Abstract
Cotton is the most important textile crop in the world due to its cellulose-enriched fibers. Sucrose synthase genes (Sus) play pivotal roles in cotton fiber and seed development. To mine and pyramid more favorable alleles for cotton molecular breeding, single nucleotide polymorphisms (SNPs) of GhSus family genes were investigated across 277 upland cotton accessions by EcoTILLING. As a result, a total of 24 SNPs in the amplified regions of eight GhSus genes were identified. These SNPs were significantly associated with at least one fiber- or seed-related trait measured in Nanjing, Anyang and Kuche in 2007–2009. Four main-effect quantitative trait nucleotides (QTNs) and five epistatic QTNs, with 0.76–3.56% of phenotypic variances explained by each QTN (PVE), were found to be associated with yield-related traits; six epistatic QTNs, with the 0.43–3.48% PVE, were found to be associated with fiber quality-related traits; and one main-effect QTN and one epistatic QTN, with the PVE of 1.96% and 2.53%, were found to be associated with seed oil content and protein content, respectively. Therefore, this study provides new information for molecular breeding in cotton.
Cotton fiber is not only the most important textile material worldwide, but also the ideal experimental system for studying the mechanism of cell development due to its single-celled profile1. Cotton fibers originate from the epidermal cells of ovules, and its growth and development is a highly regulated process with four distinct, but overlapping, stages: initiation, elongation, secondary wall synthesis, and maturation2. During the development of cotton fiber, the importation and metabolism of sucrose is considered to be a major factor in determining the sink strength of tissues3. In higher plants, sucrose is the major product of photosynthesis, the main form of translocated carbon, and the main substrate of sink metabolism4. Sucrose metabolism and accumulation in plant cells involves two key enzymes: sucrose synthase (Sus, EC 2.4.1.13) and invertase (INV, EC 3.2.1.26). Sucrose synthase catalyzes a reversible reaction, but preferentially cleaves sucrose to bond uridine diphosphate (UDP) into UDP-glucose and fructose5. Sucrose synthase plays important roles in metabolic processes, including starch storing6,7,8,9,10, cellulose synthesis, sugar import11,12, environmental stress responses13,14, and nitrogen fixation, as well as arbuscule maturation and maintenance in mycorrhizal roots of legumes15,16.
Sucrose synthase (Sus) genes have been proposed to be involved in controlling cotton fiber development17. Sus activity is responsible for cellulose synthesis by supplying the UDP-glucose substrate, which is essential for cell wall thickening and cotton fiber cell development18,19,20. Suppressing Sus activity in the ovule epidermis led to a fiberless phenotype and to fewer fiber initials or shrunken or collapsed fibers in those ovules in cotton21. Over-expressing a Sus gene from potato in cotton gave rise to enhanced leaf development, improved early seed development, and promoted fiber elongation22. Thus, a new strategy was proposed to increase cotton fiber yield by improving seed development. As demonstrated in many plants, Sus isoforms are encoded by a multiple gene family. For example, maize and pea each contain three distinct Sus family members6,23. Six different Sus genes were identified in Arabidopsis thaliana, Oryza sativa, Hevea brasiliensis, and Citrus unshiu13,24,25,26. Populus has seven Sus genes27,28. In two diploid cotton species (Gossypium arboreum L. and G. raimondii Ulbr.), eight Sus genes for each species have been found29. The tetraploid cotton, G. hirsutum L., has the largest Sus family to date, containing fifteen Sus genes29. Examination of their expression patterns indicated that Sus gene family members have different expression in various tissues or organs in cotton species. Additionally, their functional analyses and isolations were confined to a limited number of cotton accessions, their diverse functions and sequence variations have not been clarified in natural populations, implying that some favorable alleles are yet to be identified for cotton.
EcoTILLING is a fast and cost-effective method of detecting rare SNPs, small insertions, and deletions (indels) in target genes in natural populations, and was adapted from Targeting Induced Local Lesions in Genomes (TILLING)30. Compared to direct sequencing, EcoTILLING has the following advantages: high-throughput, accuracy, and cost-effectiveness. To date, EcoTILLING has been used for many purposes, including mapping31,32, candidate gene discovery33,34, evaluation of nucleotide diversity35, and population genetics36,37.
In this study, EcoTILLING technology was used to reveal sequence diversities and identify SNPs in GhSus family genes in a natural population of upland cotton accessions. The SNPs were then used for association analysis of phenotype variations in plants from nine environments over three years and three locations, and for the identification of favorable GhSus alleles that confer high yield and high fiber and seed quality traits.
Results
Nucleotide polymorphisms
Fifteen genes in the GhSus family were analyzed by EcoTILLING. As a result, eight out of 15 genes showed a total of 24 putative natural variation sites in the amplified regions across 277 upland cotton accessions, and no SNPs were detected in the other seven genes (Table 1). The minor allele frequency of polymorphic sites in the 277 accessions ranged from 0.018 to 0.755, with an average 0.214. Samples containing each of the polymorphic sites were randomly sequenced to determine the position and identity of these polymorphic sites in the targeted region of genes. Twenty four putative SNPs were confirmed by sequencing in the targeted region of the eight GhSus family members. Of the 24 SNPs, 12 were in the coding regions, including seven synonymous variations and five non-synonymous variations. Of the five non-synonymous variations, two SNPs were predicted by the SIFT program to severely affect protein function (Table 1).
