Abstract
Forkhead-box domain (Fox) containing family members are known to play a role in neocorticogenesis and have also been associated with disorders on the autism spectrum. Here we show that a single RNA-binding protein, Hu antigen R (HuR), dictates translation specificity of bound mRNAs and is sufficient to define distinct Foxp-characterized subpopulations of neocortical projection neurons. Furthermore, distinct phosphorylation states of HuR differentially regulate translation of Foxp mRNAs in vitro. This demonstrates the importance of RNA binding proteins within the framework of the developing brain and further confirms the role of mRNA translation in autism pathogenesis.
Neurodevelopment relies on specific and timely expression of the genes dictating its course. Transcriptional control is one key mechanism by which appropriate development occurs and manages the expression of fate-determining genes1,2. Specifically, the transcription factors Foxp1 and Foxp2, members of the Forkhead-box containing (Fox) family, are associated with neocortical development, sensorimotor behavior, and abnormalities such as speech disorders and autism3,4,5,6,7,8.
In addition to transcriptional control, post-transcriptional regulation has emerged as another key mechanism in the spatiotemporal dynamics of brain development. RNA binding proteins (RBPs) control every step of post-transcriptional processing and as such are emerging as fate-determining molecules in neocortical development9,10. In particular, RBPs such as Fragile X mental retardation protein (FMRP) may regulate a subset of genes associated with the autism spectrum disorders11.
Another important family of RBPs is the Hu antigens. Hu antigens (HuA/HuR, HuB, HuC, HuD, and Hel-N1) are named for their role in Hu syndrome, an autoimmune paraneoplastic neurological syndrome, and all have roles in brain development12 Mammalian HuR, homologous to ELAV1 (Embryonic Lethal Abnormal Vision) in Drosophila, is a globally expressed Hu antigen and has defined roles in brain, breast, lung, and colon13,14,15,16. Our group has previously investigated both HuR and HuD RBPs, and revealed an association with neurodevelopmental defects in each case17,18.
Within the brain, our group previously showed that HuR is an essential regulator of neocortical development and mRNA translation. We found that HuR regulates temporal mRNA association with active translation sites (polysome complexes). This suggests its crucial role in the timing of when a transcript will become a protein in the developing neocortex. However, direct mRNA targets of HuR in neocortical development are unknown and a direct link between HuR and autism has not been observed.
Here, we show that HuR associates with specific and common subsets of mRNAs during early and late phases of prenatal neocortical neurogenesis. The consequence of such differential association is the regulated translational control of Foxp1 and Foxp2 at distinct time points in neocortical development. We additionally observed that phosphorylation of distinct HuR sites dictates the dynamic regulation of Foxp1 and Foxp2 in vitro.
Results
The RBP HuR binds distinct groups of mRNAs throughout development
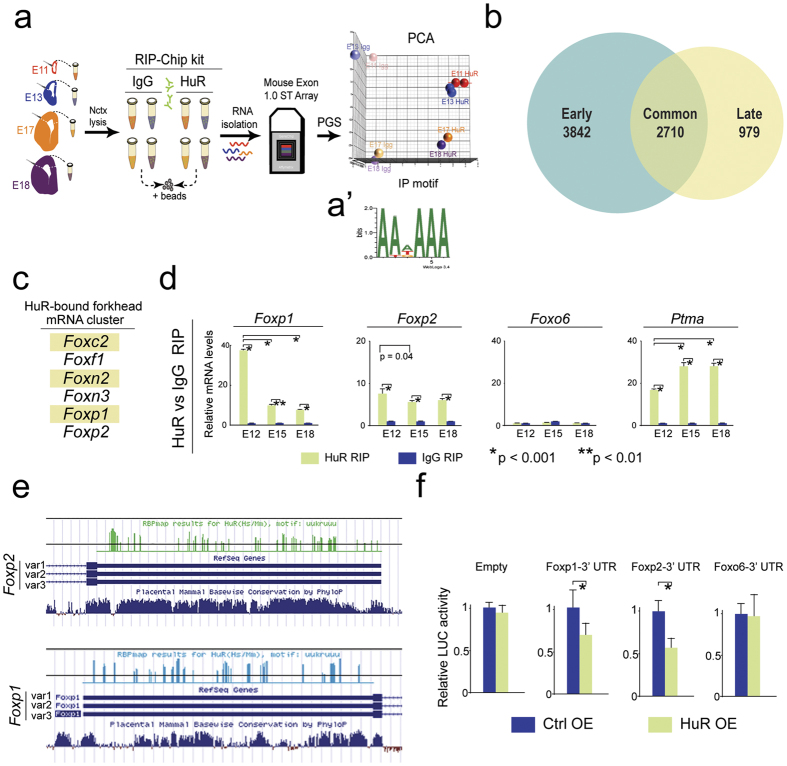
To elucidate neocortical HuR-bound mRNAs during early neurogenesis (E11 & E13) and late neurogenesis (E17 & E18), we performed RBP-immunoprecipitation coupled to a microarray assay (RIP-Chip) with an HuR RIP-Chip certified antibody and corresponding IgG (Fig. 1a; GEO accession number GSE77712). Principal component analysis (PCA) showed substantial differences in grouping between IgG and HuR pull-downs at distinct developmental stages and clustering among HuR precipitates from neocortices at early (E11 & E13) compared to later (E17 & E18) neurogenesis, substantiating the quality of the RIP experiment (Fig. 1a right).
