Abstract
Patient-matched transcriptomic studies using tumour samples before and after treatment allow inter-patient heterogeneity to be controlled, but tend not to include an untreated comparison. Here, Illumina BeadArray technology was used to measure dynamic changes in gene expression from thirty-seven paired diagnostic core and surgically excised breast cancer biopsies obtained from women receiving no treatment prior to surgery, to determine the impact of sampling method and tumour heterogeneity. Despite a lack of treatment and perhaps surprisingly, consistent changes in gene expression were identified during the diagnosis-surgery interval (48 up, 2 down; Siggenes FDR 0.05) in a manner independent of both subtype and sampling-interval length. Instead, tumour sampling method was seen to directly impact gene expression, with similar effects additionally identified in six published breast cancer datasets. In contrast with previous findings, our data does not support the concept of a significant wounding or immune response following biopsy in the absence of treatment and instead implicates a hypoxic response following the surgical biopsy. Whilst sampling-related gene expression changes are evident in treated samples, they are secondary to those associated with response to treatment. Nonetheless, sampling method remains a potential confounding factor for neoadjuvant study design.
Gene expression profiling of carcinomas has been widely used for molecular subtyping and prognostic prediction1,2,3,4,5,6,7,8,9 producing a diverse library of gene classifiers. However, these signatures may be limited by the particular dataset used to produce the signature10 and by the inherent cellular heterogeneity of tumours and the practical considerations of how samples are collected – in short the heterogeneity of the cohort and the heterogeneity of sampling.
A more informative and considered approach when performing molecular studies is to use matched biopsies from the same patient, allowing for both inter-patient variation to be controlled and changes occurring within a given tumour or organism to be more accurately modelled11,12,13. Matched sample pairs coupled with careful cohort selection should ensure that changes related to a given drug represent the largest source of variation, avoiding any unwanted contribution from confounding factors known or unknown and allow for greater statistical power with a smaller sample size14,15.
Acquisition of multiple biopsies from an individual patient has been simplified by the more common use of neoadjuvant therapy for breast cancer, an increasingly popular treatment option for initially large, inoperable or locally advanced breast tumours, as well as operable cancers susceptible to specific treatments16. Pre-operative treatment with chemotherapy, or endocrine therapy in ER+ disease, not only increases rates of breast conserving surgery17, but also allows a unique in vivo observation of tumour response to treatment12,18. This so-called ‘window of opportunity’ permits sequential biopsies of the same cancer to be taken at different time points during the course of the pre-operative treatment, allowing assessment of molecular changes in the tumour long before clinical evidence of response can be determined11,13. This has allowed the molecular effects of an administered drug to be studied and enabled biomarkers predictive of response or resistance to therapy to be identified at an increased rate. Recently our own group has demonstrated the added value of additional on-treatment measurements of gene expression to characterise and accurately predict the response to treatment13,19.
Whilst the benefits of this ‘window of opportunity’ approach are certainly attractive for translational research, matched samples have commonly been collected under the assumption that variation observed between pairs will have occurred as a result of treatment – i.e. the results apparent are due to the drug alone. A control group is often not included in these studies and those that do are commonly limited to a handful of samples (n = 820, n = 1521) or are confounded by concurrent treatments. Conversely, studies that have compared multiple biopsies from the same patient in lieu of treatment are limited to only a fraction of the total molecular repertoire, most often focussing on hormone receptor status by IHC and not full transcriptomic profiling22,23,24,25. Whilst oestrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (Her2) exhibit high concordance between sample pairs in these studies, the growing understanding of breast cancer as an increasingly heterogeneous and polygenic disease necessitates a high-throughput approach.
Previous work26 that has utilised larger-scale assays (a panel of 147 cancer related genes) investigated molecular variation under conditions of no-intervening treatment (NIT) in 21 paired core needle biopsy (CB) and excision biopsy (EB) samples. Here the diagnostic core biopsy was implicated in initiating an immune response, hypothesised to then be detected in a later surgically extracted excision biopsy. Potential stimulation of tumour-associated macrophages (TAMs) in response to CB was also reported, itself associated with poor prognosis in human breast cancer, raising concerns of taking multiple repeated biopsies from the same patient, underlining the importance of considering the full repertoire of genetic expression under conditions of no treatment.
Here we present the largest dataset to-date of untreated patient-matched breast cancer samples to determine whether, and to what extent, sample pairs exhibit molecular heterogeneity independent of treatment, and what the implications of any variation are in terms of the interpretation of patient-matched gene expression profiling studies. We explore possible causes of consistent differential expression and whether these reflect a wounding or immune response to the first biopsy, a hypoxia- or stress-induced response following blood supply interruption27 or the normal growth and evolution of tumours over time28.
Methods
Patient selection
Paired diagnostic core biopsies and surgical excision biopsies were identified from 37 patients with a primary histologically confirmed invasive breast cancer that did not receive any preoperative or neoadjuvant treatment at the Western General Hospital, Edinburgh, between 2003 and 2011. All patients gave informed consent to be included in the study, which was approved by the Lothian Regional Ethics Committee (2001/8/80 and 2001/8/81) and we confirm can that all experiments were performed in accordance with relevant guidelines and regulations. A summary and complete clinicopathological characteristics of the patients diagnostic core biopsies are given in Supplementary Tables S1 and S2.
