Abstract
Prions are formed of misfolded assemblies (PrPSc) of the variably N-glycosylated cellular prion protein (PrPC). In infected species, prions replicate by seeding the conversion and polymerization of host PrPC. Distinct prion strains can be recognized, exhibiting defined PrPSc biochemical properties such as the glycotype and specific biological traits. While strain information is encoded within the conformation of PrPSc assemblies, the storage of the structural information and the molecular requirements for self-perpetuation remain uncertain. Here, we investigated the specific role of PrPC glycosylation status. First, we developed an efficient protein misfolding cyclic amplification method using cells expressing the PrPC species of interest as substrate. Applying the technique to PrPC glycosylation mutants expressing cells revealed that neither PrPC nor PrPSc glycoform stoichiometry was instrumental to PrPSc formation and strainness perpetuation. Our study supports the view that strain properties, including PrPSc glycotype are enciphered within PrPSc structural backbone, not in the attached glycans.
Prion phenotype results from the conformational change of specific amyloidogenic proteins. This change is based on the self-sustained transfer of a structural information from a protein conformer in the prion state to the same protein in the non-prion conformation, presumably through a seeding-polymerization process. Initially formulated to explain prion diseases pathogenesis in human and animals, the prion concept has gained wider relevance in the regulation of diverse biological process and in the progression of other neurodegenerative disorders such as Alzheimer and Parkinson diseases1,2,3. Mammalian prions are primarily formed of macromolecular assemblies of PrPSc, a misfolded, ß-sheet enriched form of the ubiquitously expressed, α-helix rich, host-encoded prion glycoprotein PrPC. Within defined host species, PrPC can be transconformed in many prion variants or strains, differing in their PrPSc conformation at the level of the tertiary and/quaternary structure and in their biological properties4,5,6,7. In particular, prions maintain strain-specific stoichiometric ratios of PrPSc glycoforms on serial passaging in the same host species8,9, leading to the view that glycans may somehow participate to prion strain information encoding. Consistently, transgenic modelling suggested that PrPC glycosylation status influenced the efficacy of intra- and cross-species transmission of prions10,11,12 and prion strain properties13. However, such studies remained difficult to interpret, given that point mutations inserted to prevent N-linked glycosylation or altered trafficking of the mutant PrPC rather than N-glycans removal may be the primary cause for the observed alterations in the propagation of prions (see ref. 14 and references herein). The intrinsic convertibility of PrPC glycosylation mutants into PrPSc and the role of attached glycans in prion strainness remain thus an open question.
While the molecular mechanisms and the cellular factors potentially involved in PrPSc formation remain largely undefined, PrPC is convertible into PrPSc in a test tube after adjunction of minute amounts of PrPSc seeds by a technique designated protein misfolding cyclic amplification (PMCA)15. PMCA increases the ability of PrPSc to template the conversion of PrPC by repetitive cycles of incubation and sonication. As the main source of PrPC substrate, most of the proprietary PMCA protocols are using brain homogenate from susceptible animals or transgenic mouse models expressing the PrPC of interest. The sensitivity achieved by PMCA allows amplification of subinfectious levels of PrPSc in biological samples such as blood, urine, faeces or cerebrospinal fluid of human and animals infected with prions16,17,18,19,20. PMCA products or ‘amplicons’ are truly infectious and generally exhibit the same strain properties as the PrPSc seeds21,22,23,24. A limited number of experiments have been performed by replacing brain substrate with cell substrate25,26,27,28 despite the availability of a number of cell models expressing PrPC from different species, and permissive to prions (for review29). These cell-based PMCA assays generally yielded either low PrP conversion rates or unsatisfying sensitivity to be applied routinely in high throughput protocols. While using brain material is not a limiting step for routine use of PMCA, addressing the contribution to the prion conversion process of certain PrPC polymorphisms or mutations or post-translational modifications, such as glycosylation becomes an issue with this technique when suitable transgenic mouse models are not available.
