Abstract
We isolated Pseudomonas putida (P. putida) strain 1A00316 from Antarctica. This bacterium has a high efficiency against Meloidogyne incognita (M. incognita) in vitro and under greenhouse conditions. The complete genome of P. putida 1A00316 was sequenced using PacBio single molecule real-time (SMRT) technology. A comparative genomic analysis of 16 Pseudomonas strains revealed that although P. putida 1A00316 belonged to P. putida, it was phenotypically more similar to nematicidal Pseudomonas fluorescens (P. fluorescens) strains. We characterized the diversity and specificity of nematicidal factors in P. putida 1A00316 with comparative genomics and functional analysis, and found that P. putida 1A00316 has diverse nematicidal factors including protein alkaline metalloproteinase AprA and two secondary metabolites, hydrogen cyanide and cyclo-(l-isoleucyl-l-proline). We show for the first time that cyclo-(l-isoleucyl-l-proline) exhibit nematicidal activity in P. putida. Interestingly, our study had not detected common nematicidal factors such as 2,4-diacetylphloroglucinol (2,4-DAPG) and pyrrolnitrin in P. putida 1A00316. The results of the present study reveal the diversity and specificity of nematicidal factors in P. putida strain 1A00316.
Members of the genus Pseudomonas display remarkable physiological and metabolic versatility, which enables them to colonize diverse terrestrial and aquatic habitats such as soil1, plants2, polluted creeks3, and fresh water4. Among species of the genus, P. putida has been isolated from many different niches and has the ability to survive in soils containing high concentrations of organic contaminants and heavy metals5. They can degrade a wide variety of chemicals, including many natural and synthetic compounds6. Some P. putida strains are plant growth-promoting rhizospheric and endophytic bacteria, which make them ideal for biocontrol7. However, the effects of P. putida strains on Meloidogyne incognita (M. incognita) have seldom been reported and the mechanisms by which they control M. incognita are unclear8.
M. incognita infection can cause serious plant parasitic diseases, which greatly limits agricultural productivity and quality9. M. incognita on cucumbers and tomatoes was mainly controlled by fumigants such as methyl bromide10, metal sodium, and 1,3-dichloropropene11. Although these chemical nematicides are effective, they have been withdrawn from use or restricted owing to serious environmental safety and public health concerns12. Accordingly, it has become necessary to identify novel and environmental friendly alternatives to control plant-parasitic nematode populations.
Pseudomonas species are ubiquitous in the natural world and produce a remarkable array of secondary metabolites active against agriculturally important plant diseases13. For example, some P. fluorescens strains can produce proteins and secondary metabolites that function as biocontrol factors to kill nematodes, such as 2,4-diacetylphloroglucinol (2,4-DAPG)14,15, alkaline metalloproteinase AprA16, pyrrolnitrin (Prn)17, and hydrogen cyanide (HCN)17. Loper et al. characterized the diversity and phylogeny of plant-associated Pseudomonas spp. involved in multitrophic interactions based on the comparative genomic study18. They found that ten P. fluorescens strains exhibited a diverse spectrum of traits involved in biological control and other multitrophic interactions with plants, microbes, and insects. However, that study of secondary metabolite biosynthesis did not include functional analyses of biocontrol activity. In addition, although some other comparative genome analysis of Pseudomonas strains have also been reported, they mainly focused on the Pseudomonas strains which degraded organic pollutant or some plant growth-promoting rhizobacteria (PGPR) in Pseudomonas19,20,21,22.
In this study, we report the complete genome sequence of P. putida 1A00316 isolated from Antarctica soil. The assembled genome sequence has been deposited in NCBI Refseq database under accession number CP014343. The bacterium inhibits M. incognita in vitro and under greenhouse conditions, where it reduces symptoms up to 71.67% on tomato caused by M. incognita23. We performed comparative genomic and phenotypic analyses of 16 strains within the Pseudomonas group (five P. fluorescens, two P. protegens, and nine P. putida strains), including the newly sequenced P. putida 1A00316. We observed a diverse spectrum of genomic features, evolutionary relationships, and virulence factors among these strains. We further identified and characterized the nematicidal factors in P. putida 1A00316, and explored their distribution within other Pseudomonas strains.
Results and Discussion
Genomic features of Pseudomonas strains
The genomic features of P. putida 1A00316 and other 15 reference Pseudomonas strains are summarized in Table 1. The newly sequenced complete genome sequence of P. putida 1A00316 comprised a circular chromosome of 5,715,815 bp containing 5,039 protein-coding genes, 22 rRNA and 76 tRNA genes with an average G + C content of 64.4% (Table 1; Fig. 1A). Interestingly, P. putida 1A00316 had the highest GC content among the selected Pseudomonas strains. The genome size and number of protein-coding genes for these strains ranged from 5.72 ~ 7.07 Mb and 4,960 ~ 6,357 genes, respectively, demonstrating substantial strain-to-strain variation.
Table 1. Features of 16 Pseudomonas genomes.
