Abstract
A reverse transcription-polymerase chain reaction (RT-PCR) was used to amplify 1412 bp of the fusion protein gene (F gene) of four Newcastle disease virus (NDV) isolates; two velogenic (TY-1/90 and DIK-90) and two lentogenic isolates (Dongla 88/1 and GD.S.1). Following sequencing, nucleotide sequences were annotated and 894 bp were compared phylogenetically with those from strains previously reported in the Sudan and the virus strains published on the GenBank. It could be demonstrated that TY-1/90 and DIK-90 strains belong to the genotype VI of NDV and are in close genetic relationship to sub- genotype VIb. TY-1/90 and DIK-90 strains were observed to be genetically unrelated to the earlier Sudanese isolates of 1970/80s and the late of 2000s suggesting a different origin. The close genetic relationship to the European and African pigeon paramyxovirus type 1 (PPMV-1) suggests a common ancestor. Dongola, GD.S.1 strains were classified into genotype II that comprises non-pathogenic lentogenic NDV strains. The present genetic classification of NDV isolates of the Sudan provides valuable information on genotypes of NDV. Further molecular epidemiological investigations of the recent outbreaks of Newcastle disease in the Sudan are needed in order to improve the efficiency of control strategies and vaccine development.
Keywords: Newcastle disease virus, Phylogenetic analysis, Sudan isolates
Introduction
Newcastle disease (ND) or avian paramyxovirus 1 infection is a viral disease of domestic poultry and wild birds (Alexander, 1997). Gastrointestinal, respiratory and neurological symptoms of ND extent from subclinical to rapidly fatal based on the pathotype of the virus involved (Alexander, 2003). The disease is exceedingly contagious and over 250 species of birds of all age groups can be affected with the disease (Alexander et al., 1997). The causative virus, Newcastle disease virus (NDV), also named avian paramyxovirus type 1 (APMV-l), which has a single-stranded negative-sense RNA genome, is classified in the genus Avulavirus of the sub-family Paramyxovirinae, the family Paramyxoviridae and the order Mononegavirales (Lamb et al., 2000).
Diverse genotypes of APMV-1 circulate in various parts of the world and are genetically divided into two classes (class I and class II) and further classified into genotypes based on phylogenetic differences. Class I viruses have the longest of the APMV-1 genomes of 15,198 nucleotides, are genetically less diverse, generally present in wild waterfowl, and are of low virulence (Dimitrov et al., 2016). Viruses in this class are frequently isolated in live bird market samples and incorporate at least nine genotypes. Class II contains one genotype and its viruses are genetically more diverse, exhibit a wider range of virulence and include at least 16 genotypes (Diel et al., 2012). NDV strains of low virulence commonly used as vaccines worldwide such as LaSota and B1 are incorporated in class II, genotype II. Viruses of genotypes III, which were predominantly isolated decades ago in Japan, Taiwan and Zimbabwe are now isolated sporadically (Aldous et al., 2003; Miller et al., 2010). Viruses of genotypes V, VI, and VII, are highly versatile and have been isolated from different countries and continents. Alternately, virulent viruses of genotypes XI, XIII, XVI, XIV, XVII and XVIII appear to have a more restricted geographic distribution and have been isolated predominantly from poultry (Dimitrov et al., 2016). Another system suggested by Aldous classifies NDV into six lineages and 13 sub lineages and later, three extra sub lineages were added (Aldous et al., 2003; Snoeck et al., 2009).
In the Sudan, NDV was first reported in Khartoum in 1951 (ARSVS, 1951). Since then the disease has been regularly mentioned in all reports of the Sudan veterinary services. Diagnosis was based on the picture of disease, but the virus was isolated and identified for the first time in 1962 (Karrar and Mustafa, 1964; Eisa, 1979). According to Ballouh et al. (1983), twelve NDV isolates obtained during 1963-1979 in the Sudan were mesogenic (n=4) and velogenic (n=8). During the year 1984-1985, four virus isolates were found to be velogenic (Haroun et al., 1992). In another study, six isolates were obtained from outbreaks in the country between 1988 and 1991 and found to possess the characteristics of the viscerotropic velogenic strains of NDV [VVNDV], and concluded that the VVNDV is the most prevalent pathotype in the Sudan (Khalafalla et al., 1992).
Search of the literature revealed 3 publications containing phylogenetic analysis of NDV isolates from Sudan (Aldous et al., 2003; Ujvári et al., 2003; Hassan et al., 2010) that covered the period 1975- 2006, but without a link to classical pathotyping or the circulation and origin of this devastating virus in the country. According to Ujvári et al. (2003) analysis of 68 strains of avian paramyxovirus type 1 of pigeons (PPMV-1) showed a close genetic relationship to Sudanese viruses from the mid-1970 suggesting that PPMV-1 could be of African origin.
Here, we genetically classified additional Sudanese NDV isolates obtained between 1988 and 1999 and discussed the origin and circulation of various virus genotypes in the Sudan.
Materials and Methods
Viruses
NDV strains were obtained from the repository of Department of Microbiology, Faculty of Veterinary Medicine, University of Khartoum. TY-1/90 and Dik-90 are virulent strains isolated from ND outbreaks that occurred in south of Khartoum during July - September 1990 (Khalafalla et al., 1992). Dongola 88/1 strain is a local isolate of lentogenic NDV, which was isolated in 1988 from apparently healthy chickens in Dongola city of Northern Sudan (Khalafalla, 1994). According to Khalafalla (1994) the chickens from which Dongla 88/1 isolate was obtained were vaccinated with Komarov strain of NDV. LaSota VC is a live vaccine strain (INTERVET, The Netherlands) used to vaccinate chickens in the Sudan. I2 strain is a live lentogenic thermostable NDV vaccine strain originally isolated in the Australian Center for International Agriculture Research (ACIAR) (Bensink and Spradbrow, 1999). GD.S.1 is a field virus isolated from non-vaccinated local chickens in Gedarif city in June 1993 (Abdelaziz et al., 2004). The viruses were propagated in embryonated chicken eggs and identified as NDV by hemagglutination inhibition (HI) test according to standard techniques (OIE, 2008).