Figure 1. HuR binds Foxp1 and Foxp2, autism-associated Fox mRNAs in developing neocortices.
(a) Schematic of RIP-ChIP approach from E11, E13, E17 and E18 neocortices to discover HuR-bound mRNAs. RNAs recovered from precipitates were analyzed using Mouse Exon 1.0 ST arrays and PGS or Bioconductor. PCA analysis on the right is shown. (a’) The most prevalent motif of mRNA transcripts in HuR RIP was that of HuR. (b) HuR-bound mRNAs were identified during only early (3842 transcripts, blue), or only late (979 transcripts, yellow) neurogenesis, or present in both groups (2710 transcripts, green overlap) during neocorticogenesis. (c) HuR binds a group of mRNAs characterized by the Fox domain. (d) Additional HuR RIPs from E12, E15, and E18 neocortices followed by qRT-PCR on precipitates validated HuR binding of Foxp1 and Foxp2 across neurogenesis. Values were normalized to Gapdh and IgG. Error bars represent SD, *p < 0.05. (e) Predicted HuR binding sites in 3′ UTRs of Foxp1 and Foxp2. (f) HuR suppressed luciferase activity when 3′ UTRs of Foxp1 or Foxp2 were present, but not of Foxo6.
Using stringent criteria (fold-change > 2 and False Discovery Rate < 5%), we found 6552 transcripts bound in early neocortical neurogenesis, significantly fewer transcripts bound during late neurogenesis (3689), and an overlap of 2710 transcripts bound across these developmental periods (Figs 1b and S1a).
In order to confirm coverage of our screen, we compared our results with a study whose aim it was to determine all binding targets of HuR in HeLa cells. Out of the 1091, 1862, and 564 genes in the early, common, and late lists, respectively, there were 119 (p = 7.92 × 10−15), 273 (p = 1.59 × 10−59), and 73 (p = 5.29 × 10−13) overlaps between our screens. It should be noted that CNS-specific mRNAs which are expressed in murine neocortical tissue may not have been expressed in the HeLa line, accounting for some of the variance between the lists.
We noticed genes associated with autism (members of the FoxP family and DYRK1A) to be HuR-bound and wanted to determine if targets of HuR were enriched for genes related to autism. We did not observe significant overlap between neocortical targets of HuR and genes found in the SFARI autism database, but observed that 12 genes overlap with early, 13 with common, and 2 with late HuR RIP targets; www.sfari.org, Simons Foundation, Table S1). Four genes from this list (NF1, DYRK1A, FOXP1, and GSK3B) were found to be in common with the previous HeLa screen19. Additionally, we found a significant overlap of early-bound HuR target genes and those bound by FMRP9 (51 genes, Fig. S1b, Table S2). These data suggest that HuR may contribute to regulation of FMRP targets.
HuR can regulate Foxp1 and Foxp2 via their 3’ UTRs
HuR-bound mRNAs were clustered using functional gene annotation (WebGestalt)32, and we found a group of TFs known to be involved in neurogenesis and associated with autism (e.g., Mef2c33, Foxp14, Foxp2 4; Fig. 1c, S1b). Subsequent qPCR analyses of additional HuR-RIPs (n = 3 per developmental stage) at E12, E15, and E18 confirmed HuR binding of different members of the Fox family (Fig. 1d, S1c). Foxp1 mRNA was enriched early-on. Meanwhile, Foxp2 mRNA was bound throughout neocortical neurogenesis, similarly to the known HuR target Ptma20, and in contrast to the negative control-identified as not bound by HuR in our screen, Foxo6 (Fig. 1d). HuR did not preferentially bind any splice variant of Foxp1 or Foxp2 (Fig. S1d). Though the expression and roles of Foxp1 and Foxp2 protein in the developing brain are well documented8,23,24,25; their upstream regulation has yet to be fully understood.
HuR is known to bind the untranslated regions (UTRs) of mRNAs, which have major roles in post-transcriptional regulation22. We found that both Foxp1 and Foxp2 had putative HuR binding sites in their 3′ UTRs (Fig. 1e) using RNPmap software (rbpmap.technion.ac.il). We tested for the functional regulation of FoxP1 or Foxp2 3′ UTRs in the presence of HuR using a luciferase assay in HEK293T cells24. When normalized over Renilla luciferase, we found that over-expression (OE) of HuR had no appreciable\effect on the expression of empty firefly luciferase vector nor of Foxo6 3′ UTR-cloned firefly luciferase (Fig. 1f; n = 3; p > 0.05). However, with either Foxp1 or Foxp2 3′ UTRs cloned downstream of the luciferase open reading frame (ORF), we found a significant reduction in firefly luciferase expression in the presence of the HuR OE, but not the control vector (Fig. 1f; p < 0.05; n = 3). Collectively, these data suggest that HuR may regulate autism-associated Foxp1 and Foxp2 expression during neocorticogenesis.