Table 1. List of nucleotide polymorphisms in candidate genes.
| Gene | Nucleotide polymorphisma | Ratiob | Variation of amino acidc | SIFTd |
|---|---|---|---|---|
| GhSus1At | A918G | 0.612 | No | |
| G1783C | 0.342 | No | ||
| A2940T | 0.338 | T678= | ||
| GhSus1Dt | T650C | 0.089 | No | |
| G751A | 0.089 | V153I | 0 | |
| G2709C | 0.107 | E601Q | 0.12 | |
| GhSus3At | T881A | 0.072 | No | |
| T1320C | 0.072 | No | ||
| G1327A | 0.072 | No | ||
| T2294C | 0.018 | L504S | 0 | |
| A2472C | 0.018 | No | ||
| GhSus4Dt | A1886T | 0.025 | No | |
| T2167C | 0.025 | E388= | ||
| GhSus5Dt | G491T | 0.029 | L108F | 1 |
| C2648G | 0.025 | P660A | 1 | |
| GhSus6At | G3143T | 0.159 | No | |
| C4655T | 0.209 | No | ||
| G4697A | 0.209 | W755= | ||
| G5005A | 0.209 | No | ||
| GhSus7Dt | T192C | 0.755 | T64= | |
| C404T | 0.755 | D109= | ||
| T407C | 0.755 | E110= | ||
| GhSus8Dt | C577T | 0.09 | No | |
| T1334C | 0.069 | G298= |
aThe first letter indicates bp at this site in TM-1 sequence, followed by the position of the SNP in the TM-1 sequence, and then the nucleotide that is the rare variant at this site.
bRatio was calculated by dividing the number of similar nucleotide changes identified on the EcoTILLING gel by 277 upland cotton accessions.
cThe first letter indicates the common amino acid at this site, followed by the position of the SNP within the predicated protein sequence and the amino acid change induced by the variant nucleotide polymorphism; “=” means no change in amino acid encoded by that codon (synonymous variation); “No” means that SNP was in intron.
dA non-synonymous SNP is predicted to be damaging to the encoded protein if the SIFT score is <0.05 (bold).
Haplotype diversity analysis
The haplotype distribution for each of the amplicons is shown in Table 2. The GhSus1At amplicon had the largest number of haplotypes in the GhSus gene family, with eight haplotypes detected, while the amplicons GhSus4Dt and GhSus7Dt each had only two haplotypes. The levels of haplotype diversity (HD) varied markedly in different amplicons of the GhSus family. The highest level of HD was in amplicon GhSus1At, with 0.574, while the lowest was in amplicon GhSus4Dt, with only 0.049. One major haplotype was detected in each GhSus gene sequence except for amplicon GhSus1At, for which no haplotype exhibited a frequency higher than 0.5 (Supplementary Table S1).
Table 2. Gene length, number of SNPs, number of haplotypes and haplotype diversity (HD) for the GhSus family genes.
| Gene | Length (bp) | Number of SNP | Number of Haplotype | HD |
|---|---|---|---|---|
| GhSus1At | 3650 | 3 | 8 | 0.574 |
| GhSus1Dt | 3648 | 3 | 4 | 0.290 |
| GhSus3At | 3491 | 5 | 4 | 0.143 |
| GhSus4Dt | 4790 | 2 | 2 | 0.049 |
| GhSus5Dt | 3167 | 2 | 4 | 0.064 |
| GhSus6At | 5176 | 4 | 4 | 0.386 |
| GhSus7Dt | 3428 | 3 | 2 | 0.372 |
| GhSus8Dt | 3762 | 2 | 4 | 0.201 |
Population structure
Analysis of the population structure composed of 277 upland cotton varieties showed that the log probability of data [In P(D)] value corresponding to each hypothetical number of subpopulations (k) continued to increase with k value and did not reach a peak. An ad hoc statistic ∆k value was then calculated according to the method described by Evanno et al.38. The values showed a much higher likelihood at k = 2 than at k = 3–10, suggesting that the whole panel could be divided into two major subpopulations, SP1 and SP2, respectively. The SP1 group contained 89 accessions; 16 from America, three from Europe, seven from North regions of China, 12 from Northwest regions of China, 32 from the Yellow River region of China, 15 from the Yangtze River regions of China, and four from other Asian regions. The SP2 group consisted of 188 accessions; four from Australia, five from Africa, 34 from America, two from Europe, six from North regions of China, 3 from Northwest regions of China (Supplementary Table S2), 101 from the Yellow River region of China, 33 from the Yangtze River regions of China. The corresponding Q matrix at k = 2 was then used for the subsequent association analysis.
Complete linkage disequilibrium of SNPs was detected as follows within five single GhSus genes: GhSus1Dt-T650C and GhSus1Dt-G751A in GhSus1Dt-G751A; GhSus4Dt-A1886T and GhSus4Dt-T2167C in GhSus4Dt; GhSus3At-T881A, GhSus3At-T1320C, and GhSus3At-G1327A in GhSus3At; GhSus3At-T2294C and GhSus3At-A2472C in GhSus3At; GhSus6At-C4655T, GhSus6At-G4697A, and GhSus6At-G5005A in GhSus6At; GhSus7Dt-T192C, GhSus7Dt-C404T, and GhSus7Dt-T407C in GhSus7Dt. All SNPs in complete linkage disequilibrium were combined into a single haplotypes and those were used for further analysis (here, for easy description of the results, only one SNP name from each haplotype was used instead of haplotype). Although ten of those SNPs (for example, GhSus1At-A918G and GhSus1At-G1783C) result in synonymous variations or are located in intron regions, they may be in complete linkage disequilibrium with other non-synonymous mutations, which could have functional relevance to the organism. Therefore, these SNPs were further analyzed. Thus, a total of 15 SNPs were included in the association analysis, namely, three SNPs in GhSus1At (GhSus1At-A918G, GhSus1At-G1783C, and GhSus1At-A2940T), each two SNPs in GhSus1Dt (GhSus1Dt-G751A and GhSus1Dt-G2709C), in GhSus3At (GhSus3At-T881A and GhSus3At-T2294C), in GhSus5Dt (GhSus5Dt-G491T and GhSus5Dt-C2648G), in GhSus6At (GhSus6At-G3143T and GhSus6At-C4655T) and in GhSus8Dt (GhSus8Dt-C577T and GhSus8Dt-T1334C), and each one in GhSus4Dt (GhSus4Dt-A1886T) and in GhSus7Dt (GhSus7Dt-T192C).