Sample collection, RNA processing and microarray hybridisation
Core biopsies were taken at diagnosis in all patients using a 14-gauge automated needle device. Multiple cores were taken per tumour and combined as individual samples. Surgical excision biopsies of breast tumour were collected between 13 and 53 days later (mean interval = 27.5 days). Samples were snap frozen in liquid nitrogen and stored at −80 °C before homogenisation and RNA extraction using the RNeasy Mini Kit with RNAse Free DNAse treatment (Qiagen). RNA quantity and quality was assured using a Nanodrop 2000 c spectrophotometer (Thermo Scientific). RNA was reverse transcribed and amplified using the WT-Ovation FFPE System Version 2 (NuGEN), purified using the Qiaquick PCR purification Kit (NuGEN), biotinylated using the IL Encore Biotin Module (NuGEN), and purified using the minElute Reaction Cleanup Kit (Qiagen). At each step RNA/cDNA quantity and quality was assured by repeat assessment with the Nanodrop 2000 c prior to advancing to the next stage. Labelled cDNA was hybridized to Illumina Human HT-12 version 3 and version 4 whole-genome expression bead arrays according to the standard protocol for NuGEN labelled samples. Data was extracted using GenomeStudio software (Illumina).
Data analysis
All analysis was conducted in R (http://www.r-project.org) using software packages available via CRAN (http://cran.r-project.org/) and Bioconductor (http://www.bioconductor.org/). Data was pre-processed using the lumi package29. Log2 transformation, quantile normalisation and summarisation was performed for all Illumina probe profiles. Probe expression information was extracted and detected probes were standardised, firstly by passing a detection p-value threshold (≤0.05) and then by being called present in ≥3 samples. This was carried out three times, once each for three processing batches that comprised the dataset. A common feature list was determined by those probes common to all three dataset batches before re-mapping to Ensembl gene sequences using the biomaRt package30,31. Multiple probes-per-Ensembl ID were resolved by mean averaging. Batch correction was performed using ComBat32 to integrate processing batches for further analysis (Supplementary Fig. S2). Sample groupings were compared using Pearson product-moment correlations. All significances were calculated by a two-tailed Wilcoxon rank sum test and corrected for multiple testing (FDR), unless otherwise stated. Intrinsic subtype assignment was performed using the genefu package33. Pairwise differential gene expression was calculated using significance analysis of microarrays (SAM), part of the siggenes package34. Hierarchical clustering was performed on pairwise fold change expression values using a complete linkage method. SAM analysis of treated data was performed using a hundred 18-sample permutations, to fairly match sample size between subsets, with the intersecting significant genes being taken as a mean average. The raw and processed data from this study can be accessed from NCBI GEO under the accession GSE76728.
Results
Consistent treatment-independent gene expression changes between diagnostic and surgical paired samples
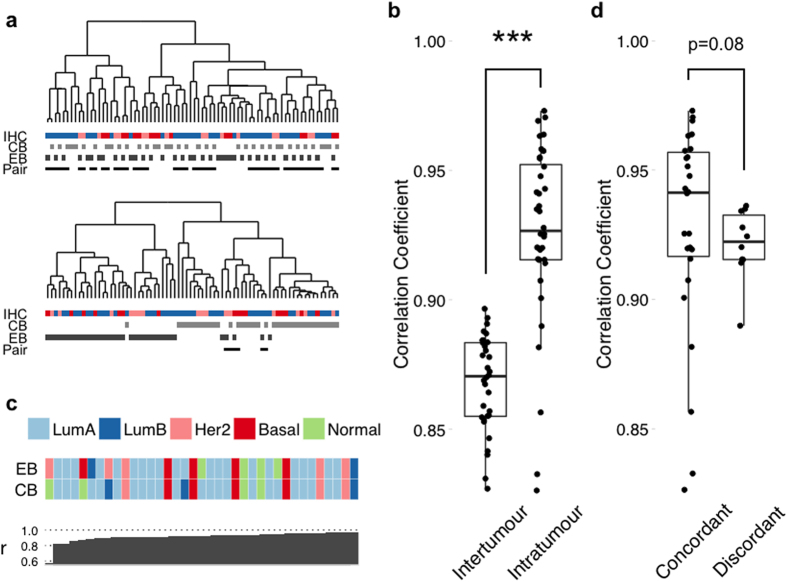
Unsupervised clustering using the 500 most variable genes across the patient-matched samples demonstrated high concordance, with 25/37 pairs observed to cluster at the first level of the dendrogram (Fig. 1a upper). In order to assess whether core biopsy and surgical excision sample pairs varied from one another, Pearson correlations were calculated between intra-tumour (paired) and inter-tumour (non-paired) samples (Fig. 1B). Intra-tumour differences were more significantly (p = 7e-11) correlated (median r = 0.92 range r = 0.60–0.97) than the mean inter-tumour variations (median = 0.87, range r = 0.77–0.90). Technical interference due to sample processing is expected to decrease correlation between samples, though only nominally35. Correlations below r ≃ 0.97 are likely due to underlying tumour heterogeneity, implicating a biological cause to the variation.