In the present study, we adapted the miniaturized-bead PMCA (mb-PMCA) protocol22 to the use, as PrPC substrate, of cell lysates from RK13 cell lines30 expressing PrPC from different species or with point mutation. We report highly efficient amplification of scrapie, hamster, human and to a lesser extent of mouse-adapted prion strains. We next addressed the question of the prion convertibility of several ovine PrP glycosylation mutants defective in glycosylation at either sites of PrP or at both sites14. At variance with earlier reports using cell14,31 or transgenic mouse11 modelling, PrPC glycosylation mutants were converted as efficiently as wild-type PrPC into bona fide prions by PMCA with two unrelated prion strains. Our study also reveals that interactions between defined stoichiometric ratios of neither PrPC nor PrPSc glycoforms are key to PrPSc formation, leading to the view that strain-specific glycotype is enciphered within PrP backbone. In ethical and practical terms, the development of a highly sensitive cell lysate-based PMCA will allow reduction and even bypassing the use of animal tissues.
Results
Cell-based mb-PMCA efficiently amplifies prions
Previously, we developed a so-called mb-PCMCA procedure allowing efficient amplification of prions from different species in a round of 48 hours and in a micro-plate format18,22,32. The experimental conditions were primarily established with brain material from transgenic tg338 mice overexpressing ovine PrPC and the 127S scrapie strain, a prototypal ‘fast’ strain killing tg338 mice within 2 months33. To progressively replace tg338 brain substrate by cell substrate, we used rabbit kidney epithelial RK13 cells expressing constitutively the VRQ allele of ovine PrPC (P2FJ6 clone), and susceptible to 127S prions32,34. At the same protein concentration, these cells express approximately 2/3 of PrPC levels compared with tg338 mouse brain, and the PrPC glycoform pattern is cell-specific (Fig. 1A and refs 30,35). The cell-based mb-PMCA (Cell-mb-PMCA) procedure was performed by seeding serial 10-fold dilutions of 127S-infected brain homogenate in P2FJ6 cell lysates, and running 96 incubation/sonication cycles for a round of 48 h in 96-well microplates. The amplicons were treated with proteinase K (PK) to eliminate PrPC before detection of PK-resistant PrPSc (PrPres) by Western-blotting.
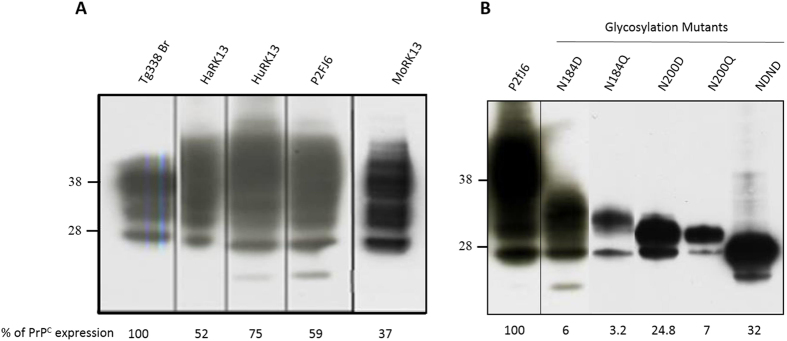
Figure 1. Electrophoretic profiles and expression levels of PrPC in the different RK13 cell lines used.
(A) RK13 cell lines expressing hamster (HaRK13), human (HuRK13), sheep (P2FJ6 clone) and mouse (MoRK13) wild-type PrP and (B) the different ovine PrP glycosylation mutants (as referenced in Table 2) were probed for the presence of PrPC by western blotting (SAF34 anti-PrP monoclonal antibody). Brain lysate of ovine PrP tg338 mice (A) or cell lysates from P2FJ6 clone (B) were used to determine the relative expression levels in the different cell lines. Migration size of standard molecular mass marker (kDa) is indicated on the left.
Cell-mb-PMCA efficiency to amplify 127S prions was tightly dependent of the total protein concentration in the cell lysate. At the concentration of 2 mg/mL, there was no significant amplification of 127S seeds (not shown). At 6 mg/mL, PrPres was detected in reaction mixtures seeded with 104-fold diluted 127S brain homogenate. Further concentration of the cell lysate to 10 mg/mL led to detection of PrPres from reaction mixtures seeded with 105–106-fold diluted inoculum (Fig. 2). Supplementation of the 10 mg/mL cell lysate with brain homogenate from PrP0/0 mice (1:1 ratio) allowed detection of PrPres in 127S brain homogenate diluted up to 107-fold (Fig. 2). PMCA performed with untransfected parental RK13 cells resulted in no detectable amplification of PrPres (Fig. S1), consistent with the non-detection of endogenous rabbit PrPC in these cells30 and the poor convertibility of rabbit PrPC by 127S-like prions36.
