, Bentley S. Screening a diverse collection of Artemisia annua germplasm accessions for the antimalarial compound, artemisinin. Plant Gen Res. 2012;10:152–154. doi: 10.1017/S1479262112000159. [DOI] [Google Scholar]
- 60.Larson TR, Branigan C, Harvey D, Penfield T, Bowles D, Graham IA. A survey of artemisinic and dihydroartemisinic acid contents in glasshouse and global field-grown populations of the artemisinin-producing plant Artemisia annua L. Ind Crops Prod. 2013;45:1–6. doi: 10.1016/j.indcrop.2012.12.004. [DOI] [Google Scholar]
- 61.Graham IA, Besser K, Blumer S, Branigan CA, Czechowski T, Elias L, Guterman I, Harvey D, Isaac PG, Khan AM, Larson TR, Li Y, Pawson T, Penfield T, Rae AM, Rathbone DA, Reid S, Ross J, Smallwood MF, Segura V, Townsend T, Vyas D, Winzer T, Bowles D. The genetic map of Artemisia annua L. identifies loci affecting yield of the antimalarial drug artemisinin. Science. 2010;327:328–331. doi: 10.1126/science.1182612. [DOI] [PubMed] [Google Scholar]
- 62.Townsend T, Segura V, Chigeza G, Penfield T, Rae A, Harvey D, Bowles D, Graham IA. The use of combining ability analysis to identify elite parents for Artemisia annua F1 hybrid production. PLoS One. 2013;8:e61989. doi: 10.1371/journal.pone.0061989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muranaka T, Saito K. Production of pharmaceuticals by plant tissue cultures. In: Mander L, Lui HW, editors. Comprehensive natural products II: chemistry and biology. Development and modification of bioactivity. Oxford: Elsevier; 2010. pp. 615–628. [Google Scholar]
- 64.Weathers PJ, Elkholy S, Wobbe KK. Artemisinin: the biosynthetic pathway and its regulation in Artemisia annua, a terpenoid-rich species. In Vitro Cell Dev Biol Plant. 2006;42:309–317. doi: 10.1079/IVP2006782. [DOI] [Google Scholar]
- 65.Liu C, Zhao Y, Wang Y. Artemisinin: current state and perspectives for biotechnological production of an antimalarial drug. Appl Microbiol Biotechnol. 2006;74:11–20. doi: 10.1007/s00253-006-0452-0. [DOI] [PubMed] [Google Scholar]
- 66.Vergauwe A, Cammaert R, Vandenberghe D, Genetelo C, Inze D, Van Montagu M, Van den Eeckhout E. Agrobacterium tumefaciens-mediated transformation of Artemisia annua L. and regeneration of transgenic plants. Plant Cell Rep. 1996;15:929–933. doi: 10.1007/BF00231590. [DOI] [PubMed] [Google Scholar]
- 67.Vergauwe A, Van Geldre E, Inzé D, Van Montagu M, Van den Eeckhout E. Factors influencing Agrobacterium tumefaciens-mediated transformation of Artemisia annua L. Plant Cell Rep. 1998;18:105–110. doi: 10.1007/s002990050540. [DOI] [PubMed] [Google Scholar]
- 68.Han JL, Wang H, Ye HC, Liu Y, Li ZQ, Zhang Y, Zhang YS, Yan F, Li GF. High efficiency of genetic transformation and regeneration of Artemisia annua L. via Agrobacterium tumefaciens-mediated procedure. Plant Sci. 2005;168:73–80. doi: 10.1016/j.plantsci.2004.07.020. [DOI] [Google Scholar]
- 69.Lualon W, De-Eknamkul W, Tanaka H, Shoyama Y, Putalun W. Artemisinin production by shoot regeneration of Artemisia annua L. using thidiazuron. Z Naturforsch. 2008;63:96–100. doi: 10.1515/znc-2008-1-218. [DOI] [PubMed] [Google Scholar]
- 70.Tian N, Liu S, Ting H, Huang J, van der Krol S, Bouwmeester H, Liu Z. An improved Agrobacterium tumefaciens mediated transformation of Artemisia annua L. by using stem internodes as explants. Czech J Genet Plant Breed. 2013;49:123–129. [Google Scholar]
- 71.Wang J, Nie J, Pattanaik S, Yuan L. Efficient Agrobacterium-mediated transformation of Artemisia annua L. using young inflorescence. In Vitro Cell Dev Biol. 2016;52:204–211. doi: 10.1007/s11627-015-9744-3. [DOI] [Google Scholar]
- 72.Kiani BH, Suberu J, Barker GC, Mirza B. Development of efficient miniprep transformation methods for Artemisia annua using Agrobacterium tumefaciens and Agrobacterium rhizogenes. In Vitro Cell Dev Biol. 2014;50:590–600. doi: 10.1007/s11627-014-9607-3. [DOI] [Google Scholar]
- 73.Nguyen KT, Towler MJ, Weathers PJ. The effect of roots and media constituents on trichomes and artemisinin production in Artemisia annua L. Plant Cell Rep. 2013;32:207–218. doi: 10.1007/s00299-012-1355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen DH, Ye HC, Li GF. Expression of a chimeric farnesyl diphosphate synthase gene in Artemisia annua L. transgenic plants via Agrobacterium tumefaciens-mediated transformation. Plant Sci. 2000;155:179–185. doi: 10.1016/S0168-9452(00)00217-X. [DOI] [PubMed] [Google Scholar]
- 75.Banyai W, Kirdmanee C, Mii M, Supaibulwatana K. Overexpression of farnesyl pyrophosphate synthase (FPS) gene affected artemisinin content and growth of Artemisia annua L. Plant Cell Tissue Organ Cult. 2010;103:255–265. doi: 10.1007/s11240-010-9775-8. [DOI] [Google Scholar]
