Abstract
Importance
High-grade serous carcinoma (HGSC) is the most prevalent and lethal form of ovarian cancer. HGSCs frequently arise in the distal fallopian tubes rather than the ovary, developing from small precursor lesions called serous tubal intraepithelial carcinomas (TICs or more specifically STICs). While STICs have been reported to harbor TP53 mutations, detailed molecular characterizations of these lesions are lacking.
Observations
We performed targeted next generation sequencing (NGS) on formalin-fixed, paraffin- embedded tissue from four women, two with HGSC and two with uterine endometrioid carcinoma (UEC) who were diagnosed with synchronous STICs. We detected concordant mutations in both HGSCs with synchronous STICs, including TP53 mutations as well as assumed germline BRCA1/2 alterations, confirming a clonal relationship between these lesions. NGS confirmed the presence of a STIC clonally unrelated to one case of UEC. NGS of the other tubal lesion diagnosed as a STIC unexpectedly supported the lesion as a micrometastasis from the associated UEC.
Conclusions and Relevance
We demonstrate that targeted NGS can identify genetic lesions in minute lesions such as TICs, and confirm TP53 mutations as early driving events for HGSC. NGS also demonstrated unexpected relationships between presumed STICs and synchronous carcinomas, suggesting potential diagnostic and translational research applications.
INTRODUCTION
Ovarian cancers will account for over 14,000 estimated deaths in the United States in 2015, with nearly two-thirds presenting at high stage with dismal five year overall survival (27%) 1. Despite advances in surgery, medicine, and imaging, ovarian cancer mortality has changed little over several decades, and the need for early detection of cancers at a curable stage remains unmet. A dualistic model of ovarian cancer pathogenesis posits a heterogeneous group of low-grade, clinically indolent, genomically stable tumors (Type I, which account for ~25% of all ovarian carcinomas) and a high-grade, clinically aggressive group with high risk for distant metastases (Type II, comprising the remaining ~75%) 2–4. These groups can also be broadly distinguished on morphologic grounds, with high-grade serous carcinoma (HGSC) comprising most Type II cancers, the most common and lethal ovarian carcinoma group. In contrast to Type I ovarian carcinomas 3,4, Type I tumors are characterized by very frequent TP53 mutations (>95%)5 and associated genomic instability.
Many, if not most ovarian HGSCs are derived from precursor lesions arising from epithelium in the fimbriated end of the fallopian tube 6–8. These precursor lesions, termed serous tubal intraepithelial carcinomas (TICs and more specifically STICs), demonstrate atypical histologic changes that are reminiscent of HGSC. Furthermore, STICs harbor clonal TP53 mutations 6,9, indicating that this alteration is an early event in the oncogenesis of HGSC. However, comprehensive sequencing based assessment of molecular alterations in TICs has not been reported in large part due to their minute size. Likewise it is unclear if presumed STICs may in fact represent metastatic tubal deposits.
To more comprehensively assess somatic alterations in TICs and assess relationships between TICs and synchronous carcinomas, we performed a pilot study of targeted next generation sequencing (NGS) on a series of four TICs (two discovered incidentally) in patients undergoing total abdominal hysterectomy/bilateral salpingo-ophorectomy (TAH/BSO) for gynecologic malignancies.
METHODS
Tissue samples
Four cases of gynecologic malignancies diagnosed in 2011–12 with co-existing TIC were selected from the University of Michigan Department of Pathology case files following Institutional Review Board approval. Review of hematoxylin and eosin stained slides by experienced gynecologic pathologists (J.N.S. and K.R.C.) confirmed the diagnosis in each case. Available demographic and clinicopathologic data were obtained from the medical record.
Immunohistochemistry
Immunohistochemistry was performed using the Ventana Benchmark System (Ventana Medical Systems; Tucson, Arizona) on formalin-fixed, paraffin-embedded (FFPE) tissue sections cut to a thickness of 4μm. Antibody clones and staining evaluation are described in the Supplement eMethods.