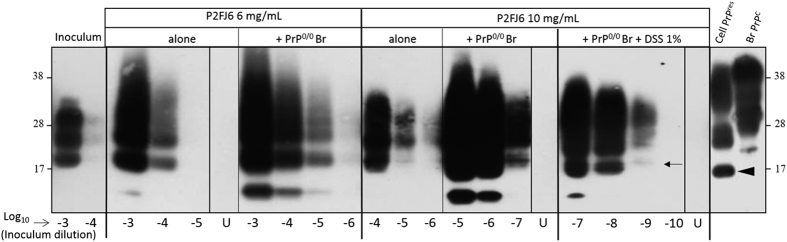
Figure 2. Endpoint titration of 127S prions by Cell lysate based mb-PMCA supplemented with PrP knockout brain.
P2FJ6 cell lysates were prepared at two protein concentrations and used either alone or mixed (1:1) with 10% PrP0/0 mouse brain (Br) lysate in the absence and the presence of 1% Dextran sulphate sodium (DSS), as indicated. The lysates were then used as PMCA substrate to amplify serial 10-fold dilutions of brain homogenate from tg338 mice infected with 127S prions. Each dilution was directly analysed by Western blotting for PrPres content (Sha31 antibody) after proteinase K treatment. For comparison purposes, the first two lanes illustrate PrPres content in non-amplified products (10−3 and 10−4 dilutions); the last two lanes PrPres and PrPC electrophoretic profiles in 127S infected P2FJ6 cell line (Cell PrPres) and normal tg338 mouse brain lysates (Br PrPC), respectively. Lanes U correspond to unseeded lysates ran on the same microplate. Note the difference in unglysosylated PrPres molecular mass between Cell-PMCA generated products (small arrow) and cell-passaged prions (arrowhead). *Low size PrPres fragments.
Various molecules reportedly enhance prion conversion and amplification, in a strain-dependent manner37,38,39,40,41,42,43. We examined whether addition of negatively charged molecules, such as dextran sulfate (DSS) would further improve Cell-mb-PMCA sensitivity. Addition of 1% DSS (>500 kDa) led to the amplification of the 127S strain up to 10−9 dilution, i.e. approx. 10–100-fold less the sensitivity routinely obtained with tg338 brain as substrate22. All the unseeded samples were negative in these experiments (Fig. 2, lanes U). The conversion yield of PrPC present in the cell lysate, as examined after thermolysin treatment of the amplified products22 was ≤30% (data not shown), as previously observed with tg338 mouse brain material22.
Compared to brain PrPres (Fig. 2, lanes Inoculum), the PrPres glycoprofile of the amplified product was cell-specific, consistent with the differences in PrPC glycans content between brain and non-neuronal cultured cell models30,35. Furthermore, the cell-mb-PMCA amplicon exhibited a higher molecular mass (~2 kDa) than that of PrPres accumulating in infected P2FJ6/Rov cell cultures (Fig. 2, compare with Cell PrPres lane). In Rov cells, biosynthetized PrPSc is naturally cleaved by cathepsin proteases, to produce the so-called C2 fragment. This fragment is more truncated than the PK-resistant core of 127S PrPSc 35. Conceivably, interactions between PrPSc and cathepsin proteases may not occur during Cell-mb-PMCA, due to disruption of endolysosomial vesicles by the detergent used in the PMCA lysis buffer.
We next determined whether the infectivity of the amplified products generated using cell lysates would correlate with the efficacy of amplification. The amplicons obtained with the 10−7 127S seed with 10 mg/mL cell lysates supplemented with PrP0/0 brain, in the presence or absence of 1% DSS, were tenfold diluted up to the 10−7 dilution and immediately inoculated intracerebrally to reporter tg338 mice (Table 1). Mice inoculated with unseeded controls did not develop any clinical disease and were euthanized healthy at 240 days post-inoculation. A 100% attack rate was observed with the cell-generated amplicons diluted up to 10−4 fold. At the 10−5 dilution, 1/5 (−DSS) and 2/5 (+DSS) tg338 mice were infected. At the 10−6 and 10−7 dilution, none of the mice developed the disease and were euthanized healthy. There was thus no significant impact of adding DSS to the PMCA reaction on the infectivity of the amplified products. Collectively, these data indicate that the cell-generated amplicons were 100-fold less infectious than the brain-generated amplicons (Table 1 and ref. 22). This value was consistent with the difference of amplification efficiency observed between cell and brain lysates.