Figure 1. Evidence of treatment independent variation between breast cancer diagnostic core biopsies and surgical excision samples.
(a) Hierarchical clustering of the 37 patient-matched diagnostic core and excision biopsy samples using the 500 most variable genes (upper) and a SAM derived signature of 50 genes consistently differentially expressed between core and excision biopsies (lower). Bars represent IHC status (ER+/Her2− = Blue; ER+/Her2+ = Pink; ER-/Her2− = Red) or biopsy method. Lower-most bar indicates where sample pairs have co-aggregated. Two-thirds (25/37) of the pairs cluster at the first level of the upper dendrogram, whereas pairwise association is lost in 31/37 cases for the lower. (b) There is a significantly stronger correlation between biopsy pairs (intra-tumour) than between different tumours (mean inter-tumour). ***p < 0.001. (c) Discordance in molecular subtype assignment between core and excision biopsies. Patients are ranked left to right by pairwise correlation. Colours represent SSP subtypes (Luminal A = Dark blue; Luminal B = Light Blue; Her2 = Pink; Basal = Red; Normal = Green). (d) Sample pairs were called discordant when Biopsy A ≠Biopsy B for at least 4/5 classifiers. Comparison of concordant vs. discordant pairwise correlations then revealed an inverse relationship between correlation and discordancy.
To determine the impact of differences in gene expression apparent between diagnostic core and excision biopsy pairs, pairwise SAM analysis identified 50 significantly differentially expressed genes (48 upregulated, 2 downregulated; FDR = 5%), between the surgical excision and diagnostic core biopsies (Supplementary Table S3). Clustering using these genes was able to separate samples by their sampling method in 31/37 cases (Fig. 1a lower). Five subtyping signatures (PAM50, Sorlie 2003, Hu 2006, Desmedt 2008, Wirapati 2008) were applied to each sample individually and concordant (subtype agreement between pairs with >3 subtype classifiers) vs. discordant (≤3 pair subtype agreements) results recorded (Fig. 1c (PAM50 only) and S5 (all methods)). Pairwise correlation between concordant and discordant assignments, revealed a trend (p = 0.08, Wilcoxon) between decreased pairwise correlation and greater discordance (Fig. 1d).
Pathway analysis of the 50 gene NIT signature revealed enrichment for MAPK signalling (DUSP1, JUN, NR4A1 and FOS), cancer specific (COX-2, PGE2, JUN and FOS), apoptosis induction (FOS and JUN) and genomic reformatting following (brain) ischaemia (EGR1 and JUN) pathways. A number of these genes are examples of ‘early’ or ‘primary’ growth response genes induced by both cell-extrinsic and cell intrinsic signals that do not require de novo protein synthesis for their expression36.
Patient-matched gene expression changes may be associated with either time or biopsy methodology
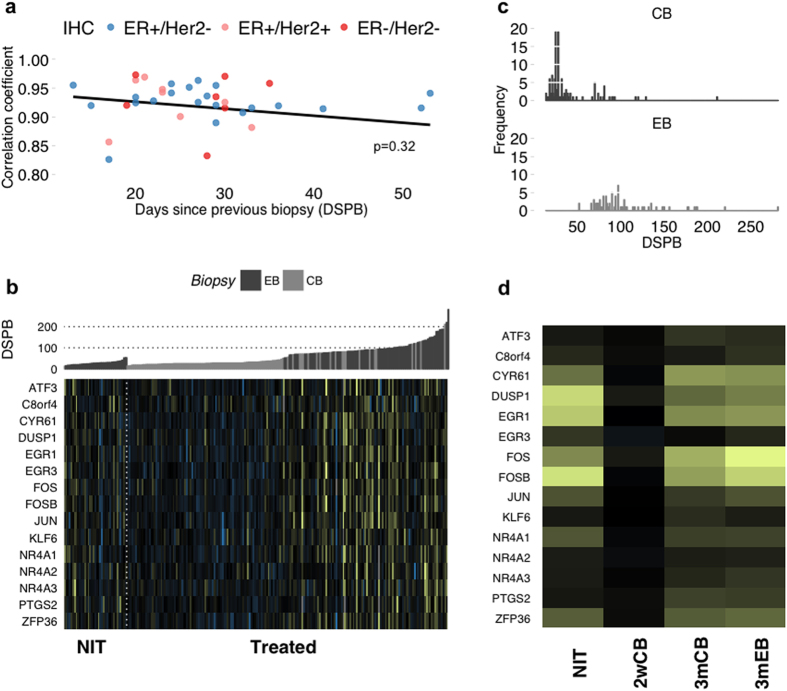
Having determined significant and consistent changes in gene expression exist between diagnostic core and surgical excision pairs we sought to identify the underlying cause. Several hypotheses were immediately apparent – greater changes may occur following a shorter time interval between sampling if consistent gene expression changes reflect a wounding/immune response to the diagnostic core biopsy26; or it may be anticipated that expression patterns diverge over time to reflect tumour evolution; this may in turn be driven by tumour subtype28. However, comparisons of pairwise correlations defined by either IHC status subtype (ER+/Her2−, ER+/HER2+ or ER-/HER2−), PAM50 subtype (cross-table comparing IHC and PAM50 subtype assignment in Supplementary Table S4) or as a function of time interval between biopsies revealed no trend associated with either factor (p = 0.43 and p = 0.32 respectively) (Fig. 2a). This suggested that a progression in breast cancer-related biological changes were unlikely to be the root cause of the observed pairwise variation. To further investigate whether breast cancer biology could be responsible for the observed pairwise variation, we compared 7 breast cancer-related expression modules37 between core