Association between SNPs and fiber- or seed-related traits
In this study, eleven SNPs were found to be significantly associated with traits (four SNPs were main-effect QTNs and eleven pairs of SNPs were involved in epistatic QTNs) (Table 3). Of them, six QTNs were associated with fiber quality traits. Three epistatic QTNs (GhSus1At-A918G × GhSus5Dt-C2648G, GhSus1At-A918G × GhSus6At-C4655T, and GhSus1Dt- G751A × GhSus3At-T881A) were associated with fiber length (FL), which explained 3.48%, 1.09%, and 1.50% of the phenotypic variation (PVE), respectively. Two epistatic QTNs (GhSus1Dt-G751A × GhSus8Dt-T1334C and GhSus1Dt-G2709C × GhSus8Dt-C577T) were associated with fiber micronaire (FM), which explained 1.27% and 0.89% of PVE, respectively. One epistatic QTN (GhSus3At-T881A × GhSus7Dt-T192C) was associated with fiber strength (FS), which explained 0.43% of PVE. Nine QTNs were associated with yield traits. One main-effect QTN (GhSus1At-A2940T) was associated with lint percentage (LP), which explained 3.43% of PVE. Two main-effect QTNs (GhSus1At-G1783C and GhSus3At-T881A) and two epistatic QTNs (GhSus1At-A2940T × GhSus1Dt-G2709C and GhSus6At-C4655T × GhSus7Dt-T192C) were associated with boll weight (BW), which explained 3.56% and 1.76%, and 2.03% and 0.76% of PVE, respectively. One main-effect QTN (GhSus3At-T881A) and three epistatic QTNs (GhSus1At-A918G × GhSus1Dt-G2709C, GhSus1At-G1783C × GhSus1Dt- G2709C, and GhSus1Dt -G751A × GhSus1Dt-G2709C) was associated with seed index (SI), which explained 1.32% and 2.10%, 2.27%, and 0.89% of PVE, respectively. One epistatic QTN (GhSus1At-A918G × GhSus1Dt-G2709C) and one main-effect QTN (GhSus6At-C4655T) were associated with protein content (PC) and oil content (OC), which explained 2.53% and 1.96% of PVE, respectively. No significant environmental interactions were found.
Table 3. Association analysis with fiber- or seed- realted traits of SNPs in the GhSus family genes.
| Trait | QTN | LOD | r2(%) | F | P | Favorable allele | |
|---|---|---|---|---|---|---|---|
| Alleles | Effect | ||||||
| LP | GhSus1At-A2940T | 16.59 | 3.43 | 15.16 | 1.01E-04** | GhSus1At-2940T | 0.95% |
| BW | GhSus1At-G1783C | 14.60 | 3.56 | 12.71 | 3.71E-04** | GhSus1At-1783C | 0.18 g |
| GhSus3At-T881A | 15.30 | 1.76 | 77.09 | 3.02E-18** | GhSus3At-881A | 0.23 g | |
| GhSus1At-A2940T × GhSus1Dt-G2709C | 10.64 | 2.03 | 21.42 | 6.03E-10** | GhSus1At-2940T × GhSus1Dt-2709C | 0.14 g | |
| GhSus6At-C4655T × GhSus7Dt-T192C | 8.44 | 0.76 | 33.00 | 1.04E-08** | GhSus6At-4655T × GhSus7Dt-192C | 0.08 g | |
| SI | GhSus3At-T881A | 10.82 | 1.32 | 65.96 | 7.25E-16** | GhSus3At-881A | 0.40 g |
| GhSus1At-A918G × GhSus1Dt-G2709C | 10.98 | 2.10 | 10.83 | 1.01E-03* | GhSus1At-918G × GhSus1Dt-2709C | 0.26 g | |
| GhSus1At-G1783C × GhSus1Dt-G2709C | 17.73 | 2.27 | 28.49 | 5.91E-13** | GhSus1At-1783C × GhSus1Dt-2709C | 0.29 g | |
| GhSus1Dt-G751A × GhSus1Dt-G2709C | 6.58 | 0.89 | 40.02 | 2.98E-10** | GhSus1Dt-751A × GhSus1Dt-2709C | 0.25 g | |
| FL | GhSus1At-A918G × GhSus5Dt-C2648G | 16.53 | 3.48 | 14.62 | 4.90E-07** | GhSus1At-918G × GhSus5Dt-2648G | 0.42 cm |
| GhSus1At-A918G × GhSus6At-C4655T | 7.98 | 1.09 | 31.51 | 2.21E-08** | GhSus1At-918A × GhSus6At-4655C | 0.23 cm | |
| GhSus1Dt-G751A × GhSus3At-T881A | 11.97 | 1.50 | 22.29 | 2.57E-10** | GhSus1Dt-751A × GhSus3At-881A | 0.39 cm | |
| FS | GhSus3At-T881A × GhSus7Dt-T192C | 4.81 | 0.43 | 14.22 | 7.24E-07** | GhSus3At-881A × GhSus7Dt-192C | 0.19 cN/tex |
| FM | GhSus1Dt-G751A × GhSus8Dt-T1334C | 5.15 | 0.89 | 14.56 | 5.17E-07** | GhSus1Dt-751A × GhSus8Dt-1334C | 0.07 |
| GhSus1Dt-G2709C × GhSus8Dt-C577T | 5.11 | 1.27 | 6.72 | 1.23E-03* | GhSus1Dt-2709G × GhSus8Dt-577C | 0.08 | |
| PC | GhSus1At-A918G × GhSus1Dt-G2709C | 9.57 | 2.53 | 11.11 | 1.62E-05** | GhSus1At-918A × GhSus1Dt-2709G | 0.49% |
| OC | GhSus6At-C4655T | 3.67 | 1.96 | 15.21 | 1.04E-04** | GhSus6At-4655C | 0.54% |
* and **at the 0.05 and 0.01 levels of significance on the genome, respectively. LP, lint percentage (LP); BW, boll weight; SI, seed index; FL, fiber length; FM, fiber micronaire; FU, fiber uniformity; FS, fiber strength; PC, protein content; OC, oil content. r2 is the phenotypic variance explained by each QTN.