HuR conditional mutants reveal time-sensitive role of post-transcriptional regulation of Foxp1 and Foxp2
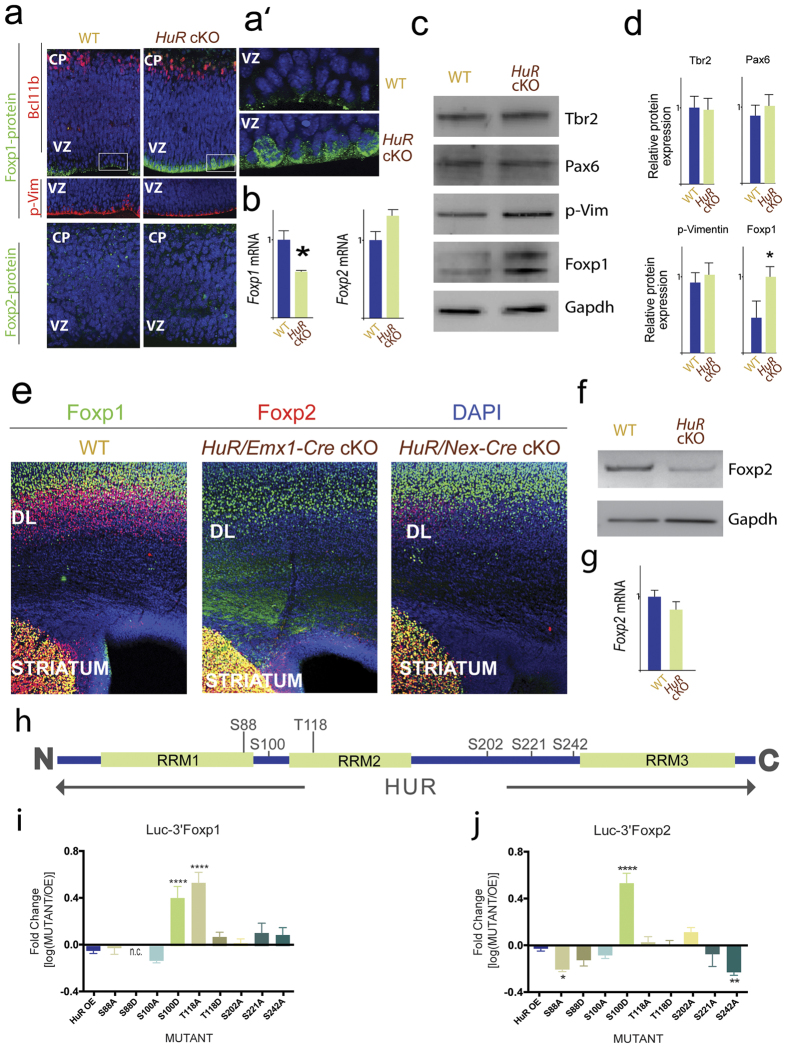
To examine the role of HuR in the post-transcriptional processing of Foxp1 and Foxp2 mRNA in the E13 neocortex in vivo, we generated an HuR conditional knock-out (HuR cKO) line by crossing HuR-floxed mice to Emx1-Cre (HuR/Emx1-Cre cKO; HuR depleted in radial glia progenitors and postmitotic neurons.). We performed qRT-PCR on total mRNA isolated from E13 WT and HuR cKO littermates and found a significant decrease in Foxp1 mRNA levels (Fig. 2b; n = 2; p < 0.05). Protein levels in HuR cKOs were assessed using immunohistochemistry (IHC) and western blotting (WB) (Fig. 2a,c,d). Surprisingly, despite lower Foxp1 mRNA levels, we found increased Foxp1-protein expression in RGs at E13, indicating HuR suppression of Foxp1 translation during early neocortical neurogenesis. When we assessed Foxp2 mRNA levels, we found them to be unchanged at E13 (Fig. 2b). Furthermore, IHC revealed that Foxp2 protein was not increased in E13 HuR/Emx1-Cre cKO neocortices (Fig. 2b), corroborated by WB (data not shown). This suggests distinct temporal regulation of Foxp1 and Foxp2 transcripts by HuR.
Figure 2. Neocortical HuR-dependent differential regulation of Foxp1 and Foxp2 mRNA translation depends on HuR phospho-states.
(a) Representative confocal images of immunostained E13 neocortex. Foxp1 protein (green, upper panels) was increased in mitotic RG in HuR cKOs at E13 and in BCl11b-expressing (red) post-mitotic neurons. Foxp2 protein (green, lower panels) showed no change. Boxed insets of VZ are shown enlarged in (a’). (b) Foxp1 mRNA levels were reduced in the E13 HuR cKO neocortex, but not Foxp2. (c) WB analysis confirmed that E13 HuR depletion increased Foxp1 protein, with no detectable changes in Tbr2 or Pax6. Quantification shown in (d). Values were normalized to Gapdh. (e) Representative confocal images of immunostained P0 neocortex. Foxp2 protein (red) was decreased in neocortices of both neocortical HuR cKOs, while being spared in basal ganglia (BG). Foxp1 (green) expression was not lost in HuR cKOs. DAPI is in blue. (f) WB analysis confirmed decreased levels of protein in HuR cKO, Nex-Cre (g) qRT-PCR revealed that levels of FoxP2 mRNA were unchanged in HuR cKO, Emx-Cre (h) Schematic of HuR protein with tested Ser/Thr sites. (i,j) Translation assay of Luc-3′Foxp1 (i) and of Luc-3′Foxp2 (j) in the presence of HuR OE or a phoshosite-mutated versions. n.c. = not completed Readings represent Luciferase (Luc) protein amount normalized to qRT-PCR measured Luciferase (Luc) mRNA levels. Graphs are presented as the log10 fold change of the HuR mutant readings over control OE for Luc/Luc. DL = deep layers. Error bars represent SEM, *p < 0.05, **p < 0.01, ****p < 0.0001.