and excision biopsies (Supplementary Fig. S3), which revealed no contrasting trend in gene expression. In conjunction to assigning cause, it remained equally important to determine whether our NIT signature genes were evident to alter in an equivalent treated data set. Using a patient-matched cohort treated with letrozole, collected and processed within our group in a manner analogous to the NIT data13, there appeared to be a strong relationship between the time interval between biopsies and a subset of our NIT gene signature (Figs 2b and S4), with treated samples biopsied after 3 months exhibiting significant differential expression of these NIT genes. In these instances however, time interval almost wholly coincided with the final biopsy method and we therefore sought to determine which factor, if either, was dominant. Dividing the treated data into 3 subsets determined by the time since previous biopsy (Fig. 2c) as 2-week CB (2wCB, n = 95), 3-month CB (3 mCB, n = 18) and 3-month EB (3 mEB, n = 70), allowed comparison of changes assumed to result from treatment by both time interval and sampling method. Biopsy time interval was apparent as the factor most associated with the genes altered in our NIT data, with 3-month samples alone exhibiting expression fold changes similar to those observed in the NIT data, implying time on treatment as the defining factor (Fig. 2d). However, in tandem, global GSEA analysis was able to demonstrate that our NIT signature could significantly define the differences between untreated and treated data only when sampling method differed (NIT vs. 2wCB, p = 0; NIT vs. 3 mCB, p = 0.02; NIT vs. 3 mEB, p = 0.25). Similarly, only in the instance of excision biopsy was SAM analysis able to recapitulate >30% of the genes differentially expressed in the NIT data (mean = 47%). More so, out of 3955 differentially expressed treated-EB genes, seven of those common to NIT differential expression were amongst the top 15 in terms of fold change magnitude. Taken together, this implies both time on treatment as well as biopsy method are able to impact upon NIT signature gene expression.
Figure 2. Factors associated with consistent gene expression changes between diagnostic core biopsy and surgical excision of breast tumours in the absence of treatment.
(a) Pairwise correlations between biopsy pairs are not explained as either a function of time between biopsy (p = 0.32) or IHC status (p = 0.43). ER+/Her2− = Blue; ER+/Her2+ = Pink; ER-/Her2− = Red. (b) Heatmap showing differential expression of NIT signature genes in NIT and letrozole treated cohorts. Colours represent gene expression fold changes (up = yellow; down = blue) between samples and their subsequent patient-matched biopsies. Samples are ordered by increasing time between biopsies and reveal a pattern associated with either extraction method - CB (grey) or EB (dark grey) - or time. (c) Frequency distribution of biopsy time intervals. For further analysis, the letrozole treated data was split into three subsets – 2-week CB (2wCB), 3-month CB (3 mCB) and 3-month EB (3 mEB) – to investigate the effects of biopsy method and/or time on gene expression. (d) Mean expression fold changes since previous biopsy for 2 week, 3 month and NIT samples. 3 month subsets closely resemble NIT expression changes, though SAM analysis revealed a greater intersection of differentially expressed genes between NIT and 3 mEB samples than between NIT and 3 mCB samples.
Multiple patient-matched datasets also demonstrate changes in NIT early growth response genes
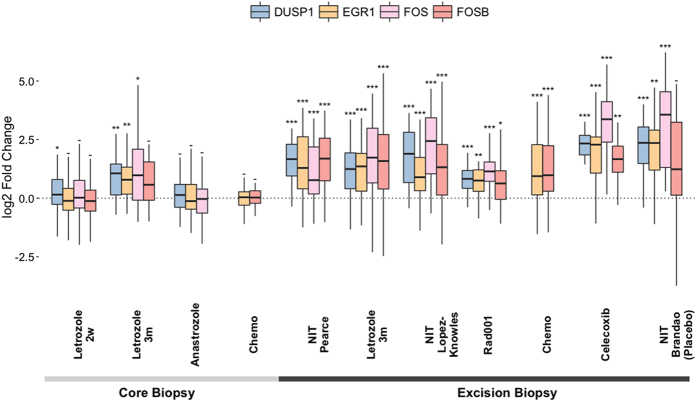
To further investigate the evident relationship between sampling method, time on treatment and NIT signature expression, we selected a panel of four genes (DUSP1, EGR1, FOS, FOSB) that were observed to be most representative of these factors, as well as being well characterised in the literature, for comparison in six external validation datasets (Table 1, Fig. 3). Significant differential gene expression was consistently observed to a greater degree only when an excision biopsy followed a previous core biopsy for these four genes.