- 76.Shen Q, Chen YF, Wang T, Wu SY, Lu X, Zhang L, Zhang FY, Jiang WM, Wang GF, Tang KX. Overexpression of the cytochrome P450 monooxygenase (cyp71av1) and cytochrome P450 reductase (cpr) genes increased artemisinin content in Artemisia annua (Asteraceae) Gen Mol Res. 2012;11:3298–3309. doi: 10.4238/2012.September.12.13. [DOI] [PubMed] [Google Scholar]
- 77.Xiang L, Zeng L, Yuan Y, Chen M, Wang F, Liu X, Zeng L, Lan X, Liao Z. Enhancement of artemisinin biosynthesis by overexpressing dxr, cyp71av1 and cpr in the plants of Artemisia annua L. Plant Omics J. 2012;5:503–507. [Google Scholar]
- 78.Yuan Y, Liu W, Zhang Q, Xiang L, Liu X, Chen M, Lin Z, Wang Q, Liao Z. Overexpression of artemisinic aldehyde Δ11(13) reductase gene-enhanced artemisinin and its relative metabolite biosynthesis in transgenic Artemisia annua L. Biotechnol Appl Biochem. 2015;62:17–23. doi: 10.1002/bab.1234. [DOI] [PubMed] [Google Scholar]
- 79.Chen Y, Shen Q, Wang Y, Wang T, Wu S, Zhang L, Lu X, Zhang F, Jiang W, Qiu B, Gao E, Sun X, Tang K. The stacked over-expression of FPS, CYP71AV1 and CPR genes leads to the increase of artemisinin level in Artemisia annua L. Plant Biotechnol Rep. 2013;7:287–295. doi: 10.1007/s11816-012-0262-z. [DOI] [Google Scholar]
- 80.Wang Y, Jing F, Yu S, Chen Y, Wang T, Liu P, Wang G, Sun X, Tang K. Co-overexpression of the HMGR and FPS genes enhances artemisinin content in Artemisia annua L. J Med Plant Res. 2011;51:3396–3403. [Google Scholar]
- 81.Alam P, Abdin MZ. Over-expression of HMG-CoA reductase and amorpha-4,11-diene synthase genes in Artemisia annua L. and its influence on artemisinin content. Plant Cell Rep. 2011;30:1919–1928. doi: 10.1007/s00299-011-1099-6. [DOI] [PubMed] [Google Scholar]
- 82.Alam P, Kamaludding Sharaf-Eldin MA, Elkholy SF, Abdin MZ. The effect of over-expression of rate limiting enzymes on the yield of artemisinin in Artemisia annua. Rend Fis Acc Lincei. 2015;27:311–319. doi: 10.1007/s12210-015-0481-7. [DOI] [Google Scholar]
- 83.Zhang L, Jin F, Li F, Li M, Wang Y, Wang G, Sun X, Tang K. Development of transgenic Artemisia annua (Chinese wormwood) plants with an enhanced content of artemisinin, an effective anti-malarial drug, by hairpin-RNA-mediated gene silencing. Biotechnol Appl Biochem. 2009;52:199–207. doi: 10.1042/BA20080068. [DOI] [PubMed] [Google Scholar]
- 84.Ferreira JFS. Nutrient deficiency in the production of artemisinin, dihydroartemisinic acid, and artemisinic acid in Artemisia annua L. J Agric Food Chem. 2007;55:1686–1694. doi: 10.1021/jf063017v. [DOI] [PubMed] [Google Scholar]
- 85.Wang ML, Jiang YS, Wei JQ, Wei X, Qi XX, Jiang SY, Wang ZM. Effects of irradiance on growth, photosynthetic characteristics, and artemisinin content of Artemisia annua L. Photosynthetica. 2007;46:17–20. doi: 10.1007/s11099-008-0004-1. [DOI] [Google Scholar]
- 86.Pan WS, Zheng LP, Tian H, Li WY, Wan JW. Transcriptome responses involved in artemisinin production in Artemisia annua L. under UV-B radiation. J Photochem Photobiol B. 2014;140:292–300. doi: 10.1016/j.jphotobiol.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 87.Marchese JA, Ferreira JFS, Rehder VLG, Rodrigues O. Water deficit effect on the accumulation of biomass and artemisinin in annual wormwood (Artemisia annua L., Asteraceae) Braz J Plant Physiol. 2010;22:1–9. doi: 10.1590/S1677-04202010000100001. [DOI] [Google Scholar]
- 88.Nguyen KT, Arsenault PR, Weathers PJ. Trichomes + roots + ROS = artemisinin: regulating artemisinin biosynthesis in Artemisia annua L. In Vitro Cell Dev Biol Plant. 2011;47:329–338. doi: 10.1007/s11627-011-9343-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pandey N, Pandey-Rai S. Updates on artemisinin: an insight to mode of actions and strategies for enhanced global production. Protoplasma. 2016;253:15–30. doi: 10.1007/s00709-015-0805-6. [DOI] [PubMed] [Google Scholar]
- 90.Han J, Wang H, Lundgren A, Brodelius PE. Effects of overexpression of AaWRKY1 on artemisinin biosynthesis in transgenic Artemisia annua plants. Phytochemistry. 2014;102:89–96. doi: 10.1016/j.phytochem.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 91.Lu X, Zhang L, Zhang F, Jiang W, Shen Q, Zhang L, Lv Z, Wang G, Tang K. AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea. New Phytol. 2013;198:1191–1202. doi: 10.1111/nph.12207. [DOI] [PubMed] [Google Scholar]
- 92.Yu ZX, Li JX, Yang CQ, Hu WL, Wang LJ, Chen XY. The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol Plant. 2012;5:353–365. doi: 10.1093/mp/ssr087. [DOI] [PubMed] [Google Scholar]
- 93.Ji Y, Xiao J, Shen Y, Ma D, Li Z, Pu G, Li X, Huang L, Liu B, Ye H, Wang H. Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua. Plant Cell Physiol. 2014;55:1592–1604. doi: 10.1093/pcp/pcu090. [DOI] [PubMed] [Google Scholar]