We next sought to determine if HuR is required for Foxp2 protein expression later in development. IHC of P0 WT, HuR/Emx1-Cre cKO, and HuR/Nex-Cre cKO (HuR depleted in postmitotic neurons.) neocortices revealed decreased Foxp2 protein expression in both HuR cKOs (Fig. 2e, red) and presence of Foxp1 protein at P0 (Fig. 2e, green); WB confirmed decreased Foxp2 protein in HuR cKO (Fig. 2f). qRT-PCR confirmed no changes in total Foxp2 mRNA levels in the HuR cKO (Emx1-Cre, Fig. 2g) nor in subcellular distribution between nucleus and cytoplasm (Emx1-Cre, Fig. S2a). These findings indicate a role for HuR in regulation of Foxp2 mRNA translation. We further determined that HuR was acting in a cell-autonomous fashion (Fig. S2c–h).
Phosphorylation sites on HuR act in post-transcriptional regulation of Foxp1 and Foxp2
Given HuR’s ability to inhibit translation of Foxp1 and promote Foxp2, we wanted to explore HuR’s capacity to both bind similar targets and temporally regulate them. HuR has multiple residues which are amenable to phosphorylative regulation (Fig. 2h, www.phosphosite.org) and these have been shown to influence HuR’s function as an RBP28,29. To test this hypothesis, we transfected HuR phosphomutants and phosphomimics along with 3′ UTR Foxp1- or Foxp2-luciferase constructs in HEK293T cells and performed a translation assay24.
For the Foxp1-luciferase transfected cells, the phosphomimic S100D and the phosphomutant T118A had significantly higher translation of the luciferase gene than the wild type HuR overexpression (Fig. 2i). In Foxp2-luciferase transfected cells, the phosphomimic S100D, as in Foxp1, had higher translation than wild type HuR (Fig. 2j). However, in contrast to Foxp1, the phosphomutants S88A and S242A had significantly lower levels of Foxp2 translation than the wild type. This suggests that the S100 site is important to the regulation of translation of both Foxp1 and Foxp2 mRNA transcripts, while the S88, T118, and S242 sites contribute to the differential regulation of these two related transcripts.
Discussion
While there is much known about the downstream targets and effects of Foxp228 few studies have identified the mechanisms behind Foxp2 or Foxp1 mRNA regulation. Recently, we identified WNT3 as a positive post-transcriptional regulator of Foxp2, but not Foxp124. Another study examining post-transcriptional regulation of Foxp2 found it to be a target of the miRNAs miR-9 and miR-13222. One transcriptional regulator of Foxp2, lef1, was previously determined in Danio rerio (Zebrafish) to bind an enhancer element of Foxp2 in tectum and hindbrain, but not the telencephalon, a precursory structure to the cerebral cortex29. Though these results have yet to be confirmed in higher taxonomic organisms, this is particularly interesting evidence of regulation from an evolutionary perspective. Given this dearth, our study provides needed insight into the regulation of Foxp2.
What is particularly compelling about the regulation of Foxp1 and Foxp2 protein synthesis is the dependence on time. Early neocortical neurogenesis has been known to be susceptible to teratogenic influence that then results in autism-like phenotypes, further underscoring the importance of timing and timed control in brain development30,31. Our data show the existence of groups of transcripts bound by an RBP, HuR, during early neurogenesis, late neurogenesis, and throughout. Collectively, these data support the idea that some RBP-bound mRNAs form RBP-defined operons10,18 which are functionally and structurally related mRNAs poised for translation in developing systems, allowing for prompt and rapid response to developmental needs and specification signals. While Foxp2 mRNA levels did not change in HuR cKO, we unexpectedly found decreased Foxp1 mRNA paralleled by increased Foxp1 protein at E13. This unusual finding may be the consequence of a misbalanced steady state of Foxp1 mRNA. For example, not enough Foxp1 mRNA is being transcribed and to compensate for the absence of HuR binding, this mRNA is now being prematurely translated and decayed. As one of many options, this intriguing finding should be further characterized in future studies.
While many studies have shown that HuR can be regulated via its phosphorylation sites, we provide the first evidence that differential HuR phosphorylation states may regulate mRNA translation of the bound autism-associated genes Foxp1 and Foxp2. Sole phospho-sites have been involved in either inhibiting or promoting HuR activity26,34,35,36. In addition to the work presented here, one other study showed that tandem phosphorylation of HuR at S221 and S318 was necessary for mRNA binding (COX-2, cyclin-A, and cyclin-D) and nuclear translocation37. Our data agree with the hypothesis that the HuR-phosphorylation pattern is responsible for differential translational regulation of bound mRNAs. However, we cannot exclude the possibility that the distinct combinations of phosphorylated sites are further involved in the dynamic temporal regulation of Foxp1 and Foxp2 expression levels, with certain sites activated at certain time points in development. While there are some known kinases targeting HuR, including Cdk1(for S202) and PKC α/δ (for S221), no kinase has yet been identified for S24226. The precise spatiotemporal mechanism of HuR-phosphosite-mediated regulation as well as which sites are active when in vivo are key questions that remain to be answered.
Within the neocortex, Foxp1 and Foxp2 define functionally distinct subpopulations of neocortical glutamatergic projection neurons23 and mutations in each are associated with autism3,4,5,6,7,8. This novel mechanism of post-transcriptional regulation demonstrates how intrinsic HuR can differentially regulate translation of bound mRNAs during the intricate steps of neocorticogenesis. In conclusion, this study demonstrates the importance of RBPs within the framework of the developing brain and further confirms that altered mRNA translation contributes to autism pathogenesis.