Targeted Next Generation Sequencing
Ten μm FFPE sections (10 per sample) were cut from representative blocks from the primary tumor and the tubal lesion from each case. Tumor tissue containing high estimated tumor content (ranging from 60–80% tumor nuclei) was macrodissected. DNA isolation, next generation sequencing using the DNA component of the Oncomine Comprehensive Panel (OCP), and data analysis to identify prioritized non-synonymous mutations was performed essentially as described10,11. We have extensively validated this OCP workflow performance using molecular standards and routine tissue samples11, and detailed information is provided in the Supplement eMethods.
RESULTS
Clinicopathologic Characteristics
We identified four cases from 2011–2012 in which TICs (presumed serous were identified in TAH/BSO specimens resected for gynecologic cancer (TABLE 1 and FIGURE 1A&B). Patients 1 and 2 were diagnosed with HGSC (stage IIC and IIIC, respectively) and patients 3 and 4 demonstrated uterine endometrioid carcinoma (UEC), FIGO grade 1 (stage IIIA and IB, respectively). Additional clinical information is provided in the Supplement eResults. Immunohistochemistry for p53 was performed for each presumed STIC, showing strong diffuse nuclear expression in patients 2–4, and a total lack of expression in patient 1 (TP53 immunohistochemistry for patient 4 shown in FIGURE 1C).
Table 1.
Clinicopathologic and coverage data for sequenced samples
| Patient | Age | Primary Tumor Type (Stage) | Site | gDNA isolated (ng) | Mapped Reads | % reads on target | Average coverage depth | Target bases at 100X coverage | Total Called variants1 | Variants passing filtering2 | Prioritized variants3 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 53 | HGSC (IIC) | Tube | 89 | 1,599,999 | 93% | 621x | 89% | 192 | 2 | 2 |
| Ovary | 86 | 806,616 | 93% | 333x | 82% | 169 | 3 | 3 | |||
| 2 | 48 | HGSC (IIIC) | Tube | 176 | 1,940,650 | 94% | 752x | 90% | 232 | 2 | 2 |
| Ovary | 1,864 | 859,438 | 97% | 354x | 89% | 192 | 3 | 2 | |||
| 3 | 53 | UEC, FIGO 1 (IIIA) | Tube | 228 | 1,844,577 | 94% | 690x | 84% | 210 | 1 | 1 |
| Ovary | 452 | 803,820 | 98% | 331x | 83% | 160 | 6 | 5 | |||
| 4 | 62 | UEC, FIGO 1 (IB) | Tube | 16 | 1,376,506 | 93% | 510x | 82% | 514 | 3 | 3 |
| Ovary | 436 | 736,811 | 93% | 293x | 85% | 164 | 3 | 2 | |||
|
|
|||||||||||
| Median | 202 | 1,117,972 | 94% | 432x | 85% | 192 | 3 | 2 | |||
HGSC= High grade serous carcinoma, UEC= Uterine endometrioid carcinoma, gDNA= genomic DNA.
Variants called by automated low stringency variant calling.
Variants passing filtering of technical artifacts, poorly supported variants, germline SNPs and synonymous/non-coding variants.