Table 1. Composition of patient-matched neoadjuvant breast tumour datasets used within our study.
| Neoadjuvant treatment | n | Biopsy Time Interval/days |
Extraction Method |
Dataset/Reference | |||
|---|---|---|---|---|---|---|---|
| median | range | 1st | On- | Final | |||
| None | 37 | 27 | 13–53 | CB | – | EB | This study |
| Letrozole | 122 | 107 | 13–884 | CB | CB | CB EB | Turnbull et al.13 |
| Celecoxib/none | 22 15 | unspecified | 14–21 | CB | – | EB | (Brandão et al.21 |
| Anastrozole | 81 On-18 Final | 14 | 14–112 | CB | CB | CB | (Smith et al.57 |
| RAD001 | 21 | 14 | 14 | CB | – | EB | (Sabine et al.58 |
| Anthracycline-based Chemotherapy | 69 | unspecified | unspecified | CB | CB | EB | (Magbanua et al.59 |
| None | 56 | 14 | 14 | CB | EB | Lopez-Knowles et al.47 | |
Figure 3. Multiple patient-matched datasets demonstrate shared changes in NIT early growth response genes.
Pairwise analysis of four early growth response genes among the NIT signature in six validation datasets. These genes potentially represent an association between gene expression and sampling method, with surgically excised samples (EB) showing greater expression fold-change than their core biopsied (CB) counterparts. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; − = not significant.
Tumour sampling appears independent of an immune or wound-healing response
It remained important to determine whether gene expression changes in lieu of treatment were able to elicit either an immune or wound-healing response that may be detrimental to a patient’s health or directly confound gene expression profiling results. Evaluation of both a published immune-related 9 gene panel, suggested to be upregulated in response to a diagnostic CB26 and a 589 gene signature representative of a wound-healing response38 failed to show any association with sampling method or time between biopsy, with sample expression unlikely to have been affected in terms of these biological categories (Supplementary Fig. S2).
Discussion
Our study reports gene expression changes in the largest cohort of sequential samples from patients receiving no-intervening treatment yet assembled to demonstrate the molecular variation that occurs independent of treatment in the neoadjuvant and preoperative setting. Significant pairwise changes in gene expression were observed and a 50 gene signature identified comprised of genes associated with a number of cell growth, cell stress and cancer related signalling pathways, including ATF3, EGR1, FOS, FOSB and JUN, each of which have been previously implicated in prognostic discrimination and pathogenesis of breast cancer39,40,41,42,43 as well as other cancers44.
We report that sampling method, specifically core versus excision biopsy, has a direct impact on gene expression and has the potential to introduce a confounding factor to downstream analysis. The most probable explanation of this expression variation between sample pairs is the technical issue of warm ischemia before newly biopsied samples are processed, when the cellular metabolic machinery attempts to mount a survival or apoptotic response before all metabolic activity ceases. Tissue ischemia may result from exclusion of the vascular supply or simply from handling by the surgeon, scrub nurse, pathologist and tumour bank personnel before the sample is frozen in liquid nitrogen. This time delay is likely (on average) to be significantly longer for surgical excision specimens than core biopsy samples. The effects of ischemia on gene expression has been described previously45 and warm ischemia associated with the surgical extirpation of human tissues has significant effects on gene expression. These data support the careful monitoring of ischemic time for tissues harvested for the purpose of gene profiling. Similarly, Dash et al.46 demonstrated significant changes in the expression of FOS, JUN, ATF3 in a study to examine the effect of processing time on prostate cancer samples.
Similar conclusions are proposed in a recent study published during the review process47, where CB and EB pairs were also analysed in terms of correlation and differential gene expression. Variation between sample pairs was found to be evident though modest and those genes found to be differentially expressed intersected with those highlighted in this study (34% of our NIT signature genes). Of particular interest, the overlapping genes included the four genes highlighted in Fig. 3 as well as several others indicative of a stress or early growth response (RGS1, RGS2, ATF3, JUN), similarly proposed as a reaction to surgery-associated ischaemia. Indeed, the rationale of the study stemmed from a desire to investigate post-operative processing time and its effect on gene expression, citing two smaller studies which both highlighted ischaemia as a potential source of molecular variation48,49. Again, key molecules including FOSB were highlighted as being demonstrative of this effect. Importantly, the investigators additionally compared excision biopsies placed into RNAlater either immediately post-surgery or following an interval, and again observed early growth and stress response associated genetic expression to increase.
In a second analogous study, of 147 breast cancer-related genes measured by Nanostring in 21 patients, Jeselsohn et al.26 proposed that sequential breast cancer biopsies reveal activation of an immune response, characterised by a panel of 9 immune-related genes, of which CD68 is known to activate tumour-associated macrophages and implicated in increasingly severe prognosis50, as well as being present in the clinically available Oncotype DX® breast cancer assay8. However, none of these genes were found to be significantly differentially expressed in our larger, whole genome cohort (Supplementary Fig. S1). Conversely, a number of studies have demonstrated a high degree of concordance between classical IHC markers for breast cancer, namely oestrogen, progesterone and HER2 receptors24,25 between diagnostic core and excision biopsies, suggesting discordance may be limited to the level of transcription. Our study demonstrates that pairwise variation at the transcriptome level is not limited to the classical markers of breast cancer, though in some cases paired samples may be classified differently using molecular signatures (Figs 1c and S5). The causative factor behind this variation is most likely due to sampling method in our NIT cohort, with surgical resection resulting in gene expression in response to stress. It remains unclear whether this effect translates to samples following treatment, with time on treatment being observed to mediate molecular changes against the background of the sampling method.