Methods
Animals
All procedures and animal husbandry were approved by and carried out in accordance with the Rutgers University Institutional Animal Care and Use Committee (IACUC) guidelines (protocol number: I12-065). Isolation of wild type (WT) embryonic cortices and in utero electroporation experiments were performed in pregnant CD-1 dams (Charles River). Generation of HuR conditional deletion and WT littermate control animals was accomplished using Jackson laboratory Emx1-Cre (Strain Name: B6.129S2-Emx1tm1(cre)Krj/J, Stock Number#: 005628) and Nex-Cre (kind gift from M. Schwab). To generate embryonic HuR deletion mice, we produced timed pregnancies by placing a male with females overnight and checking for plugs the next day. The presence of plugs was considered embryonic day 0.5 (E0.5). Neocortical dissections were performed under RNAse free conditions on ice in PBS or Hank’s Balanced Salt Solution (HBSS) supplemented with D-glucose and HEPES38. Neocortices were either used immediately or stored at −80 °C until use. At least eight neocortices per the genotype were analyzed in each of the experimental approaches.
Immunocytochemistry
Brains were dissected in PBS and immediately submerged in 4% paraformaldehyde (PFA) (Sigma-Aldrich #P6148). Three ten-minute washes were carried out at room temperature while agitating and brains were subsequently left agitating in fresh PFA at 4 °C overnight. Fixed brains were washed once with PBS and placed in 30% sucrose (J.T. Baker #4072-05) and stored at 4 °C. Fixed brains were embedded in 3% agarose (Lab Express #2001) and sectioned (Leica Vibratome VT100S) at 70 μm. Sections were stored in 30% sucrose.
For immunostaining, sections were first subjected to three five-minute PBS washes while gently agitating. Next, sections were blocked in PBS with 5% normal donkey serum [Jackson Immuno, 1% bovine serum albumin (BioMatik #A2134), 0.1% glycine (VWR #BDH4156)] containing 0.1% L-lysine (Sigma-Aldrich 100999242) and 0.4% Triton-X (Fisher #BP151) pH 7.5] for 45 minutes at room temperature. After blocking, sections were placed in their respective primary antibodies (Table S3) in blocking solution with 0.4% Triton-X 100 overnight or up to two nights with gentle shaking at 4 °C. Sections were washed in PBS three times and placed in Triton-free blocking solution with secondary antibodies (Table S4) for two hours while gently shaking at room temperature. Sections were then washed three times in PBS for five minutes with gentle shaking and placed in PBS with DAPI (Roche diagnostics #10-236-276-001) for five minutes shaking at room temperature. They were washed again in PBS for five minutes two times. Sections were mounted in VectaShield (Vector Laboratories, Inc. H-1000), cover-slipped, and sealed with nail polish. Imaging was carried out using an Olympus multi-photon/confocal microscope FV1000MPE.
Western Blot
Protein lysates were prepared using T-PER reagent (Thermo Scientific #78510), Lysates were cleared by centrifugation for 10 minutes at approximately 13,000 × g. For loading volume adjustment, lysates were analyzed for total RNA content using a spectrophotometer. We used the Invitrogen SureLock Western blot system with Invitrogen Bis-Tris 4–12% gradient gels and followed the manufacturer’s protocol for running and transfer onto nitrocellulose membranes (BioRad #162-0214).
After transfer, membranes were washed in PBS solution with 0.04% Triton-X 100 (PBS-T), shaking at room temperature for five minutes. Membranes were blocked (blocking solution: 5% milk, 10% FBS, PBS-T) for one hour at room temperature and placed in probing solution (PBS-T and 10% FBS) with primary antibody (see Table S3) overnight or up to two nights, shaking at 4 °C. The blots were washed in PBS solution for five minutes three times, and placed in probing solution with secondary antibody (Table S4) forone1 hour. Blots were washed three times in PBS-T and once in PBS for 5 minutes, and developed (Protein Simple ChemiGlow West #60-12596-00) and imaged with Genesnap software and Syngene G:Box imager. Densitometry quantifications were completed in ImageJ (version:2.0.0-rc-30/1.49u) software.
Quantitative Real Time PCR (qRT-PCR) and Subcellular Distribution
RNA was isolated using the PARIS kit for cortical and nuclear/cytoplasmic fractionation or Trizol-LS (Life Technologies #10296028), following the manufacturer’s protocol. RNA samples were DNAsed (Ambion #AM1907) to remove contaminating DNA. RT-PCR was performed using Applied Biosystems StepOne Real-Time PCR system with Step-one v2.1 software (#4376373) using RNA-Ct 1-Step Taqman kit (#4392653) and Taqman probes (Table S5) according to the manufacturer’s protocol with reactions adjusted to 10-μl total volume. Each qRT-PCR was performed with more than three technical replicates within more than four biological samples per developmental stage or experimental condition.
In utero Electroporation
E13 neocortices were in utero electroporated as previously described17,38. Briefly, E13 pregnant CD-1 dams (Charles river) were anesthetized, embryos removed, and 1 ug total DNA was injected into the ventricular space using a pulled glass pipette (prepared with Sutter Instrument Co. P-87 pipette puller). Five electric pulses were delivered (BTX Harvard apparatus ECM 830; voltage: 37 V, pulse length: 50 ms, interval between pulses: 950 ms) to embryos by placing the plus electrode above the head to drive DNA into the neocortex. The surgeries lasted approximately twenty-five minutes. Pups were placed back into the mother, muscles and skin were sutured (Ethicon, Silk, 5-0), and gestation was allowed to continue until the appropriate age for subsequent analyses. At least two or three transfected neocortices were analyzed per experimental condition.