| Features | Chromosome size (Mb) | G + C (%) | CDSs | Average CDSs length (nt) | Coding (%) | rRNA genes | tRNA genes | Scaffolds | Accession number | Virulence factors |
|---|---|---|---|---|---|---|---|---|---|---|
| Pf_A506 | 5.96 | 60.0 | 5267 | 993 | 87.7 | 19 | 69 | 1 | NC_017911.1 | 87 |
| Pps_CHA0 | 6.87 | 63.4 | 6115 | 996 | 88.6 | 15 | 68 | 1 | NC_021237.1 | 101 |
| Pf_F113 | 6.85 | 60.8 | 5862 | 1011 | 86.6 | 16 | 66 | 1 | NC_016830.1 | 110 |
| Pf_Pf0-1 | 6.44 | 60.5 | 5722 | 1008 | 89.6 | 19 | 73 | 1 | NC_007492.2 | 77 |
| Pps_Pf-5 | 7.07 | 63.3 | 6108 | 1013 | 87.5 | 16 | 71 | 1 | NC_004129.6 | 106 |
| Pf_Q2–87 | 6.37 | 60.6 | 5597 | 996 | 87.6 | 19 | 68 | 1 | NZ_CM001558.1 | 103 |
| Pf_SBW25 | 6.72 | 60.5 | 5921 | 1000 | 88.1 | 16 | 66 | 1 | NC_012660.1 | 91 |
| Pp_1A00316 | 5.72 | 64.4 | 5039 | 987 | 87.2 | 22 | 76 | 1 | CP014343 | 92 |
| Pp_BIRD-1 | 5.73 | 61.7 | 4960 | 1002 | 86.7 | 22 | 64 | 1 | NC_017530.1 | 64 |
| Pp_F1 | 5.96 | 61.9 | 5250 | 1005 | 88.5 | 19 | 76 | 1 | NC_009512.1 | 66 |
| Pp_GB-1 | 6.08 | 61.9 | 5408 | 1003 | 89.2 | 22 | 74 | 1 | NC_010322.1 | 66 |
| Pp_H8234 | 6.87 | 61.6 | 6357 | 934 | 86.4 | 18 | 69 | 1 | NC_021491.1 | 72 |
| Pp_HB3267 | 5.88 | 62.6 | 5292 | 966 | 87.0 | 22 | 70 | 2 | NC_019905.1 | 69 |
| Pp_KT2440 | 6.18 | 61.5 | 5350 | 1000 | 86.5 | 22 | 74 | 1 | NC_002947.3 | 63 |
| Pp_S16 | 5.98 | 62.3 | 5218 | 971 | 84.7 | 19 | 70 | 1 | NC_015733.1 | 65 |
| Pp_W619 | 5.77 | 61.4 | 5182 | 988 | 88.7 | 22 | 75 | 1 | NC_010501.1 | 62 |
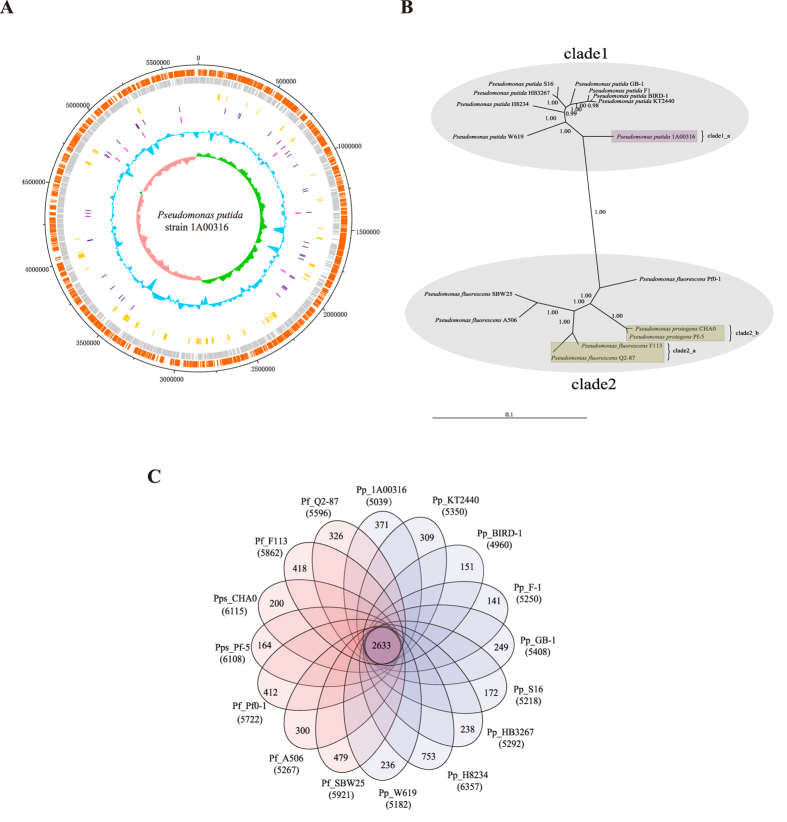
Figure 1. Genomic features and a comparative genomic analysis of Pseudomonas strains.
(A) Circular plot of the P. putida 1A00316 chromosome. Circles are numbered from 1 (outermost) to 8 (innermost). Circle 1 represents the whole chromosome; Circles 2 and 3 show the locations of predicted CDSs on the positive and negative strands, respectively; Circle 4, genomic islands; Circle 5, tRNA genes; Circle 6, rRNA genes; Circle 7, %G + C; Circle 8, GC skew ((G−C)/(G + C)). (B) Phylogenetic tree depicting the relationships among 16 Pseudomonas strains. The values shown at interior nodes represent clade credibility, which is the likelihood of the clade based on the posterior probability values generated using MrBayes. (C) Genomic diversity of 16 Pseudomonas strains. Each strain is represented by an oval. The number of orthologous coding sequences (CDSs) shared by all strains (i.e., the core genome) is shown in the center. Overlapping regions show the number of CDSs conserved only within the specified genomes. Numbers in non-overlapping portions show the number of CDSs unique to each strain. The total number of protein-coding genes within each genome is listed below the strain name.
Phylogenetic tree
We constructed a phylogenetic tree of the 16 Pseudomonas strains based on 16 S rRNA and five conserved genes (dnaE, guaA, gyrB, recA, and rpoB)24 (Fig. 1B). The 16 Pseudomonas strains clustered into two major clades. The P. fluorescens group clearly formed a single, large clade composed of four distinct sub-clades. P. fluorescens Q2–87 (producing 2,4-DAPG, which may have nematicidal activity)25 and nematicidal strain P. fluorescens F_11326 were in clade2_a, and clade2_b was composed of two other closely related nematicidal strains, P. protegens CHA014,15,16 and Pf-527. The other large clade was composed of nine P. putida strains and was clearly divided into two sub-clades. Clade1_a contained only P. putida 1A00316, indicating that it was distinct from other P. putida strains. We also observed similar patterns between the above-described phylogenetic tree and a maximum likelihood phylogeny inferred from 2,408 single-copy protein-coding genes conserved in these Pseudomonas strains (see Supplementary Figure S1).
Analysis and comparison of virulence factors
To further explore the relationships among the Pseudomonas strains, we predicted virulence factors in each strain using the virulence factor database (VFDB)28 and performed a comprehensive comparative analysis of these factors. As summarized in Table 1, the number of virulence factors in strains of the P. fluorescens group (77–110) was generally higher than that in P. putida group (62–72), except for P. putida 1A00316 (92). We speculated that P. putida 1A00316 differs from other P. putida strains with respect to virulence metabolism and is more similar to P. fluorescens group. To evaluate this conjecture, we compared the types of virulence factors between P. putida 1A00316 and other P. fluorescens and P. putida strains. P. putida 1A00316 had 30 unique virulence factors with respect to P. putida strains, but only eleven unique virulence factors with respect to the nematicidal P. fluorescens strains. These results support our observation that at least with respect to virulence genes, P. putida 1A00316 is more similar to P. fluorescens than P. putida group.
