Abstract
Various herbal medicines have been developed and used in various parts of the world for thousands of years. Although locally grown indigenous plants were originally used for traditional herbal preparations, Western herbal products are now becoming popular in Japan with the increasing interest in health. At the same time, there are growing concerns about the substitution of ingredients and adulteration of herbal products, highlighting the need for the authentication of the origin of plants used in herbal products. This review describes studies on Cimicifuga and Vitex products developed in Europe and Japan, focusing on establishing analytical methods to evaluate the origins of material plants and finished products. These methods include a polymerase chain reaction-restriction fragment length polymorphism method and a multiplex amplification refractory mutation system method. A genome-based authentication method and liquid chromatography-mass spectrometry-based authentication for black cohosh products, and the identification of two characteristic diterpenes of agnus castus fruit and a shrub chaste tree fruit-specific triterpene derivative are also described.
Electronic supplementary material
The online version of this article (doi:10.1007/s11418-016-1006-0) contains supplementary material, which is available to authorized users.
Keywords: Herbal product, Quality control, DNA identification, LC/MS, Cimicifuga, Vitex
Introduction
Herbal medicines have been used for thousands of years to treat illness in various parts of the world. Kampo medicines (traditional Japanese medicines) were developed in Japan using herbal plants native to Asia, whereas Western herbal plants native to Europe have traditionally been used as crude drugs. The raw materials and extracts are now standardized to be used for medicines in many countries. Thus, closely related plants are used worldwide for different pharmaceuticals with different treatment applications, depending on their background. For example, the flowering aerial part of Leonurus cardiac L. is used to relieve symptoms of nervous tension under the name ‘motherwort’ in Europe, whereas the flowering aerial part of L. japonicas Houtt. and L. sibiricus L. is used as a crude drug named Leonurus Herb (益母草 in Japanese) as a component of Kampo formulations for the treatment of women’s disorders in Japan (Table 1). When herbal medicines are made from locally grown plants there is less concern, but misidentification and confusion may occur when both Western and the Asian herbal medicines are used in the same area.
Table 1.
Examples of the Western herbs and crude drugs in Japan derived from plants of the same genus
| Genus | Western herba | Crude drug in Japan | |||||
|---|---|---|---|---|---|---|---|
| Name | Botanical origin | Part of use | Name | Japanese name | Botanical origin | Part of use | |
| Artemisia | Wormwood | A. absinthium L. | Leaf, flowering top | Artemisia capillaris flower | 茵蔯蒿 | A. capillaris Thunb. | Flower head |
| Cimicifuga | Black cohosh | C. racemosa (L.) Nutt | Rhizome, root | Cimicifuga Rhizome | 升麻 |
C. simplex (DC.) Turcz. C. dahurica (Trucz.) Maxim. C. heracleifolia Kom. C. foetida L. |
Rhizome |
| Leonurus | Motherwort | L. cardiac L. | Flowering aerial part | Leonurus herb | 益母草 |
L. japonicas Houtt. L. sibiricus L. |
Flowering aerial part |
| Quercus | Oak bark |
Q. robur L. Q. petraea (Matt.) Liebl. Q. pubescens Willd |
Bark | Quercus bark | 樸樕 |
Q. acutissima Carruth. Q. serrata Murray, Q. mongolica Fisch. ex Ledeb. Q. uariabilis Blume |
Bark |
| Rosa | Dog rose |
R. cania L., R. pendulina L. and other Rosa species |
Receptacle and sepal | Rose fruit | 営実 | R. multiflora Thunnb | Fruit, pseudocarp |
| Vitex | Agnus castus fruit | V. agnus-castus L. | Fruit | Shrub chaste tree fruit | 蔓荊子 |
V. rotundifolia L. f. V. trifolia L. |
Fruit |
aHerbal substances defined in the European Pharmacopoeia
In Japan, Western herbal preparations have only been used as health food products because of the lack of usage experience but they now are becoming more popular due to the changes in cause-specific morbidity and the increasing interest in self-medication. After guidelines for the approval of pharmaceuticals including Western herbs were established based on a notification from the Japanese Ministry of Health, Labour and Welfare [1], two products prepared from Western herbs have been approved as over-the-counter (OTC) drugs in Japan, and more products are expected to become available to the Japanese market in the near future. Practical methods for verifying the botanical sources of herbal products are thus required to ensure the safety and efficacy of such products [2–4]. This review describes some of our recent studies on the genetic and chemical authentication of the original plant species in herbal products.
Studies on the botanical origins of Cimicifuga products
Black cohosh (Cimicifuga racemosa (L.) Nutt.) is one of the most commonly used Western herbs today. Its roots and rhizomes have traditionally been used for the treatment of women’s disorders in North America and Europe, and it is now used globally to alleviate menopausal symptoms, being one of the top 10 best-selling herbs in the USA. Black cohosh extracts or materials are officially prescribed in the European Pharmacopoeia (EP) and standardized as an active ingredient of OTC drugs in Europe. In the USA, black cohosh extracts and materials are prepared for use as dietary supplements, and The United States Pharmacopeia and The National Formulary (USP−NF) provides the standards.
Because the regulation of herbal products as dietary supplements in the USA is less strict than for OTC drugs in Europe, the substitution and adulteration of black cohosh dietary supplements with related Cimicifuga plants has frequently occurred in North America [5–7]. Additional reports have described hepatotoxicity associated with the use of black cohosh products, especially from products derived from the wrong plant parts instead of the roots and rhizomes or different Cimicifuga species [8–12]. The Ministry of Health, Labour and Welfare of Japan as well as other national ministries have updated their notifications about the risk of adverse liver reactions caused by the use of products containing black cohosh.
One possible reason why Cimicifuga plants native to Asia have been found in dietary supplements in the USA is that Chinese crude drugs were mistakenly recognized as black cohosh and were mixed with it. Cimicifuga plants are also used as traditional medicines in East Asia including Japan; the rhizome of C. dahurica (Turcz.) Maxim., C. foetida L. and C. heracleifolia Kom. is defined in the Chinese Pharmacopoeia (CP) as a crude drug named Cimicifuga Rhizome with heat-clearing and detoxifying effects, whereas the Japanese Pharmacopoeia (JP) describes C. simplex (DC.) Wormsk. in addition to the above three species as the botanical origin of Cimicifuga Rhizome (or “升麻” in Japanese) [13]. It is illegal in Japan for any health foods to contain materials or extracts prepared from C. simplex, C. dahurica, C. foetida or C. heracleifolia instead of C. racemosa, because Cimicifuga Rhizome is a pharmaceutical ingredient used exclusively for drugs.
Because of these problems, we developed genetic and chemical methods to simultaneously discriminate C. racemosa from other related species for the evaluation of black cohosh products that are commercially available in Japan.
Genome-based authentication of black cohosh products
Molecular biological techniques are useful and powerful methods to detect genetic differences among closely related plant species [14–17]. For Cimicifuga plants, a random amplified fragment length polymorphism fingerprinting method was developed to identify the four sympatric species distributed in eastern North America (C. racemosa, C. americana Michx., C. pachypoda Elliott and C. rubifolia Kearney) [18], and a multiplex amplification refractory mutation system (ARMS) method was developed to identify four Asian species (C. dahurica, C. foetida, C. heracleifolia, and C. simplex) [19]. To the best of our knowledge, there has been no report about the simultaneous identification of both North American and Asian species. To establish a DNA analytical method that can discriminate C. racemosa from possible contaminated species, we prepared six Cimicifuga plants, identified each species by sequencing, and attempted to develop polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and ARMS methods [20].