Cell culture conditions and transfections
N2A cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO #31053-028) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich #F6178), 20 mM HEPES (GIBCO #15630-080), sodium pyruvate (GIBCO #11360-070), glutamax (GIBCO #35050-061), and penicillin/streptomycin (Cellgro #30-001-C1). HEK293T cells were cultured in DMEM supplemented with 5% fetal bovine serum (FBS; Sigma-Aldrich #F6178), 20 mM HEPES (GIBCO #15630-080), sodium pyruvate (GIBCO #11360-070), glutamax (GIBCO #35050-061), and penicillin/streptomycin (Cellgro #30-001-C1). Both cell lines were fed by aspirating media, washing with PBS 1X and replacing with fresh media. Passage of cells was performed in TrypLE select (GIBCO #12563-029) for five minutes, after which supplemented DMEM media was added at two times the volume of TrypLE and cells were pelleted at 300 rpm for three minutes (Beckman Coulter Allegra 21 centrifuge) and re-suspended in fresh media or lysis buffer for downstream protein/RNA isolation. Transfections were performed using Lipofectamine 2000 (Invitrogen #52758) according to the manufacturer’s protocol.
To prepare for primary neuronal cultures, one day before neocortical dissections, cover slips were treated with 1M HCl and then washed and coated with 15 ug/ml poly-L-ornithine (Sigma-Aldrich #P3655) for forty-five minutes at room temperature. Poly-L-ornithine was aspirated from cover slips that were then treated with 4 μg/ml of mouse laminin (GIBCO #1094706) for 10–15 hours at 37 °C. To isolate primary neurons, neocortical dissections were performed in HBSS supplemented with 0.5% D-Glucose (American Analytical #AB00715-00500) and 25 mM HEPES (GIBCO #15630-080). Dissected neocortices were minced and dissociated in HBSS supplemented with 0.5% D-Glucose, 25 mM HEPES, 0.5 mg/ml DNase I (Sigma-Aldrich #D5025), 20 U/ml Papain (Calbiochem #5125), and 5.0 mg/ml Dispase II (Sigma-Aldrich #D4693-1G). Cells were incubated for five minutes, pelleted, and re-suspended in supplemented HBSS three times. Primary dissociated cells were re-suspended in primary cell culture media including Neurobasal medium (GIBCO #1097077), Sodium Pyruvate (GIBCO #11360-070), 2 mM glutamax (GIBCO #35050-061), penicillin/streptomycin (Cellgro #30-001-C1), and B-27 supplement (GIBCO #17504). All cells were cultured at 37 °C with 5% CO2. For immunostaining, cells were fixed with 4% PFA for twenty minutes.
HuR RIP
HuR-associated RNAs from neocortices at distinct ages (E11, E12, E13, E15, E17, E18, P0) were isolated using the RIP kit (MBL international #RN1001) and rabbit anti-HuR (MBL international #RN004P) RIP certified antibody according to manufacturer’s instructions. HuR function was shown to react to UV39,40 used in HITS CLIP, which is avoided by the RIP-ChIP kit.
Previously, targets of HuR were identified in a human HeLa cell line using the PAR-CLIP method. To compare our findings, we first converted their list of target human gene symbols (Supplementary Table 2) to mouse gene symbols. Starting with 1216 human genes, we used the biomaRt tool within R to query the Ensembl database and found the mouse homolog associated gene name for each. With some imprecision in matching for species, this produced 1214 results, but only 1192 unique mouse gene symbols. From these, we then used the GeneOverlap package in R to apply Fisher’s exact test for significance of the overlap between lists of gene symbols from our RIP-ChIP screen and the PAR-CLIP one.
Microarray analysis
Total RNA from developing neocortices and RIPs were sent for exon microarray hybridization at the UMDNJ-RWJMS facility using the GeneChip Mouse Exon 1.0 ST Array (Affymetrix; n = 2/developmental stage or experimental condition). The Array image (CEL) files were read into either Partek’s Genome Suite (Affymetrix) or R/BioConductor using the oligo package41 RMA normalized, and assembled into core transcript models using NetAffx-supplied annotations. A linear model was fit with the limma package42 and enrichment contrasts selected based on a log2 fold-change of +1 or greater and a Benjamini-Hochberg-adjusted p value of 0.05 or less. Early (E11 and E13) and late (E17 and E18) samples were grouped separately. Of the 4,092 and 2,604 transcripts found to be significantly enriched by HuR RIP at early and late times, respectively, there were 2,244 and 1,803 unique, assigned Entrez Gene IDs and so these were selected for functional analysis using WebGestalt32. These transcripts were consolidated into gene symbols for testing overlaps with FMRP-binding mRNAs or autism-associated mRNAs (Fig. S1) using the GeneOverlap package in Bioconductor. Probeset and exon plots were produced using the GenomeGraphs package in Bioconductor.