Identification and comparison of protein families
Using orthoMCL29, we identified a core genome containing 2,633 protein-coding genes shared among the 16 Pseudomonas strains (Fig. 1C). The core genome accounted for only 41–53% of the proteome of each strain, indicating a high degree of genomic diversity in the Pseudomonas group. To examine the genomic diversity among the nematicidal strains, we determined the size of the core genome and the pan-genome of four nematicidal strains (P. putida 1A00316, and P. fluorescens F_113 and two P. protegens strains), which contained 3,261 and 8,666 protein-coding genes, respectively. We also characterized the core genome and pan-genome of four additional strains, including the three nematicidal P. fluorescens strains and one randomly selected P. putida strain that lacks nematicidal ability. Their core genomes and pan-genomes contained 3,136 ~ 3,217 and 9,108 ~ 10,003 genes, respectively. These results demonstrated that the genomic complexity among the four nematicidal strains is less than that of other taxon combinations (not all strains in this group can kill M. incognita).
P. putida 1A00316 contained a nematicidal factor encoding alkaline metalloproteinase AprA
P. putida 1A00316 exhibits strong inhibition against M. incognita in vitro and under greenhouse conditions23, while all of the other selected P. putida strains lack the ability to kill M. incognita. Thus, additional studies of this important bacterium, especially its nematicidal factors, are worthwhile. Several factors active against M. incognita have been detected in other nematicidal bacteria26,27,30,31,32, including Prn, 2,4-DAPG, HCN, and alkaline metalloproteinase AprA. To detect such factors and identify their functions in P. putida 1A00316, we performed a series of biological experiments and bioinformatics analyses.
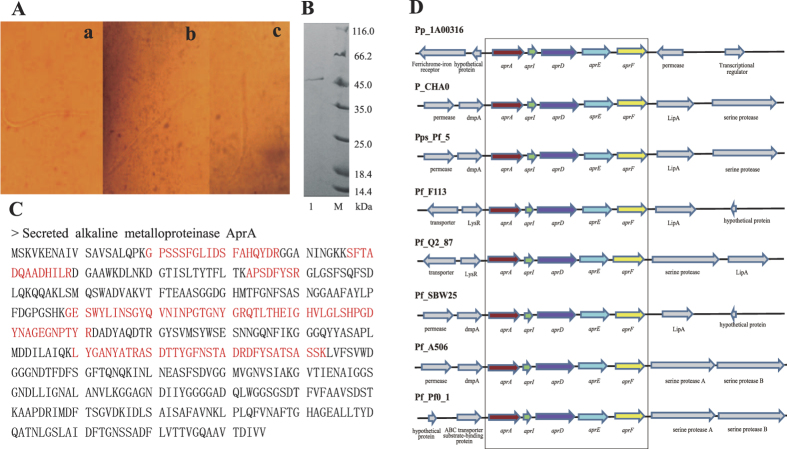
We examined the toxicity of the crude protein extract from P. putida 1A00316 against M. incognita under an inverted microscope. Initially, the cuticles and intestinal tissues of M. incognita were smooth and intact. A vacuole began to form and the intestinal tissue became ambiguous after 12 h, but the body wall was still integrated. After 24 h, the intestinal tissue disappeared completely (Fig. 2A). High temperature and proteinase K inactivated the preparations, confirming that the active fraction was a protein. The crude protein was then purified by ammonium sulfate precipitation, Sephadex G75 chromatography, cation-exchange CM52 chromatography and assayed for nematicidal activity and protein content. After the cation-exchange CM52 column step, we observed a mortality rate of the active fraction of 41.6%. SDS-PAGE showed a prominent band of approximately 48.0 kDa (Fig. 2B), in reasonable agreement with the previously determined molecular mass of 49.9 kDa for alkaline metalloprotease AprA from a different P. fluorescens strain16.
Figure 2. Characterization of the alkaline metalloproteinase AprA in P. putida 1A00316.
(A) The light microscope results for M. incognita processed with the P. putida 1A00316 protein. ‘a’, ‘b’ and ‘c’ processed with the nematicidal protein for 0 h, 12 h and 24 h respectively. (B) The purified fraction exhibiting nematicidal activity after Sephadex G75 chromatography was assayed by SDS-PAGE. M represents the protein marker, line 1 represents the P. putida 1A00316 purified fraction exhibiting nematicidal activity after Sephadex G75 chromatography. (C) Protein bands detected by MALDI-TOF MS. The six red amino acid fragments were peptides, and can all be matched to a protein annotated alkaline metalloproteinase AprA. (D) aprA gene cluster in different Pseudomonas strains.
The 48-kDa band identified by SDS-PAGE was excised and analyzed by MALDI-TOF MS. Six peptides were detected for the protein, all of which matched to the gene encoding alkaline metalloproteinase AprA in P. putida 1A00316 genome (Fig. 2C). We detected the aprA gene cluster in all of the selected P. fluorescens strains, but only in P. putida 1A00316 within the P. putida group (Fig. 2D), suggesting that AprA protein may have an important function in this peculiar bacterium. The upstream and downstream genes of aprA gene cluster were conserved in the P. fluorescens strains, but were quite different in P. putida 1A00316. We detected genes encoding a ferrochrome-iron receptor and a hypothetical protein upstream of the aprA gene cluster in P. putida 1A00316, and genes encoding a permease and a transcriptional regulator downstream of the cluster. In addition to the comparative analysis of the genetic organization of the region surrounding aprA gene cluster, we compared AprA sequences between P. putida 1A00316 and the P. fluorescens group strains. Based on the sequence alignment, the identity of AprA protein between P. putida 1A00316 and four nematicidal strains was 70%, and the ZnMc_serralysin-like structural domain and active sites were highly conserved (Supplementary Figure S2). Mutations in AprA and GacS/GacA signal transduction pathway lead to a sharp decline in nematicidal activity in P. fluorescens CHA016, indicating that both are involved in the process of killing M. incognita. Based on the above results, we proved that AprA protein in P. putida 1A00316 contributes to the biocontrol of M. incognita.