- 94.Zhang F, Fu X, Lv Z, Lu X, Shen Q, Zhang L, Zhu M, Wang G, Sun X, Liao Z, Tang K. A basic leucine zipper transcription factor, AabZIP1, connects abscisic acid signaling with artemisinin biosynthesis in Artemisia annua. Mol Plant. 2015;8:163–175. doi: 10.1016/j.molp.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 95.Pu GB, Ma DM, Chen JL, Ma LQ, Wang H, Li GF, Ye HC, Liu BY. Salicylic acid activates artemisinin biosynthesis in Artemisia annua L. Plant Cell Rep. 2009;25:1127–1135. doi: 10.1007/s00299-009-0713-3. [DOI] [PubMed] [Google Scholar]
- 96.Maes L, Van Nieuwerburgh FCW, Zhang Y, Reed DW, Pollier J, Vande Casteele SRF, Inzé D, Covello PS, Deforce DLD, Goossens A. Dissection of the phytohormonal regulation of trichome formation and biosynthesis of the antimalarial compound artemisinin in Artemisia annua plants. New Phytol. 2011;189:176–189. doi: 10.1111/j.1469-8137.2010.03466.x. [DOI] [PubMed] [Google Scholar]
- 97.Caretto S, Quarta A, Durante M, Nisi R, De Paolis A, Blando F, Mita G. Methyl jasmonate and miconazole differently affect artemisinin production and gene expression in Artemisia annua suspension cultures. Plant Biol. 2011;13:51–58. doi: 10.1111/j.1438-8677.2009.00306.x. [DOI] [PubMed] [Google Scholar]
- 98.Xiang L, Zhu S, Zhao T, Zhang M, Liu W, Chen M, Lan X, Liao Z. Enhancement of artemisinin content and relative expression of genes of artemisinin biosynthesis in Artemisia annua by exogenous MeJA treatment. Plant Growth Regul. 2015;75:435–441. doi: 10.1007/s10725-014-0004-z. [DOI] [Google Scholar]
- 99.Zhang L, Lu X, Shen Q, Chen Y, Wang T, Zhang F, Wu S, Jiang W, Liu P, Zhang L, Wang Y, Tang K. Identification of putative Artemisia annua ABCG transporter unigenes related to artemisinin yield following expression analysis in different plant tissues and in response to methyl jasmonate and abscisic acid treatments. Plant Mol Biol Rep. 2012;30:838–847. doi: 10.1007/s11105-011-0400-8. [DOI] [Google Scholar]
- 100.Lu X, Lin X, Shen Q, Zhang F, Wang Y, ChenY Wang T, Wu S, Tang K. Characterization of the jasmonate biosynthetic gene allene oxide cyclase in Artemisia annua L., source of the antimalarial drug artemisinin. Plant Mol Biol Rep. 2011;29:489–497. doi: 10.1007/s11105-010-0252-7. [DOI] [Google Scholar]
- 101.Zhang F, Lu X, Lv Z, Zhang L, Zhu M, Jiang W, Wang G, Sun X, Tang K. Overexpression of the Artemisia orthologue of ABA receptor, AaPYL9, enhances ABA sensitivity and improves artemisinin content in Artemisia annua L. PLoS One. 2013;8:e56697. doi: 10.1371/journal.pone.0056697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Singh ND, Kumar S, Daniell H. Expression of β-glucosidase increases trichome density and artemisinin content in transgenic Artemisia annua plants. Plant Biotechnol J. 2016;14:1034–1045. doi: 10.1111/pbi.12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dilshad E, Cusido RM, Palazon J, Estrada KR, Bonfill M, Mirza B. Enhanced artemisinin yield by expression of rol genes in Artemisia annua. Malar J. 2015;14:424. doi: 10.1186/s12936-015-0951-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arora M, Saxena P, Choudhary DK, Abdin MZ, Varma A. Dual symbiosis between Piriformospora indica and Azotobacter chroococcum enhances the artemisinin content in Artemisia annua L. World J Microbiol Biotechnol. 2016;32:19. doi: 10.1007/s11274-015-1972-5. [DOI] [PubMed] [Google Scholar]
- 105.Awasthi A, Bharti N, Nair P, Singh R, Shukla AK, Gupta MM, Darokar MP, Kalra A. Synergistic effect of Glomus mosseae and nitrogen fixing Bacillus subtilis strain Daz26 on artemisinin content in Artemisia annua L. Appl Soil Ecol. 2011;49:125–130. doi: 10.1016/j.apsoil.2011.06.005. [DOI] [Google Scholar]
- 106.van Herpen TWJM, Cankar K, Nogueira M, Bosch D, Bouwmeester HJ, Beekwilder J. Nicotiana benthamiana as a production platform for artemisinin precursors. PLoS One. 2010;5:e14222. doi: 10.1371/journal.pone.0014222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Saxena B, Subramaniyan M, Malhotra K, Bhavesh NS, Potlakayala SD, Kumar S. Metabolic engineering of chloroplasts for artemisinic acid biosynthesis and impact on plant growth. J Biosci. 2014;39:33–41. doi: 10.1007/s12038-013-9402-z. [DOI] [PubMed] [Google Scholar]
- 108.Marienhagen J, Bott M. Metabolic engineering of microorganisms for the synthesis of plant natural products. J Biotechnol. 2013;163:166–178. doi: 10.1016/j.jbiotec.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 109.Martin VJJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 110.Newman JD, Marshall J, Chang M, Nowroozi F, Paradise E, Pitera D, Newman KL, Keasling JD. High-level production of amorpha-4,11-diene in a two-phase partitioning bioreactor of metabolically engineered Escherichia coli. Biotechnol Bioeng. 2006;95:684–691. doi: 10.1002/bit.21017. [DOI] [PubMed] [Google Scholar]