Table 1. Incubation time of tg338 mice inoculated with serial tenfold dilutions of Cell-based mb-PMCA-generated 127S prions.
| Dilution | Incubation time in days ± SEM (n/n0) | ||
|---|---|---|---|
| PMCA substrate | |||
| Cell lysate − DSS | Cell lysate + DSS | tg338 brain | |
| 10−1 | 75 ± 1 (5/5) | 93 ± 1 (5/5) | 60 ± 1 (4/4) |
| 10−3 | 91 ± 2 (5/5) | 100 ± 4 (5/5) | 72 ± 2 (5/5) |
| 10−4 | 108 ± 6 (5/5) | ND | |
| 10−5 | 114 (1/5)* | 98; 124 (2/5)* | 98 ± 3 (5/5) |
| 10−6 | >240 (0/5)* | ND | |
| 10−7 | >240 (0/5)* | >240 (0/5)* | 124 (1/5) |
| Unseeded | >240 (0/5)* | >240 (0/5)* | |
Amplicons obtained from a 10−7 127S seed mixed with 10 mg/mL cell lysates supplemented with PrP0/0 brain, in the absence (−DSS) or presence (+DSS) of 1% dextran sulfate, were tenfold diluted up to the 10−7 dilution and immediately inoculated intracerebrally to reporter tg338 mice. n/n0: number of mice with neurological disease and positive for PrPres in the brain by immunoblotting/number of inoculated tg338 mice.
*Non-affected mice euthanized healthy at 240 dpi. Data in italic are from22. ND: not done.
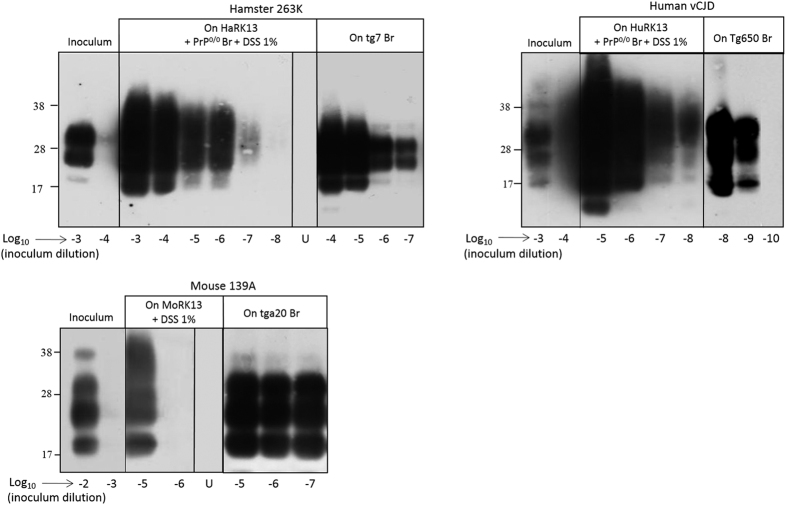
We next examined whether the Cell-mb-PMCA protocol (10 mg/mL protein concentration, addition of 1% DSS and PrP0/0 brain) would amplify prion from other species. We seeded RK13 cell lysates expressing hamster (HaRK13), human (methionine at codon 129, HuRK13) or mouse (MoRK13) PrPC (Fig. 144 and unpublished data) with serial dilutions of 263K, vCJD and 139A prions, respectively and compared the sensitivity achieved with that obtained with transgenic mouse brain as substrate. The results are summarized in Fig. 3, which is representative of more than 4 independent experiments. PrPres from hamster 263 K and human vCJD prions was amplified from 10−7- and 10−8-diluted input seeds by using HaRK13 and HuRK13 cell substrate, respectively. This sensitivity was close to that obtained with transgenic mouse brain (Fig. 3 and ref. 22). In contrast, 139A prions were less efficiently amplified, as two PMCA rounds without PrP0/0 brain supplementation were necessary for PrPres detection from the 10−5 dilution, compared to the 10−7 dilution amplified in one round with tga20 brain lysate (Fig. 3 and ref. 22). More sensitive cell clones expressing higher levels of mouse PrPC (Fig. 1) or Mo RK13 cell-specific conditions are to be found to improve the amplification of 139A prions.
Figure 3. Comparison of the efficacy of brain-based or cell-based PMCA to amplify hamster, vCJD and mouse prions.