Comparative analysis of small-molecule nematicidal factors in Pseudomonas strains
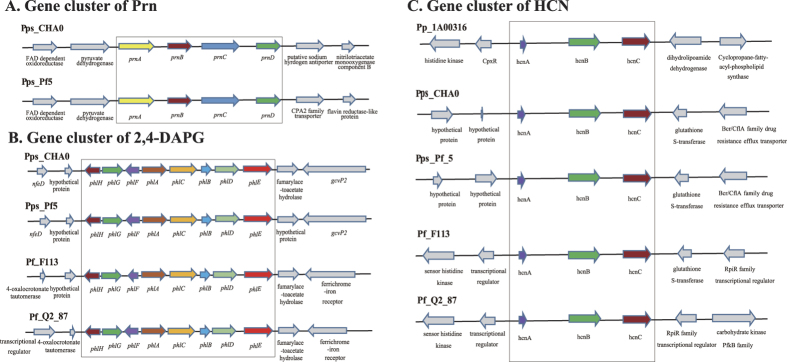
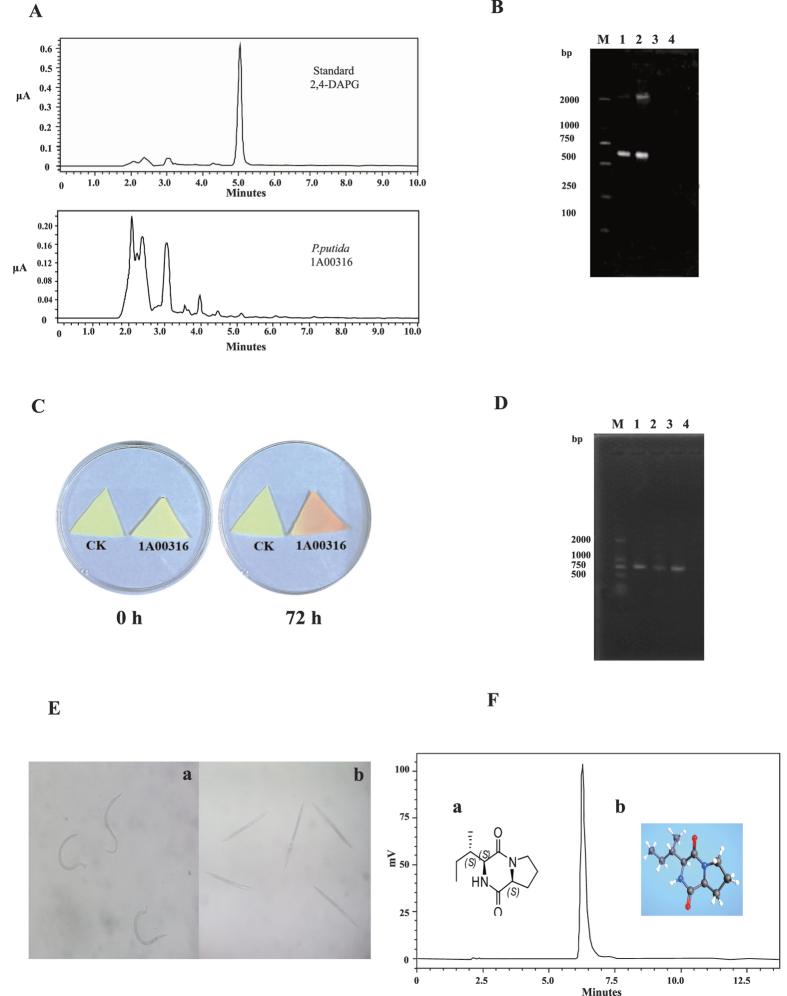
Nematicidal strains typically contain several biocontrol factors. The death of M. incognita is usually not caused by a single factor, but by the synergistic effects of several factors15,33. AprA was not the only nematicidal factor in P. putida 1A00316, and we believe that small molecular metabolites also act as biocontrol factors, such as in other nematicidal bacteria17. Figure 3 summarizes the diversity of small molecular nematicidal factors in the selected bacterial strains examined in this study. We only observed the biosynthetic Prn gene cluster in two nematicidal strains, P. protegens CHA0 and Pf-5 (Fig. 3A), and detected 2,4-DAPG biosynthesis genes in three nematicidal P. fluorescens strains and P. fluorescens Q2–87, but not in P. putida 1A00316 (Fig. 3B). We further compared the retention times of standard 2,4-DAPG and crude extract from the culture supernatant of P. putida 1A00316 by high-performance liquid chromatography (HPLC) (Fig. 4A), and confirmed the absence of 2,4-DAPG. We further PCR-amplified the locus of phlD gene in P. putida 1A00316, P. fluorescens CHA0, and P. protegens Pf-534 (Fig. 4B). As shown in Fig. 4A,B, we were unable to amplify phlD in P. putida 1A00316 and had not detected 2,4-DAPG in the crude extract of this bacterium. Based on these analyses, we determined that P. putida 1A00316 could not produce 2,4-DAPG. Accordingly, 2,4-DAPG was not associated with nematicidal activity in this strain.
Figure 3. Genetic organization of the region containing small-molecular-weight nematicidal factors.
(A) Prn gene cluster. (B) Cluster of genes encoding 2,4-DAPG. (C) HCN gene cluster. The genes included in the box are the synthetic gene cluster related to each nematicidal factor, while genes outside the box are located upstream/downstream of each gene cluster.
Figure 4. Experimental analysis of small molecule of nematicidal factors in P. putida 1A00316.
(A) HPLC chromatogram of standard 2,4-DAPG and crude extracts of P. putida 1A00316 fermentation broth. (B) PCR amplification of the phlD gene fragment. M represents DL 2000, lines 1–3 represent the DNA of P. fluorescens CHA0, P. fluorescens Pf-5, and P. putida 1A00316, respectively, and line 4 is the water control. (C) HCN detection with biochemical methods. HCN produced by P. putida 1A00316 in microtiter plates with indicator paper at 0 and 72 h. Yellow paper indicated that P. putida 1A00316 could not produce HCN, while orange indicated that it could produce HCN. (D) PCR amplification of the hcnB gene fragment. M represents the marker of DL 2000, lines 1–3 represent the DNA of P. fluorescens CHA0, P. fluorescens Pf-5, and P. putida 1A00316, respectively, and line 4 is the water control. (E) The light microscopy results for M. incognita treated with the small metabolite cyclo-(l-Ile-l-Pro) produced by P. putida 1A00316. ‘a’ and ‘b’ treated by cyclo-(l-Ile-l-Pro) for 0 h and 24 h respectively. (F) Structures of cyclo-(l-Ile-l-Pro) and HPLC results for purified cyclo-(l-Ile-l-Pro) from P. putida 1A00316. ‘a’ and ‘b’ represent the two-dimensional and three-dimensional structures of cyclo-(l-Ile-l-Pro), respectively.
However, we did detect hcn biosynthetic genes in the P. putida 1A00316 genome, and these loci were virtually identical in sequence and organization to those of four other nematicidal strains (Fig. 3C). The sequences of genes located upstream and downstream of the hcn gene cluster in four P. fluorescens strains were highly similar or identical but differed from those present in P. putida 1A00316, in which the two-component system response regulator CpxR and a signal transduction histidine kinase were located upstream and dihydrolipoamide hydrogenase and cyclopropane-fatty-acyl-phospholipid synthase were downstream of the hcn gene cluster. We also confirmed the presence of hcn gene cluster in P. putida 1A00316 genome using a biochemical experiment. As shown in Fig. 4C,D, we detected a color change from yellow to orange on the picric acid indicator paper after 72 h, indicating the production of HCN by 1A00316, and we successfully PCR-amplified the gene from the strain. According to previous studies, HCN is toxic to Caenorhabditis elegans35 and M. incognita36, suggesting that HCN contributes to the biocontrol of M. incognita in P. putida 1A00316 as well as other nematicidal Pseudomonas strains.