Cloning and Constructs
The full-length of Foxp2 3′ UTR was amplified by PCR using the forward primer 5′-TCGCGACTAGTGAACGAACTTGTGACACCTCAGTG-3′ and reverse primer 5′-CATATGCGGCCGTGTACTTCAGAAATGTAACCAACTG-3′. The Foxp1 3′ UTR was amplified with the forward primer 5′-TCGCGACTAGTACATGGAGTGAACCTCTGGGC-3′ and reverse primer 5′-CATATGCGGCCGCATTTAAGAATGCGCTCATGTCAG-3′. Restriction enzyme sites Spe I and Eag I were added to each of the forward primers and reverse primers, respectively, and used for cloning the Foxp2 and Foxp1 3′ UTRs downstream of a firefly Luciferase cassette in the pcDNA3.1-Luciferase plasmid which digested with Xba I and Eag I24.
HuR wild type, phosphomimics, and phosphomutants were cloned with TAP tags (Generously shared by Myriam Gorospe tested mutants are indicated in Fig. 2G 34,43). Mutants (S88, S100, T118, S200, S202, and S242) were generated by site-directed mutagenesis of serine/threonine residues to alanine (S/T → A); mimics (S88, S100, and T118) were generated with their serine/threonine residues mutated to aspartic acid (S/T → D).
Translational Luciferase Assay
To test the effects of these phosphorylation sites on Foxp1 and Foxp2 translation, we performed double transfections with the phosphomutants, phosphomimics, and Foxp1 or Foxp2 with firefly luciferase proteins in their 3′ UTR regions IN HEK293T cells24. At least six biological replicates and two technical replicates were performed for each sample; HuR-TAP was biologically replicated at least twenty-four times. Luciferase readings were taken to determine protein expression of the Foxp1/2 constructs (Luc) on a TD-20/20 Luminometer (Turner Designs) and using the Dual-Luciferase Reporter Assay (Promega E1910). Renilla readings were taken as a control to ensure comparable cell counts. Luciferase reagents were made fresh and came from the same kit44.
Additionally, RNA was isolated from these samples and quantified with qRT using a luciferase probe (luc) (Table S5). Using these two readings (Luc/luc), protein and mRNA levels in each sample, we were able to generate a proxy for translation by normalizing protein levels to levels of mRNA24. A metric for measuring translation is useful, as levels of mRNA and protein are both informative points of information to the actual state of the cell. For example, in relativity to each other, high levels of mRNA and low protein levels indicate low translation whereas low levels of mRNA and high levels of protein indicate high translation. Data is presented in Fig. 2h,i as their log10 values, as we are generating a ratio of mutant translation levels over wild-type translation levels. Outlier analysis was determined by the ROUT test with Q = 1% and the data was subsequently analyzed without the outlier. 6 outliers were removed from the Foxp1 assay (Fig. 2h) out of 160 data points (3.8%) and 10 outliers were removed from the Foxp2 assay (Fig. 2i) out of 172 data points (5.8%).
Statistical analysis
One-way ANOVA and SPSS software were used for statistical analysis when multiple conditions were compared, while t-test was used when only two experimental conditions within a developmental stage were compared, unless otherwise indicated. N of each experiment and the respective p-values are indicated in either figure legend, results, methods, or in the main text. A Kruskal-Wallis Multiple Comparisons test was performed on data represented in Fig. 2h,i and was calculated in GraphPad’s Prism 6.0 software. A two-way ANOVA was used to compare cultured neurons represented in Fig. S2f (n = 975 neurons, 4 slides, 8 coverslips) and S2g (n = 1082 neurons, 4 slides, 8 coverslips). Scoring and statistical analysis was performed by a blinded experimenter.
Additional Information
How to cite this article: Popovitchenko, T. et al. The RNA binding protein HuR determines the differential translation of autism-associated FoxP subfamily members in the developing neocortex. Sci. Rep. 6, 28998; doi: 10.1038/srep28998 (2016).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants (NS064303 and NS075367) to M.R.R. We would like to thank Dr. Mengqing Xiang and Dr. Hui Wang for assistance with the luminometer and luciferase assay. Dr. Erik DeBoer, Waleed Adawi, Akshay Thaper, and Nicholas Page for their assistance with data collection.
Footnotes
Author Contributions T.P. and M.R.R. wrote the main manuscript text; D.L.K., J.X. and M.K. contributed critical reagents; R.P.H. performed bioinformatics analysis; T.P., K.T., B.V. and M.R.R. performed experiments; T.P. and M.R.R. prepared all Figures; All authors reviewed manuscript.