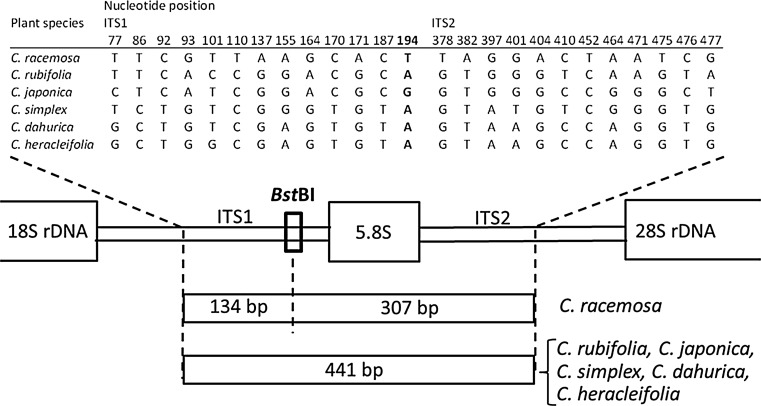
Eight referential Cimicifuga plants were kindly provided by the Research Center for Medicinal Plant Resources, National Institute of Biomedical Innovation, Health and Nutrition, and were identified as six species (one C. racemosa, three C. simplex and one each of C. japonica (Thunb.) Spreng, C, dahurica, C. heracleifolia and C. rubifolia) by DNA sequencing on the ITS and trnL−F regions (Table 2). Based on these sequences and sequences obtained from the DNA databases, we first developed a novel PCR-RFLP method for the identification of C. racemosa targeting on its unique BstBI restriction site (5ʹ TT*CGAA 3′) on the ITS1 region (Fig. 1). When the 441 bp of a DNA fragment on the ITS1 region was amplified by PCR from referential plant samples and digested with BstBI, two unique fragments of 134 and 307 bp were observed only from the referential C. racemosa plants (Fig. 2).
Table 2.
Genetic identification of referential Cimicifuga plant samples
| Sample ID | Plant species | ITS region | Registered GenBank accession no. | trnL-F region | Registered GenBank accession no. | ||
|---|---|---|---|---|---|---|---|
| GenBank accession no. | Sequence identity (%) | GenBank accession no. | Sequence identity (%) | ||||
| C1 | C. rubifolia | Z98297 | 99 | AB987680 | AJ223004 | 98 | AB987688 |
| C2 | C. japonica | Z98291 | 98 | AB987681 | AJ222999 | 98 | AB987689 |
| C3 | C. simplex | Z98299 | 99 | AB987682 | AJ223008 | 98 | AB987690 |
| C4 | C. simplex | Z98299 | 99 | AB987683 | AJ223008 | 98 | AB987691 |
| C5 | C. simplex | AB044408 | 96 | AB987684 | AJ223008 | 98 | AB987692 |
| C6 | C. dahurica | Z98284 | 99 | AB987685 | AJ222992 | 98 | AB987693 |
| C7 | C. heracleifolia | Z98290 | 97 | AB987686 | EU186382 | 97 | AB987694 |
| C8 | C. racemosa | Z98296 | 99 | AB987687 | AJ223003 | 98 | AB987695 |
Fig. 1.
Partial ITS sequences observed in referential Cimicifuga plants. Sequence data are deposited in GenBank under accession nos. AB987680–AB987687. Bold the sequence recognized by BstBI
Fig. 2.
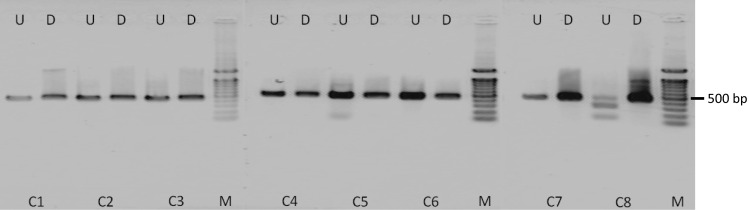
PCR-RFLP analysis of Cimicifuga plants showing the restriction enzyme BstBI on a partial ITS region. Data taken from reference 20. The forward primer: 5′-ATCATCCATTGTCGGGTCAT-3′, the reverse primer: 5′-GTTTACAACCACCGCAGACC-3′. Sample IDs are shown in Table 2. M 100 bp DNA ladder, D digested, U undigested
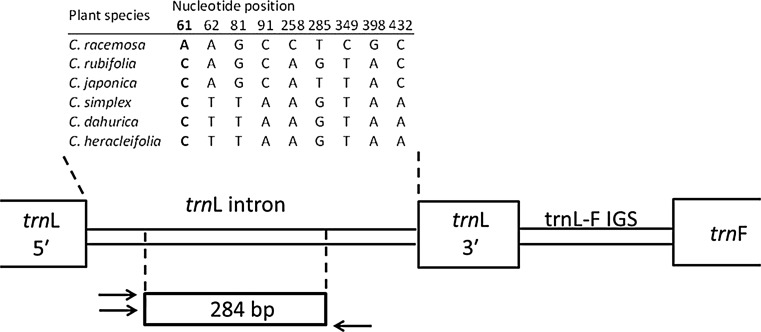
Although we found this to be a convenient and useful method to identify species of plant samples and to detect the presence of C. racemosa in black cohosh products, we could not detect contaminated or adulterated products by this method because undigested fragments remained. Therefore, we next developed an ARMS method for discriminating C. racemosa from other related species, using two types of specific sense primers targeting the nucleotide substitution at position 61 on the trnL gene (Fig. 3). The species-specific and the common fragments (284 bp) were successfully observed from C. racemosa and the other five species when the PCR was performed with DNAs from referential plants using the specific primer sets for C. racemosa and those for others, respectively (Fig. 4). Our results indicated that the ARMS analysis was a more accurate method than the PCR-RFLP method to identify C. racemosa from the related species, although the ARMS method takes a longer time.
Fig. 3.
Partial trnL-F gene sequences observed in referential Cimicifuga plants. The sequence data were deposited in GenBank under accession nos. AB987688–AB987695
Fig. 4.
ARMS analysis of Cimicifuga plants in trnL−trnF IGS region. Data taken from reference 20. a PCR amplification using a specific primer set for C. racemosa. b PCR amplification using a primer set for other Cimicifuga species. The sequences of a species-specific sense primer for C. racemosa and another five Cimicifuga species: 5′-CCTGAGCCAAATCCTGTTCTA-3′ and 5′-CCTGAGCCAAATCCTGTTCTC-3′. The sequence of a common anti-sense primer: 5′-TCAAATCAGCCACTCCATAGTATGA-3′. Sample IDs are shown in Table 2. M 100 bp DNA ladder
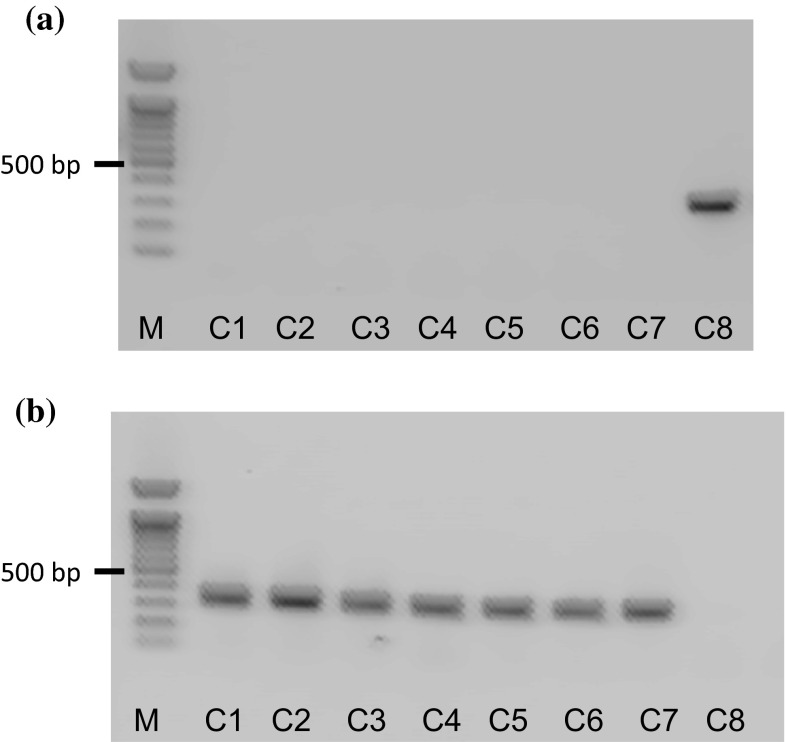
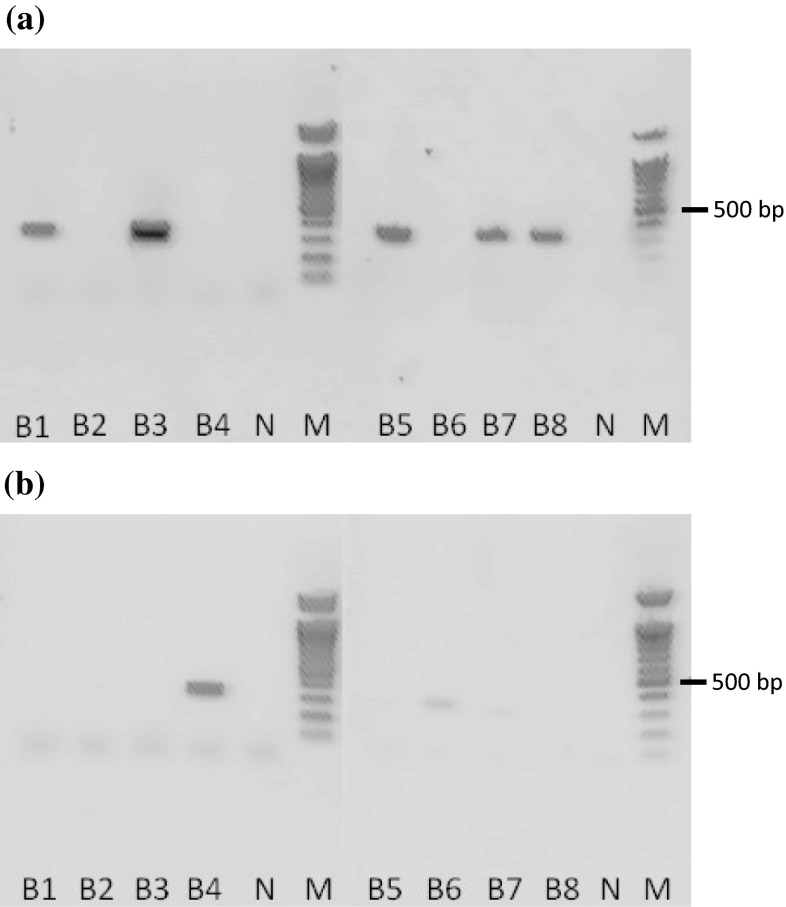
We then applied the ARMS method to test the original plants of eight black cohosh products available on the Japanese market, and the results demonstrated that three of the eight products were not derived from C. racemosa (Fig. 5; Table S1). We also performed TLC and HPLC analyses for detecting two marker compounds—actein (1) and 23-epi-26-deoxyactein (2)—(Fig. 6) from black cohosh products, and we found that the three of the eight products did not contain marker compounds, as was also shown by the results of the ARMS analysis (data not shown). These TLC and HPLC results completely followed the results of the ARMS analysis and strongly demonstrated that the ARMS analysis is a useful method for authenticating the botanical source of black cohosh products.
Fig. 5.