A nematicidal factor in P. putida 1A00316 is a small molecular metabolite, cyclo-(l-Ile-l-Pro)
We observed substantial nematicidal activity in n-butyl alcohol extracts from the culture filtrate of P. putida 1A00316 (Fig. 4E), and the samples were loaded for HPLC. Finally, the chromatogram of P24-P36 was eluted and we detected a single peak at 210 nm with a retention time of 6.0 min (Fig. 4F).
Based on an electron ionization mass spectrometry (EI/MS) analysis, we confirmed that the compound had a molecular ion at m/z 211.1449 with a formula of C11H18N2O2. We also determined its two-dimensional and three-dimensional structures (Fig. 4F). Based on the characteristics of 1H nuclear magnetic resonance (NMR) spectrum and other two-dimensional NMR spectra (Table 2), the compound was cyclic dipeptide cyclo-(l-Ile-l-Pro) (Fig. 4F). Cyclic dipeptides (also known as 2,5-dioxopiperazines) are formed by two amino acids with peptide bond cyclization, which are the smallest cyclo(peptide) in nature and can be formed by alpha amino acids (often l-amino acids) to generate a relatively stable six-member ring, and acts as an important pharmacophore in medicinal chemistry. A variety of cyclic dipeptides with different compositions and structures have been discovered. Generally, cyclo(dipeptide)s and their corresponding synthases differ among species. For example, Streptomyces noursei37, Mycobacterium tuberculosis H37Rv38, and Staphylococcus haemolyticus JCSC143539 have cyclo-(l-Phe-l-Leu) (synthase, gi: 323463044), cyclo-(l-Tyr-l-Tyr) (synthase, gi: 614130859), and cyclo-(l-Leu-l-Leu) (synthase, gi: 123658993), respectively. Interestingly, though some species share the same cyclo(dipeptide)s, their biosynthetic pathways are highly different39,40,41. We compared the synthases responsible for cyclo(dipeptide)s in other bacteria to all the encoding genes of P. putida 1A00316, and observed a very low amino acid sequence identity, suggesting that the synthases in P. putida 1A00316 are unique. Although the compound cyclo-(l-Ile-l-Pro) has been reported previously and shows antimicrobial activity against Vibrio anguillarum (minimal inhibitory concentration: 0.03 to 0.03 μg/mL)42, its nematicidal activity had not reported previously.
Table 2. NMR results for the nematicidal compound in P. putida 1A00316.
| Peak | 1H | Multiplicity | J-value | C13 |
|---|---|---|---|---|
| 1 | – | – | – | 172.4 |
| 2 | 4.2 | m | – | 60.2 |
| 3 | 2.33 | m | – | 29.8 |
| 1.93 | m | – | ||
| 4 | 1.94 | m | 23.3 | |
| 2.03 | m | |||
| 5 | 3.55 | m | – | 46.4 |
| 1’ | – | – | – | 167.7 |
| 2’ | 4.07 | t | 2.21 | 61.6 |
| 3’ | 2.16 | m | – | 37.4 |
| – | ||||
| 4’ | 1.32 | m | – | 25.7 |
| 1.44 | m | – | ||
| 5’ | t | 7.46 | 12.6 | |
| 6’ | d | 7.18 | 15.7 |
To comprehensively analyze nematicidal activity, we treated M. incognita with various concentrations of cyclo-(l-Ile-l-Pro) (0.13, 0.33, and 0.50 mg/mL). As summarized in Table 3, as the cyclo-(l-Ile-l-Pro) concentration and incubation time increased, the mortality of stage 2 juveniles (J2) increased.
Table 3. Nematicidal activity for cyclo-(l-isoleucyl-l-proline) at different concentrations.
| Concentration (μg/mL) | Mortality (%) |
||
|---|---|---|---|
| 48 h | 72 h | 96 h | |
| 130 | 12.07 ± 2.12 | 28.98 ± 4.03 | 29.90 ± 1.34 |
| 330 | 28.01 ± 4.76 | 34.17 ± 1.52 | 43.00 ± 2.86 |
| 500 | 37.50 ± 2.29 | 46.19 ± 2.76 | 50.00 ± 3.03 |
| CK | 2.05 ± 0.36 | 5.70 ± 0.76 | 7.90 ± 1.02 |
CK (control check): sterile water.
Conclusions
P. putida strain 1A00316 was isolated from Antarctica and displayed distinct traits compared with other P. putida strains. In a comparative genomic analysis of 16 Pseudomonas genomes, we observed high diversity of nematicidal factors. P. putida 1A00316 and P. fluorescens F_113 had three nematicidal factors, P. protegens strains CHA0 and Pf-5 had five nematicidal factors, and the other strains did not have nematicidal factors. We have not detected common nematicidal factors such as 2,4-DAPG and Prn in P. putida 1A00316, but we detected the synthetic gene clusters encoding for HCN and AprA, which are related to nematicidal activity. The expression of these two factors was detected using biochemical methods, and nematicidal activity of AprA protein was confirmed. In addition, we isolated and characterized a new nematicidal factor cyclo-(l-Ile-l-Pro) in P. putida 1A00316. Our study combines a comparative genomic analysis with functional identification and active substance analyses to explore the diversity and specificity of nematicidal factors in P. putida strain 1A00316, and also provides a new nematicidal factor for the control of M. incognita population.
Material and Methods
P. putida 1A00316 genome sequence and CDS annotation
The P. putida 1A00316 genome was sequenced using the PacBio single molecule real-time (SMRT) technology (Wuhan Institute of Biotechnology, Wuhan, China)43. A 3 ~ 20 kb genomic DNA library was prepared suitable for P6/C4 chemistry. Using one SMRT cell on the PacBio RSII sequencing platform, 108,842 reads with a mean read length of 7,363 bp were obtained. The reads were assembled using Hierarchical Genome Assembly Process 3 (HGAP3) within the SMRT Analysis version 2.3.0 software with the default parameters44. The structure and functional annotation of protein-coding genes, including tRNA genes, were predicted using the RAST automatic annotation pipeline45. rRNA genes were identified with RNAmmer46.
Selection and characterization of Pseudomonas strains
Sixteen Pseudomonas strains were selected for a comparative genome analysis based on their reported biological control properties. The newly sequenced P. putida 1A00316 was selected based on its extreme habitat and ability to kill M. incognita. The other 15 Pseudomonas strains were selected, in part, owing to their complete genome sequences and annotation information. In addition, among the seven P. fluorescens group strains, P. fluorescens F_113, P. protegens CHA0, and P. protegens Pf-5 are effective biological control agents against M. incognita. The genome sequences and annotation information for the 15 Pseudomonas strains were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/, Table 1).