Lysates from RK13 cell lines expressing hamster, human and mouse PrPC (supplemented with 1% DSS and eventually PrP0/0 brain (Br)) or brain homogenates from hamster PrP (tg7), human PrP (tg650) and mouse PrP (tga20) mice were seeded with serial dilutions of brain homogenates containing hamster 263K prions, human vCJD prions or mouse 139A prions and submitted to a single round (263K, vCJD) or 2 rounds (139A) of PMCA. Unamplified inoculums (two first lanes of each panel), unseeded controls (lanes U) and the amplified samples were digested with PK before western blotting (Sha31 antibody) analysis for PrPres content.
Collectively, our data indicate that Cell-based PMCA, as mouse brain based mb-PMCA22, is a versatile protocol, allowing amplification of minutes amounts of prions from different species, including human.
Cell lysates with high protein concentration avoid the use of PrP knock-out mouse brain material and DSS for efficient prion amplification
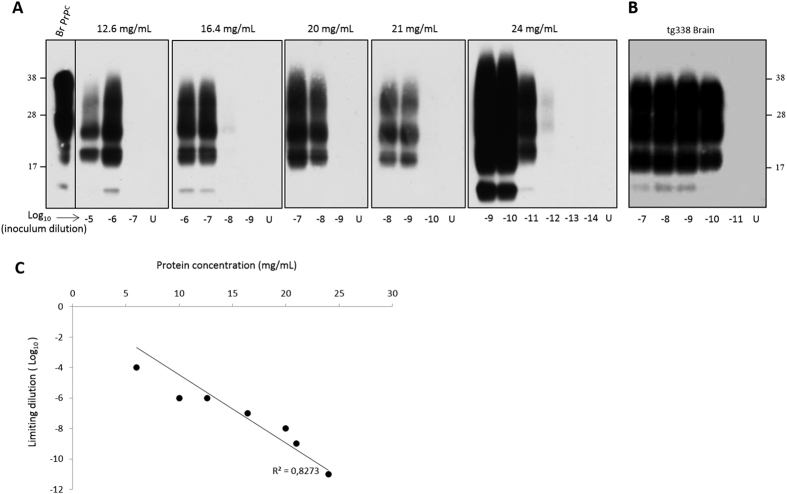
The positive correlation between the protein concentration in the cell lysate used as substrate and the PMCA sensitivity to detect 127S prions led us to reason that increasing further the total protein concentration over 10 mg/mL in the cell lysates might allow efficient prion amplification without additives. To obtain highly concentrated cell lysates, we cultivated P2FJ6 cells in multilayer preparative flasks. The cell lysates were then used ‘crude’ in Cell-mb-PMCA reactions. As shown in Fig. 4A, concentrating cell lysates from 12 mg/mL to 24 mg/mL increased the sensitivity of 127S detection by 5 Log10. At that concentration, the sensitivity achieved was similar or even higher than that obtained with tg338 brain substrate run in the same micro-plate (Fig. 4B). Reporting the limiting dilution achieved to the total protein concentration in the cell lysate showed a strong correlation between total protein concentration and the efficacy of PMCA amplification (Fig. 4C). Of note, a 10% brain homogenate would provide a 10–12 mg/mL protein concentration. Thus, the use of highly concentrated cell lysate with regard to protein content allows amplifying minute amounts of 127S prions in a brain-free context.
Figure 4. Highly efficient amplification of 127S prions using concentrated cell lysate alone by a single round of Cell-mb-PMCA.
(A) P2FJ6 cell lysates were concentrated with regard to the total protein concentration and were directly seeded with serial 10-fold dilutions of tg338 brain homogenate containing 127S prions for a single round Cell-mb-PMCA. (B) In parallel, the same dilutions were submitted to mb-PMCA by using tg338 brain homogenate as substrate. Unseeded controls (lanes U) and the amplified samples were digested with PK before western blotting analysis for PrPres content (Sha31 antibody). The first lane contains undigested normal tg338 mouse brain. (C) Regression analysis of 127S endpoint dilution achieved by cell-mb-PMCA versus total protein concentration in the P2FJ6 cell lysate.