ARMS analysis of black cohosh products using the partial trnL gene. Data taken from reference 20. a PCR amplification using a specific primer set for C. racemosa. b PCR amplification using a primer set for other Cimicifuga species. B1, 2, 3, 4 and 6 contained powdered dried material within the capsules, whereas B5, 7, and 8 contained both powdered material and dry extract within the capsules. N no template control, M 100 bp DNA ladder
Fig. 6.
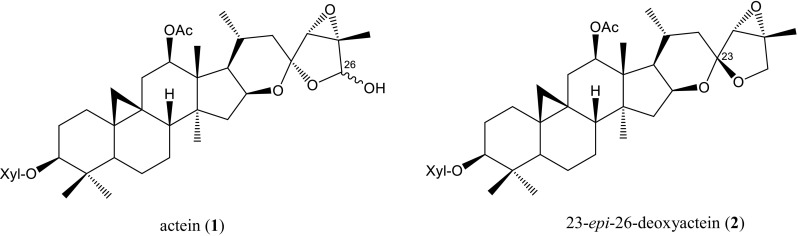
Chemical structures of actein and 23-epi-26-deoxyactein. Xyl β-D-xylopyranosyl
LC/MS-based authentication of black cohosh products
Although the ARMS analysis was a very precise and straightforward way to evaluate the botanical origin of black cohosh products as described above, it had a disadvantage in that hardly any DNA was obtained from the products consisting of dry extracts. On the other hand, chemical analyses using marker compounds were also difficult to use for evaluating the botanical origin because most of the metabolites from closely related plant species overlap. The EP sets an identification test for black cohosh to confirm the presence of actein (1) and 23-epi-26-deoxyactein (2) by TLC. However, since these compounds are present in other Cimicifuga species as well as C. racemosa, the EP sets three additional TLC tests for substitution and adulteration to compare the chromatogram obtained from the reference solution with each chromatogram from C. americana, C. foetida, and the mixture of C. dahurica and C. heracleifolia [21].
In the course of our studies on the botanical origins of black cohosh products, we found and reported a simple way to detect substitution and adulteration [22]. We prepared 25 black cohosh products including six OTC drugs purchased in Europe (Table 3: BC-1 to -19, BC-M1 to –M6) and five Cimicifuga Rhizome crude drugs of JP grade (Table 3: CR-1 to -5), and we genetically identified their origins by DNA sequencing and analyzed their constituents by liquid chromatography-mass spectrometry (LC/MS). The sequencing analysis of the ITS region revealed that five of the 10 tested health food products containing powdered plant material contained undeclared C. dahurica, and that Cimicifuga Rhizome products were derived from C. dahurica except for CR-3, which contained both C. dahurica and C. heracleifolia (Table 3).
Table 3.
The Cimicifuga products applied for LC/MS analysis
| Sample ID | Classa | Content | Source plantb | XIC pattern | Predicted species |
|---|---|---|---|---|---|
| BC-1 | A | Powdered plant material | C. racemosa | I | |
| BC-2 | A | Dry extract | I | C. racemosa | |
| BC-3 | A | Powdered plant material | C. racemosa | I | |
| BC-4 | A | Powdered plant material and dry extract | C. racemosa | I | |
| BC-5 | A | Dry extract | IV | Others | |
| BC-6 | A | Powdered plant material | C. dahurica | II | |
| BC-7 | A | Powdered plant material and dry extract | C. dahurica | III | |
| BC-8 | A | Dry extract | I | C. racemosa | |
| BC-9 | A | Powdered plant material and dry extract | C. racemosa | I | |
| BC-10 | A | Dry extract | I | C. racemosa | |
| BC-11 | A | Dry extract | I | C. racemosa | |
| BC-12 | A | Powdered plant material | C. dahurica | II | |
| BC-13 | A | Dry extract | I | C. racemosa | |
| BC-14 | A | Dry extract | I | C. racemosa | |
| BC-15 | A | Powdered plant material | C. racemosa | I | |
| BC-16 | A | Powdered plant material and dry extract | C. dahurica | III | |
| BC-17 | A | Dry extract | I | C. racemosa | |
| BC-18 | A | Dry extract | IV | Others | |
| BC-19 | A | Powdered plant material and dry extract | C. dahurica | III | |
| BC-M1 | B | Dry extract | I | C. racemosa | |
| BC-M2 | B | Dry extract | I | C. racemosa | |
| BC-M3 | B | Dry extract | I | C. racemosa | |
| BC-M4 | B | Dry extract | I | C. racemosa | |
| BC-M5 | B | Dry extract | I | C. racemosa | |
| BC-M6 | B | Dry extract | I | C. racemosa | |
| CR-1 | C | Cut crude drug | C. dahurica | II | |
| CR-2 | C | Cut crude drug | C. dahurica | II | |
| CR-3 | C | Cut crude drug | C. dahurica, C. heracleifolia | II | |
| CR-4 | C | Cut crude drug | C. dahurica | II | |
| CR-5 | C | Cut crude drug | C. dahurica | II |
a A health food, B OTC drug of EP grade, C crude drug of JP grade
bThe species of plant materials were identified by DNA sequencing
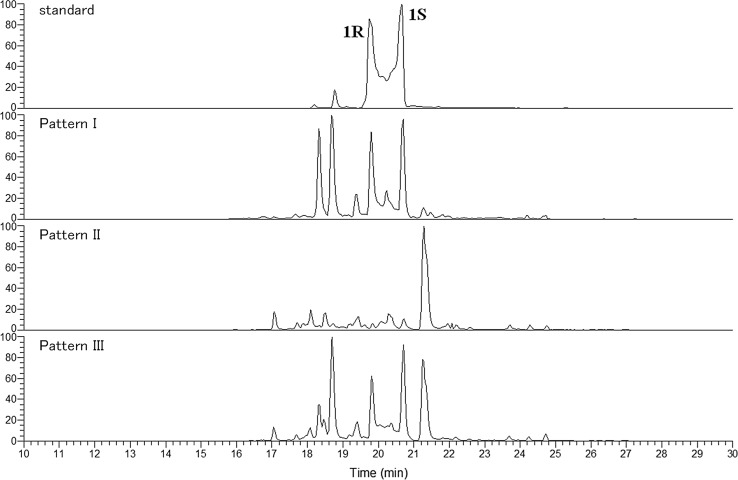
When these 15 samples were analyzed by LC/MS for the detection of actein (1) and 23-epi-26-deoxyactein (2), they showed three chromatographic patterns (I, II and III) based on the extracted ion chromatograms (XIC) at m/z 735.4 corresponding to actein (Fig. 7). All of the black cohosh products derived from C. racemosa (BC-1, -3, -4, -9, and -15) definitely contained marker compounds and showed pattern I. Of the 10 products that contained plant materials of C. dahurica, all of the Cimicifuga Rhizome samples (BC-6 and BC-12) showed pattern II, whereas the chromatograms of the other black cohosh samples (BC-7, -16, and -19) showed pattern III.