Phylogenetic tree construction
A phylogenetic tree of 16 Pseudomonas strains was generated based on the concatenated sequences of 16S rRNA and five conserved housekeeping genes (dnaE, guaA, gyrB, recA, and rpoB) for each strain, and a multi-sequence alignment was generated using MUSCLE47. Gblocks was employed to identify conserved regions from which to generate the phylogenetic tree48. The phylogeny was constructed using MrBayes (GTR Substitution model, mcmc method, discard first 250 trees sampled) and visualized using TreeDyn49. In addition, 2,408 conserved single-copy proteins common to the 16 Pseudomonas genomes were identified with orthoMCL. After concatenation, cluastal omega50 was used to generate a multi-sequence alignment for these single-copy proteins and a phylogenetic tree was generated using Phylip (version 3.696) based on the maximum likelihood method.
Identification of virulence factors
Virulence factors were identified based on the core dataset in VFDB database using BLASTP with an E-value cut-off of 1e−5 and an identity threshold of 60%28.
Detection of protein families
An all-versus-all BLASTP search was used to identify similar proteins. After filtering proteins with low sequence quality (The length of these proteins is less than 10 and the percentage of stop codon is higher than 20%), homologous proteins and clustered protein families were identified using orthoMCL with the default parameters29.
Purification and identification of a nematicidal factor as the alkaline metalloproteinase AprA
P. putida 1A00316 fermentation liquid (200 mL) was collected and centrifuged at 4225 × g for 15 min at 4 °C. The resultant supernatant was precipitated with ammonium sulfate to 60% saturation with a stirrer (4 °C). The precipitate was suspended in a minimum amount of sodium phosphate buffer (pH 7.4) and dialyzed against 50 mM phosphate buffer at 4 °C for 24 h with a stirrer.
The effect of temperature on nematicidal activity was examined after boiling the crude protein for 60 min, and cooling to room temperature. The effect of proteinase K was determined by mixing the crude protein with proteinase K (1 mg/mL) at 40 °C (pH 8.0). As a control, the crude protein was inactivated by heating at 100 °C and mixed with protease. Finally, the activity was quantitatively assayed as described below.
To further purify the crude protein, the solution was filtered through a membrane (0.22 μm), loaded onto a Sephadex G75 column connected to an ultraviolet detector, and eluted with ultrapure water. Fractions were collected and dried using a vacuum freeze dryer and then assayed by SDS-PAGE and by a nematicidal activity test as described below. Fractions with nematicidal activity were then loaded onto a CM52 column connected to an ultraviolet detector and the column was eluted with 10 mM phosphate buffer (pH 7.5) containing 0.5 M NaCl. The fractions were collected, dried as above, and then assayed again by SDS-PAGE and for nematicidal activity.
Nematicidal activity bioassays in vitro
To examine nematicidal activity in vitro, 200 μL crude filtrate or fractions of the eluant from Sephadex or CM52 column were transferred to 96-well plates and then the wells were filled with freshly hatched juvenile suspension (approximately 30 M. incognita/μL). Each treatment was replicated three times. The plates were incubated at 20 °C for 48 h, and dead M. incognita was counted after exposure under an inverted microscope. M. incognita was considered dead when no movement was observed for 2 s after mechanical touching with a needle. The percentages of dead nematodes observed were corrected by eliminating the natural death in a negative control according to the Schneider-Orelli formula51.
SDS-PAGE and MALDI-TOF MS
SDS-PAGE was performed with the Mini-PROTEAN III Gel System (Bio-Rad, Hercules, CA, USA) using 1.0-mm-thick slab gels of 12% (v/v) polyacrylamide. The proteins were stained with Coomassie Brilliant Blue G-250. Protein bands identified by Coomassie Brilliant Blue were excised and subjected to MALDI-TOF MS analyses.
Analysis of key genes encoding 2,4-DAPG and HCN in P. putida 1A00316
The retention times of 2,4-DAPG in crude extracts of P. putida 1A00316 were compared by HPLC (Shimadzu, Kyoto, Japan). In addition, the phlD gene encoding 2,4-DAPG was PCR-amplified. The forward primer for phlD was 5′-ACCCACCGCAGCATCGTTTATGAGC-3′ and the reverse primer was 5′-CCGCCGGTATGGAAGATGAAAAAGTC-3′. The key gene encoding HCN was also confirmed by PCR amplification. The forward primer for the gene was 5′-GCCTGCTCGTTCAACCGTA-3′ and the reverse primer was 5′-CGCAGCCAGCCCACGTC-3′.
Biochemical identification of HCN from P. putida 1A00316
HCN was detected according to the method in reference52. The indicator solution contained 38.46 mL of saturated picric acid solution (1.3%) and 61.54 mL of sodium carbonate solution (3.25%), mixed evenly. Filter paper was cut into triangles, dried after sterilization, and immersed in the indicator solution until saturated. After drying, the paper was used as a HCN indicator. P. putida 1A00316 was inoculated onto a Petri dish, the indicator paper was placed on the inside of the Petri dish cover, and the Petri dish was sealed. A Petri dish without P. putida 1A00316 was regarded as a negative control. After culturing at 28 °C for 3 to 4 days, the color of the indicator paper was observed. If the color of the indicator paper changed from yellow to orange or red, it was confirmed that P. putida 1A00316 can produce HCN.
Extraction and purification of a new nematicidal compound in P. putida 1A00316
P. putida 1A00316 was cultivated for 48 h at 28 °C with shaking (180 rpm) in 500 mL Erlenmeyer flasks containing 200 mL of 2216E medium. The fermentation was centrifuged at 4225 × g for 10 min at 4 °C. The supernatant was extracted with equal volumes (100 mL) of n-butyl alcohol three times. The organic phase was evaporated, and the residue was dissolved in 2 mL of methanol and filtered through a 0.22 μm filter.
The crude extract was analyzed by chromatography using a silica gel column (5 × 120 cm) containing 160 g silica gel (200–300 mesh) eluted with a stepwise ethyl acetate/MeOH gradient of increasing polarity. The fractions were monitored by thin-layer chromatography (TLC, ethyl acetate/MeOH 6:1, spraying with iodine), and similar fractions were combined and tested against J2 M. incognita. Fractions that showed high nematicidal activity were loaded on a silica gel column, eluted with CH2Cl2:MeOH at various ratios (80:1, 50:1, and 30:1), screened by TLC (CH2Cl/MeOH 15:1, spraying with iodine) and again tested against J2 M. incognita. The active fraction was further purified by column (90 × 1.8 cm) chromatography on a Sephadex LH-20 (stepwise gradient of 50–100% methanol).