It is important that results from preoperative window or neoadjuvant studies are carefully scrutinised, as a study by Morrogh et al.51 - using a 502 cancer-related gene panel to examine 16 paired patient samples, 8 of which were untreated controls - claimed that window trials are influenced by the wound-healing process. They proposed that upregulation of MLL and FOSB was evidence of this, irrespective of treatment. This mirrored our own findings, though it is necessary to note that Morrogh et al. were limited by sample size and inconsistencies in the effect direction of proliferation markers (increased Ki67, but decreased PCNA). Nonetheless it remains likely that the overall pathway level message within our study - upregulation of proliferation - was a consequence of an early growth response to the biopsy itself. It is crucial to determine whether there is a genuine immune response associated with biopsy type as inflammatory signatures have been associated with poor anti-proliferative responses to aromatase inhibitors13,52. Clear evidence of both an immune or wound response signature53 was completely absent in our untreated samples, implying that any contribution of biopsy methodology to an inflammatory response is likely to be minor.
With the potential for pairwise variation irrespective of treatment, our study raises potential concerns of the suitability of the neoadjuvant window in gene expression profiling studies. Recent results of the ALTTO (Adjuvant Lapatinib And/Or Trastuzumab Treatment Optimisation) clinical trial were, however, found to be consistent with the predicted benefits from the neoALTTO trial. This supported the utility of the neoadjuvant setting as a suitable and important window for evaluating promising new targeted agents54, as well as the continued use of patient-matched samples to assess intervention studies for translational research. Nonetheless it remains critical to understand that whilst patient-matched samples reduce variation due to individuals, all possible sources of variation must be considered for an optimal experimental design. For example, in an intervention study to assess dietary changes on normal breast tissue from pre-menopausal women it was considered optimal to schedule the sequential biopsies one menstrual cycle apart, rather than using a fixed window of time55, as there is clear evidence that menstrual changes in oestrogen levels caused significant changes in gene expression56. Underlying tumour heterogeneity is an inevitable variable when performing neoadjuvant or window studies, however our study suggests that the method of sample collection should be considered along with treatment, time interval and clinicopathological features as an important potential confounding factor. These considerations are of particular importance if a study’s purpose is the development of a prognostic/predictive classifier or identification of a biomarker, with genes present in our NIT signature excluded from the analysis.
Our study demonstrates that consistent molecular changes arise in tumours in the absence of treatment and these can impact upon classification. These changes appear to be an artefactual ischemic response resulting from the sampling methodology itself, rather than reflecting the effects of a previous biopsy. Careful consideration should be given in future studies that seek to illustrate molecular changes between paired biopsies in the neoadjuvant setting for breast cancer and likely other cancers that make use of the same experimental design.
Additional Information
Accession Code: The raw and processed data from this study can be accessed from NCBI GEO under the accession GSE76728.
How to cite this article: Pearce, D. A. et al. Tumour sampling method can significantly influence gene expression profiles derived from neoadjuvant window studies. Sci. Rep. 6, 29434; doi: 10.1038/srep29434 (2016).
Supplementary Material
Acknowledgments
We are grateful for funding from Breast Cancer Now and to CRUK for a PhD stipend for DAP.
Footnotes
Author Contributions A.H.S., J.M.D. and J.M.S.B. conceived the study and designed the experiments. L.R., J.S.T. and J.M.D. collected and reviewed samples. V.S.S., L.M.A. and A.K.T. carried out experiments. D.A.P., L.M.A., A.K.T. and A.H.S. performed the analysis. D.A.P. and A.H.S. drafted the paper. All authors approved the final manuscript.