Fig. 7.
Representative XIC of black cohosh samples classified as patterns I, II, and III. Data taken from reference 22. 1R: (R)-actein, 1S: (S)-actein
Pattern III appears to overlap with patterns I and II, and the products of pattern III consisted of powdered materials and dry extracts; it was thus strongly suggested that BC-7, -16 and -19 consisted of powdered materials of C. dahurica and an extract prepared from C. racemosa. Taking these results together with our above-described findings, we speculated that patterns I, II, III represent their botanical origins as C. racemosa, C. dahurica and the above-described mixture, respectively. We therefore established a simple method to detect substitution and adulteration of black cohosh products by monitoring a specific mass range on LC/MS.
We then analyzed the other black cohosh samples (which consisted only of dry extracts) by LC/MS to evaluate their botanical origins, and we found that 13 products including OTC drugs correctly used the dry extract of C. racemosa, and we observed that the XIC from BC-5 and BC-18 were out of synchronization with all three patterns (Table 3). Our finding that all the OTC drugs of EP grade were classified in the pattern I group strongly supports the usefulness of this method. The use of our new method could thus be a possible solution to the problem of adulteration of black cohosh products available in Japan.
Studies of the botanical origins of Vitex products
The dried ripe fruit of Vitex agnus-castus L. is defined under the names of agnus castus fruit (ACF) and chaste tree, respectively, in the EP and USP−NF. ACF is used to treat symptoms of premenstrual syndrome and climacteric disorder, and it is becoming popular in Japan as the active ingredient of OTC drugs and health food products. On the other hand, the fruit of its congenic species, V. rotundifolia L. f. and V. trifolia L., has been used as a crude drug named shrub chaste tree fruit (SCTF) (蔓荊子 in Japanese) for its pain relief, sedative and anti-inflammatory effects.
Although the EP sets an identification test for ACF to confirm the presence of aucubin and agnuside by TLC as well as a purity test by macroscopic analysis based on the size of the fruit to distinguish V. agnus-castus from other Vitex species [23], the chemical components and morphological characteristics of Vitex species are too similar to discriminate between them. Considering the internationalization of the supply chain of herbal plants, this situation could lead to misidentification of the botanical origin of these crude drugs, resulting in unexpected adverse reactions as well as a decline in their pharmaceutical efficacy. We therefore attempted to identify the marker compounds that distinguish ACF and SCTF with the goal of establishing quick methods to evaluate their origins.
Identification of characteristic diterpenes of ACF
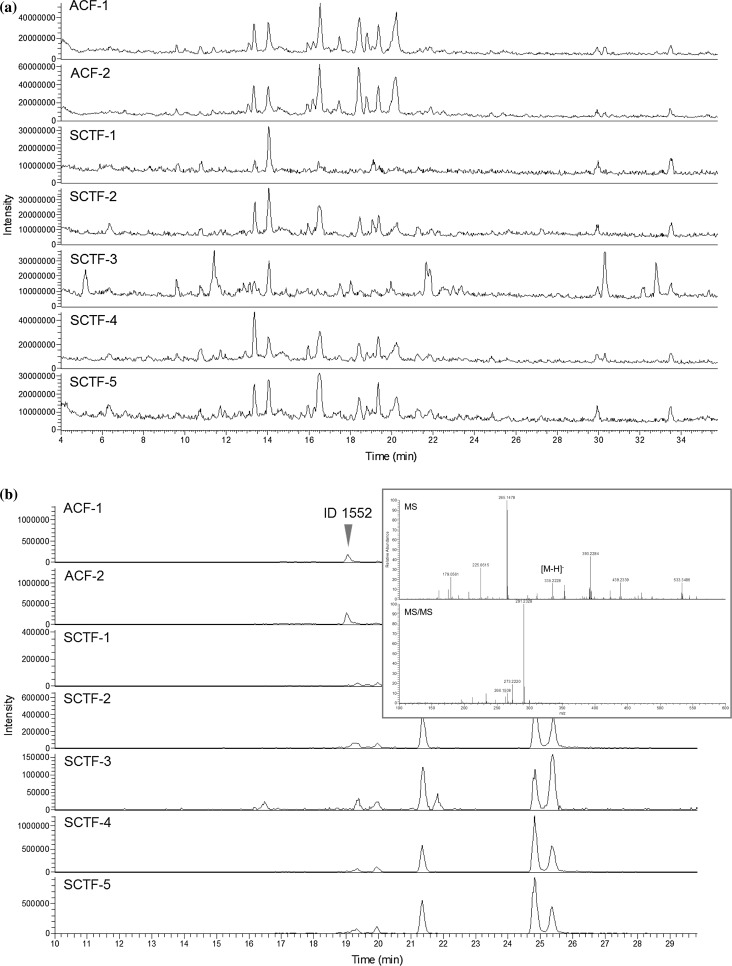
We first prepared crude drugs of ACF of EP grade and SCTF of Japanese standards for non-pharmacopoeial crude drugs (non-JP) grade to confirm their origins by DNA sequencing, and we applied them in a differential analysis using LC/MS data [24]. The sequencing analysis of the LFY/FLO gene revealed that two samples of ACF available in Germany and five samples of SCTF in Japan were derived from V. agnus-castus and V. rotundifolia or its mixture with V. trifolia, respectively (Table S2). To identify candidate marker compounds for ACF against SCTF, we performed a high-resolution LC/MS analysis of crude drug samples in an automated differential analysis, and we obtained 23 peaks as candidates. We then filtered these candidates on the basis of each formula and intensity, and seven peaks were finally picked up as characteristic compounds for ACF.