RT-PCR
Primer design
Primers were designed from conserved nucleotide regions of the NDV fusion protein gene (F-gene). First, sequences of the NDV F-gene of various isolates of NDV were retrieved from the GenBank® (http://www.ncbi.nlm.nih.gov/Genbank/index.html) and were used for multiple sequence alignment to determine conserved regions in the F-gene. Second, conserved regions in the sequences were selected and primers were manually designed. The primers were NCD_F (5` CAG GCC TCT TGC AGC TGC 3`) and NCD_R (5` TAT AGG TAA TGA GAG CAG ACG 3`).
RNA extraction
NDV RNA was extracted from harvested virus in chicken embryonic allantoic fluids using Roti®-Quick-Kit (Roth, Germany) according to the manufactures’ instructions. RNA pellet was dissolved in 15-20 µl DEPC-treated autoclaved distilled water and was immediately used in RT-PCR.
OneStep RT-PCR
RT-PCR was performed using QIAGEN® OneStep RT-PCR Kit (Qiagen, Germany). Primer stock solutions of 100 µM were first prepared and then working dilutions of 10 µM were used in the RT-PCR reactions. Briefly, NDV RNA samples, primers, dNTP mix, 5x OneStep RT-PCR Buffer, Q-solution and RNase-free water were thawed and placed on ice. All components required for RT-PCR were contained in the typical master mix except the template RNA. Five microliters of RNA were added to each 20 µl of mastermix in a PCR tube. A negative control (without template RNA) was included in every experiment. The tubes were incubated in a thermal cycler (Primus96, Peqlab Biotech, Germany) and the RT-PCR program was carried out with the following steps: reaction mixtures were subjected to a 30-min reverse transcription step at 50°C, then heating for 15 min at 95°C to activate the Hot Start Taq DNA polymerase, followed by 35 cycles of 95°C for 30 s, 60°C for 32 s and 72°C for 32 s and a final extension at 72°C for 2 min.
Analysis of PCR product
The PCR products were separated electrophoretically in 2% agarose gel (Sigma, UK) in Tris/Boric acid/EDTA running buffer (TBE buffer). Agarose gel was prepared by dissolving 1 g of agarose in 50 ml of TBE buffer. Ethidium bromide (1µl/40 ml agrose) was added and the gel was cast into the tray, combs were placed and the gel inside the tray was allowed to solidify for 30 min. Five µl of the PCR products were mixed with one µl of 6X loading dye and transferred into the wells. Two microliters of one Kb DNA ladder (GeneRuler™ 1kb DNA Ladder Plus #SM1331, Fermentas, Germany) was loaded into the first well of the gel. The gel was allowed to electrophorese for 45 min (120V and 30 mA), then DNA was visualized under UV light and the picture was documented using a gel documentation system (Bio-Rad, England).
Nucleotide sequencing and phylogenetic analysis
PCR products were sent for commercial sequencing to MWG-Biotech AG Company, Germany). Editseq program was used to edit the sequences before forward and reverse sequences of each strain were joined into one sequence using the sequence assembly software Seqman (Lasergen, DNAstar, US). Multiple sequence alignment was performed using ClustalW (http://clustalw.ddbj.nig.ac.jp/top-e.html) and phylogenetic and molecular evolutionary analyses were conducted using MEGA 6 (Tamura et al., 2013). The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The significance of all deduced phylogenetic trees was verified by bootstrap analysis of 1000 replicates.
The nucleotide sequences of isolates TY-1/90 and DIK-90 were compared with nucleotide sequence in the GenBank database using the online BLASTN program on the NCBI website (Altschul et al., 1997).
To assess the genetic lineage of the NDV strains used in this study, either phylogenetic analysis was carried out using NDV fusion protein gene sequences, which were obtained during this study, or reference sequences obtained from the GenBank. The strain name, host, region and year of isolation, genotype assignment and accession numbers, for all sequences used in the analysis are shown in Table 1.
Table 1.
Referential Newcastle disease virus (NDV) strains used in this study for phylogenetic analysis of the fusion protein gene.