The supernatant was filtered with a 0.22 μm filter, and 10 μL samples were loaded for HPLC analysis by injection onto an Agilent TC-C18 column (4.6 × 250 mm, Santa Clara, CA, USA). Elution was performed with acetonitrile/sterile distilled water (SDW) (2:8, v/v). A variable-wavelength recorder was set at 210 nm to detect the compounds eluted from the column at a flow rate of 1.1 mL min−1.
Identification of a new nematicidal compound in P. putida 1A00316
The chemical structures of isolated compounds were determined by electrospray ionization mass spectrometry (ESI) and 1H and 13C NMR. Chromatographic separation was performed on an Agilent 6540 UHD Accurate-Mass Q-TOF LC/MS, and chromatographic analysis was achieved on a C18 column (particle size 5 mm, 100 × 2.1 mm, Agilent Technology) with an injection volume of 1 μL. The mobile phase was acetonitrile/SDW (2:8, v/v) at a flow rate of 0.3 mL/min. Approximately 5 mg of the purified compound was dissolved in methanol-d4 (CD3OD) and subjected to a spectral analysis. NMR spectra were recorded on a Bruker DRX 500 NMR instrument, operated at 500 MHz for 1H NMR, and 125 MHz for 13C NMR, both at room temperature. 1H and 13C NMR assignments were supported by 1H-1H correlation spectroscopy (COSY), heteronuclear multiple-quantum coherence (HMQC), nuclear Overhauser effect spectrometry (NOESY), and heteronuclear multiple-bond correlation (HMBC) experiments.
Determination of the absolute configuration of the nematicidal compound in P. putida 1A00316
The absolute configurations of the amino acids in compounds were determined using Marfey’s FDAA (1-fluoro-2, 4-dinitrophenyl-5-l-alanine amide) derivatization method53. A sample (1 mg) of the compound was heated with 0.1 mL of 6N HCl at 120 °C for 20 h. The hydrolysate was evaporated until dry and dissolved in H2O. The retention times of all derivatives obtained from the hydrolysis of test compounds were compared with the derivatized standard d-amino and l-amino acids.
Additional Information
How to cite this article: Guo, J. et al. Comparative genomic and functional analyses: unearthing the diversity and specificity of nematicidal factors in Pseudomonas putida strain 1A00316. Sci. Rep. 6, 29211; doi: 10.1038/srep29211 (2016).
Supplementary Material
Acknowledgments
We thank Marine Culture Collection of China (MCCC) for providing strain Pseudomonas putida 1A00316. This work was supported by grants (2013CB127504 and 2015CB150506) from the National Basic Research Program of China (973), the National Natural Science Foundation of China (31271406) and the program for New Century Excellent Talents in University (NCET-13-0807).
Footnotes
Author Contributions J.Z. and L.-L.C. conceived the project and designed the experiment. J.G., W.-L.P. and R.-R.W. designed and performed the molecular modeling and comparative genome analysis. X.J. and Q.N. designed and performed nematicidal protein and small molecules isolation, structure identification and function analysis. Y.Z., Z.S., L.Z., M.C., G.L., H.Z., Z.Z., D.H., W.C. and Z.Y. take part in the different experiments and analyzed the experimental data. All authors contributed to writing and review of the manuscript. All authors read and approved the final manuscript.
References
- Nakazawa T. Travels of a Pseudomonas, from Japan around the world. Environ. Microbiol. 4, 782–786 (2002). [DOI] [PubMed] [Google Scholar]
- Taghavi S. et al. Horizontal gene transfer to endogenous endophytic bacteria from poplar improves phytoremediation of toluene. Appl. Environ. Microb. 71, 8500–8505 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E. N., Cho M. C., Kim Y., Kim C. K. & Lee K. Expansion of growth substrate range in Pseudomonas putida F1 by mutations in both cymR and todS, which recruit a ring-fission hydrolase CmtE and induce the tod catabolic operon, respectively. Microb. 149, 795–805 (2003). [DOI] [PubMed] [Google Scholar]
- Rosson R. A. & Nealson K. H. Manganese binding and oxidation by spores of a marine Bacillus. J. Bacteriol. 151, 1027–1034 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K. E., Weinel C. & Paulsen I. T. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4, 799–808 (2002). [DOI] [PubMed] [Google Scholar]
- Wu X. et al. Comparative genomics and functional analysis of niche-specific adaptation in Pseudomonas putida. FEMS Microbiol. Rev. 35, 299–323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghavi S. et al. Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar trees. Appl. Environ. Microbiol. 75, 748–57 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. et al. Isolation and characterisation of rhizosphere bacteria active against Meloidogyne incognita, Phytophthora nicotianae and the root knot-black shank complex in tobacco. Pest Manag. Sci. 71, 415–422 (2015). [DOI] [PubMed] [Google Scholar]
- Kayani M. Z., Mukhtar T. & Hussain M. A. Evaluation of nematicidal effects of Cannabis sativa L. and Zanthoxylum alatum Roxb. against root-knot nematodes, Meloidogyne incognita. Crop Prot. 39, 52–56 (2012). [Google Scholar]
- Giannakou I. O., Karpouzas D. G. & Prophetou-Athanasiadou D. A novel non-chemical nematicide for the control of root-knot nematodes. Appl. Soil Ecol. 26, 69–79 (2004). [Google Scholar]
- Ntalli N. G. & Caboni P. Botanical nematicides: a review. J. Agric. Food Chem. 60, 9929–9940 (2012). [DOI] [PubMed] [Google Scholar]
- Schneider S. M. et al. Research on alternatives to methyl bromide: pre-plant and post-harvest. Pest Manag. Sci. 59, 814–826 (2003). [DOI] [PubMed] [Google Scholar]
- Gross H. & Loper J. E. Genomics of secondary metabolite production by Pseudomonas spp. Nat. Prod. Rep. 26, 1408–1446 (2009). [DOI] [PubMed] [Google Scholar]
- Siddiqui I. A. & Shaukat S. S. Suppression of root-knot disease by Pseudomonas fluorescens CHA0 in tomato: importance of bacterial secondary metabolite, 2, 4-diacetylpholoroglucinol. Soil Biol. Biochem. 35, 1615–1623 (2003). [Google Scholar]
- Neidig N., Paul R. J., Scheu S. & Jousset A. Secondary metabolites of Pseudomonas fluorescens CHA0 drive complex non-trophic interactions with bacterivorous nematodes. Microb. Ecol. 61, 853–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui I. A., Haas D. & Heeb S. Extracellular protease of Pseudomonas fluorescens CHA0, a biocontrol factor with activity against the root-knot nematode Meloidogyne incognita. Appl. Environ. Microbiol. 71, 5646–9 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi M. et al. Pyrrolnitrin and Hydrogen Cyanide Production by Pseudomonas chlororaphis Strain PA23 Exhibits Nematicidal and Repellent Activity against Caenorhabditis elegans. PLoS One 10, e0123184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper J. E. et al. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 8, e1002784 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. Z. et al. Genomic analysis of Pseudomonas putida: genes in a genome island are crucial for nicotine degradation. Sci. Rep. 2, 377 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki R. et al. Comparative genome analysis of Pseudomonas knackmussii B13, the first bacterium known to degrade chloroaromatic compounds. Environ. Microbiol. 17, 91–104 (2014). [DOI] [PubMed] [Google Scholar]