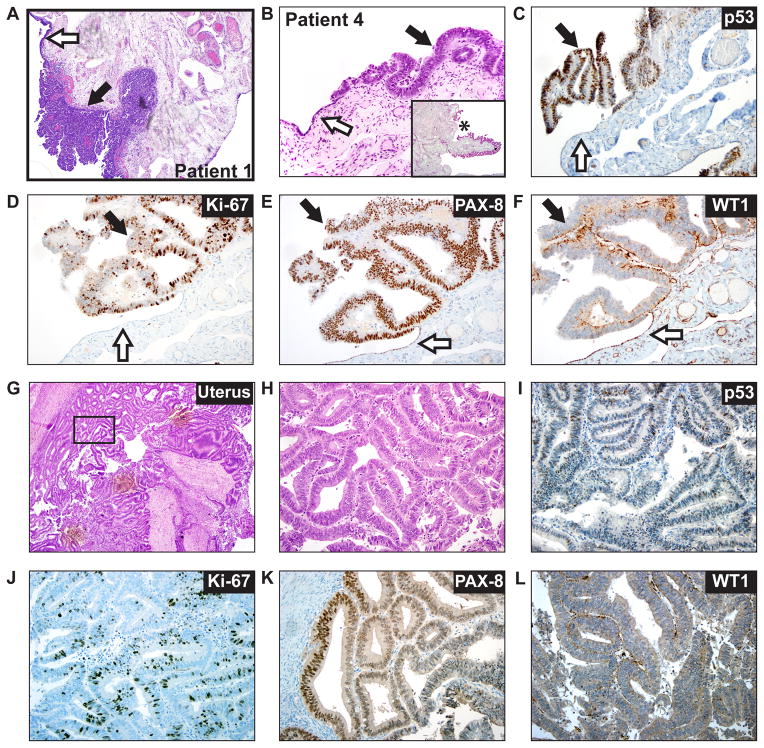
Figure 1. Intraepithelial tubal metastasis mimicking a STIC in a patient with uterine endometrial carcinoma.
A. Low power (40X) photomicrograph of a hematoxylin and eosin (H&E) stained slide from the fimbriated end of the distal fallopian tube of patient 1. The black arrow indicates the presence of STIC (noninvasive into the underlying stroma) and white arrow identifies normal adjacent tubal epithelium. B. High power (400x) photomicrograph of a H&E stained slide from the fimbriated end of the distal fallopian tube of patient 4. Inset panel shows low power (40x) micrograph of same specimen with asterisk marking area of magnification. The black arrow identifies area of significant cytologic atypia, morphologically consistent with a tubal intraepithelial carcinoma (TIC). The white arrow indicates adjacent normal tubal epithelium. C. Immunohistochemical staining (IHC) for p53 in the TIC with clonal type overexpression (black arrow). The white arrow indicates a wild type p53 staining pattern. D. IHC for Ki-67 in the TIC (black arrow) shows a mitotic index of approximately 40%. The white arrow shows low staining in the normal epithelium. E. IHC for PAX8 in the TIC (black arrow) shows strong nuclear reactivity. White arrow indicates normal epithelium. F. IHC for WT1 in the TIC (black arrow) shows absent nuclear expression. White arrow indicates WT expression in normal epithelium. G. Low power (40x) photomicrograph of an H&E stained slide from patient 4’s primary UEC. H. High power (400x) photomicrograph of the area indicated by the rectangle in panel G. I. IHC for p53 in the primary UEC shows a wild type staining pattern. J. IHC for Ki-67 in the primary UEC shows a mitotic index of approximately 20%. K. IHC for PAX-8 in the primary UEC shows patchy nuclear positivity. L. IHC for WT1 in the primary UEC shows absent nuclear expression. Next generation sequencing detected the presence of a TP53 mutation exclusively in the tubal lesion from patient 4, consistent with IHC results (see C vs. I), supporting the tubal lesion from patient 4 as a mucosal micrometastasis/implant from the patient’s primary UEC, rather than a STIC.
Next Generation Sequencing Results
Manual macrodissection was used to isolate TICs and matched invasive carcinomas from each case (see eFigure 1 in the Supplement). Targeted NGS was performed on 5–20ng of genomic DNA, using a custom multiplexed PCR-based Ion Torrent Ampliseq panel comprising 2,462 amplicons covering 135 cancer related genes (DNA component of the OCP, genes given in eTable 1 in the Supplement) with sequencing performed on the IonTorrent PGM Sequencer. Detailed information on DNA yield and sequencing statistics, including germ line SNP concordance in paired cases (≥92% per paired sample), is provided in TABLE 1, and eFigure 2 and eResults in Supplement.