Efficient Cell-based mb-PMCA conversion of ovine PrP glycosylation mutants by two distinct prion strains
PrPC has two variably occupied glycosylation sites at amino acid N184 (site 1) and N200 (site 2) (ovine PrP sequence numbering). Modelling in RK13 cells previously suggested that unglycosylated double PrP mutant failed to be converted by 127S prions, even after being properly expressed at the cell surface by an ectopic glycosylation site in the N-terminus of PrPC 14. Prion convertibility of monoglycosylated mutant was site-dependent with mutants at site 2 all being convertible and mutants at site 1 being convertible only when the N184D amino acid substitution was performed. These negative results opened the possibility that some mutants were intrinsically not convertible into prions, due to point mutation or to N-glycans removal. We examined this possibility by seeding the different cell lysate mutants (Fig. 5 and Table 2) with serially diluted seeds of 127S prions and running Cell-mb-PMCA (10 mg/mL cell lysate supplemented with 10% PrP0/0 brain homogenate, no DSS).
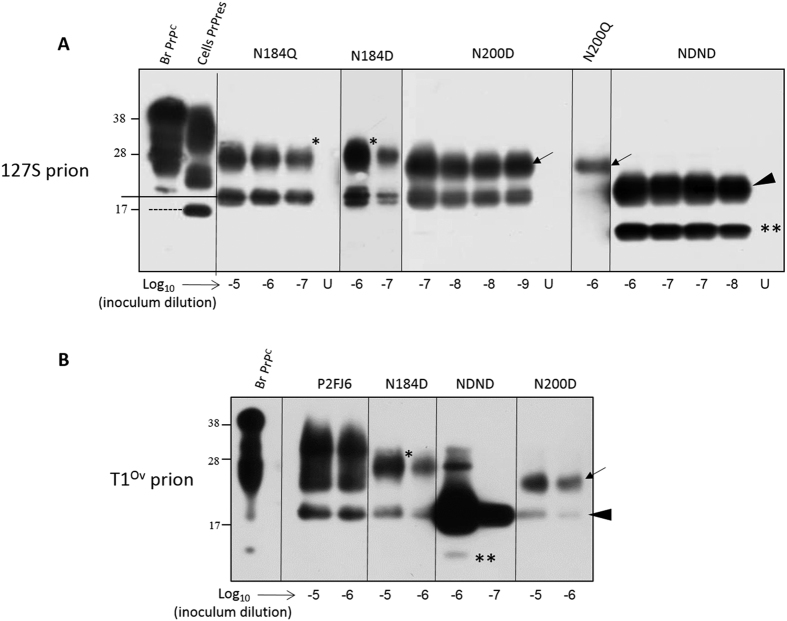
Figure 5. Efficient conversion of PrPC glycosylation mutants by two distinct prions in Cell-mb-PMCA reactions.
Lysates from RK13 cells expressing ovine PrPC mutated on the first N-glycosylation site at residue 184 (N184Q, N184D), on the second glycosylation site at residue 200 (N200D, N200Q) or at both residue (NDND) glycosylation sites were mixed with PrP0/0 brain lysate (1:1 dilution), seeded with serial 10-fold dilutions of tg338 brain homogenate containing either 127S (A) or T1Ov (B) prions and submitted to 2 rounds of Cell-mb-PMCA. Lanes U contains unseeded controls. All samples were PK-treated before western blot analysis (Sha31 antibody). For comparison, the electrophoretic profiles of tg338 brain PrPC (lane Br. PrPC) and P2FJ6 PrPres (lane Cell PrPres) are shown. To facilitate the interpretation of the western blot, PrPres monoglycosylated at residue 200 or 184 are indicated by asterisks and arrows, respectively. Full-length unglycosylated PrPres is indicated by an arrowhead. **Indicates low size PrPres fragments, highly abundant following conversion of unglycosylated PrPC by 127S but not by T1Ov prions.
Table 2. Convertibility of the PrP glycosylation mutants in cell culture and by cell PMCA.
| Glycotype | Genotype | Cell culture conversion* | Cell-PMCA conversion | Incubation time of tg338 mice inoculated with Cell-PMCA amplicons in days ± SEM (n/n0) |
|---|---|---|---|---|
| Wild type (VRQ) | Wild type | + | + | 70 ± 1 (5/5) |
| Monoglycosylated (Site 1) | N184D | + | + | 68 ± 1 (5/5) |
| N184Q | _ | + | 77 ± 1 (5/5) | |
| Monoglycosylated (Site 2) | N200D | + | + | 69 ± 1 (5/5) |
| N200Q | + | + | nd | |
| Unglysosylated | N184D-N200D | − | + | 68 ± 1 (5/5) |
| Unglysosylated mutants → tg338 brain PMCA# | + | 65 ± 1 (5/5) | ||
| Unseeded Wild type (VRQ) | none | − | >250 (0/5) |
n/n0: number of mice with neurological disease and positive for PrPres in the brain by immunoblotting/number of inoculated tg338 mice. The seeds used to infect mice were from a 127S 10−7 seed amplified over 2 rounds, so as to avoid any residual input.