References
- Martynoga B., Drechsel D. & Guillemot F. Molecular control of neurogenesis: a view from the mammalian cerebral cortex. Cold Spring Harbor Perspect in Biol 4, a008359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoer E. M., Kraushar M. L., Hart R. P. & Rasin M.-R. Post-transcriptional regulatory elements and spatiotemporal specification of neocortical stem cells and projection neurons. Neuroscience 248, 499–528 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski Rosenfeld & Blumenthal. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell 149, 525–537 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon C. & Rappold G. A. The distinct and overlapping phenotypic spectra of FOXP1 and FOXP2 in cognitive disorders. Human Genetics 131, 1687–1698 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszer M. et al. Impaired synaptic plasticity and motor learning in mice with a point mutation implicated in human speech deficits. Curr. Bio. 18, 354–362 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. S. et al. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413, 519–523 (2001). [DOI] [PubMed] [Google Scholar]
- Araujo D. J. et al. FoxP1 orchestration of ASD-relevant signaling pathways in the striatum. Genes Dev. 29, 2081–2096 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. et al. Foxp1 regulates cortical radial migration and neuronal morphogenesis in developing cerebral cortex. PLoS ONE 10, e0127671 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell R. B. RNA protein interaction in neurons. Annu Rev Neurosci. 36, 243–270 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J. D. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 8, 533–543 (2007). [DOI] [PubMed] [Google Scholar]
- Darnell J. C. et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano H. J. & Darnell R. B. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J Neuro. 17, 3024–3037 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy N. S., Chung S., Furneaux H. & Levy A. P. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem. 273, 6417–6423 (1998). [DOI] [PubMed] [Google Scholar]
- Leandersson K., Riesbeck K. & Andersson T. Wnt-5a mRNA translation is suppressed by the Elav-like protein HuR in human breast epithelial cells. Nucleic Acids Res. 34, 3988–3999 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbán S. et al. Influence of the RNA-binding protein HuR in pVHL-regulated p53 expression in renal carcinoma cells. Mol Cell Biol. 23, 7083–7095 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silanes I. et al. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene 22, 7146–7154 (2003). [DOI] [PubMed] [Google Scholar]
- DeBoer E. M. et al. Prenatal deletion of the RNA-binding protein HuD disrupts postnatal cortical circuit maturation and behavior. J Neurosci. 34, 3674–3686 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraushar M. et al. Temporally defined neocortical translation and polysome assembly are determined by the RNA-binding protein Hu antigen R. Proceedings of the National Academy of Sciences 111, E3815–E3824 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva S. et al. Transcriptome-wide Analysis of Regulatory Interactions of the RNA-Binding Protein HuR. Mol Cell 43, 340–352 (2011). [DOI] [PubMed] [Google Scholar]
- de Silanes I. L. et al. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci USA. 101, 2987–2992 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisaoka T., Nakamura Y., Senba E. & Morikawa Y. The forkhead transcription factors, Foxp1 and Foxp2, identify different subpopulations of projection neurons in the mouse cerebral cortex. Neuroscience 166, 551–563 (2010). [DOI] [PubMed] [Google Scholar]
- Clovis Y. M. et al. Convergent repression of Foxp2 3′ UTR by miR-9 and miR-132 in embryonic mouse neocortex: implications for radial migration of neurons. Development 139, 3332–3342 (2012). [DOI] [PubMed] [Google Scholar]
- Ferland R. J. et al. Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. J Comp Neurol. 460, 266–279 (2003). [DOI] [PubMed] [Google Scholar]
- Kraushar M. L. et al. Thalamic WNT3 Secretion Spatiotemporally Regulates the Neocortical Ribosome Signature and mRNA Translation to Specify Neocortical Cell Subtypes. J. Neurosci. 35, 10911–10926 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura S. et al. Expression pattern of the winged-helix/forkhead transcription factor Foxp1 in the developing central nervous system. Gene Expr Patterns 3, 193–197 (2003). [DOI] [PubMed] [Google Scholar]
- Kim H. H., Yang X., Kuwano Y. & Gorospe M. Modification at HuR (S242) alters HuR localization and proliferative influence. Cell Cycle. 7, 3371–3377 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. H. et al. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 22, 1804–1815 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes S. et al. Foxp2 Regulates Gene Networks Implicated in Neurite Outgrowth in the Developing Brain. Plos Genet. 7, e1002145 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonkowsky J. et al. Domain-specific regulation of foxP2 CNS expression by lef1. BMC Dev Biol. 8, 103 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G. C., Reuhl K. R., Cheh M., McRae P. & Halladay A. K. A new neurobehavioral model of autism in mice: pre-and postnatal exposure to sodium valproate. J Autism Dev Disord. 36, 779–793 (2006). [DOI] [PubMed] [Google Scholar]
- Ardinger H. et al. Verification of the fetal valproate syndrome phenotype. Am J Med Genet. 29, 171–185 (1988). [DOI] [PubMed] [Google Scholar]
- Zhang B., Kirov S. & Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic acids research, 10.1093/nar/gki475 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai N.-P. et al. Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell 151, 1581–1594 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. H. & Gorospe M. Phosphorylated HuR shuttles in cycles. Cell Cycle 7, 3124–3126 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K. et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell. 25, 543–557 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova N., Yang X., King P. & Nabors B. Phosphoregulation of the RNA-binding protein Hu antigen R (HuR) by Cdk5 affects centrosome function. J Biol Chem. 287, 32277–32287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doller A. et al. Tandem phosphorylation of serines 221 and 318 by protein kinase Cdelta coordinates mRNA binding and nucleocytoplasmic shuttling of HuR. Mol Cell Biol. 30, 1397–1410 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rašin M. R. et al. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci. 10, 819–827 (2007). [DOI] [PubMed] [Google Scholar]
- Mazan-Mamczarz K. et al. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci USA 100, 8354–8359 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. et al. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 19, 2340–2350 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho B. S. & Irizarry R. A. A. Framework for oligonucleotide microarray preprocessing. Bioinformatics 26, 2363–2367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K. Limma: linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor. pp 397–420 (2005). [Google Scholar]
- Lal A. et al. Antiapoptotic function of RNA‐binding protein HuR effected through prothymosin α. EMBO J. 24, 1852–1862 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. & Matise M. In Vivo Dual Luciferase Reporter Assay with Chick Neural Tube In Ovo Electroporation System. Methods Mol Biology. 1018, 211–217 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.