NGS variant calls were filtered using predefined criteria (see Supplement) to nominate potential somatic driving alterations. Somatic TP53 mutations were present in all four tubal lesions (TABLE 2), with the two HGSC patients showing evidence of a clonal relationship between the TICs and the primary serous ovarian carcinomas (details provided in the Supplement). For the two patients who presented with incidental TICs in the context of UEC, NGS demonstrated that the two lesions in patient 3 were genetically heterogeneous, with the TIC harboring a single TP53 mutation and the UEC showing somatic MTOR, PTEN, KRAS, PIK3CA, and ATM mutations (TABLE 2), consistent with a genetically distinct STIC and UEC. In patient 4, who harbored a TIC and a UEC (histology shown in FIGURE 1B & G,H), the tubal lesion surprisingly demonstrated somatic PTEN and CTNNB1 mutations in addition to a TP53 mutation; the matched UEC also demonstrated concordant PTEN and CTNNB1 mutations, but no TP53 mutation was noted (despite over 200 covering reads at that position). Correspondingly, IHC for p53 showed a clonal staining pattern in the TIC, with wild type staining observed in the primary tumor (FIGURE 1C vs. I). Additional IHC stains (FIGURE 1D–F&J–L) demonstrate both the primary tumor and TIC to be positive for PAX8 and negative for WT1. The TIC showed a mitotic index around 40% (as assessed by Ki-67 IHC), while the primary tumor demonstrated 20–30% Ki-67 staining. As described below, this genomic profile and immunophenotype supports the tubal lesion as a micrometasis/tube implant from the UEC, rather than a STIC.
Table 2.
Prioritized mutations from next generation sequencing of STICs and synchronous gynecologic malignancies
| Patient | Site | Gene | Exonic Function | Location | Nucleotide | Amino Acid | Variant Allele Frequency | Coverage at Variant Allele |
|---|---|---|---|---|---|---|---|---|
| 1 | Tube | BRCA2 | frameshift deletion | chr13:32914209 | c.5718_5719del | p.1906_1907del | 72% | 137 |
| Tube | TP53 | stopgain SNV | chr17:7578263 | c.C190T | p.R64X | 48% | 85 | |
|
| ||||||||
| Ovary | BRCA2 | stopgain SNV | chr13:32972626 | c.A9976T | p.K3326X | 50% | 87 | |
| Ovary | BRCA2 | frameshift deletion | chr13:32914209 | c.5718_5719del | p.1906_1907del | 54% | 122 | |
| Ovary | TP53 | stopgain SNV | chr17:7578263 | c.C190T | p.R64X | 11% | 8 | |
|
| ||||||||
| 2 | Tube | BRCA1 | stopgain SNV | chr17:41244787 | c.C2620T | p.Q874X | 52% | 464 |
| Tube | TP53 | nonsynon. SNV | chr17:7577580 | c.A305G | p.Y102C | 10% | 118 | |
|
| ||||||||
| Ovary | TP53 | nonsynon. SNV | chr17:7577580 | c.A305G | p.Y102C | 69% | 225 | |
| Ovary | BRCA1 | stopgain SNV | chr17:41244787 | c.C2620T | p.Q874X | 88% | 351 | |
|
| ||||||||
| 3 | Tube | TP53 | nonsynon. SNV | chr17:7577535 | c.G350C | p.R117T | 15% | 90 |
|
| ||||||||
| Uterus | MTOR | nonsynon. SNV | chr1:11184573 | c.C6644A | p.S2215Y | 18% | 36 | |
| Uterus | PTEN | nonsynon. SNV | chr10:89685308 | c.A203T | p.Y68F | 15% | 41 | |
| Uterus | KRAS | nonsynon. SNV | chr12:25398284 | c.G35T | p.G12V | 20% | 81 | |
| Uterus | PIK3CA | nonsynon. SNV | chr3:178916876 | c.G263A | p.R88Q | 21% | 85 | |
| Uterus | ATM | nonsynon. SNV | chr11:108142000 | c.C2944T | p.R982C | 20% | 81 | |
|
| ||||||||
| 4 | Tube | TP53 | nonsynon. SNV | chr17:7577548 | c.G337C | p.G113R | 12% | 60 |
| Tube | PTEN | stopgain SNV | chr10:89692904 | c.C388T | p.R130X | 18% | 36 | |
| Tube | CTNNB1 | nonsynon. SNV | chr3:41266137 | c.C134T | p.S45F | 14% | 151 | |
|
| ||||||||
| Uterus | PTEN | stopgain SNV | chr10:89692904 | c.C388T | p.R130X | 34% | 73 | |
| Uterus | CTNNB1 | nonsynon. SNV | chr3:41266136 | c.T133C | p.S45P | 33% | 133 | |
nonsynon. SNV = nonsynonmous single nucleotide variant.