*Cell conversion was assessed in ref. 14.
#The PMCA product inoculated was obtained by seeding tg338 brain lysate with unglycosylated PrPSc seed (10−7 dilution), itself obtained by seeding the NDND double mutant cell lysate with 10−8 diluted 127S seed. nd: not done.
Figure 1B illustrates PrPC electrophoretic pattern and expression level in the different glycosylation mutant cell lysates relative to the wild type P2FJ6 cells. Overall, PrPC expression level in the different PrP glycosylation mutants was low, ranging from ~3% (N184Q) to 32% (NDND double mutant). All the PrPC glycosylation mutants were converted into PrPSc by Cell-mb-PMCA. In one round, the limiting dilution of the 127S input seed established at 10−5 for N184Q and N200Q and 10−6 for N200D and NDND mutants. Despite low expression levels of mutant PrPC in the cell lysates, the sensitivity achieved was thus only 100- (N184Q) and 10-fold less (N200D, NDND) than that observed by using wild-type cell lysates. After a second round (Fig. 5A), the limiting dilution established between 10−7 and 10−9 for all but the N200Q mutants, which established at 10−6. Taken together, these results demonstrate that unglycosylated and monoglycosylated PrPC mutants are intrinsically convertible into PrPSc by 127S prions, independently of their non-convertibility once expressed in RK13 cells.
We next examined whether the absence of PrPC glycosylation requirement for in vitro prion conversion would apply to another prion strain designated T1Ov, and obtained after adaption to tg338 mice of prions responsible for a rare cortical, MM2 form of sporadic Creutzfeldt-Jakob disease45. T1Ov prions have no strain properties in common with 127S prions but share similar efficacy to be amplified by PMCA using tg338 brain as substrate45. In two rounds, the limiting dilution of the T1Ov input seed established at 10−6 for all the N184D and N200D mutants, as for wild-type PrP, and 10−7 for the NDND mutant (Fig. 5B). It can be noted that the proportion of low-size PrPres fragments in the lowly-glycosylated amplicons markedly differed between the two strains (Fig. 5), further differentiating the two agents.
Cell-based mb-PMCA lowly glycosylated prions are highly infectious
We finally addressed whether the monoglycosylated or unglycosylated PrPSc products generated by glycosylation mutant Cell mb-PMCA were infectious and retained strain-specific biochemical and neuropathological properties. Amplicons generated from reaction mixtures seeded with 10−7 127S brain material, -and amplified over 2 rounds to exclude any residual infectivity of the input seed-, were inoculated by intracerebral route to reporter tg338 mice. As shown in Table 2, the amplified products generated with the glycosylation mutants induced disease in mice with similar efficacy as the products generated on the wild-type PrP cell substrate. Mean incubation time to disease was the shortest after inoculation of the non-glycosylated amplicons (68 days) compared to wild-type generated amplicons (70 days). N184Q-derived amplicons were the less efficient, inducing disease in 77 days. Reporting the incubation time values to 127S dose-response curve34 and quantifying the amount of PrPres in the amplicons allowed calculating specific infectivity values, that is the amount of infectivity per molecule of PrPres generated by the PMCA reaction. Assuming a straight correlation between PrPres content in the PMCA amplicons and infectivity, the specific infectivity per unit PrPres appeared 10 to 80-fold higher for the lowly –glycosylated amplicons than for the wild-type amplicons (Fig. S2).