| Sno | Strain | Host | Year | Country | Genotype | Accession number |
|---|---|---|---|---|---|---|
| 1 | Ulster 2C/67 | Fowl | 1967 | N. Ireland | I | D00243 |
| 2 | V4 Queensland/66 | Fowl | 1966 | Australia | I | M24693 |
| 3 | Beaudette C/45 | Fowl | 1945 | USA | II | X04719 |
| 4 | La Sota/46 | Fowl | 1946 | USA | II | M24696 |
| 5 | Texas G.B 48 | Fowl | 1948 | USA | II | M24698 |
| 6 | B1 | Fowl | 2000 | USA | II | AF309418 |
| 7 | V4 | Fowl | 2003 | China | II | AY225110 |
| 8 | Clone 30 | Fowl | 2005 | _ | II | Y18898 |
| 9 | LaSota | Fowl | 1999 | Netherlands | II | AF077761 |
| 10 | Miyadera/51 | Fowl | 1951 | Japan | III | M18456 |
| 11 | Miyadera/51/1 | Fowl | 1951 | Japan | III | M24701 |
| 12 | Herts/33 | Fowl | 1933 | UK | IV | M24702 |
| 13 | Italien/45 | Fowl | 1945 | Italy | IV | M17710 |
| 14 | Chicken/Sudan/Obied/1987 | Fowl | 1987 | Sudan | IV | GQ258674 |
| 15 | Cormorant/US(CA)D9704285 | Cormorant | 1997 | USA | V | GQ288381 |
| 16 | Cormorant/US(CA)92-23071 | Cormorant | 1992 | USA | V | GQ288388 |
| 17 | VRDC/NDV/F/N9 | Fowl | 2012 | India | V | KJ621049 |
| 18 | Iraq AG68 | Fowl | 1998 | Iraq | VI | AF001108 |
| 19 | Q-GB 506/97 | Fowl | 1998 | Denmark | VI | AF001129 |
| 20 | Pigeon/Guangdong/GZ293 | Pigeon | 2014 | China | VI | KT381592 |
| 21 | TX3503/04 | Pigeon | 2008 | USA | VI | KT381592 |
| 22 | SD-2/75 | Fowl | 1975 | Sudan | VI | AY151384 |
| 23 | SD-3/75 | Fowl | 1975 | Sudan | VI | AY151383 |
| 24 | SD-4/75 | Fowl | 1975 | Sudan | VI | AY151385 |
| 25 | Kuwait 256 | Fowl | 1968 | Kuwait | VIa | AF001109 |
| 26 | Lebanon 70 | Fowl | 1970 | Lebanon | VIa | AF001110 |
| 27 | Ch/98-1 | Pigeon | 1998 | China | VIb | AF358785 |
| 28 | VRD07-163/2007 | Dove | 2012 | Nigeria | VIb | JQ039385 |
| 29 | DK-6/95 | Ostrich | 1995 | Denmark | VId | AF001130 |
| 30 | A-24/96 | Fowl | 1996 | Austria | VId | AF001133 |
| 31 | Warwick/66 | Fowl | 1966 | Great Britain | VIe | Z12111 |
| 32 | JX-1/94 | Fowl | 1994 | China | VIf | AF458021 |
| 33 | ZhJ-2/86 | Fowl | 1986 | China | VIg | AF458016 |
| 34 | Sh-1/97 | Fowl | 1997 | China | VIg | AF458018 |
| 35 | TW-C69-2-9 | Fowl | 1969 | Taiwan | VIh | AY372129 |
| 36 | TW-C81-4-3 | Fowl | 1981 | Taiwan | VIh | AY372133 |
| 37 | JS-2/98(Go) | Goose | 1998 | China | VII | AF456439 |
| 38 | AE 232/1/96 | Partridge | 1996 | UAE | VIIb | AF109884 |
| 39 | ZW3422/95 | Fowl | 1997 | Zimbabwe | VIIb | AF109877 |
| 40 | CH-A7/96 | Fowl | 1996 | China | VIIc | AY028995 |
| 41 | JS-3/00 | Fowl | 2000 | China | VIIc | AF458010 |
| 42 | Chicken/Sudan/02/2005 | Fowl | 2005 | Sudan | VIId | GQ258669 |
| 43 | Chicken/Sudan/08/2004 | Fowl | 2004 | Sudan | VIId | GQ258675 |
| 44 | Chicken/Sudan/05/2004 | Fowl | 2004 | Sudan | VIId | GQ258672 |
| 45 | Chicken/Sudan/04/2003 | Fowl | 2003 | Sudan | VIId | GQ258671 |
| 46 | Chicken/Sudan/03/2003 | Fowl | 2003 | Sudan | VIId | GQ258670 |
| 47 | Chicken/Sudan/06/2006 | Fowl | 2006 | Sudan | VIId | GQ258673 |
| 48 | Taiwan 95 | Fowl | 1995 | Taiwan | VIIe | U62620 |
| 49 | Nd/chicken/TR/53 | Fowl | 2014 | Turkey | VIIi | KT585633 |
| 50 | Nd/chicken/TR/19-2 | Fowl | 2012 | Turkey | VIIi | KT585617 |
| 51 | F48E9 | Fowl | 1948 | China | IX | AF163440 |
| 52 | FJ-1/85 | Fowl | 1985 | China | IX | AF458009 |
| 53 | TW-C69-10-36 | Fowl | 1969 | Taiwan | X | AY372163 |
| 54 | TW-C81-4-6 | Fowl | 1981 | Taiwan | X | AY372137 |
Results
Reverse transcription polymerase chain reaction (RT-PCR)
Following RNA isolation, RNA from all the strains was reverse transcribed then amplified in a OneStep RT-PCR reaction to confirm NDV identity and to achieve molecular characterization analysis. The primer set used in this study was able to amplify fragments with the expected sizes of 1412 bp, from the NDV fusion protein gene. Examination of the amplified PCR products following electrophoresis on agarose gels revealed the expected sizes of amplicons for NDV isolates (Fig. 1).
Fig. 1.
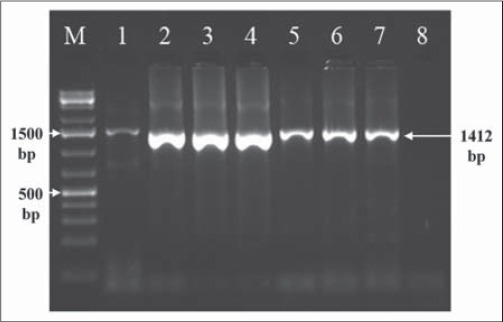
RT-PCR amplification of the Newcastle disease virus fusion protein gene using F1 (forward) and R (reverse) primer combination, which gave a product size of 1412 bp. The amplicons were electrophoresed in 2% agarose gel. Lanes: M, molecular size marker; Lanes 1-7 are NDV strains I2, TY-1/90, C30, Dik-90, GD.S.1, Lasota and Dongola, respectively; Lane 8, –ve control (H2O).