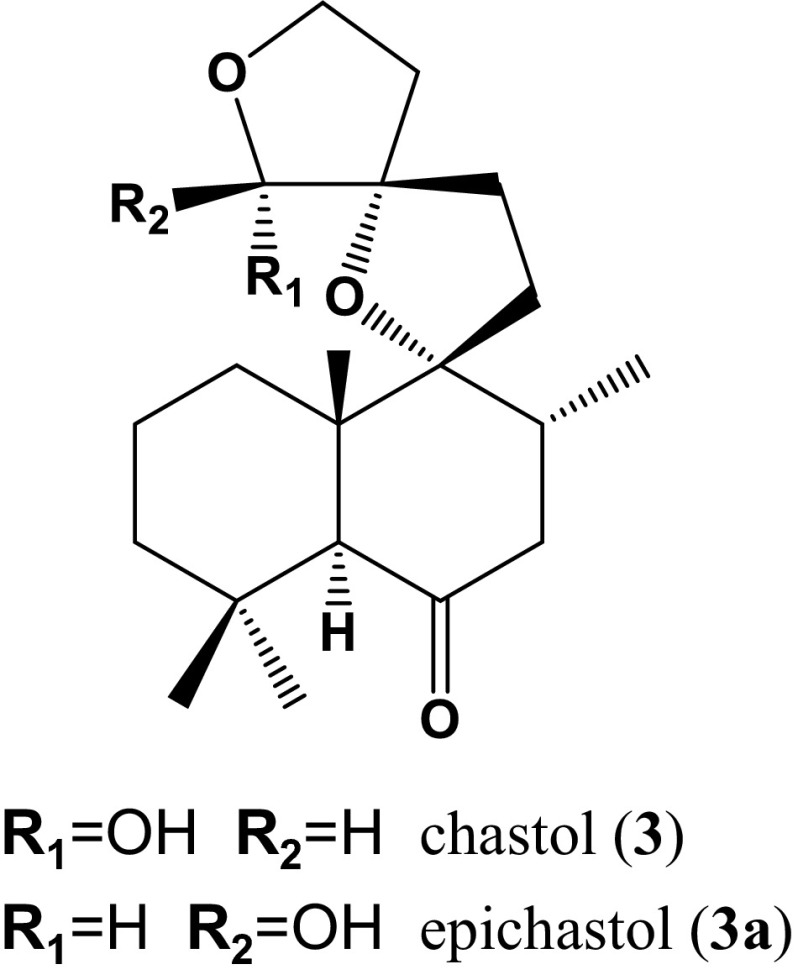
The chemical formulas calculated based on the accurate mass and MS fragmentation pattern of these peaks were collated with the phytochemical metabolite database, and we selected a putative diterpene with a novel structure—ID 1552 (RT 19.02 min, m/z 335.2228, formula C20H31O4 for [M-H]−)—as the first target to identify the chemical structure (Fig. 8). An ACF health food product was used as a source material of peak ID 1552. The repeated column chromatography and preparative TLC of a CHCl3 extract of the ACF health food product afforded compound 3/3a as a mixture. Compound 3/3a was obtained as a white powder. The 1H-NMR and 13C-NMR spectroscopic data of 3/3a (Table 4) were very similar to those of known compounds, leucasperols A and B [25], and the complete structures of 3/3a were determined on the basis of the spectroscopic data from HMBC, COSY, NOESY and differential NOE NMR experiments (Fig. 9). Since there have been no reports on the isolation of compounds bearing this structure other than leucasperols A and B from Leucas aspera L., we proposed the trivial names chastol and epichastol for compounds 3 and 3a, respectively.
Fig. 8.
Chromatograms of ACF and SCTF samples. Data taken from reference 24. Two samples of ACF available in Germany and five samples of SCTF in Japan were derived from V. agnus-castus and V. rotundifolia or their mixture with V. trifolia, respectively. a TIC of ACF and SCTF samples. b XIC of ACF and SCTF samples at m/z 335.22–335.23. The MS and MS/MS spectra of peak ID 1552 from ACF-2 are shown in the upper-right window
Table 4.
Comparison of 1H- and 13C-NMR data between Ref. and compounds 3/3a in chloroform-d (800 and 200 MHz, δ in ppm, J in Hz)
| Position | LA/LB*1 (5:2 mixture) | 3/3a (4:1 mixture) |
|---|---|---|
| δ | δ | |
| 1 | 1.86 (1H, m) 1.44 (1H, dt, 12.5, 3.7) CH2 (29.9/28.7) |
1.56 (overlapping) 1.41 (1H, dd,10.4, 4.8) CH2 (30.4/30.3) |
| 2 | 1.64-1.68 (1H, m) 1.59 (1H, dd, 9.1, 4.3) CH2 (26.5/26.5) |
1.57 (overlapping) 1.50 (1H, m) CH2 (17.2/17.3) |
| 3 | 3.11 (1H, dd, 11.6, 4.3)/3.07 (1H, dd, 11.3, 4.3) – CH2 (78.2/78.4) |
1.20 (overlapping) 0.97 (1H, ddd-like, 12.8, 6.3, 4.0) CH2 (41.5/41.5) |
| 4 | C (37.4/37.4) | C (31.2/30.9) |
| 5 | 2.63 (1H, s)/2.64 (1H, s) CH (58.5/58.1) |
2.51 (1H, s)/2.62 (1H, s) CH (58.2/57.6) |
| 6 | C (210.3/210.6) | C (210.5/210.6) |
| 7 | 2.41 (1H, td, 13.1, 0.9)/2.36 (1H, td, 13.1, 1.0) 2.06 (1H, dd, 13.1, 4.3)/2.01 (1H, overlapping) CH2 (48.7/48.9) |
2.43 (1H, m) 2.00 (1H, td, 12.8, 4.8) CH2 (47.6/47.8) |
| 8 | 2.10-2.19 (1H, m) CH (38.6/37.8) |
2.09 (m) CH (37.4/36.6) |
| 9 | C (92.7/93.0) | C (91.9/91.8) |
| 10 | C (47.3/47.5) | C (46.4/46.8) |
| 11 | 2.14 (1H, dd, 9.4, 3.6) 1.83 (1H, m) CH2 (29.0/31.2) |
2.17 (1H, ddd, 13.0, 8.8, 3.2) 1.83 (1H, ddd, 13.0, 9.6, 4.8) CH2 (28.0/27.6) |
| 12 | 2.02 (1H dd, 9.1, 3.6) 1.92 (1H, dd, 9.1, 4.3) CH2 (36.6/37.2) |
1.99 (1H ddd, 12.8, 8.8, 3.2) 1.92 (1H, m) CH2 (35.4/35.4) |
| 13 | C (91.3/92.5) | C (90.3/91.7) |
| 14 | 2.43 (1H, dt, 11.9, 9.8) 1.92 (1H, ddd, 11.9, 7.3, 2.7) CH2 (35.5/31.1) |
2.43 (1H, ddd, 12.5, 9.6, 2.4) 1.94 (1H, ddd, 12.5, 7.2, 3.2) CH2 (34.6/35.9) |
| 15 | 4.11 (1H, ddd, 9.8, 8.7, 2.7)/4.04 (1H, td, 8.5, 3.9) 3.79 (1H, td, 8.7, 7.3)/3.97 (1H, td, 8.5, 7.0) CH2 (64.8/66.3) |
4.13 (1H, ddd, 9.6, 8.8, 3.2)/4.06 (1H, m) 3.77 (1H, ddd, 8.8, 7.2, 2.4)/4.00 (1H, m) CH2 (64.0/65.5) |
| 16 | 4.81 (1H, s)/5.33 (1H, s) CH (99.0/101.6) |
4.78 (1H, s)/5.37 (1H, s) CH (98.1/100.6) |
| 17 | 0.95 (3H, d, 6.4)/0.96 (3H, d, 6.4) CH3 (16.9/17.6) |
0.93 (3H, d, 6.4) CH3 (16.0/16.7) |
| 18 | 1.08 (3H, s)/1.07 (3H, s) CH3 (27.7/27.6) |
0.91 (3H, s)/0.89 (3H, s) CH3 (31.7/31.6) |
| 19 | 1.16 (3H, s)/1.15 (3H, s) CH3 (14.9/15.0) |
1.14 (3H, s)/1.18 (3H, s) CH3 (20.8/21.7) |
| 20 | 0.88 (3H, s)/0.87 (3H, s) CH3 (20.0/20.2) |
0.81 (3H, s)/0.86 (3H, s) CH3 (19.0/19.2) |
| 16-OH | 3.18 (1H, br s)*2 |
*1 LA leucasperol A, LB leucasperol B; a pair of signals from tautomers is shown as ‘LA/LB’
*2 Disappeared upon treatment with heavy water
Fig. 9.