References
- Dekker T. J. A. et al. Reliability of core needle biopsy for determining ER and HER2 status in breast cancer. Ann. Oncol. 24, 931–937 (2013). [DOI] [PubMed] [Google Scholar]
- van de Vijver M. J. et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347, 1999–2009 (2002). [DOI] [PubMed] [Google Scholar]
- Filipits M. et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin. Cancer Res. 17, 6012–6020 (2011). [DOI] [PubMed] [Google Scholar]
- Ma X.-J. et al. A five-gene molecular grade index and HOXB13:IL17BR are complementary prognostic factors in early stage breast cancer. Clin. Cancer Res. 14, 2601–2608 (2008). [DOI] [PubMed] [Google Scholar]
- Jankowitz R. C. et al. Prognostic utility of the breast cancer index and comparison to Adjuvant! Online in a clinical case series of early breast cancer. Breast Cancer Res. 13, R98 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C. et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 98, 262–272 (2006). [DOI] [PubMed] [Google Scholar]
- Toussaint J. et al. Improvement of the clinical applicability of the Genomic Grade Index through a qRT-PCR test performed on frozen and formalin-fixed paraffin-embedded tissues. BMC Genomics 10, 424 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik S. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 351, 2817–2826 (2004). [DOI] [PubMed] [Google Scholar]
- Sorlie T. et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci.USA 100, 8418–8423 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ein-Dor L., Kela I., Getz G., Givol D. & Domany E. Outcome signature genes in breast cancer: is there a unique set? Bioinformatics 21, 171–178 (2005). [DOI] [PubMed] [Google Scholar]
- Sabine V. S. et al. Gene expression profiling of response to mTOR inhibitor everolimus in pre-operatively treated post-menopausal women with oestrogen receptor-positive breast cancer. Breast Cancer Res Treat 122, 419–428 (2010). [DOI] [PubMed] [Google Scholar]
- Sims A. H. & Bartlett J. M. Approaches towards expression profiling the response to treatment. Breast Cancer Res. 10, 115 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull A. K. et al. Accurate Prediction and Validation of Response to Endocrine Therapy in Breast Cancer. J. Clin. Oncol. 33, 2270–2278 (2015). [DOI] [PubMed] [Google Scholar]
- Sims A. H. Bioinformatics and breast cancer: what can high-throughput genomic approaches actually tell us? J Clin Pathol 62, 879–885 (2009). [DOI] [PubMed] [Google Scholar]
- Turnbull A. K. et al. Direct integration of intensity-level data from Affymetrix and Illumina microarrays improves statistical power for robust reanalysis. BMC Med. Genomics 5, 35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untch M., Konecny G. E., Paepke S. & von Minckwitz G. Current and future role of neoadjuvant therapy for breast cancer. Breast 23, 526–537 (2014). [DOI] [PubMed] [Google Scholar]
- Mamounas E. P. Facilitating breast-conserving surgery and preventing recurrence: aromatase inhibitors in the neoadjuvant and adjuvant settings. Ann. Surg. Oncol. 15, 691–703 (2008). [DOI] [PubMed] [Google Scholar]
- Macaskill E. J. & Dixon J. M. Neoadjuvant use of endocrine therapy in breast cancer. Breast J. 13, 243–250 (2007). [DOI] [PubMed] [Google Scholar]
- Arthur L. M. et al. Molecular Changes in Lobular Breast Cancers in Response to Endocrine Therapy. Cancer Res. 74, 5371–5376 (2014). [DOI] [PubMed] [Google Scholar]
- Morrogh M. et al. Differentially expressed genes in window trials are influenced by the wound-healing process: lessons learned from a pilot study with anastrozole. J Surg Res 176, 121–132 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandão R. D. et al. A randomised controlled phase II trial of pre-operative celecoxib treatment reveals anti-tumour transcriptional response in primary breast cancer. Breast Cancer Res. 15, R29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnedos M. et al. Discordance between core needle biopsy (CNB) and excisional biopsy (EB) for estrogen receptor (ER), progesterone receptor (PgR) and HER2 status in early breast cancer (EBC). Ann. Oncol. 20, 1948–1952 (2009). [DOI] [PubMed] [Google Scholar]
- Badoual C. et al. Pathological prognostic factors of invasive breast carcinoma in ultrasound-guided large core biopsies-correlation with subsequent surgical excisions. Breast 14, 22–27 (2005). [DOI] [PubMed] [Google Scholar]
- Cahill R. A., Walsh D., Landers R. J. & Watson R. G. Preoperative profiling of symptomatic breast cancer by diagnostic core biopsy. Ann. Surg. Oncol. 13, 45–51 (2006). [DOI] [PubMed] [Google Scholar]
- Loubeyre P. et al. Concordance between core needle biopsy and surgical excision specimens for tumour hormone receptor profiling according to the St. Gallen Classification, in clinical practice. Breast J. 19, 605–610 (2011). [DOI] [PubMed] [Google Scholar]
- Jeselsohn R. M. et al. Digital Quantification of Gene Expression in Sequential Breast Cancer Biopsies Reveals Activation of an Immune Response. PLos One 8, e64225 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger R. H., Rolfs A., Marti H. H., Bauer C. & Gassmann M. Hypoxia, a novel inducer of acute phase gene expression in a human hepatoma cell line. J. Biol. Chem. 270, 27865–27870 (1995). [DOI] [PubMed] [Google Scholar]
- Geyer F. C. et al. Molecular analysis reveals a genetic basis for the phenotypic diversity of metaplastic breast carcinomas. J. Pathol. 220, 562–573 (2010). [DOI] [PubMed] [Google Scholar]
- Du P., Kibbe W. A. & Lin S. M. lumi: a pipeline for processing Illumina microarray. Bioinformatics 24, 1547–1548 (2008). [DOI] [PubMed] [Google Scholar]
- Durinck S. et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440 (2005). [DOI] [PubMed] [Google Scholar]
- Durinck S., Spellman P. T., Birney E. & Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W. E., Li C. & Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- Haibe-Kains B., Schroeder M., Bontempi G., Sotiriou C. & Quackenbush J. genefu: Relevant Functions for Gene Expression Analysis, Especially in Breast Cancer. R package version 1.8.0 (2012). [Google Scholar]
- Schwender, H. siggenes: Multiple testing using SAM and Efron’s empirical Bayes approaches. (2012).