Sequencing and sequence alignment
Sequencing of the fusion protein gene of TY-1/90, DIK-90, GD.S.1, and Dongola 88/1 strains using the NCD_F forward and NCD_R reverse primers resulted in readable sequences that varied in length between 906-1002 nucleotides. For the purpose of accurate alignment data, all the sequences were reduced to an equal length of 894 nucleotides. This length covered the region between nucleotide numbers 588 and 1481 of the complete coding sequence (1662 nucleotides) of the fusion protein gene.
The sequence alignment performed for TY-1/90, DIK-90, GD.S.1, and Dongola 88/1 showed that all the sequences were identical in 759 nucleotides (84.9%), while differed from each other in 135 nucleotides (15.1%). The alignment showed that the sequences were grouped into two groups according to the number of identity a group contained strains TY-1/90 and Dik-90 (Velogenic strains) and a group contained the rest of the sequences (Lentogenic strains). Strains TY-1/90 and Dik-90 showed a high degree of identity among themselves as 885 out of 894 (98.99%) nucleotides were similar.
BLAST search and phylogenetic analysis of partial fusion gene
Results of the BLAST search for TY-1/90 partial F gene nucleotide sequences revealed 96% identity with NDV strain APMV1/chicken/Japan/Osaka/2440/1969 (AB853926.2) isolated in Japan in 1969, strain IS 744/99 (JF795620.1) isolated in Israel in 2011 and strain ETH10065 (KC205475.1) isolated in Ethiopia in 2014. Strain DIK-90 demonstrated 96% nucleotide identity with strain APMV1/chicken/Japan/Osaka/2440/1969 (AB853926.2), strain Warwick (Z12111.1) isolated in Great Britain in 1966, Pigeon/Nigeria/VRD07-173/2007 (JQ039395.1) isolated in Nigeria in 2007, strain dove/Nigeria/VRD07-163/2007 (JQ039385.1) isolated in Nigeria in 2007 and strain 2011_Ethiopia_ETH10065 (KC205475.1) isolated in Ethiopia in 2014.
In light of the percent identity matrix created by Clustal 2.1 (Table 2), Sudan isolates of NDV of the 2000s (chicken/Sudan/02-06) showed 96.18-100% nucleotide sequence homology between them, while Sudan isolates of 1970s in addition to isolate Chicken/Sudan/Obeid isolated in 1987 shared 95.99- 98.41% identity. The Sudan 1970/80s shared 87.58-89.81% identity with the Sudan 2000s isolates. On the other hand, isolates of 1990s analyzed in this study (TY-1/90 and DIK-90) showed only 41.61-43.62% nucleotide homology with both the Sudan 1970/80s and 2000s isolates (Table 2).
Table 2.
Percent Identity Matrix created by Clustal 2.1 for published velogenic Sudanese isolates and representative VI genotypes of Newcastle disease virus from different regions of the world based on nucleotide sequence of partial F gene.
| Virus isolate | (1) | (2) | (3) | (4) | (5) | (6) | (7) | (8) | (9) | (10) | (11) | (12) | (13) | (14) | (15) | (16) | (17) | (18) | (19) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (1) Warwick | 100 | 93.8 | 86.8 | 85.3 | 89.4 | 92.6 | 92.8 | 44.1 | 42.3 | 42.3 | 42.9 | 44.5 | 44.0 | 44.0 | 43.7 | 42.7 | 43.3 | 43.0 | 43.0 |
| (2) Sh-1/97 | 93.8 | 100. | 92.0 | 91.4 | 94.0 | 94.1 | 94.4 | 41.9 | 42.3 | 42.3 | 43.0 | 43.4 | 43.6 | 43.4 | 42.6 | 41.6 | 43.2 | 41.9 | 41.9 |
| (3) TX3503/04 | 86.8 | 92.0 | 100. | 94.8 | 94.0 | 93.9 | 94.0 | 43.6 | 43.0 | 43.0 | 43.6 | 44.3 | 43.4 | 43.4 | 43.2 | 42.5 | 43.3 | 42.7 | 42.5 |
| (4) Pigeon/Gdg/GZ293 | 85.3 | 91.4 | 94.8 | 100. | 92.7 | 92.4 | 92.5 | 44.6 | 44.6 | 43.3 | 44.0 | 44.8 | 44.0 | 44.0 | 44.7 | 43.0 | 44.0 | 43.3 | 43.0 |