DISCUSSION
Here, using targeted NGS on macrodissected routine FFPE archival tissue, we report a comprehensive investigation of somatic driving mutations associated with fallopian tube TICs and their relationship to synchronous gynecologic malignancies. In all tubal lesions, somatic TP53 mutations were identified, consistent with previous reports that TP53 mutation occurs early in the pathogenesis of HGSC 9.9 and the common use of TP53 immunostaining in STIC diagnosis. The genetic concordance between the STICs and HGSCs in patients 1 and 2 supports a clonal relationship between the two lesions. In contrast, the mutational discordance between the STIC and UEC in patient 3 implies separate and independent neoplastic processes in the fallopian tube and uterus. Patients 1 and 2 harbored germline BRCA1/2 mutations, and STICs are frequently found in the fallopian tubes of BRCA mutated patients with concomitant HGSC and in ~5% of those undergoing prophylactic TAH/BSO for HGSC risk-reduction 12. The STIC in patient 3 is potentially sporadic as no germline cancer predisposing mutations in BRCA1, BRCA2 or MSH2 were identified, however additional cancer predisposing loci were not assessed as the OCP was designed to interrogate somatic driving alterations.
Patient 4 demonstrated a small tubal lesion that resembled a STIC by morphology and immunophenotype (clonal TP53); however, NGS sequencing demonstrated PTEN and CTNNB1 mutations in addition to TP53. PTEN and CTNNB1 are characteristic alterations of UEC (reviewed in 13), but are uncommon in HGSC 4,14. Given that the TIC and primary UEC present in patient 4 harbored concomitant PTEN and CTNNB1 mutations, we propose that the fallopian tube lesion represents a mucosal UEC micrometastasis mimicking a STIC, rather than an independent STIC. The TP53 mutation present in the tubal lesion as supported by both NGS and IHC, but not in the endometrial primary, provides further support that the tube lesion represents a micrometastasis. These findings highlight that although STICs may be precursors to HGSC, not all high grade TICs are of tubal origin. Likewise, the fallopian tube can harbor metastases from other sites, even in an apparent intraepithelial fashion, and the possibility that a TIC could represent a metastasis should be considered even in the context of serous histologies (e.g., some “STICs” may represent metastases from primary peritoneal HGSCs). Reliance on clonal p53 expression by IHC for the diagnosis of STIC, as with this patient, can be a potential pitfall complicating the identification of fallopian tube metastases.
This pilot study demonstrates the feasibility of targeted NGS on very small epithelial lesions such as TICs, with analysis performed on as little as 5ng of input genomic DNA isolated by macrodissection from FFPE tissue. Although macrodissection in this context is challenging, the clonal TP53 IHC expression in each TIC supports the use of the TP53 variant allele frequency (detected by NGS) to estimate tumor content and supports our approach. Further studies including larger cohorts of synchronous and asynchronous TIC/HGSC cases using this methodology or laser capture microdissection and more comprehensive sequencing may be useful in identifying possible lesions driving progression and potential biomarkers to assist with early detection or minimal residual disease monitoring. Lastly, as shown in case 4, characterization of additional cases diagnosed as STIC may help identify micrometastases to the tubal mucosa that can morphologically mimic true STICs.