- 111.Pitera DJ, Paddon CJ, Newman JD, Keasling JD. Balancing a heterologous mevalonate pathway for improved isoprenoid production in Escherichia coli. Metab Eng. 2007;9:193–207. doi: 10.1016/j.ymben.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 112.Anthony JR, Anthony LC, Nowroozi F, Kwon G, Newman JD, Keasling JD. Optimization of the mevalonate-based isoprenoid biosynthetic pathway in Escherichia coli for production of the anti-malarial drug precursor amorpha-4,11-diene. Metab Eng. 2009;11:13–19. doi: 10.1016/j.ymben.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 113.Tsuruta H, Paddon CJ, Eng D, Lenihan JR, Horning T, Anthony LC, Regentin R, Keasling JD, Renninger NS, Newman JD. High-level production of amorpha-4,11-diene, a precursor of the antimalarial agent artemisinin, in Escherichia coli. PLoS One. 2009;4:e4489. doi: 10.1371/journal.pone.0004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang C, Chen X, Zou R, Zhou K, Stephanopoulos G, Too HP. Combining genotype improvement and statistical media optimization for isoprenoid production in E. coli. PLoS One. 2013;8:e75164. doi: 10.1371/journal.pone.0075164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang C, Zou R, Chen X, Stephanopoulos G, Too HP. Experimental design-aided systematic pathway optimization of glucose uptake and deoxyxylulose phosphate pathway for improved amorphadiene production. Appl Microbiol Biotechnol. 2015;99:3825–3837. doi: 10.1007/s00253-015-6463-y. [DOI] [PubMed] [Google Scholar]
- 116.Wang JF, Xiong ZQ, Li SY, Wang Y. Enhancing isoprenoid production through systematically assembling and modulating efflux pumps in Escherichia coli. Appl Microbiol Biotechnol. 2013;97:8057–8067. doi: 10.1007/s00253-013-5062-z. [DOI] [PubMed] [Google Scholar]
- 117.Zhang C, Chen X, Stephanopoulos G, Too HP. Efflux transporter engineering markedly improves amorphadiene production in Escherichia coli. Biotechnol Bioeng. 2016 doi: 10.1002/bit.25943. [DOI] [PubMed] [Google Scholar]
- 118.Chang MCY, Eachus RA, Trieu W, Ro DK, Keasling JD. Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat Chem Biol. 2007;3:274–277. doi: 10.1038/nchembio875. [DOI] [PubMed] [Google Scholar]
- 119.Dietrich JA, Yoshikuni Y, Fisher KJ, Woolard FX, Ockey D, McPhee DJ, Renninger NS, Chang MCY, Baker D, Keasling JD. A novel semi-biosynthetic route for artemisinin production using engineered substrate-promiscuous P450BM3. ACS Chem Biol. 2009;4:261–267. doi: 10.1021/cb900006h. [DOI] [PubMed] [Google Scholar]
- 120.Lindahl AL, Olsson ME, Mercke P, Tollbom O, Schelin J, Brodelius M, Brodelius PE. Production of the artemisinin precursor amorpha-4,11-diene by engineered Saccharomyces cerevisiae. Biotechnol Lett. 2006;28:571–580. doi: 10.1007/s10529-006-0015-6. [DOI] [PubMed] [Google Scholar]
- 121.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MCY, Withers ST, Shiba Y, Sarpong R, Keasling JD. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 122.Ro DK, Ouellet M, Paradise EM, Burd H, Eng D, Paddon CJ, Newman JD, Keasling JD. Induction of multiple pleiotropic drug resistance genes in yeast engineered to produce an increased level of anti-malarial drug precursor, artemisinic acid. BMC Biotechnol. 2008;8:83. doi: 10.1186/1472-6750-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lenihan JR, Tsuruta H, Diola D, Renninger NS, Regentin R. Developing an industrial artemisinic acid fermentation process to support the cost-effective production of antimalarial artemisinin-based combination therapies. Biotechnol Prog. 2008;24:1026–1032. doi: 10.1002/btpr.27. [DOI] [PubMed] [Google Scholar]
- 124.Westfall PJ, Pitera DJ, Lenihan JR, EngD Woolard FX, Regentin R, Horning T, Tsuruta H, Melis DJ, Owens A, Fickes S, Diola D, Benjamin KR, Keasling JD, Leavell MD, McPhee DJ, Renninger NS, Newman JD, Paddon CJ. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc Natl Acad Sci USA. 2012;109:E111–E118. doi: 10.1073/pnas.1110740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schenkman JB, Jansson I. The many roles of cytochrome b5. Pharmacol Ther. 2003;97:139–152. doi: 10.1016/S0163-7258(02)00327-3. [DOI] [PubMed] [Google Scholar]
- 126.Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, Polichuk DR, Teoh KH, Reed DW, Treynor T, Lenihan J, Fleck M, Bajad S, Dang G, Diola D, Dorin G, Ellens KW, Fickes S, Galazzo J, Gaucher SP, Geistlinger T, Henry R, Hepp M, Horning T, Iqbal T, Jiang H, Kizer L, Lieu B, Melis D, Moss N, Regentin R, Secrest S, Tsuruta H, Vazquez R, Westblade LF, Xu L, Yu M, Zhang Y, Zhao L, Lievense J, Covello PS, Keasling JD, Reiling KK, Renninger NS, Newman JD. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 127.Paddon CJ, Keasling JD. Semi-synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development. Nat Rev. 2014;12:355–367. doi: 10.1038/nrmicro3240. [DOI] [PubMed] [Google Scholar]
- 128.Horváth Z, Horosanskaia E, Lee JW, Lorenz H, Gilmore K, Seeberger PH, Seidel-Morgenstern A. Recovery of artemisinin from a complex reaction mixture using continuous chromatography and crystallization. Org Process Res Dev. 2015;19:624–634. doi: 10.1021/acs.oprd.5b00048. [DOI] [Google Scholar]