| (5) 5Ch/98-1 | 89.4 | 94.0 | 94.0 | 92.7 | 100. | 94.2 | 94.5 | 45.3 | 45.3 | 44.0 | 44.3 | 45.6 | 44.8 | 44.8 | 44.5 | 43.6 | 44.0 | 43.9 | 43.6 |
| (6) TY-1/90 | 92.6 | 94.2 | 93.9 | 92.4 | 94.2 | 100. | 99.0 | 42.4 | 42.6 | 42.3 | 43.0 | 43.4 | 43.6 | 42.6 | 42.6 | 41.3 | 42.0 | 41.6 | 41.6 |
| (7) Dik_90 | 92.8 | 94.4 | 94.0 | 92.5 | 94.5 | 99.0 | 100. | 42.8 | 42.6 | 42.3 | 43.0 | 43.4 | 42.6 | 42.6 | 42.4 | 41.3 | 42.0 | 41.6 | 41.6 |
| (8) TW-C69-2-9 | 44.1 | 41.9 | 43.6 | 44.6 | 45.3 | 42.4 | 42.7 | 100. | 88.5. | 89.0 | 89.0 | 96.6 | 97.4 | 97.9 | 97.4 | 95.0 | 96.8 | 96.3 | 96.3 |
| (9) Chicken/Sudan/06 | 42.3 | 42.3 | 42.9 | 43.3 | 44.0 | 42.6 | 42.6 | 88.5 | 100. | 96.2 | 96.2 | 97.8 | 87.9 | 87.6 | 87.3 | 88.2 | 88.5 | 88.2 | 88.2 |
| (10) Chicken/Sudan/08 | 42.3 | 42.3 | 43.0 | 43.3 | 43.6 | 42.3 | 42.3 | 89.0 | 96.2 | 100. | 98.4 | 86.3 | 87.9 | 87.6 | 87.3 | 87.6 | 87.9 | 88.2 | 88.2 |
| (11) Chicken/Sudan/03* | 43.0 | 43.0 | 43.6 | 44.0 | 44.3 | 43.0 | 43.0 | 89.0 | 97.8 | 98.4 | 100. | 87.9 | 89.5 | 89.2 | 88.9 | 89.2 | 89.5 | 89.8 | 89.8 |
| (12) DK-6/95 | 44.5 | 43.4 | 44.2 | 44.8 | 45.6 | 43.4 | 43.4 | 96.6 | 86.3 | 87.9 | 86.3 | 100. | 95.1 | 95.4 | 94.9 | 92.5 | 94.6 | 93.6 | 93.3 |
| (13) Lebanon 70 | 44.0 | 42.6 | 43.4 | 44.0 | 44.8 | 42.6 | 42.6 | 97.4 | 87.9 | 87.9 | 89.5 | 95.1 | 100. | 99.2 | 98.7 | 96.8 | 97.5 | 97.6 | 97.3 |
| (14) Kuwait 256 | 44.0 | 42.6 | 43.4 | 44.0 | 44.8 | 42.6 | 42.6 | 97.9 | 87.6 | 87.6 | 89.2 | 95.4 | 99.2 | 100. | 99.5 | 96.5 | 97.1 | 97.3 | 97.1 |
| (15) Iraq AG68 | 43.7 | 42.4 | 43.1 | 43.7 | 44.5 | 42.4 | 42.4 | 97.4 | 87.3 | 87.3 | 88.9 | 94.9 | 98.7 | 99.5 | 100. | 96.0 | 96.8 | 96.8 | 96.5 |
| (16) SD-4/75 | 42.7 | 41.6 | 42.5 | 43.0 | 43.6 | 41.3 | 41.3 | 95.0 | 88.2 | 87.6 | 89.2 | 92.5 | 96.8 | 96.5 | 96.0 | 100. | 98.4 | 97.6 | 97.3 |
| (17) Chicken/SD/Obied | 43.3 | 42.0 | 43.3 | 44.0 | 44.0 | 42.0 | 42.0 | 96.8 | 88.5 | 87.9 | 89.5 | 94.6 | 97.5 | 97.1 | 96.8 | 98.4 | 100. | 98.4 | 98.4 |
| (18) SD-2/75 | 43.0 | 42.0 | 42.7 | 43.3 | 43.9 | 41.6 | 41.6 | 96.3 | 88.2 | 88.2 | 89.8 | 93.6 | 97.6 | 97.3 | 96.8 | 97.6 | 98.4 | 100. | 99.7 |
| (19) SD-3/75 | 43.0 | 41.9 | 42.5 | 43.0 | 43.6 | 41.6 | 41.6 | 96.3 | 88.2 | 88.2 | 89.8 | 93.3 | 93.6 | 97.0 | 96.5 | 97.3 | 98.4 | 99.7 | 100. |
Based on the phylogenetic analysis performed in this study, all studied NDV strains can be divided into two clusters, which represent the genotypes II (strains I-2, Clone 30, GD.S.1, LaSota VC and Dongola 88/1) and VI (TY-1/90 and DIK-90). TY-1/90 and DIK-90 strains clustered close to sub-genotypes b, f, g and e of genotype VI at one branch of the tree, while the other Sudanese strains of the 1970s, 1980s and 2000s clustered close to sub-genotypes a, d and h of genotype VI at the other branch (Fig. 2).
Fig. 2.
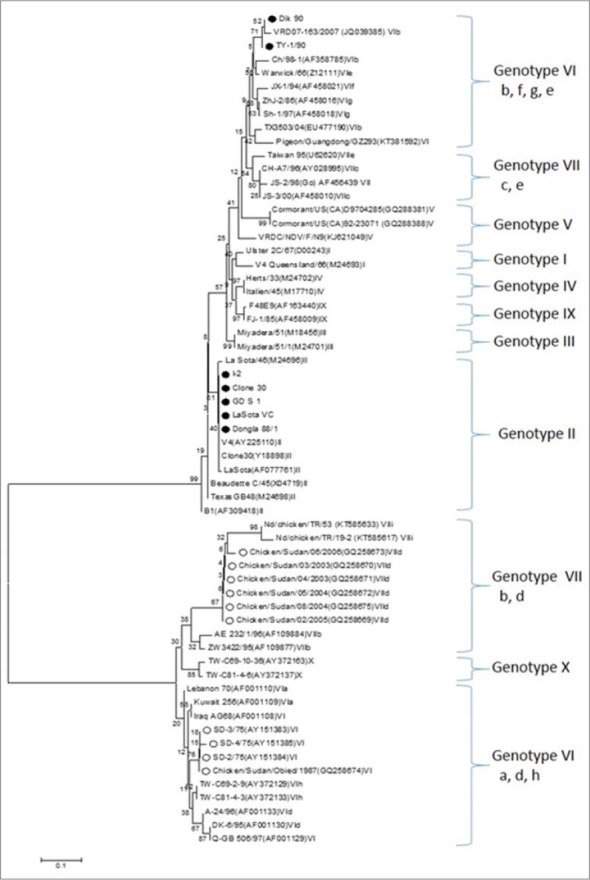
Rooted phylogenetic tree of the nucleotide sequences of the fusion protein gene of Newcastle disease virus strains. The evolutionary history was inferred using the Neighbor-Joining method. The optimal tree with the sum of branch length = 2.20497769 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. The analysis involved 61 nucleotide sequences. Evolutionary analyses were conducted in MEGA7. Isolates analyzed in this study are marked with filled circle, whereas those reported earlier from the Sudan are marked with empty circles.