Using epistatic association analyses, seven pairs of GhSus family genes, namely, GhSus1At × GhSus1Dt (BW, PC, and three for SI), GhSus6At × GhSus7Dt (BW), GhSus1At × GhSus5Dt (FL), GhSus1At × GhSus6At (FL), GhSus1Dt × GhSus3At (FL), GhSus3At × GhSus7Dt (FS), GhSus1Dt × GhSus8Dt (two for FM) were found to have epistatic interactions (Table 3). Among all the seven epistatic interactions, two-way ANOVA was used to further detect the corresponding interactions of these gene haplotypes. As a result, six pairs of gene haplotypes were found to have epistatic interactions except for GhSus3At × GhSus7Dt (FS) (Supplementary Table S3).
Favorable QTN alleles mined in upland cotton accessions
The phenotypic effects of each QTN allele were estimated by the method of Lü et al.39, and a total of 17 desirable alleles were identified. Phenotypic effects for each favorable allele are shown in Table 3. Among the favorable alleles, GhSus3At-881A had the most positive phenotypic effects for BW and SI, and increased BW and SI by 0.23 g and 0.40 g, respectively. One epistatic QTN, between GhSus1At-918G and GhSus5Dt-2648G, increased FL by 0.42 mm; another epistatic QTN, between GhSus1Dt-2709G and GhSus8Dt-577C, decreased FM by 0.08; GhSus1At-2940T increased LP by 0.95%; the epistatic QTN between GhSus3At-881A and GhSus7Dt-192C increased FS by 0.19 cN/tex; the epistatic QTN between GhSus1At-918A and GhSus1Dt-2709G increased PC by 0.49%; and GhSus6At-4655C increased OC by 0.54%.
Expression analysis of GhSus gene at different developmental stages
To better understand the potential functions of GhSus isoforms, their expression patterns were analyzed in fiber and ovule at different developmental stages using upland cotton TM-1 published RNA-seq data40. Most of GhSus genes were expressed (fragments per kilobase of transcript per million reads sequence (FPKM) > 1) in both fiber and ovule except for GhSus4A and GhSus4D which were not expressed in either tissue. The expression levels of GhSus1A and GhSus1D were both high in fiber at 5 DPA (days post-anthesis), then declined notably at 20 DPA (Fig. S1). The transcripts of GhSus5D and GhSus8D showed relatively higher in fiber from 5 to 25 DPA, and were very weak in ovule (Fig. S1).
Discussion
To understand genetic variation and mine more favorable alleles, 277 upland cotton accessions from China and another 5 geographic areas were analyzed for allele diversity in fifteen GhSus family genes by EcoTILLING. The results revealed 24 SNPs among the family genes after analysis of 14,957,446 bp sequences. Of the 24 SNPs, half (12/24) were in coding regions, including seven synonymous variations and five nonsynonymous variations. Of the five nonsynonymous variations, two SNPs were predicted by the SIFT program to severely affect protein function. Although SIFT has a 20% false-positive error and some mutations predicted to be deleterious may be functionally neutral, these scores may be useful in prioritizing mutations for further study and for the analysis of possible contributions of the GhSus family gene members to stress tolerance.
Sus is a key enzyme in plant sucrose metabolism. In many plants, Sus isoforms are encoded by a small multi-gene family13,24. Fifteen Sus genes have been detected in G. hirsutum so far; previous reports showed that some of these genes were related to the initiation, elongation, and secondary cell wall deposition of the single-celled fibers41,42,43,44. However, little is known about the underlying genetics and molecular biology of the Sus genes and their physiological functions during fiber growth and development.
In the present study, eleven SNPs in the GhSus family gene members were significantly associated with at least one agronomic trait. Five main-effect QTNs and twelve epistatic QTNs containing these eleven SNPs were detected for eight traits in upland cotton. Although most of the QTNs were associated with SNPs that are expected to result in synonymous mutations or be located in intron sequences, these SNPs may be in complete linkage disequilibrium with other non-synonymous mutations, which could have functional relevance to the organism, or these SNPs could become the targets of other genes, leading to effects on agronomic traits. All of these are waiting for further experimental confirmation.
Using 2,878 QTLs mapped in a previous study40, we found that only five out of eleven SNPs in our study were located in the same regions as previous reports. For example, the locus GhSus1Dt-G751A associated with FL on chromosome (Chr.) 19 was located in the region of qFL-D5-1 identified by Hu et al.45. The loci GhSus1Dt-G751A and GhSus1Dt-G2709C, both associated with FM on Chr. 19, were located in the region of qFM-C19-1 identified by Sun et al.46. The locus GhSus1Dt-G2709C associated with PC on Chr. 19 was located in the region of qPro2-c19-1 identified by Yu et al.34. The locus GhSus5Dt-C2648G associated with FL on chromosome 25 was located in the regions of qFL-C25-1 and qFL-C25-2 identified by Sun et al.46. The locus GhSus7Dt-T192C associated with BW on chromosome 16 was located in the region of qBW-C16-1 identified by Wu et al.47. The locus GhSus7Dt-T192C was also associated with FS, which was located in the regions of F2:3-qFS-c16-1 and F2:3-qFS-c16-2 identified by Yu et al.48. This comparison demonstrates that our association analysis further confirmed the previous results of QTL mapping suggesting that these QTLs are stable, repeatable, and reliable.
The quality and productivity of cotton fiber is mainly affected by two biological processes: fiber initiation and fiber elongation49. The fiber elongation process (~3–23 days post anthesis, DPA) determines the fiber final length and strength. Fiber strength also depends on formation of the secondary cell wall (~16–40 DPA), which contains >95% of the dry weight of cellulose in the mature cotton fiber. As GhSus genes are vital for fiber cell development, suppression experiments result in a disruption of fiber and seed development43. Due to the severity of the phenotypic effects, it was difficult to analyze the impact of different GhSus family genes on fiber quality from pleiotropic developmental effects on fiber growth. As a reverse genetic research method, EcoTILLING technology provided a new approach for GhSus family gene function analysis.