Supplementary Material
Acknowledgments
JNS, SAT, and KRC developed the study concept and design; ASM, JNS, DHH, AKC, CJL, SAT, and KRC participated in the acquisition, analysis, and/or interpretation of data; ASM, SAT, and KRC drafted and critically reviewed the manuscript; and SAT and KRC obtained funding. This work was supported in part by the A. Alfred Taubman Medical Research Institute (SAT). SAT has a sponsored research agreement with ThermoFisher Scientific that provided access to the NGS panel used herein. ThermoFisher Scientific had no other role in the study design, collection of data, analysis, drafting/review of the manuscript, or decision to submit for publication. ASM and SAT had access to all data and were responsible for primary data analysis.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Kurman RJ, Shih Ie M. Pathogenesis of ovarian cancer: lessons from morphology and molecular biology and their clinical implications. Int J Gynecol Pathol. 2008;27(2):151–160. doi: 10.1097/PGP.0b013e318161e4f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurman RJ, Shih Ie M. The origin and pathogenesis of epithelial ovarian cancer: a proposed unifying theory. Am J Surg Pathol. 2010;34(3):433–443. doi: 10.1097/PAS.0b013e3181cf3d79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurman RJ, Shih Ie M. Molecular pathogenesis and extraovarian origin of epithelial ovarian cancer--shifting the paradigm. Hum Pathol. 2011;42(7):918–931. doi: 10.1016/j.humpath.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed AA, Etemadmoghadam D, Temple J, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol. 2010;221(1):49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kindelberger DW, Lee Y, Miron A, et al. Intraepithelial carcinoma of the fimbria and pelvic serous carcinoma: Evidence for a causal relationship. Am J Surg Pathol. 2007;31(2):161–169. doi: 10.1097/01.pas.0000213335.40358.47. [DOI] [PubMed] [Google Scholar]
- 7.Piek JM, van Diest PJ, Zweemer RP, et al. Dysplastic changes in prophylactically removed Fallopian tubes of women predisposed to developing ovarian cancer. J Pathol. 2001;195(4):451–456. doi: 10.1002/path.1000. [DOI] [PubMed] [Google Scholar]
- 8.Piek JM, Verheijen RH, Kenemans P, Massuger LF, Bulten H, van Diest PJ. BRCA1/2-related ovarian cancers are of tubal origin: a hypothesis. Gynecol Oncol. 2003;90(2):491. doi: 10.1016/s0090-8258(03)00365-2. [DOI] [PubMed] [Google Scholar]
- 9.Lee Y, Miron A, Drapkin R, et al. A candidate precursor to serous carcinoma that originates in the distal fallopian tube. J Pathol. 2007;211(1):26–35. doi: 10.1002/path.2091. [DOI] [PubMed] [Google Scholar]
- 10.Cani AK, Hovelson DH, McDaniel AS, et al. Next-Gen Sequencing Exposes Frequent MED12 Mutations and Actionable Therapeutic Targets in Phyllodes Tumors. Mol Cancer Res. 2015;13(4):613–619. doi: 10.1158/1541-7786.MCR-14-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hovelson DH, McDaniel AS, Cani AK, et al. Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia. doi: 10.1016/j.neo.2015.03.004. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conner JR, Meserve E, Pizer E, et al. Outcome of unexpected adnexal neoplasia discovered during risk reduction salpingo-oophorectomy in women with germ-line BRCA1 or BRCA2 mutations. Gynecol Oncol. 2014;132(2):280–286. doi: 10.1016/j.ygyno.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Hara AJ, Bell DW. The genomics and genetics of endometrial cancer. Adv Genomics Genet. 2012;2012(2):33–47. doi: 10.2147/AGG.S28953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.