Discussion
Four strains of NDV, 2 velogenic (TY-1/90 and DIK-90) and two lentogenic (Dongola and GD.S.1), were selected for this study. A number of 894 nucleotides of the fusion protein gene of each of the strains were sequenced and analyzed before a phylogenetic tree was constructed.
The phylogenetic analysis showed that TY-1/90 and DIK-90 strains belonged to genotype VI within a sub-group containing sub-genotypes b, f, g and e. The BLAST search revealed a 96% nucleotide homology with VI genotype NDVs mostly from Africa and Asia that were characterized as belonging to sub genotypes a, b, f and e (Van Borm et al., 2012; de Almeida et al., 2013; Umali et al., 2013). As the partial sequence of the F gene does not allow precise delineation of NDV viruses at the sub-genotype level (de Almeida et al., 2013), it is not possible to designate a sub-genotype for isolates TY-1/90 and DIK-90. These two strains originated from natural severe outbreaks of ND that occurred in Tayba, 30 Km south of Khartoum, in July 1990 (TY-1/90) and Dikheinat, 15 Km south of Khartoum, in September 1990 (DIK-90) (Khalafalla et al., 1992), respectively. Affected chickens exhibited symptoms characterized by profuse diarrhea and paralysis of wings and legs demonstrating the involvement of digestive and nervous systems. According to the classical pathotyping, TY-1/90 and DIK-90 strains were described as viscerotropic velogenic NDV in light of the fact that they kill chicken embryos quickly and produce haemorrhagic lesions in the intestinal tract in infected chickens (Khalafalla et al., 1992). The mean death time (MDT) and intracerebral pathogenicity index (ICPI) values used in the pathotyping were similar to those described by Hanson et al. (1973) for the viscerotropic pathotypes of NDV.
Genotype VI has been shown to be characterized by frequent branching and repeated epizootics caused by molecular variants separated by only short evolutionary intervals (Lomniczi et al., 1998). The genotype was divided into eight subtypes (a-h) (Liu et al., 2003; Tsai et al., 2004; Lien et al., 2007). Genotype VIb is comprised mostly of viruses associated with panzootic infection in pigeons (Columba livia) (Aldous et al., 2003) and isolates studied so far usually belonged to mAb group P (Alexander et al., 1997). Pigeon isolates were further divided into two genetic lineages (Ujvári et al., 2003). The majority of isolates clustered into a single genetic lineage, termed VIb/1, within the genotype VI of NDV strains of chickens, whereas a small number of isolates that originated in Croatia after 1995, are grouped in a highly diverged lineage, termed VIb/2. Our phylogenetic analysis showed that the Sudanese strains TY-1/90 and DIK-90 studied in this study cluster close to genotype VIb that included four strains isolated from pigeons and doves. This finding supports the assumption that the pigeon PMV-1 (PPMV-1) could be of African origin (Ujvári et al., 2003).
The mid-1970s recorded outbreaks of ND occurred in chicken in Sudan were caused by Genotype VI including SD-2/75, SD-3/75 and SD-4/75 strains (Ujvári et al., 2003). The aforementioned Sudanese isolates clustered together, in this study within the VI genotype group (containing sub-genotypes a, d and h) viruses in a separate branch that likewise contains the Middle Eastern isolates of subtype VIa. The previously described Sudanese isolates of 1970/80s and 2000s isolates shared a relatively high (87.58-89.81%) sequence homology, while isolates of 1990s analyzed in this study (TY-1/90 and DIK-90) shared only 41.61-43.62% nucleotide homology with them. Furthermore, the results of the phylogenetic analysis affirm that TY-1/90 and DIK-90 are distinct from previously reported NDV isolates of the Sudan as each group cluster in different branches of the tree. Accordingly, the origin of these two groups of NDVs seems to be different.
Our results demonstrated that the Sudanese strains Dongola 88/1 and GD.S.1 fit in with genotype II. Genotype II consists primarily of viruses that were isolated from 1945 to 2000 worldwide, varying from virulent to mild isolates (Aldous et al., 2003). Generally, genotype II has two subclusters a velogenic and a lentogenic (Qin et al., 2008). The Beaudette C/45 and Texas48 isolates, which have a polybasic-F0 cleavage site motif typical for velogenic NDV strain fall into one subcluster, while the reference vaccine strains (LaSota, B1, Clone 30, La Sota/46 and V4), which have a motif typical of lentogenic NDV strain fall into the other subcluster. All the above-mentioned isolates analyzed in this study fell into the lentogenic cluster of genotype II.
For Dongola 88/1 strain, this result is in accordance with our previous findings of Khalafalla (1994). Khalafalla described Dongola strain, which was isolated in 1988 from apparently healthy chickens in Dongola town of northern Sudan, as a lentogenic pathotype of NDV, causing a mild or inapparent infection of the respiratory tract. The study of the mAb binding pattern placed Dongola strain in group E of paramyxoviruses (Khalafalla, 1994). The results obtained for GD.S.1, which was isolated from Gedarif in June 1999, are in contrast with the previous description of the strain as velogenic (Abdelaziz et al., 2004). These two isolates are probably originated lentogenic strains of NDV used as vaccines. Further whole genome sequencing is expected to unveil whether these isolates were vaccine or field lentogenic strains.