- 129.Mannan A, Shaheen N, Arshad W, Qureshi RA, Zia M, Mirza B. Hairy roots induction and artemisinin analysis in Artemisia dubia and Artemisia indica. Afr J Biotechnol. 2008;7:3288–3292. [Google Scholar]
- 130.Mannan A, Ahmed I, Arshad W, Asim MF, Qureshi RA, Hussain I, Mirza B. Survey of artemisinin production by diverse Artemisia species in northern Pakistan. Malar J. 2010;9:310. doi: 10.1186/1475-2875-9-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mannan A, Ahmed I, Arshad W, Hussain I, Mirza B. Effects of vegetative and flowering stages on the biosynthesis of artemisinin in Artemisia species. Arch Pharm Res. 2011;34:1657–1661. doi: 10.1007/s12272-011-1010-6. [DOI] [PubMed] [Google Scholar]
- 132.Arab HA, Rahbari S, Rassouli A, Moslemi MH, Khosravirad F. Determination of artemisinin in Artemisia sieberi and anticoccidial effects of the plant extract in broiler chickens. Trop Anim Health Prod. 2006;38:497–503. doi: 10.1007/s11250-006-4390-8. [DOI] [PubMed] [Google Scholar]
- 133.Zia M, Mannan A, Chaudhary MF. Effect of growth regulators and amino acids on artemisinin production in the callus of Artemisia absinthium. Pak J Bot. 2007;39:799–805. [Google Scholar]
- 134.Dilshad E, Cusido RM, Estrada KR, Bonfill M, Mirza B. Genetic transformation of Artemisia carvifolia Buch with rol genes enhances artemisinin accumulation. PLoS One. 2015;10:e0140266. doi: 10.1371/journal.pone.0140266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nibret E, Wink M. Volatile components of four Ethiopian Artemisia species extracts and their in vitro antitrypanosomal and cytotoxic activities. Phytomedicine. 2009;17:369–374. doi: 10.1016/j.phymed.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 136.Kraft C, Jenett-Siems K, Siems K, Jakupovic J, Mavi S, Bienzle U, Eich E. In vitro antiplasmodial evaluation of medicinal plants from Zimbabwe. Phytother Res. 2003;17:123–128. doi: 10.1002/ptr.1066. [DOI] [PubMed] [Google Scholar]
- 137.Gathirwa JW, Rukunga GM, Njagi ENM, Omar SA, Guantai AN, Muthaura CN, Mwitari PG, Kimani CW, Kirira PG, Tolo FM, Ndunda TN, Ndiege IO. In vitro anti-plasmodial and in vivo anti-malarial activity of some plants traditionally used for the treatment of malaria by the Meru community in Kenya. J Nat Med. 2007;61:261–268. doi: 10.1007/s11418-007-0140-0. [DOI] [Google Scholar]
- 138.Ramazani A, Sardari S, Zakeri S, Vaziri B. In vitro antiplasmodial and phytochemical study of five Artemisia species from Iran and in vivo activity of two species. Parasitol Res. 2010;107:593–599. doi: 10.1007/s00436-010-1900-4. [DOI] [PubMed] [Google Scholar]
- 139.Komori A, Suzuki M, Seki H, Nishizawa T, Meyer JJM, Shimizu H, Yokoyama S, Muranaka T. Comparative functional analysis of CYP71AV1 natural variants reveals an important residue for the successive oxidation of amorpha-4,11-diene. FEBS Lett. 2013;587:278–284. doi: 10.1016/j.febslet.2012.11.031. [DOI] [PubMed] [Google Scholar]
- 140.Muangphrom P, Suzuki M, Seki H, Fukushima EO, Muranaka T. Functional analysis of orthologous artemisinic aldehyde Δ11(13)-reductase reveals potential artemisinin-producing activity in non-artemisinin-producing Artemisia absinthium. Plant Biotechnol. 2014;31:483–491. doi: 10.5511/plantbiotechnology.14.0807a. [DOI] [Google Scholar]
- 141.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwing KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NPJ, Lindegardh N, Socheat D, White NJ. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Witkowski B, Khim N, Chim P, Kim S, Ke S, Kloeung N, Chy S, Duong S, Leang R, Ringwald P, Dondorp AM, Tripura R, Benoit-Vical F, Berry A, Gorgette O, Ariey F, Barale JC, Mercereau-Puijalon O, Menard D. Reduced artemisinin susceptibility of Plasmodium falciparum ring stages in western Cambodia. Antimicrob Agents Chemother. 2013;57:914–923. doi: 10.1128/AAC.01868-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han KT, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, Rahman MR, Hasan MM, Islam A, Miotto O, Amato R, MacInnis B, Stalker J, Kwiatkowski DP, Bozdech Z, Jeeyapant A, Cheah PY, Sakulthaew T, Chalk J, Intharabut B, Silamut K, Lee SJ, Vihokhern B, Kunasol C, Imwong M, Tarning J, Taylor WJ, Yeung S, Woodrow CJ, Flegg JA, Das D, Smith J, Venkatesan M, Plowe CV, Stepniewska K, Guerin PJ, Dondorp AM, Day NP, White NJ. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Amaratunga C, Sreng S, Suon S, Phelps ES, Stepniewska K, Lim P, Zhou C, Mao S, Anderson JM, Lindegardh N, Jiang H, Song J, Su XZ, White NJ, Dondorp AM, Anderson TJC, Fay MP, Mu J, Duong S, Fairhurst RM. Artemisinin-resistant Plasmodium falciparum in Pursat province, western Cambodia: a parasite clearance rate study. Lancet Infect Dis. 2012;12:851–858. doi: 10.1016/S1473-3099(12)70181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Afonso A, Hunt P, Cheesman S, Alves AC, Cunha CV, do Rosário V, Cravo P. Malaria parasites can develop stable resistance to artemisinin but lack mutations in candidate genes atp6 (encoding the sarcoplasmic and endoplasmic reticulum Ca2+ ATPase), tctp, mdr1, and cg10. Antimicrob Agents Chemother. 