The expression patterns of GhSus family genes during fiber cell development were analyzed using RNA-seq data from G. hirsutum TM-144. All seven genes associated with the fiber-related traits (Table 3) were highly expressed in fiber cells during the fiber elongation stage or secondary cell wall formation stage. Five GhSus genes (GhSus1At, GhSus1Dt, GhSus5Dt, GhSus6At, and GhSus3At) associated with fiber length were found to be highly expressed during the fiber elongation process (Supplementary Fig. S1). Two genes (GhSus3At and GhSus7At) associated with fiber strength had relatively high expression in the secondary cell wall formation stage (~16–40 DPA) (Supplementary Fig. S1). Sucrose synthase (Susy) is the major enzyme of Sucrose (Suc) hydrolysis to UDP-glucose that could be used as a substrate for cellulose synthesis in cotton (G. hirsutum) fibers50. The combination of GhSus gene expression patterns and QTL analysis in our study further confirmed that GhSus family genes play important roles in fiber development.
Six SNPs (GhSus1At-A918G, GhSus1At-G1783C, GhSus1Dt-G751A, GhSus1Dt-G2709C, GhSus3At-T881A, and GhSus6At-C4655T) were found to be associated with seed-related traits (SI, PC, and OC) in addition to the SNPs associated with fiber-related traits. In a previous study, Sus was found to be abundant in transfer cells located at the innermost layer of the seed coat and in developing filial tissues, where Sus is involved in transfer cell wall ingrowth3 and endosperm cellularization51. Suppressing the expression of Sus in cotton seed coats leads to a fiberless phenotype43, and silencing its expression in the filial tissue results in stunted and unviable seeds and loss of transfer cells43,51. Xu22 overexpressed the potato Sus gene in cotton, and revealed that increased Sus activity in cotton could improve seed development, leading to enhanced seed weight and number. Overexpression of Sus could also enhance sink strength and sucrose supply by expanding leaves22. Jiang42 overexpressed GhSusA1 in transgenic cotton and found this gene increased fiber length and strength. These findings provide an indication that it is valuable to increase fiber yield by enhancing fiber length and seed number. The new GhSus alleles found in our study may be used to increase fiber yield.
Materials and Methods
Plant materials
A set of 277 Upland cotton varieties possessing diverse fiber qualities, kindly provided by the Cotton Research Institute, Chinese Academy of Agricultural Sciences (CRI-CAAS), were subjected to EcoTILLING analysis. Of the 277 upland cotton accessions, four were from Australia, five from Africa, five from Europe, 50 from America, and 213 from Asia. Of the 213 accessions from Asia, two were from Pakistan, two from Vietnam, and 209 from China. The 209 accessions from China were from four geographic regions (the North region, Yellow River region, Yangtze River region, and northwest inland region). Detailed information on all accessions is shown in Supplementary Table S2.
Phenotypic data
All 277 accessions were sown in three consecutive growing seasons (2007–2009) simultaneously at three main ecological cotton-growing areas of China: Nanjing in Jiangsu Province (31°14′~32°37′N, 118°22′~119°14′E; the average annual frost free period lasts 227–278 days, the average active accumulated temperature was 3500–5500 °C, annual rainfall was about 1000–1600 mm; Yangtze River valley cotton growing region), Anyang in Henan Province (35°12′~36°22′N, 113°37′~114°58′E; the average annual frost free period lasts 180–230 days, the average active accumulated temperature was 3800–4900 °C, annual rainfall was about 500–1000 mm; Yellow River valley cotton growing region) and Kuche in Xinjiang Uygur Autonomous Region (40°46′~42°35′N, 82°35′~84°17′E; the average annual frost free period lasts 170–230 days, the average active accumulated temperature was 3000–5400 °C, annual rainfall was about 15–380 mm; Northwest cotton growing region). A randomized complete block design with three replications was employed in the field trials, and each block was settled within a single-row plot at 5 m long and 0.7 m wide. Conventional field production management techniques that were adjusted to local practice were used.
Fiber samples were collected from bolls at the internal middle parts of plants, ginned by roller and sent to the Cotton Quality Supervision, Inspection and Testing Center of the Ministry of Agriculture, China to test fiber length (FL in mm), fiber strength (FS in cN/tex), fiber micronaire (FM), and fiber uniformity (FU in %) using an Uster HVI 900. Four yield traits, namely, lint percentage (LP in %), Seed index (g/100 seeds), boll weight (BW in g), and boll number per plant (BN), were also calculated. Seed kernel protein content (PC in %) and oil content (OC in %) were measured in 277 accessions. Phenotypic scores from field trials were shown in Supplementary Tables S6 and S7.
DNA extraction
Equal quantities of fresh young leaves from each cotton variety were collected and immediately brought to the laboratory where total genomic DNA was extracted according to the protocol described by Paterson et al.52. DNA from all samples was quantified using a spectrophotometer and normalized to a concentration of 20~60 ng/μl.
Designation and verification of subgenome-specific primers for duplicated gene amplification
To amplify target fragments of GhSus gene family sequences, gene-specific primers were designed according to the published DNA sequences of GhSus gene family members29,53 (Supplementary Tables S4 and S5). The expression patterns of GhSus family genes were analyzed from the public RNA-seq data40.
Since cultivated Upland cotton is allotetraploid, where many single nucleotide diversities exist between subgenomes, subgenome-specific primers were designed for GhSus gene family members to avoid errors caused by SNPs derived from duplicated genes in the A- and D-subgenomes. SNAPER software (http://ausubellab.mgh.harvard.edu/) was employed to design the subgenome- specific primers. All primers were evaluated for their subgenome specificity using PCR. Firstly, DNA templates from two diploid species, G. raimondii (D genome) and G. herbaceum var. africanum (A genome), and one tetraploid species, G. hirsutum acc. TM-1 (AD genome), were amplified. The PCR products were then subjected to polyacrylamide gel electrophoresis (PAGE) to verify the primer specificity. When the PCR products only existed in two species, G. herbaceum var. africanum and G. hirsutum acc. TM-1, but not in G. raimondii, the primers were considered to be At subgenome-specific primers. When the PCR products only existed in G. raimondii and G. hirsutum acc. TM-1, but not in G. herbaceum var. africanum, the primers were considered to be Dt-subgenome specific primers. After subgenome-specific primers were acquired, semi-nested PCR primers were designed to further improve the PCR specificity and increase the product concentration.