A limitation of this study is that we used partial F gene sequences to characterize genetically our NDV isolates instead of the full F gene as proposed by Diel et al. (2012).
It can be concluded that both lentogenic and velogenic NDV exist in Sudan. Genotype VI of NDV has been maintained since 1970s and until at least the 1990s. This genotype has probably given origin to PPMV-1 strains of the 1980s. Amid the same period, other genotypes of NDV existed in Sudan. Genotype IV was isolated from domestic fowls on 2 occasions; in 1987 (Hassan et al., 2010) and in 1991 (Aldous et al., 2003). More recently, genotype VIId was isolated from vaccinated immature and adult layer flocks in Khartoum State and eastern Sudan (Gadarif) between 2003 and 2006 (Hassan et al., 2010). Along these lines, further studies using phylogenetic analysis are necessary to enable a full view on the evolving status of ND in Sudan, to trace the sources of infection, and to assist in taking decisions to improve the disease control efforts.
Conflict of interest
The authors of this article declare that there is no conflict of interest.
Acknowledgements
Thanks are to the staff of the Virology Research Laboratory, Department of Microbiology, Faculty of Veterinary Medicine, University of Khartoum. We are also grateful to Prof. Abdelrahim Mohamed Elhussein and the staff of the Central Laboratory, Ministry of Science and Technology, for their support and help. Special thanks are due to Dr. Abdel Gaffar Elfahal.
References
- Abdelaziz S.A, Khalafalla A.I, Ali A.S, Elhassan S.M. Newcastle disease in village chickens in the Sudan. Survey of disease incidence and Isolation of the causative virus. J. Anim. Vet. Adv. 2004;3(1):36–38. [Google Scholar]
- Aldous E.W, Mynn J.K, Banks J, Alexander D.J. A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene. Avian Pathol. 2003;32(3):239–256. doi: 10.1080/030794503100009783. [DOI] [PubMed] [Google Scholar]
- Alexander D.J. Newcastle disease and other avian Paramyxoviridae infections. In: Calneck B.W, editor. Diseases of Poultry. 10th edn. Ames, IA: Iowa State University Press; 1997. pp. 541–569. [Google Scholar]
- Alexander D.J, Manvell R.J, Lowings J.P, Frost K.M, Collins M.S, Russell P.H, Smith J.E. Antigenic diversity and similarities detected in avian paramyxovirus type 1 (Newcastle disease virus) isolates using monoclonal antibodies. Avian Pathol. 1997;26(2):399–418. doi: 10.1080/03079459708419222. [DOI] [PubMed] [Google Scholar]
- Alexander D.J. Newcastle disease, other paramyxoviruses and pneumovirus infections. In: Saif Y.M, Barnes H.J, Glisson J.R, Fadly A.M, McDougald D.J, Swayne D.E, editors. Diseases of Poultry. 11th edn. Ames: Iowa State Press; 2003. pp. 63–100. [Google Scholar]
- Altschul S.F, Madden T.L, Schäffer A.A, Zhang J, Zhang Z, Miller W, Lipman D.J. Gapped BLAST and PSI-BLAST:a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARSVS. Annual Reports of the Sudan Veterinary Services (ARSVS) 1950-1951. Khartoum, Sudan: 1951. [Google Scholar]
- Ballouh A, Nayil A.A, Ali B.H. Pathotypes of Newcastle disease virus from the Sudan. Sudan J. Vet. Sci. Anim. Husb. 1983;24(1):69–78. [Google Scholar]
- Bensink Z, Spradbrow P. Newcastle disease virus strain I2--a prospective thermostable vaccine for use in developing countries. Vet. Microbiol. 1999;68(1-2):131–139. doi: 10.1016/s0378-1135(99)00069-3. [DOI] [PubMed] [Google Scholar]
- de Almeida R.S, Hammoumi S, Gil P, Briand F.X, Molia S, Gaidet N, Cappelle J, Chevalier V, Balança G, Traoré A, Grillet C, Maminiaina O.F, Guendouz S, Dakouo M, Samaké K, Bezeid Oel.M, Diarra A, Chaka H, Goutard F, Thompson P, Martinez D, Jestin V, Albina E. New avian paramyxoviruses type I strains identified in Africa provide new outcomes for phylogeny reconstruction and genotype classification. PLoS One. 2013;8(10):e76413. doi: 10.1371/journal.pone.0076413. http://dx.doi.org/10.1371/journal.pone.0076413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diel D.G, da Silva L.H, Liu H, Wang Z, Miller P.J, Afonso C.L. Genetic diversity of avian paramyxovirus type 1:proposal fora unified nomenclatureand classification system of Newcastle disease virus genotypes. Infect. Genet. E. 2012;12(8):1770–1779. doi: 10.1016/j.meegid.2012.07.012. [DOI] [PubMed] [Google Scholar]
- Dimitrov K.M, Ramey A.M, Qiu X, Bahl J, Afonso C.L. Temporal, geographic, and host distribution of avian paramyxovirus 1 (Newcastle disease virus) Infect. Genet. E. 2016;39:22–34. doi: 10.1016/j.meegid.2016.01.008. [DOI] [PubMed] [Google Scholar]
- Eisa M. The isolation and partial characterization of a Newcastle disease virus. Sudan J. Vet. Sci. Anim. Husb. 1979;20(1):1–10. [Google Scholar]