- Shen X., Hu H., Peng H., Wang W. & Zhang X. Comparative genomic analysis of four representative plant growth-promoting rhizobacteria in Pseudomonas. BMC Genomics 14, 271 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen R. L. et al. Unearthing the genomes of plant-beneficial Pseudomonas model strains WCS358,WCS374 and WCS417. BMC Genomics 16, 539 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J. P. et al. Mechanism of antagonistic bacteria Pseudomonas putida 1A00316 from the South Pole soil against Meloidogyne incognita. Chin. J. Appl. Environ. Biol. 20, 1046–1051 (2014). [Google Scholar]
- Ronquist F. et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangera M. G. & Thomashow L. S. Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2,4-diacetylphloroglucinol from Pseudomonas fluorescens Q2–87. J. Bacteriol. 181, 3155–3166 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimecombe M. J., De Leij F. A. & Lynch J. M. Effect of Introduced Pseudomonas fluorescens Strains on Soil Nematode and Protozoan Populations in the Rhizosphere of Wheat and Pea. Microb. Ecol. 38, 387–397 (1999). [DOI] [PubMed] [Google Scholar]
- Loper J. & Gross H. Genomic analysis of antifungal metabolite production by Pseudomonas fluorescens Pf-5. Eur. J. Plant Pathol. 119, 265–278 (2007). [Google Scholar]
- Chen L., Xiong Z., Sun L., Yang J. & Jin Q. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 40, D641–645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C. J. Jr. & Roos D. S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers J. M., De Bruijn I., Nybroe O. & Ongena M. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol. Rev. 34, 1037–1062 (2010). [DOI] [PubMed] [Google Scholar]
- Khan Z. et al. A plant growth promoting rhizobacterium, Paenibacillus polymyxa strain GBR-1, suppresses root-knot nematode. Bioresour Technol. 99, 3016–3023 (2008). [DOI] [PubMed] [Google Scholar]
- Oka Y. J., Chet I. & Spiegel Y. Control of the root-k not nematode Meloidogyne javanica by Bacillus cereus. Biocontrol Sci. Technol. 3, 115–126 (1993). [Google Scholar]
- Tian B., Yang J. & Zhang K. Q. Bacteria used in the biological control of plant-parasitic nematodes: populations, mechanisms of action, and future prospects. FEMS Microbiol. Ecol. 61, 197–213 (2007). [DOI] [PubMed] [Google Scholar]
- Mavrodi O. V. et al. Genetic Diversity of phlD from 2,4-Diacetylphloroglucinol-Producing Fluorescent Pseudomonas spp. Phytopathology 91, 35–43 (2001). [DOI] [PubMed] [Google Scholar]
- Gallagher L. A. & Manoil C. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183, 6207–6214 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui I. A., Shaukat S. S., Sheikh I. H. & Khan A. Role of cyanide production by Pseudomonas fluorescens CHA0 in the suppression of root-knot nematode, Meloidogyne javanica in tomato. World J. Microb. Biot. 22, 641–650 (2006). [Google Scholar]
- Sauguet L. et al. Cyclodipeptide synthases, a family of class-I aminoacyl-tRNA synthetase-like enzymes involved in non-ribosomal peptide synthesis. Nucleic Acids Res. 39, 4475–4489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetting M. W., Hegde S. S. & Blanchard J. S. The structure and mechanism of the Mycobacterium tuberculosis cyclodityrosine synthetase. Nat. Chem. Biol. 6, 797–799 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi F. et al. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187, 7292–7308 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondry M. et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond–forming enzymes. Nat. Chem. Biol. 5, 414–420 (2009). [DOI] [PubMed] [Google Scholar]
- Bonnefond L. et al. (2011) Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog. Proc. Natl. Acad. Sci. USA. 108, 3912–3917 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fdhila F., Vázquez V., Sánchez J. L. & Riguera R. dd-diketopiperazines: Antibiotics active against Vibrio anguillarum isolated from marine bacteria associated with cultures of Pecten maximus. J. Nat. Prod. 66, 1299–1301 (2003). [DOI] [PubMed] [Google Scholar]
- Li Y., Li Q., Li X., Song J. & Sun C. Complete chloroplast genome sequence of Fritillaria unibracteata var. wabuensis based on SMRT Sequencing Technology. Mitochondrial DNA 15, 1–2 (2015). [DOI] [PubMed] [Google Scholar]
- Liao Y. C., Lin S. H. & Lin H. H. Completing bacterial genome assemblies: strategy and performance comparisons. Sci. Rep. 5, 8747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K. et al. The RAST Server: rapid annotations using subsystems technology. BMC genomics 9, 75 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K. et al. RNAmmer, consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasar A., Mulet M., Lalucat J. & García-Valdés E. PseudoMLSA: a database for multigenic sequence analysis of Pseudomonas species. BMC Microbiol. 10, 118 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G. & Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577 (2007). [DOI] [PubMed] [Google Scholar]
- Chevenet F., Brun C., Bañuls A. L., Jacq B. & Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC bioinformatics 7, 439 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F. & Higgins D. G. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 1079, 105–116 (2014). [DOI] [PubMed] [Google Scholar]
- Ntalli N. G., Ferrari F., Giannakou I. & Menkissoglu-Spiroudi U. Synergistic and antagonistic interactions of terpenes against Meloidogyne incognita and the nematicidal activity of essential oils from seven plants indigenous to Greece. Pest Manag. Sci. 67, 341–351 (2011). [DOI] [PubMed] [Google Scholar]
- Cardoso A. P., Ernesto M., Cliff J., Egan S. V. & Bradbury J. H. Cyanogenic potential of cassava flour: field trial in Mozambique of a simple kit. Int. J. Food Sci. Nutr. 49, 93–99 (1998). [DOI] [PubMed] [Google Scholar]
- Nishanth Kumar S., Dileep C., Mohandas C., Nambisan B. & Ca J. Cyclo (d-Tyr-d-Phe): a new antibacterial, anticancer, and antioxidant cyclic dipeptide from Bacillus sp. N strain associated with a rhabditid entomopathogenic nematode. J. Pept. Sci. 20, 173–185 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.