PCR amplification and EcoTILLING assays
Celery juice extract (CEL I enzyme) was extracted from celery following the method described by Till et al.54. CEL I enzymatic activity was evaluated by the digestion of heteroduplexes formed by control substances C and G, which were both supplied in the Surveyor Mutation Discovery kit. The enzyme digestion conditions followed those described in the Surveyor Mutation Discovery kit protocol (Transgenomics).
To identify nucleotide diversities by EcoTILLING, gene- and subgenome- specific primers were designed based on the GhSus family gene sequences and used in a semi-nested PCR. To avoid base mismatch during PCR, an ExTaq (TAKARA, China) enzyme with high fidelity was used in three replicates. The first PCR was performed with gene- and subgenome- specific primer pairs. The PCR amplified products were then used as templates for the second semi-nested PCR reaction. Both steps of the PCR reaction used the same forward primer but distinct reverse primers (Supplementary Table S6).
After PCR, an EcoTILLING protocol was employed for SNP screening, as described by Kadaru et al.55 with some modifications. First, the semi-nested PCRs were performed, and then the PCR products of each cotton variety were mixed with that of TM-1 and incubated at 99 °C for denaturing 10 min, followed by annealing with 28 cycles of 20 sec at 72 °C with a decrement of 0.3 °C per cycle and then cooling to 42 °C with a decrement of 1 °C per cycle, before finally being cooled to 24 °C for 1 min. Finally, if SNPs existed between the cotton variety and TM-1, base-mismatched DNA heteroduplexes would be formed in the mixture of the two PCR products and could be digested specifically at mismatched base position by endonuclease, CEL I. The digestion of heteroduplexes generated several small-sized DNA bands that were visualized using PAGE, implying that SNPs existed between the cotton variety and TM-1 gene sequences. Otherwise, if no SNPs existed between the cotton variety and TM-1, heteroduplexes would not be formed and could not be digested by CEL I. No small-sized DNA fragments generated. Once a polymorphic band was identified, the corresponding DNA samples were amplified using gene- and subgenome- specific primers. The resulting PCR fragments were directly sequenced for each polymorphic site from at least three accessions to confirm that only two alleles segregated at any specific site.
Nomenclature of SNP
Here, SNP was named as “gene + subgenome + dash + base in TM-1 + base position + base in the accession analyzed. For example, the SNP, GhSus1Dt-G751A, demonstrates that G base on the position of Dt subgenome in TM-1 is substituted by A base in the accession analyzed (Supplementary Table S8).
DNA sequencing and statistical analysis
Once polymorphisms were found for a targeted region of the GhSus family genes by EcoTILLING, accessions that showed polymorphisms on the gel were randomly selected for sequencing by a commercial company (Genscript Biotech Co. China) to confirm the polymorphisms. ClustalX software was used to find the SNP sites in the obtained sequences. The SIFT (Sorting Intolerant from Tolerant) method56 was used to predict deleterious effects of SNPs to protein function. Haplotype diversity (HD) was analyzed using DnaSP v5.157.
Calculation of gene expression levels
Genomic (as reference) and RNA-seq data of upland cotton TM-1 was obtained from the NCBI SRA database (accession codes: PRJNA248163)40. RNA-seq reads were mapped to the TM-1 genome (Version 1.1) using Tophat (Version 2.0.8)58. To measure gene expression level in fiber and ovule, we calculated the expression of each gene using FPKM (Fragments per Kilobase of exon model per Million mapped reads) with Cufflinks (Version 2.1.1)59.
Association analysis between SNPs and agronomic traits
A possible population structure was depicted using 258 simple sequence repeat (SSR) markers by STRUCTURE software version 260. The length of the burn-in period and the number of Markov chain Monte Carlo replications after burn-in were all assigned at 100,000 with an admixture and allele frequencies correlated model. The hypothetical number of subpopulations (k) ranged from 1 to 10 was performed at five independent run iterations. The log probability of data from the STRUCTURE output was used to estimate the k according to the method described by Evanno et al.38. Based on the k, the population structure matrix (Q) was generated for further marker-trait association analysis.
The tentative association between nucleotide variations in the Sus gene family with agronomic traits was performed by an epistatic association mapping (EAM) approach proposed by Lü et al.39. The genetic model extended was as follows:
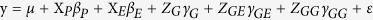 |
where y is the phenotypic value of agronomic traits, Xp is the Q matrix for the population structure, XE, ZG, ZGE, and ZGG are the design matrices of the environment effect, QTL effect, QTL-by-environment interaction effect, and QTL-by-QTL interaction effect, respectively, and βP, βE, γG, γGE, and γGG are the corresponding effects. Note that γG includes additive and dominant effects, γG includes additive-by-environment and dominant-by-environment effects, and γGG includes additive-by-additive, additive-by-dominant, dominant-by-additive and dominant-by-dominant effects. All the effects in the above model were estimated by empirical Bayes61. Results from Monte Carlo simulation studies showed that these estimates were unbiased61,62,63. The significance threshold of the logarithm of odds (LOD) score was set at 3, which is equivalent to the significance level of 0.000263.
Additional Information
How to cite this article: Zeng, Y.-D. et al. EcoTILLING revealed SNPs in GhSus genes that are associated with fiber- and seed-related traits in upland cotton. Sci. Rep. 6, 29250; doi: 10.1038/srep29250 (2016).
Supplementary Material
Acknowledgments
This work was financially supported in part by the National Natural Science Foundation of China (30971824), the National Key Technology Support Program of China during the Twelfth Five-year Plan Period [grant number 2013BAD01B03-04], Jiangsu Collaborative Innovation Center for Modern Crop Production. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions B.-L.Z., X.-M.D. and T.-Z.Z. conceived and designed the experiments, Y.-D.Z. and J.-L.S. performed the experiments, Y.-D.Z., S.-H.B. and Y.-M.Z. analyzed the data, Y.-D.Z., K.-S.D. and T.T. contributed reagents/materials/analysis tool, J.-L.S. performed the phenotype investigation, B.-L.Z. and Y.-D.Z. wrote the manuscript.