2006;50:480–489. doi: 10.1128/AAC.50.2.480-489.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, Lim P, Mao S, Sopha C, Sam B, Anderson JM, Duong S, Chuor CM, Taylor WRJ, Suon S, Mercereau-Puijalon O, Fairhurst RM, Menard D. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in vitro and ex vivo drug-response studies. Lancet Infect Dis. 2013;13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mok S, Ashley EA, Ferreira PE, Zhu L, Lin Z, Yeo T, Chotivanich K, Imwong M, Pukrittayakamee S, Dhorda M, Nguon C, Lim P, Amaratunga C, Suon S, Hien TT, Htut Y, Faiz MA, Onyamboko MA, Mayxay M, Newton PN, Tripura R, Woodrow CJ, Miotto O, Kwiatkowski DP, Nosten F, Day NPJ, Preiser PR, White NJ, Dondorp AM, Fairhurst RM, Bozdech Z. Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science. 2015;347:431–435. doi: 10.1126/science.1260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Takala-Harrison S, Clark TG, Jacob CG, Cummings MP, Miotto O, Dondorp AM, Fukuda MM, Nosten F, Noedl H, Imwong M, Bethell D, Se Y, Lon C, Tyner SD, Saunders DL, Socheat D, Ariey F, Phyo AP, Starzengruber P, Fuehrer HP, Swoboda P, Stepniewska K, Flegg J, Arze C, Cerqueira GC, Silva JC, Ricklefs SM, Porcella SF, Stephens RM, Adams M, Kenefic LJ, Campino S, Auburn S, MacInnis B, Kwiatkowski DP, Su XZ, White NJ, Ringwald P, Plowe CV. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc Natl Acad Sci USA. 2012;110:240–245. doi: 10.1073/pnas.1211205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cheeseman IH, Miller BA, Nair S, Nkhoma S, Tan A, Tan JC, Saai SA, Phyo AP, Moo CL, Lwing KM, McGready R, Ashley E, Imwong M, Stepniewska K, Yi P, Dondorp AM, Mayxay M, Newton PN, White NJ, Nosten F, Ferdig MT, Anderson TJC. A major genome region underlying artemisinin resistance in Malaria. Science. 2012;336:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Miotto O, Almagro-Garcia J, Manske M, MacInnis B, Campino S, Rockett KA, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Duong S, Nguon C, Chuor CM, Saunders D, Se Y, Lon C, Fukuda MM, Amenga-Etego L, Hodgson AVO, Asoala V, Imwong M, Takala-Harrison S, Nosten F, Su XZ, Ringwald P, Ariey F, Dolecek C, Hien TT, Boni MF, Thai CQ, Amambua-Ngwa A, Conway DJ, Djimdé AA, Doumbo OK, Zongo I, Ouedraogo JB, Alcock D, Drury E, Auburn S, Koch O, Sanders M, Hubbart C, Maslen G, Ruano-Rubio V, Jyothi D, Miles A, O’Brien J, Gamble C, Oyola SO, Rayner JC, Newbold CI, Berriman M, Spencer CCA, McVean G, Day NP, White NJ, Bethell D, Dondorp AM, Plowe CV, Fairhurst RM, Kwiatkowski DP. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet. 2013;45:648–655. doi: 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Ménard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale JC, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Ménard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Isozumi R, Uemura H, Kimata I, Ichinose Y, Logedi J, Omar AH, Kaneko A. Novel mutations in K13 propeller gene of artemisinin-resistant Plasmodium falciparum. Emerg Infect Dis. 2015;21:490–492. doi: 10.3201/eid2103.140898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Straimer J, Gnädig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, Dacheux M, Khim N, Zhang L, Lam S, Gregory PD, Urnov FD, Mercereau-Puijalon O, Benoit-Vical F, Fairhurst RM, Ménard D, Fidock DA. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nyunt MH, Hlaing T, Oo HW, Tin-Oo LLK, Phway HP, Wang B, Zaw NN, Han SS, Tun T, San KK, Kyaw MP, Han ET. Molecular assessment of artemisinin resistance markers, polymorphisms in the K13 propeller, and a multidrug-resistance gene in the eastern and western border areas of Myanmar. Clin Infect Dis. 2015;60:1208–1215. doi: 10.1093/cid/ciu1160. [DOI] [PubMed] [Google Scholar]
- 155.Bayih AG, Getnet G, Alemu A, Getie S, Mohon AN, Pillai DR. A unique Plasmodium falciparum Kelch 13 gene mutation in northwest Ethiopia. Am J Trop Med Hyg. 2016;94:132–135. doi: 10.4269/ajtmh.15-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mbengue A, Bhattacharjee S, Pandharkar T, Liu H, Estiu G, Stahelin RV, Rizk SS, Njimoh DL, Ryan Y, Chotivanich K, Nguon C, Ghorbal M, Lopez-Rubio JJ, Pfrender M, Emrich S, Mohandas N, Dondorp AM, Wiest O, Haldar K. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520:683–687. doi: 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mita T, Tachibana S, Hashimoto M, Hirai M. Plasmodium falciparum kelch 13: a potential molecular marker for tackling artemisinin-resistant malaria parasites. Expert Rev Anti-Infect Ther. 2016;14:125–135. doi: 10.1586/14787210.2016.1106938. [DOI] [PubMed] [Google Scholar]