- Kitchen R. R. et al. Correcting for intra-experiment variation in Illumina BeadChip data is necessary to generate robust gene-expression profiles. BMC Genomics 11, 134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler T., Sen R. & Roy A. L. Regulation of Primary Response Genes. Mol. Cell 44, 348–360 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmedt C. et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin. Cancer Res. 14, 5158–5165 (2008). [DOI] [PubMed] [Google Scholar]
- Chang H. Y. et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc. Natl. Acad. Sci. USA 102, 3738–3743 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland K. I., Konstadoulakis M. M., Vezeridis M. P. & Wanebo H. J. Oncogene protein co-expression. Value of Ha-ras, c-myc, c-fos, and p53 as prognostic discriminants for breast carcinoma. Ann. Surg. 221, 706–718, discussion 718–20 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronski K. et al. Early growth response gene 1 (EGR1) is deleted in estrogen receptor-negative human breast carcinoma. Cancer 104, 925–930 (2005). [DOI] [PubMed] [Google Scholar]
- Langer S. et al. Jun and Fos family protein expression in human breast cancer: correlation of protein expression and clinicopathological parameters. Eur. J. Gynaecol. Oncol. 27, 345–352 (2006). [PubMed] [Google Scholar]
- Vleugel M. M., Greijer A. E., Bos R., van der Wall E. & van Diest P. J. c-Jun activation is associated with proliferation and angiogenesis in invasive breast cancer. Hum. Pathol. 37, 668–674 (2006). [DOI] [PubMed] [Google Scholar]
- Wolford C. C. et al. Transcription factor ATF3 links host adaptive response to breast cancer metastasis. J. Clin. Invest. 123, 2893–2906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka F. et al. EGRI and FOSB gene expressions in cancer stroma are independent prognostic indicators for epithelial ovarian cancer receiving standard therapy. Genes. Chromosomes Cancer 51, 300–312 (2012). [DOI] [PubMed] [Google Scholar]
- Huang J. et al. Effects of ischemia on gene expression. J. Surg. Res. 99, 222–227 (2001). [DOI] [PubMed] [Google Scholar]
- Dash A. et al. Changes in Differential Gene Expression because of Warm Ischemia Time of Radical Prostatectomy Specimens. Am. J. Pathol. 161, 1743–1748 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Knowles E. et al. Heterogeneity in global gene expression profiles between biopsy specimens taken peri-surgically from primary ER-positive breast carcinomas. Breast Cancer Res. 18, 39 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgan E. et al. Ischemia caused by time to freezing induces systematic microRNA and mRNA responses in cancer tissue. Mol. Oncol. 5, 564–576 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktas B. et al. Global gene expression changes induced by prolonged cold ischemic stress and preservation method of breast cancer tissue. Mol. Oncol. 8, 717–727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M. J. et al. Proliferating macrophages associated with high grade, hormone receptor negative breast cancer and poor clinical outcome. Breast Cancer Res. Treat. 128, 703–711 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrogh M. et al. Differentially Expressed Genes in Window Trials are Influenced by the Wound-Healing Process: Lessons Learned from a Pilot Study with Anastrozole. J. Surg. Res. 176, 121–132 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbier A. K. et al. Molecular profiling of aromatase inhibitor-treated post-menopausal breast tumors identifies immune-related correlates of resistance. Clin Cancer Res 19, 2775–2786 (2013). [DOI] [PubMed] [Google Scholar]
- Chang H. Y. et al. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol. 2, E7 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMichele A. et al. The Neoadjuvant Model Is Still the Future for Drug Development in Breast Cancer. Clin. Cancer Res. 21, 2911–2915 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong K. R. et al. Biomarkers of dietary energy restriction in women at increased risk of breast cancer. Cancer Prev Res (Phila Pa) 2, 720–731 (2009). [DOI] [PubMed] [Google Scholar]
- Wilson C. L., Sims A. H., Howell A., Miller C. J. & Clarke R. B. Effects of oestrogen on gene expression in epithelium and stroma of normal human breast tissue. Endocr Relat Cancer 13, 617–628 (2006). [DOI] [PubMed] [Google Scholar]
- Smith I. E. et al. A phase II placebo-controlled trial of neoadjuvant anastrozole alone or with gefitinib in early breast cancer. J. Clin. Oncol. 25, 3816–3822 (2007). [DOI] [PubMed] [Google Scholar]
- Sabine V. S. et al. Gene expression profiling of response to mTOR inhibitor everolimus in pre-operatively treated post-menopausal women with oestrogen receptor-positive breast cancer. Breast Cancer Res. Treat. 122, 419–428 (2010). [DOI] [PubMed] [Google Scholar]
- Magbanua M. J. M. et al. Serial expression analysis of breast tumors during neoadjuvant chemotherapy reveals changes in cell cycle and immune pathways associated with recurrence and response. Breast Cancer Res. 17, 73 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.