- Hanson R.P, Spalatin J, Jacobson G.S. The viscerotropic pathotype of Newcastle disease virus. Avian Dis. 1973;17(2):354–361. [PubMed] [Google Scholar]
- Haroun M, Khalafalla A.I, Hajer I. Some properties of Newcastle disease virus field isolates in Sudan. Bull. Anim. Health Prod. Africa. 1992;40:107–110. [Google Scholar]
- Hassan W, Khair S.A, Mochotlhoane B, Abolnik C. Newcastle disease outbreaks in the Sudan from 2003 to 2006 were caused by viruses of genotype 5d. Virus Genes. 2010;40(1):106–110. doi: 10.1007/s11262-009-0424-4. [DOI] [PubMed] [Google Scholar]
- Karrar G, Mustafa A. Newcastle Disease in the Sudan. Bulletin de l'Office International des Epizooties. 1964;62:890–896. [Google Scholar]
- Khalafalla A.I. Isolation and characterization of lentogenic NDVs from apparently healthy chicken in the Sudan. Bull. Anim. Health Prod. Africa. 1994;42:179–182. [Google Scholar]
- Khalafalla A.I, Fadol M.A, Hameid O.A, Hussein Y.A, el Nur M. Pathogenic properties of Newcastle disease virus isolates in the Sudan. Acta Vet. Hung. 1992;40(4):329–333. [PubMed] [Google Scholar]
- Lamb R.A, Collins P.L, Kolakofsky D, Melero Y, Nagai Y, Oldstone M.B.A, Pringle C.R, Rima B.K. Family Paramyxoviridae. In: van Regenmortel M.V.H, editor. Virus Taxonomy, Seventh report of the International Committee on Taxonomy of Viruses. New York: Academic Press; 2000. pp. 549–561. [Google Scholar]
- Lien Y.Y, Lee J.W, Su H.Y, Tsai H.J, Tsai M.C, Hsieh C.Y, Tsai S.S. Phylogenetic characterization of Newcastle disease viruses isolated in Taiwan during 2003-2006. Vet. Microbiol. 2007;123(1-3):194–202. doi: 10.1016/j.vetmic.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Liu X.F, Wan H.Q, Ni X.X, Wu Y.T, Liu W.B. Pathotypical and genotypical characterization of strains of Newcastle disease virus isolated from outbreaks in chicken and goose flocks in some regions of China during 1985-2001. Arch. Virol. 2003;148(7):1387–1403. doi: 10.1007/s00705-003-0014-z. [DOI] [PubMed] [Google Scholar]
- Lomniczi B, Wehmann E, Herczeg J, Ballagi-Pordány A, Kaleta E.F, Werner O, Meulemans G, Jorgensen P.H, Manté A.P, Gielkens A.L, Capua I, Damoser J. Newcastle disease outbreaks in recent years in Western Europe were caused by an old (VI) and a novel genotype (VII) Arch. Virol. 1998;143(1):49–64. doi: 10.1007/s007050050267. [DOI] [PubMed] [Google Scholar]
- Miller P.J, Decanini E.L, Afonso C.L. Newcastle disease:evolution of genotypes and the related diagnostic challenges. Infect. Genet. 2010;10(1):26–35. doi: 10.1016/j.meegid.2009.09.012. [DOI] [PubMed] [Google Scholar]
- OIE. Newcastle disease. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. Sixth edition. Paris: OIE; 2008. pp. 576–589. [Google Scholar]
- Qin Z, Sun L, Ma B, Cui Z, Zhu Y, Kitamura Y, Liu W. F gene recombination between genotype II and VII Newcastle disease virus. Virus Res. 2008;131(2):299–303. doi: 10.1016/j.virusres.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method:A new method for reconstructing phylogenetic trees. Mol. Biol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Snoeck C.J, Ducatez M.F, Owoade A.A, Faleke O.O, Alkali B.R, Tahita M.C, Tarnagda Z, Ouedraogo J.B, Maikano I, Mbah P.O, Kremer J.R, Muller C.P. Newcastle disease virus in West Africa: new virulent strains identified in non-commercial farms. Arch. Virol. 2009;154(1):47–54. doi: 10.1007/s00705-008-0269-5. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6:Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H.J, Chang K.H, Tseng C.H, Frost K.M, Manvell R.J, Alexander D.J. Antigenic and genotypical characterization of Newcastle disease viruses isolated in Taiwan between 1969 and 1996. Vet. Microbiol. 2004;104(1-2):19–30. doi: 10.1016/j.vetmic.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Umali D.V, Ito H, Suzuki T, Shirota K, Katoh H, Ito T. Molecular epidemiology of Newcastle disease virus isolates from vaccinated commercial poultry farms in non-epidemic areas of Japan. Virol. J. 2013;9:10. doi: 10.1186/1743-422X-10-330. 330. doi:10.1186/1743-422X-10-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujvári D, Wehmann E, Kaleta E.F, Werner O, Savić V, Nagy E, Zifra G, Lomniczi B. Phylogenetic analysis reveals extensive evolution of avian paramyxovirus type 1 strains of pigeons (Columba livia) and suggests multiple species transmission. Virus Res. 2003;96(1-2):63–73. doi: 10.1016/s0168-1702(03)00173-4. [DOI] [PubMed] [Google Scholar]
- Van Borm S, Obishakin E, Joannis T, Lambrecht B, van den Berg T. Further evidence for the widespread co-circulation of lineages 4b and 7 velogenic Newcastle disease viruses in rural Nigeria. Avian Pathol. 2012;41(4):377–382. doi: 10.1080/03079457.2012.696311. [DOI] [PubMed] [Google Scholar]