References
- Ruan Y. L. Rapid cell expansion and cellulose synthesis regulated by plasmodesmata and sugar: insights from the single-celled cotton fibre. Funct. Plant Biol. 34, 1–10 (2007). [DOI] [PubMed] [Google Scholar]
- Basra A. S. & Malik C. P. Development of the cotton fiber. Int. Rev. Cytol. 89, 65–113 (1984). [Google Scholar]
- Pugh D. A., Offler C. E., Talbot M. J. & Ruan Y. L. Evidence for the role of transfer cells in the evolutionary increase in seed and fiber biomass yield in cotton. Mol. Plant 3, 1075–1086 (2010). [DOI] [PubMed] [Google Scholar]
- Farrar J., Pollock C. & Gallagher J. Sucrose and the integration of metabolism in vascular plants. Plant Sci. 154, 1–11 (2000). [DOI] [PubMed] [Google Scholar]
- Geigenberger P. & Stitt M. Sucrose synthase catalyses a readily reversible reaction in vivo in developing potato tubers and other plant tissues. Planta 189, 329–339 (1993). [DOI] [PubMed] [Google Scholar]
- Barratt D. H. P. et al. Multiple, distinct isoforms of sucrose synthase in pea. Plant Physiol. 127, 655–664 (2001). [PMC free article] [PubMed] [Google Scholar]
- Chourey P. S., Taliercio E. W., Carlson S. J. & Ruan Y. L. Genetic evidence that the two isozymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Mol. Gen. Genet. 259, 88–96 (1998). [DOI] [PubMed] [Google Scholar]
- Fu H. & Park W. D. Sink- and vascular-associated sucrose synthase functions are encoded by different gene classes in potato. Plant Cell 7, 1369–1385 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G. Q. & Sturm A. Antisense repression of sucrose synthase in carrot (Daucus carota L.) affects growth rather than sucrose partitioning. Plant Mol. Biol. 41, 465–479 (1999). [DOI] [PubMed] [Google Scholar]
- Zrenner R., Salanoubat M., Willmitzer L. & Sonnewald U. Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L). Plant J. 7, 97–107 (1995). [DOI] [PubMed] [Google Scholar]
- Klotz K. L., Finger F. L. & Shelver W. L. Characterization of two sucrose synthase isoforms in sugarbeet root. Plant Physiol. Biochem. 41, 107–115 (2003). [Google Scholar]
- Sun J., Loboda T., Sung S. J. & Black C. C. Sucrose Synthase in Wild Tomato, Lycopersicon chmielewskii, and Tomato Fruit Sink Strength. Plant Physiol. 98, 1163–1169 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniawska Z. et al. Analysis of the sucrose synthase gene family in Arabidopsis. Plant J. 49, 810–828 (2007). [DOI] [PubMed] [Google Scholar]
- Harada T., Satoh S., Yoshioka T. & Ishizawa K. Expression of sucrose synthase genes involved in enhanced elongation of pondweed (Potamogeton distinctus) turions under anoxia. Ann. Bot. 96, 683–692 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M. C. et al. Knockdown of the symbiotic sucrose synthase MtSucS1 affects arbuscule maturation and maintenance in mycorrhizal roots of Medicago truncatula. Plant Physiol. 152, 1000–1014 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohnjec N., Perlick A. M., Puhler A. & Kuster H. The Medicago truncatula sucrose synthase gene MtSucS1 is activated both in the infected region of root nodules and in the cortex of roots colonized by arbuscular mycorrhizal fungi. Mol. Plant Microbe In. 16, 903–915 (2003). [DOI] [PubMed] [Google Scholar]
- Lee J. J., Woodward A. W. & Chen Z. J. Gene expression changes and early events in cotton fibre development. Ann. Bot. 100, 1391–1401 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht G. & Mustroph A. Localization of sucrose synthase in wheat roots: increased in situ activity of sucrose synthase correlates with cell wall thickening by cellulose deposition under hypoxia. Planta 217, 252–260 (2003). [DOI] [PubMed] [Google Scholar]
- Fujii S., Hayashi T. & Mizuno K. Sucrose synthase is an integral component of the cellulose synthesis machinery. Plant Cell physiol. 51, 294–301 (2010). [DOI] [PubMed] [Google Scholar]
- Nolte K. D., Hendrix D. L., Radin J. W. & Koch K. E. Sucrose synthase localization during initiation of seed development and trichome differentiation in cotton ovules. Plant Physiol. 109, 1285–1293 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y. L., Llewellyn D. J., Furbank R. T. & Chourey P. S. The delayed initiation and slow elongation of fuzz-like short fibre cells in relation to altered patterns of sucrose synthase expression and plasmodesmata gating in a lintless mutant of cotton. J. Exp. Bot. 56, 977–984 (2005). [DOI] [PubMed] [Google Scholar]
- Xu S. M., Brill E., Llewellyn D. J., Furbank R. T. & Ruan Y. L. Overexpression of a potato sucrose synthase gene in cotton accelerates leaf expansion, reduces seed abortion, and enhances fiber production. Mol. Plant 5, 430–441 (2012). [DOI] [PubMed] [Google Scholar]
- Duncan K. A., Hardin S. C. & Huber S. C. The three maize sucrose synthase isoforms differ in distribution, localization, and phosphorylation. Plant Cell Physiol. 47, 959–971 (2006). [DOI] [PubMed] [Google Scholar]
- Hirose T., Scofield G. N. & Terao T. An expression analysis profile for the entire sucrose synthase gene family in rice. Plant Sci. 174, 534–543 (2008). [Google Scholar]
- Islam M. Z., Hu X. M., Jin L. F., Liu Y. Z. & Peng S. A. Genome-wide identification and expression profile analysis of citrus sucrose synthase genes: investigation of possible roles in the regulation of sugar accumulation. PLoS One 9, e113623 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X. et al. Structure and expression profile of the sucrose synthase gene family in the rubber tree: indicative of roles in stress response and sucrose utilization in the laticifers. FEBS J. 281, 291–305 (2014). [DOI] [PubMed] [Google Scholar]
- An X. et al. Identification and characterization of the populus sucrose synthase gene family. Gene 539, 58–67 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang D., Xu B., Yang X., Zhang Z. & Li B. The sucrose synthase gene family in Populus: structure, expression, and evolution. Tree Genet. Genomes 7, 443–456 (2010). [Google Scholar]