- 158.Wang Z, Wang Y, Cabrera M, Zhang Y, Gupta B, Wu Y, Kemirembe K, Hu Y, Liang X, Brashear A, Shrestha S, Li X, Miao J, Sun X, Yang Z, Cui L. Artemisinin resistance at the China–Myanmar border and association with mutations in the K13 propeller gene. Antimicrob Agents Chemother. 2015;59:6952–6959. doi: 10.1128/AAC.01255-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Humphreys GS, Merinopoulos I, Ahmed J, Whitty CJM, Mutabingwa TK, Sutherland CJ, Hallett RL. Amodiaquine and artemether–lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother. 2007;51:991–997. doi: 10.1128/AAC.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.van der Velden M, Rijpma SR, Russel FGM, Sauerwein RW, Koenderink JB. PfMDR2 and PfMDR5 are dispensable for Plasmodium falciparum asexual parasite multiplication but change in vitro susceptibility to anti-malarial drugs. Malar J. 2015;14:76. doi: 10.1186/s12936-015-0581-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dogovski C, Xie SC, Burgio G, Bridgford J, Mok S, McCaw JM, Chotivanich K, Kenny S, Gnädig N, Straimer J, Bozdech Z, Fidock DA, Simpson JA, Dondorp AM, Foote S, Klonis N, Tilley L. Targeting the cell stress response of Plasmodium falciparum to overcome artemisinin resistance. PLoS Biol. 2015;13:e1002132. doi: 10.1371/journal.pbio.1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Elfawal MA, Towler MJ, Reich NG, Weathers PJ, Golenbock D, Rich SM. Dried whole plant Artemisia annua as an antimalarial therapy. PLoS One. 2012;7:e52746. doi: 10.1371/journal.pone.0052746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Onimus M, Carteron S, Lutgen P. The surprising efficiency of Artemisia annua powder capsules. Med Aromat Plants. 2013;2:3. doi: 10.4172/2167-0412.1000125. [DOI] [Google Scholar]
- 164.Weathers PJ, Elfawal MA, Towler MJ, Acquaah-Mensah GK, Rich SM. Pharmacokinetics of artemisinin delivered by oral consumption of Artemisia annua dried leaves in healthy vs. Plasmodium chabaudi-infected mice. J Ethnopharmacol. 2014;153:732–736. doi: 10.1016/j.jep.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Weathers PJ, Jordan NJ, Lasin P, Towler MJ. Simulated digestion of dried leaves of Artemisia annua consumed as a treatment (pACT) for malaria. J Ethnopharmacol. 2014;151:858–863. doi: 10.1016/j.jep.2013.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Weathers PJ, Towler MJ. Changes in key constituents of clonally propagated Artemisia annua L. during preparation of compressed leaf tablets for possible therapeutic use. Ind Crops Prod. 2014;62:173–178. doi: 10.1016/j.indcrop.2014.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Weathers PJ, Towler MJ, Hassanali A, Lutgen P, Engeu PO. Dried-leaf Artemisia annua: a practical malaria therapeutic for developing countries? World J Pharmacol. 2014;3:39–55. doi: 10.5497/wjp.v3.i4.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Towler MJ, Weathers PJ. Variations in key artemisinic and other metabolites throughout plant development in Artemisia annua L. for potential therapeutic use. Ind Crops Prod. 2015;67:185–191. doi: 10.1016/j.indcrop.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Elfawal MA, Towler MJ, Reich NG, Weathers PJ, Rich SM. Dried whole-plant Artemisia annua slows evolution of malaria drug resistance and overcomes resistance to artemisinin. Proc Natl Acad Sci USA. 2015;112:821–826. doi: 10.1073/pnas.1413127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Giganti D, Bouillon A, Tawk L, Robert F, Martinez M, Crublet E, Weber P, Girard-Blanc C, Petres S, Haouz A, Hernandez JF, Mercereau-Puijalon O, Alzari PM, Barale JC. A novel Plasmodium-specific prodomain fold regulates the malaria drug target SUB1 subtilase. Nat Commun. 2014;5:4833. doi: 10.1038/ncomms5833. [DOI] [PubMed] [Google Scholar]
- 171.Guggisberg AM, Park J, Edwards RL, Kelly ML, Hodge DM, Tolia NH, Odom AR. A sugar phosphatase regulates the methylerythritol phosphate (MEP) pathway in malaria parasites. Nat Commun. 2014;5:4467. doi: 10.1038/ncomms5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wright MH, Clough B, Rackham MD, Rangachari K, Brannigan JA, Grainger M, Moss DK, Bottrill AR, Heal WP, Broncel M, Serwa RA, Brady D, Mann DJ, Leatherbarrow RJ, Tewari R, Wilkinson AJ, Holder AA, Tate EW. Validation of N-myristoyltransferase as an antimalarial drug target using an integrated chemical biology approach. Nat Chem. 2013;6:112–121. doi: 10.1038/nchem.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Allen SM, Lim EE, Jortzik E, Preuss J, Chua HH, MacRae JI, Rahlfs S, Haeussler K, Downton MT, McConville MJ, Becker K, Ralph SA. Plasmodium falciparum glucose-6-phosphate dehydrogenase 6-phosphogluconolactonase is a potential drug target. FEBS J. 2015;282:3808–3823. doi: 10.1111/febs.13380. [DOI] [PubMed] [Google Scholar]