Chemical structures of chastol and epichastol
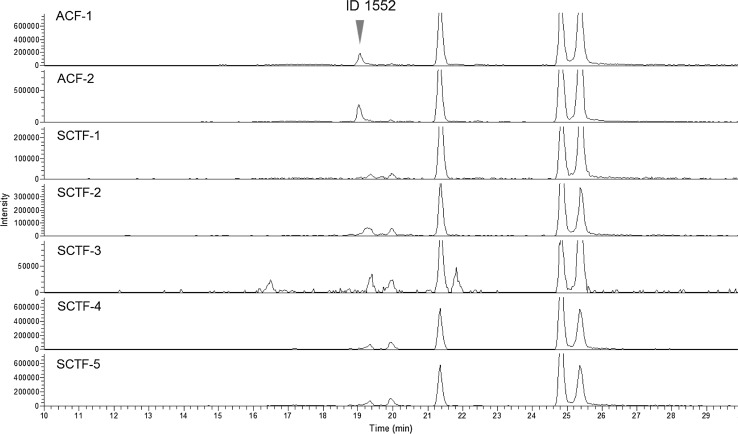
We analyzed additional ACF products by LC/MS to verify the usefulness of 3/3a as marker compounds, and we successfully confirmed their detection (Fig. 10; Table S3). Since avoiding contamination from closely related species is a significant requirement for pharmaceuticals of natural origin, this information will be valuable for the quality control of both ACF and SCTF products from the viewpoint of regulatory science.
Fig. 10.
Extracted ion chromatograms of the processed ACF samples at m/z 335.22–335.23. Data taken from reference 24. ACF-11, -12 and -13 were available in Europe as OTC drugs of EP grade and ACF-21 and -22 produced in the USA were available in Japan as health food products
Identification of an SCTF-specific triterpene derivative
Metabolomics have recently been adopted for the classification of variables and the identification of marker compounds based on the characteristics of their metabolic profiles in phytochemical investigations. For example, chemometric analyses such as a principal component analysis, a partial least-squares discriminant analysis, and an orthogonal partial least-squares discriminant analysis (OPLS-DA) are frequently used as multivariate tools for discrimination analyses [26–29]. In order to identify marker compounds for SCTF against ACF, we analyzed SCTF and ACF products by LC/MS to apply to multivariate analyses [30].
When the LC/MS data set was subjected to OPLS-DA, the clusters of SCTF and ACF were clearly separated on the score plots, and 28 ions were picked up as the most influential variables that contribute to the segregation of SCTF and ACF samples on the S-plots (Fig. 11; Table S4). We then filtered these candidates on the basis of each formula and intensity, and seven peaks were finally picked up as specific compounds for SCTF. The chemical formulas calculated based on the accurate mass and MS fragmentation pattern of these peaks were collated with the phytochemical metabolite database, and we selected a putative triterpene—ID 6436 (RT 4.59 min, m/z 663.3920, formula C40H55O8 for [M-H]−)—as the first target to identify the chemical structure.
Fig. 11.
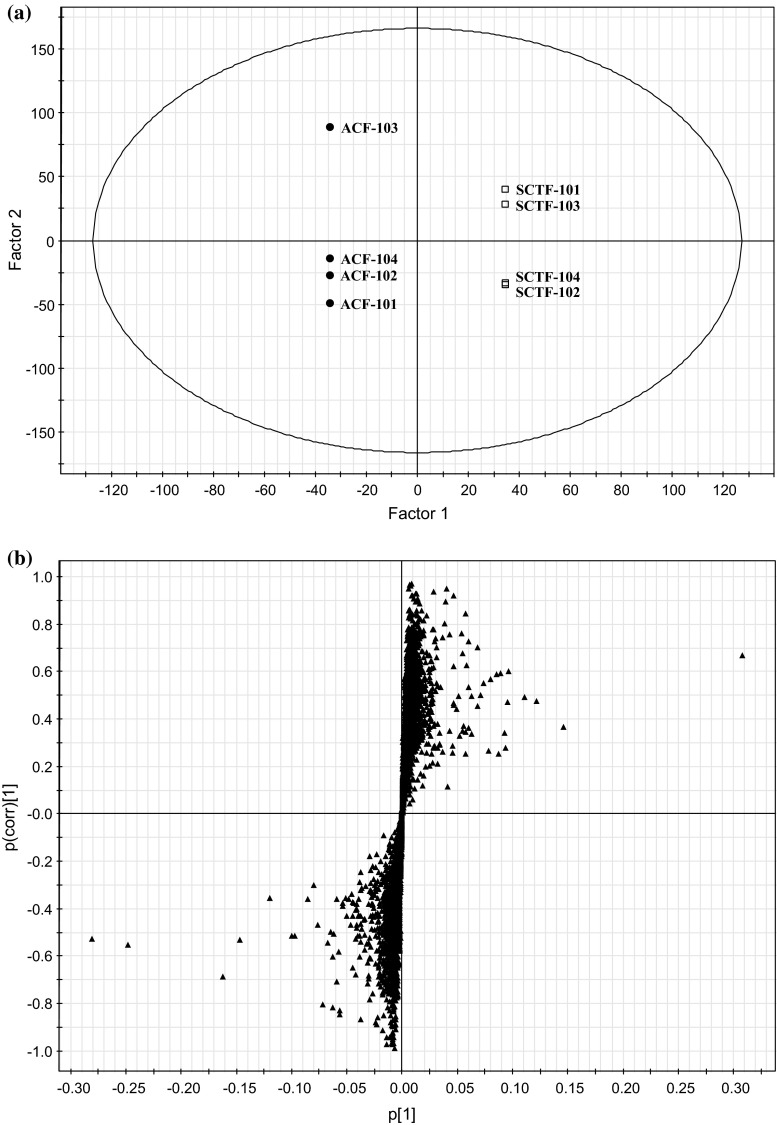
OPLS-DA/O2PLS plots based on the LC/MS data of the SCTF and ACF products. Data taken from reference 30. Four samples of SCTF were crude drugs of non-JP grade with dried fruit form. ACF-101 and -102 were crude drugs of EP grade with dried fruit form. ACF-103 and -104 were health food with capsule form. a Score plot of SCTF and ACF products. Black circles ACF. Open squares SCTF. b S-plot of SCTF and ACF products
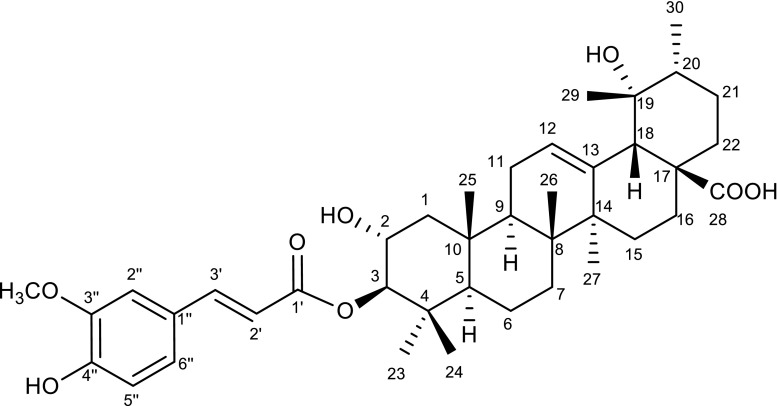
The SCTF crude drug was extracted with CHCl3 and fractionated using normal and reversed-phase column chromatography and prep-HPLC, monitoring the marker compound on LC/MS as a guide to obtain compound 4. Compound 4 was isolated as an amorphous colorless powder, and its molecular formula was determined as C40H56O8 by high-resolution electrospray ionization mass spectrometry, with 13 degrees of unsaturation. Its complete structure was determined as 3-O-trans-feruloyl tormentic acid on the basis of spectroscopic data from 1H-NMR, 13C-NMR, and HMQC and HMBC analyses (Tables 5, 6; Fig. 12); this is the first time that this compound has been isolated from a Vitex plant.
Table 5.