- Zou C. et al. Genome-wide analysis of the Sus gene family in cotton. J. Integr. Plant Biol. 55, 643–653 (2013). [DOI] [PubMed] [Google Scholar]
- Comai L. et al. Efficient discovery of DNA polymorphisms in natural populations by Ecotilling. Plant J. 37, 778–786 (2004). [DOI] [PubMed] [Google Scholar]
- Raghavan C. et al. Rapid method for detecting SNPs on agarose gels and its application in candidate gene mapping. Mol. Breed. 19, 87–101 (2006). [Google Scholar]
- Rungis D. et al. Efficient genetic mapping of single nucleotide polymorphisms based upon DNA mismatch digestion. Mol. Breed. 16, 261–270 (2005). [Google Scholar]
- Xia Y. et al. Allelic variations of a light harvesting chlorophyll a/b-binding protein gene (Lhcb1) associated with agronomic traits in barley. PLos One 7, e37573 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S. et al. Identification of rice transcription factors associated with drought tolerance using the Ecotilling method. PLos One 7, e30765 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro G., Eliott F. G. & Henry R. J. An optimized ecotilling protocol for polyploids or pooled samples using a capillary electrophoresis system. Anal. Biochem. 355, 145–147 (2006). [DOI] [PubMed] [Google Scholar]
- Gilchrist E. J. et al. Use of Ecotilling as an efficient SNP discovery tool to survey genetic variation in wild populations of Populus trichocarpa. Mol. Ecol. 15, 1367–1378 (2006). [DOI] [PubMed] [Google Scholar]
- Rakshit S. et al. Large-scale DNA polymorphism study of Oryza sativa and O. rufipogon reveals the origin and divergence of Asian rice. Theor. Appl. Genet. 114, 731–743 (2007). [DOI] [PubMed] [Google Scholar]
- Evanno G., Regnaut S. & Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620 (2005). [DOI] [PubMed] [Google Scholar]
- Lü H., Liu X., Wei S. & Zhang Y. Epistatic association mapping in homozygous crop cultivars. PLos One 6, e17773 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T. et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotech. 33, 531–537 (2015). [DOI] [PubMed] [Google Scholar]
- Bai W. Q. et al. Gibberellin overproduction promotes sucrose synthase expression and secondary cell wall deposition in cotton fibers. PLos One 9, e96537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill E. et al. A novel isoform of sucrose synthase is targeted to the cell wall during secondary cell wall synthesis in cotton fiber. Plant Physiol. 157, 40–54 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Guo W., Zhu H., Ruan Y. L. & Zhang T. Overexpression of GhSusA1 increases plant biomass and improves cotton fiber yield and quality. Plant Biotech. J. 10, 301–312 (2012). [DOI] [PubMed] [Google Scholar]
- Ruan Y. L., Llewellyn D. J. & Furbank R. T. Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 15, 952–964 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Zhang X., Zhang T. & Guo W. Molecular tagging and source analysis of QTL for elite fiber quality in upland cotton. Acta Agron. Sin. 34, 578–586 (2008). [Google Scholar]
- Sun F. et al. QTL mapping for fiber quality traits across multiple generations and environments in upland cotton. Mol. Breed. 30, 569–582 (2011). [Google Scholar]
- Wu J., Gutierrez O. A., Jenkins J. N., McCarty J. C. & Zhu J. Quantitative analysis and QTL mapping for agronomic and fiber traits in an RI population of upland cotton. Euphytica 165, 231–245 (2008). [Google Scholar]
- Yu J. et al. Identification of quantitative trait loci across interspecific F2, F2:3 and testcross populations for agronomic and fiber traits in tetraploid cotton. Euphytica 191, 375–389 (2013). [Google Scholar]
- Shi Y. H. et al. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18, 651–664 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L. et al. Transcriptomic analysis of fiber strength in upland cotton chromosome introgression lines carrying different Gossypium barbadense chromosomal segments. PLos One 9, e94642 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y. L. et al. Expression of sucrose synthase in the developing endosperm is essential for early seed development in cotton. Funct. Plant Biol. 35, 382 (2008). [DOI] [PubMed] [Google Scholar]
- Paterson A. H., Brubaker C. L. & Wendel J. F. A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol. Biol. Rep. 11, 122–127 (1993). [Google Scholar]
- Zhu H. et al. Structure, expression differentiation and evolution of duplicated fiber developmental genes in Gossypium barbadense and G. hirsutum. BMC Plant Biol. 11, 40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till B. J., Zerr T., Comai L. & Henikoff S. A protocol for TILLING and Ecotilling in plants and animals. Nat. Protoc. 1, 2465–2477 (2006). [DOI] [PubMed] [Google Scholar]
- Kadaru S. B., Yadav A. S., Fjellstrom R. G. & Oard J. H. Alternative ecotilling protocol for rapid, cost-effective single-nucleotide polymorphism discovery and genotyping in rice (Oryza sativa L.). Plant Mol. Biol. Rep. 24, 3–22 (2006). [Google Scholar]
- Ng P. C. & Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P. & Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009). [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L. & Salzberg S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M. & Donnelly P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S. An expectation-maximization algorithm for the Lasso estimation of quantitative trait locus effects. Heredity 105, 483–494 (2010). [DOI] [PubMed] [Google Scholar]
- Bu S. H. et al. Interacted QTL mapping in partial NCII design provides evidences for breeding by design. PLos One 10, e0121034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. B. et al. Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci Rep. 6, 19444 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