- 174.Li H, van der Linden WA, Verdoes M, Florea BI, McAllister FE, Govindaswamy K, Elias JE, Bhanot P, Overkleeft HS, Bogyo M. Assessing subunit dependency of the Plasmodium proteasome using small molecule inhibitors and active site probes. ACS Chem Biol. 2014;9:1869–1876. doi: 10.1021/cb5001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yuthavong Y, Tarnchompoo B, Vilaivan T, Chitnumsub P, Kamchonwongpaisan S, Charman SA, McLennan D, White KL, Vivas L, Bongard E, Thongphanchang C, Taweechai S, Vanichtanankul J, Rattanajak R, Arwon U, Fantauzzi P, Yuvaniyama J, Charman WN, Matthews D. Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target. Proc Natl Acad Sci USA. 2012;109:16823–16828. doi: 10.1073/pnas.1204556109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mokmak W, Chunsrivirot S, Hannongbua S, Yuthavong Y, Tongsima S, Kamchonwongpaisan S. Molecular dynamics of interactions between rigid and flexible antifolates and dihydrofolate reductase from pyrimethamine-sensitive and pyrimethamine-resistant Plasmodium falciparum. Chem Biol Drug Des. 2014;84:450–461. doi: 10.1111/cbdd.12334. [DOI] [PubMed] [Google Scholar]
- 177.Chitnumsub P, Jaruwat A, Riangrungroj P, Ittarat W, Noytanom K, Oonanant W, Vanichthanankul J, Chuankhayan P, Maenpuen S, Chen CJ, Chaiyen P, Yuthavong Y, Leartsakulpanich U. Structures of Plasmodium vivax serine hydroxymethyltransferase: implications for ligand-binding specificity and functional control. Acta Crystallogr D Biol Crystallogr. 2014;70:3177–3186. doi: 10.1107/S1399004714023128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pinthong C, Maenpuen S, Amornwatcharapong W, Yuthavong Y, Leartsakulpanich U, Chaiyen P. Distinct biochemical properties of human serine hydroxymethyltransferase compared with the Plasmodium enzyme: implications for selective inhibition. FEBS J. 2014;281:2570–2583. doi: 10.1111/febs.12803. [DOI] [PubMed] [Google Scholar]
- 179.Chitnumsub P, Ittarat W, Jaruwat A, Noytanom K, Amornwatcharapong W, Pomthanakasem W, Chaiyen P, Yuthavong Y, Leartsakulpanich U. The structure of Plasmodium falciparum serine hydroxymethyltransferase reveals a novel redox switch that regulates its activities. Acta Crystallogr D Biol Crystallogr. 2014;70:1517–1527. doi: 10.1107/S1399004714005598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Maenpuen S, Amornwatcharapong W, Krasatong P, Sucharitakul J, Palfey BA, Yuthavong Y, Chitnumsub P, Leartsakulpanich U, Chaiyen P. Kinetic mechanism and the rate-limiting step of Plasmodium vivax serine hydroxymethyltransferase. J Biol Chem. 2015;290:8656–8665. doi: 10.1074/jbc.M114.612275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Henrich PP, O’Brien C, Sáenz FE, Cremers S, Kyle DE, Fidock DA. Evidence for pyronaridine as a highly effective partner drug for treatment of artemisinin-resistant malaria in a rodent model. Antimicrob Agents Chemother. 2014;58:183–195. doi: 10.1128/AAC.01466-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sun W, Tanaka TQ, Magle CT, Huang W, Southall N, Huang R, Dehdashti SJ, McKew JC, Williamson KC, Zheng W. Chemical signatures and new drug targets for gametocytocidal drug development. Sci Rep. 2014;4:3743. doi: 10.1038/srep03743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Singh C, Verma VP, Hassam M, Singh AS, Naikade NK, Puri SK. New orally active amino- and hydroxy-functionalized 11-azaartemisinins and their derivatives with high order of antimalarial activity against multidrug-resistant Plasmodium yoelii in Swiss mice. J Med Chem. 2014;57:2489–2497. doi: 10.1021/jm401774f. [DOI] [PubMed] [Google Scholar]
- 184.Oliveira R, Newton AS, Guedes RC, Miranda D, Amewu RK, Srivastava A, Gut J, Rosenthal PJ, O’Neill PM, Ward SA, Lopes F, Moreira R. An endoperoxide-based hybrid approach to deliver falcipain inhibitors inside malaria parasites. ChemMedChem. 2013;8:1528–1536. doi: 10.1002/cmdc.201300202. [DOI] [PubMed] [Google Scholar]
- 185.Oliveira R, Miranda D, Magalhães J, Capela R, Perry MJ, O’Neill PM, Moreira R, Lopes F. From hybrid compounds to targeted drug delivery in antimalarial therapy. Bioorg Med Chem. 2015;23:5120–5130. doi: 10.1016/j.bmc.2015.04.017. [DOI] [PubMed] [Google Scholar]
- 186.Witschel MC, Rottmann M, Schwab A, Leartsakulpanich U, Chitnumsub P, Seet M, Tonazzi S, Schwertz G, Stelzer Mietzner T, McNamara C, Thater F, Freymond C, Jaruwat A, Pinthong C, Riangrungroj P, Oufir M, Hamburger M, Mäser P, Sanz-Alonso LM, Charman S, Wittlin S, Yuthavong Y, Chaiyen P, Diederich F. Inhibitors of plasmodial serine hydroxymethyltransferase (SHMT): cocrystal structures of pyrazolopyrans with potent blood- and liver-stage activities. J Med Chem. 2015;58:3117–3130. doi: 10.1021/jm501987h. [DOI] [PubMed] [Google Scholar]