Comparison of 1H-NMR data between compound 4 and Ref. in methanol-d 4 (800 MHz, δ in ppm, J in Hz)
| Position | 4 | Ref.*1 | ||
|---|---|---|---|---|
| δ | δ | |||
| 2 | 3.84 | (1H, ddd, J = 4.0, 10.4, 11.2) | 3.85 | (1H, ddd, J = 4.4, 9.9, 11.0) |
| 3 | 4.64 | (1H, d, J = 10.4) | 4.64 | (1H, d, J = 9.9) |
| 12 | 5.29 | (1H, t, J = 3.0) | 5.30 | (1H, br s) |
| 18 | 2.51 | (1H, s) | 2.51 | (1H, br s) |
| 23 | 0.90 | (3H, s) | 0.90 | (3H, s) |
| 24 | 0.95 | (3H, s) | 0.96 | (3H, s) |
| 25 | 1.05 | (3H, s) | 1.06 | (3H, s) |
| 26 | 0.82 | (3H, s) | 0.81 | (3H, s) |
| 27 | 1.36 | (3H, s) | 1.97*2 | (3H, s) |
| 29 | 1.20 | (3H, s) | 1.20 | (3H, s) |
| 30 | 0.93 | (3H, d, J = 6.4) | 0.93 | (3H, d, J = 6.6) |
| 2′ | 6.43 | (1H, d, J = 16.0) | 6.43 | (1H, d, J = 16) |
| 3′ | 7.62 | (1H, d, J = 16.0) | 7.63 | (1H, d, J = 16) |
| 2′’ | 7.20 | (1H, d, J = 1.6) | 7.21 | (1H, br s) |
| 5′’ | 6.81 | (1H, d, J = 8.0) | 6.81 | (1H, d, J = 7.7) |
| 6′’ | 7.07 | (1H, dd, J = 1.6, 8.0) | 7.08 | (1H, br d, J = 7.7) |
| OCH3 | 3.89 | (3H, s) | 3.90 | (3H, s) |
Data taken from reference 30
*1 Zhao Q-C, Cui C-B, Cai B, Yao X-S, Osada H, Molbank, M328 (2003)
*2 May be mistyped
Table 6.
13C-NMR data of compound 4 in methanol-d 4 (200 MHz, δ in ppm)
| Position | 4 | Ref.*1 | Position |
|---|---|---|---|
| δ | DEPT | δ | |
| 1 | 48.2 | CH2 | 48.55 |
| 2 | 67.8 | CH | 67.65 |
| 3 | 85.8 | CH | 85.64 |
| 4 | 40.8 | C | 40.65 |
| 5 | 56.6 | CH | 56.46 |
| 6 | 19.8 | CH2 | 19.58 |
| 7 | 34.2 | CH2 | 34.03 |
| 8 | 42.9 | C | 42.68 |
| 9 | 48.8 | CH | 8.71*2 |
| 10 | 39.4 | C | 39.00 |
| 11 | 24.9 | CH2 | 24.75 |
| 12 | 129.2 | CH | 129.14 |
| 13 | 140.5 | C | 140.16 |
| 14 | 41.3 | C | 41.14 |
| 15 | 29.8 | CH2 | 29.59 |
| 16 | 26.8 | CH2 | 26.60 |
| 17 | 49.3 | C | 48.72 |
| 18 | 55.3 | CH | 55.09 |
| 19 | 73.8 | C | 73.57 |
| 20 | 43.3 | CH | 43.09 |
| 21 | 27.5 | CH2 | 27.30 |
| 22 | 39.3 | CH2 | 39.22 |
| 23 | 29.4 | CH3 | 29.25 |
| 24 | 18.5 | CH3 | 18.32 |
| 25 | 17.3*3 | CH3 | 16.60 |
| 26 | 17.7*3 | CH3 | 17.07 |
| 27 | 25.0 | CH3 | 24.83 |
| 28 | 182.9 | C | 182.29 |
| 29 | 27.2 | CH3 | 27.05 |
| 30 | 16.8*3 | CH3 | 17.50 |
| 1′ | 169.8 | C | 169.61 |
| 2′ | 116.3 | CH | 116.16 |
| 3′ | 146.7 | CH | 146.50 |
| 1″ | 128.1 | C | 127.90 |
| 2″ | 111.8 | CH | 111.58 |
| 3″ | 149.6 | C | 149.40 |
| 4″ | 150.7 | C | 150.53 |
| 5″ | 116.7 | CH | 116.49 |
| 6″ | 124.3 | CH | 124.04 |
| OCH3 | 56.7 | CH3 | 56.51 |
Data taken from reference 30
*1 Zhao Q-C, Cui C-B, Cai B, Yao X-S, Osada H, Molbank, M328 (2003)
*2 May be mistyped
*3 Assigned from HMBC correlations
Fig. 12.
Chemical structure of 3-O-trans-feruloyl tormentic acid
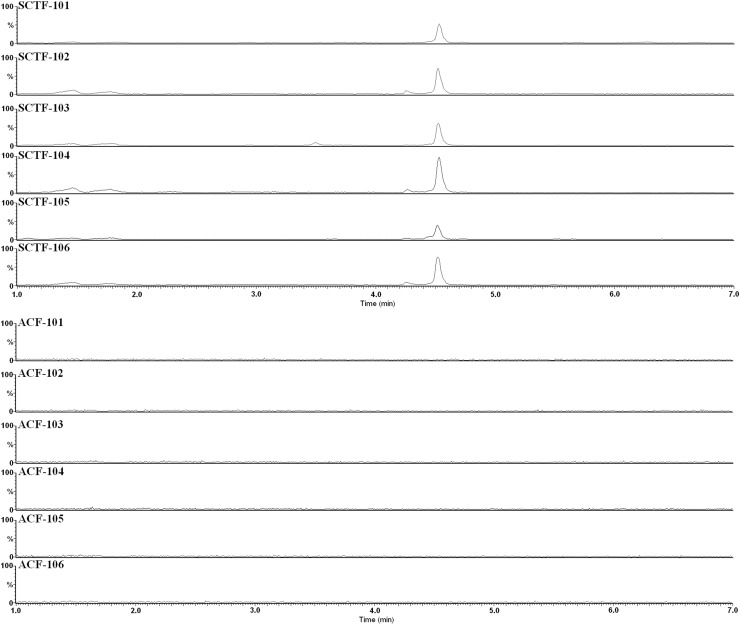
We also analyzed additional SCTF and ACF products by LC/MS to evaluate the usefulness of compound 4 as a marker compound for SCTF and successfully verified its specificity (Fig. 13; Table S5). The limit of detection of compound 4 in SCTF raw materials was 0.02 mg/ml. These results demonstrated that monitoring a marker compound for SCTF—3-O-trans-feruloyl tormentic acid—should help prevent the use of adulterated materials in ACF products, and would also be a useful means of identifying the origin of raw materials in SCTF products.
Fig. 13.
Extracted ion chromatograms of SCTF and ACF samples at m/z 663.38–663.40. Data taken from reference 30. Six samples of SCTF were crude drugs of non-JP grade with dried fruit form. ACF-101 and -102 were crude drugs of EP grade with dried fruit form. ACF-103, -104, -105 and -106 were health food with capsule form. The vertical axes represent the relative signal intensity to 60,000 and 6,000 for SCTF and ACF, respectively
Future perspective
With the changes in cause-specific morbidity and the increasing interest in self-medication, Western herbs are becoming popular in Japan. Because herbal products and their ingredients contain thousands of constituents, the authentication of original species from herbal products is the first and most important task to ensure their quality and thus their safety and efficacy. This review described the importance of quality control in the regulation of herbal products, using Cimicifuga and Vitex products as examples. There are many other Western herbs being developed for OTC drugs, foods with functional claims and health foods, and the proper use of the correct origin for each herb is necessary. It is important to continue to work on the quality control of herbal products from the viewpoint of regulatory science.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
I am deeply indebted to Dr. Yukihiro Goda and Dr. Takashi Hakamatsuka at the National Institute of Health Sciences for their helpful suggestions on this research. I also sincerely thank the researchers and students who collaborated on this project; their names are cited in the references. This work was supported by a grant for ‘Research on the Development of New Drugs’ from the Japan Health Sciences Foundation, a Health and Labour Sciences Research Grant, and a grant for ‘Research on the Development of New Drugs’ from Japan’s Agency for Medical Research and Development (AMED).
References
- 1.PFSB/ELD Notification No. 0322001 (2007) Handling in application for marketing approval of crude drug products, which have been widely used as OTC drugs in foreign countries
- 2.Goda Y. Labeling and contents of “Health Foods”. Farumashia. 2006;42:905–907. [Google Scholar]
- 3.Goda Y. The safety of health foods and importance of their origin. Yakugaku Zasshi. 2008;128:837–838. doi: 10.1248/yakushi.128.837. [DOI] [PubMed] [Google Scholar]
- 4.Cordell GA. Sustainable medicines and global health care. Planta Med. 2011;77:1129–1138. doi: 10.1055/s-0030-1270731. [DOI] [PubMed] [Google Scholar]
- 5.He K, Pauli GF, Zheng B, Wang H, Bai N, Peng T, Roller M, Zheng Q. Cimicifuga species identification by high performance liquid chromatography–photodiode array/mass spectrometric/evaporative light scattering detection for quality control of black cohosh products. J Chromatogr A. 2006;1112:241–254. doi: 10.1016/j.chroma.2006.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang B, Kronenberg F, Nuntanakorn P, Qiu M-H, Kennelly EJ. Evaluation of the botanical authenticity and phytochemical profile of black cohosh products by high-performance liquid chromatography with selected ion monitoring liquid chromatography-mass spectrometry. J Agric Food Chem. 2006;54:3242–3253. doi: 10.1021/jf0606149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang B, Ma C, Motley T, Kronenberg F, Kennelly EJ. Phytochemical fingerprinting to thwart black cohosh adulteration: a 15 Actaea species analysis. Phytochem Anal. 2011;22:339–351. doi: 10.1002/pca.1285. [DOI] [PubMed] [Google Scholar]
- 8.Whiting PW, Clouston A, Kerlin P. Black cohosh and other herbal remedies associated with acute hepatitis. Med J Aust. 2002;177:432–435. doi: 10.5694/j.1326-5377.2002.tb04886.x. [DOI] [PubMed] [Google Scholar]
- 9.Borrelli F, Ernst E. Black cohosh (Cimicifuga racemosa): a systematic review of adverse events. Am J Obstet Gynecol. 2008;199:455–466. doi: 10.1016/j.ajog.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Betz JM, Anderson L, Avigan MI, Barnes J, Farnsworth NR, Gerden B, Henderson L, Kennelly EJ, Koetter U, Lessard S, Dog TL, McLaughlin M, Naser B, Osmers RGW, Pellicore LS, Senior JR, van Breemen RB, Wuttke W, Cardellina JH. Black cohosh: considerations of safety and benefit. Nutr Today. 2009;44:155–162. doi: 10.1097/NT.0b013e3181af63f9. [DOI] [Google Scholar]
- 11.Teschke R, Bahre R, Genthner A, Fuchs J, Schmidt-Taenzer W, Wolff A. Suspected black cohosh hepatotoxicity—Challenges and pitfalls of causality assessment. Maturitas. 2009;63:302–314. doi: 10.1016/j.maturitas.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Painter D, Perwaiz S, Murty M. Black cohosh products and liver toxicity: update. Canadian Adverse Reaction Newsletter. 2010;20:1–2. [Google Scholar]
- 13.The Ministry of Health, Labour and Welfare (2011) The Japanese Pharmacopeia 16th edn. Tokyo, pp 1526–1527
- 14.Sasaki Y, Fushimi H, Cao H, Cai SQ, Komatsu K. Sequence analysis of Chinise and Japanese Curucuma Drugs on the 18S rRNA gene and trnK gene and the application of amplification-refractory mutation system analysis for their authentication. Biol Pharm Bull. 2002;25:1593–1599. doi: 10.1248/bpb.25.1593. [DOI] [PubMed] [Google Scholar]
- 15.Guo Y, Kondo K, Terabayashi S, Yamamoto Y, Shimada H, Fujita M, Kawasaki T, Maruyama T, Goda Y, Mizukami H. DNA authentication of So-jutsu (Atractylodes lancea rhizome) and Byaku-jutsu (Atractylodes rhizome) obtained in the market based on the nucleotide sequence of the 18S-5.8S rDNA internal transcribed spacer region. J Nat Med. 2006;60:149–156. doi: 10.1007/s11418-006-0032-8. [DOI] [Google Scholar]
- 16.Sasaki Y, Komatsu K, Nagumo S. Rapid detection of Panax ginseng by loop-mediated isothermal amplification and its application to authentication of Ginseng. Biol Pharm Bull. 2008;31:1806–1808. doi: 10.1248/bpb.31.1806. [DOI] [PubMed] [Google Scholar]
- 17.Sucher NJ, Carles MC. Genome-based approaches to the authentication of medicinal plants. Planta Med. 2008;74:603–623. doi: 10.1055/s-2008-1074517. [DOI] [PubMed] [Google Scholar]
- 18.Zerega NJC, Mori S, Lindqvist C, Zheng Q, Motley TJ. Using amplified fragment length polymorphisms (AFLP) to identify Black Cohosh (Actaea racemosa) Econ Bot. 2002;56:154–164. doi: 10.1663/0013-0001(2002)056[0154:UAFLPA]2.0.CO;2. [DOI] [Google Scholar]
- 19.Jigden B, Wang H, Kim YJ, Noh JH, Gotov C, Lee JI, Yang DC. Development of a multiplex polymerase chain reaction method for simultaneous detection of four Cimicifuga Species. Crop Sci. 2010;50:1961–1966. doi: 10.2135/cropsci2009.07.0412. [DOI] [Google Scholar]
- 20.Masada-Atsumi S, Onuma M, Suenaga E, Maruyama T, Hishida A, Kiuchi F, Kobayashi S, Goda Y, Hakamatsuka T. Genome-based authentication of black cohosh (Cimicifuga racemosa; Ranunculaceae) supplements available in the Japanese markets. Jpn J Food Chem Safety. 2013;20:178–188. [Google Scholar]
- 21.The European Directorate for the Quality of Medicines (2014) The European Pharmacopoeia (8th edition 8.0). Council of Europe, Strasbourg, pp 1182–1185
- 22.Masada-Atsumi S, Kumeta Y, Takahashi Y, Hakamatsuka T, Goda Y. Evaluation of the botanical origin of black cohosh products by genetical and chemical analyses. Biol Pharm Bull. 2014;62:454–460. doi: 10.1248/bpb.b13-00844. [DOI] [PubMed] [Google Scholar]
- 23.The European Directorate for the Quality of Medicines. The European Pharmacopoeia (8th edition 8.0). Council of Europe, Strasbourg, pp 1137–1138
- 24.Oshima N, Masada S, Suzuki R, Yagi K, Matsufuji H, Takahashi Y, Yahagi T, Watanabe M, Yahara S, Iida O, Kawahara N, Maruyama T, Goda Y, Hakamatsuka T. Identification of new diterpenes as marker compounds distinguishing Agnus Castus Fruit (Chaste Tree) from Shrub Chaste Tree Fruit (Viticis Fructus) Planta Med. 2016;82:147–153. doi: 10.1055/s-0035-1558089. [DOI] [PubMed] [Google Scholar]
- 25.Sadhu SK, Okuyama E, Fujimoto H, Ishibashi M. Diterpenes from Leucas aspera inhibiting prostaglandin-induced contractions. J Nat Prod. 2006;69:988–994. doi: 10.1021/np058118m. [DOI] [PubMed] [Google Scholar]
- 26.Gad HA, El-Ahmady SH, Abou-Shoer MI, Al-Azizi MM. Application of chemometrics in authentication of herbal medicines: a review. Phytochem Anal. 2013;24:1–24. doi: 10.1002/pca.2378. [DOI] [PubMed] [Google Scholar]
- 27.Wakana D, Maruyama T, Zaima K, Takeda H, Sugimura K, Anjiki N, Iida O, Kawahara N, Goda Y, Hosoe T. Standardization of ginger extracts using 1H-NMR-metabolomics. Jpn J Food Chem Safety. 2014;21:135–138. [Google Scholar]
- 28.Zhu J, Fan X, Cheng Y, Agarwal R, Moore CM, Chen ST, Tong W. PLoS One. 2014;9:e87462. doi: 10.1371/journal.pone.0087462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duan LX, Qi X, Chen M, Huang LQ. Plant metabolomics. Netherlands: Springer; 2015. Application of metabolomics in the identification of chinese herbal medicine; pp. 227–244. [Google Scholar]
- 30.Yahagi T, Masada S, Oshima N, Suzuki R, Yagi K, Matsufuji H, Suenaga E, Takahashi Y, Watanabe M, Yahara S, Iida O, Kawahara N, Maruyama T, Goda Y, Hakamatsuka T. Determination and identification of specific marker compound for discriminating Shrub Chaste Tree Fruit from Agnus Castus Fruit based on the LC/MS metabolic analysis. Chem Pharm Bull (Tokyo) 2016;64:305–310. doi: 10.1248/cpb.c15-00831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.