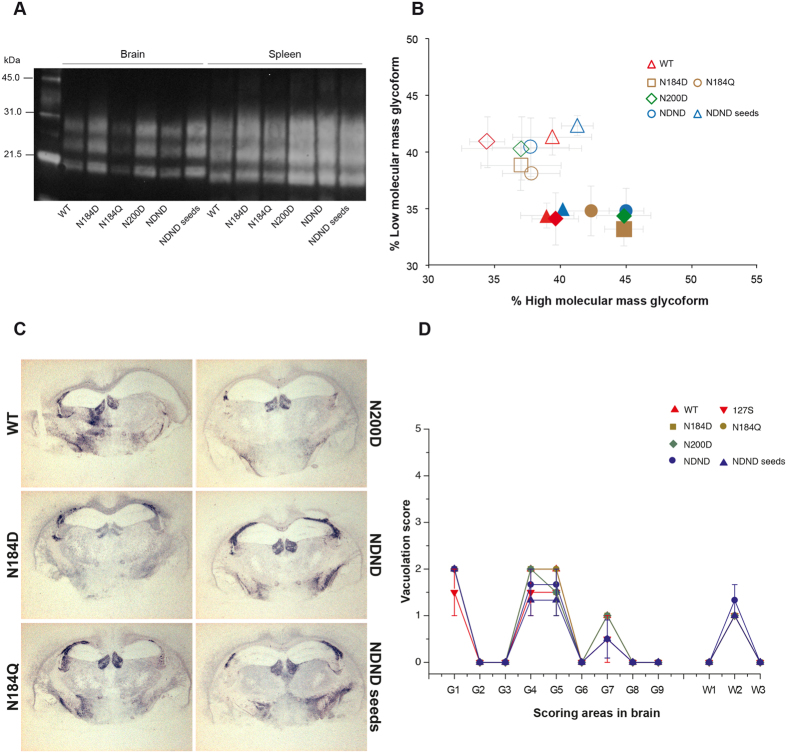
Remarkably, PrPres electrophoretic pattern in brain and spleen tissue (Fig. 6A,B), and neuroanatomical distribution of PrPres (Figs 6C and S3) and of vacuolar degeneration (Fig. 6D) in the reporter tg338 mice inoculated with unglycosylated or monoglycosylated 127S amplicons was reminiscent of 127S prions, passaged (Fig. 6) or not19,33,46,47 by Cell-mb-PMCA. Prominent PrPres deposition in the lateral hypothalamic area, in the corpus callosum, in the habenula (Fig. 6C) in the raphe nuclei of the brain stem (Fig. S3), and marked vacuolar degeneration in the dorsal medulla, hypothalamus and white matter of the mesencephalic tegmentum (Fig. 6D) were typical of 127S prions33. PrPres staining and vacuolation in the affected brain regions were sometimes less intense, as observed with infection of diluted 127S-infected tg338 brain homogenate33 or on reisolation of 127S prions to tg338 mice19,46,47.
Figure 6. Strain properties of Cell-mb-PMCA lowly-glycosylated 127S prions in tg338 mice.
Lysates from RK13 cells expressing wild-type (WT) ovine PrPC or ovine PrPC mutated on the first N-glycosylation site at residue 184 (N184D, N184Q), on the second glycosylation site at residue 200 (N200D) or at both residue (NDND) glycosylation sites were mixed with PrP0/0 brain lysate (1:1 dilution), seeded with serial 10-fold dilutions of tg338 brain homogenate containing 127S prions and submitted to 2 rounds of Cell-mb-PMCA before inoculation to tg338 mice. Seeds generated from NDND cells were also submitted to another round of PMCA using tg338 mouse brain as substrate (wild type brain PrPC). The amplicon obtained at 10−8 dilution was then used for inoculation (NDND seeds). (A) PrPres banding pattern and (B) ratios of high- and low-molecular mass PrPres glycoforms in the brain (filled symbol) and spleen (open symbol) tissue of tg338 mice inoculated with Cell-mb-PMCA products. (C) Neuroanatomical distribution of PrPres in tg338 mice inoculated with the Cell-mb-PMCA products. Representative histoblot (12F10 antibody) of brain coronal section (hippocampus level). Deposition in standardized anterio-posterior sections can be vizualized in Supplementary Figure S1. (D) Distribution of vacuolar degeneration (lesion profile) in tg338 mouse brain inoculated with the Cell-mb-PMCA products, as above. The intensity of vacuolation was scored as means standard errors of the means (error bars) in standard gray (G1 to G9) and white (W1 to W3) matter areas. These areas are as follows: G1, dorsal medulla; G2, cerebellar cortex; G3, superior colliculus; G4, hypothalamus; G5, medial thalamus; G6, hippocampus; G7, septum; G8, medial cerebral cortex at the level of the thalamus; G9, medial cerebral cortex at the level of the septum; W1, cerebellar white matter; W2, white matter of the mesencephalic tegmentum; and W3, pyramidal tract.
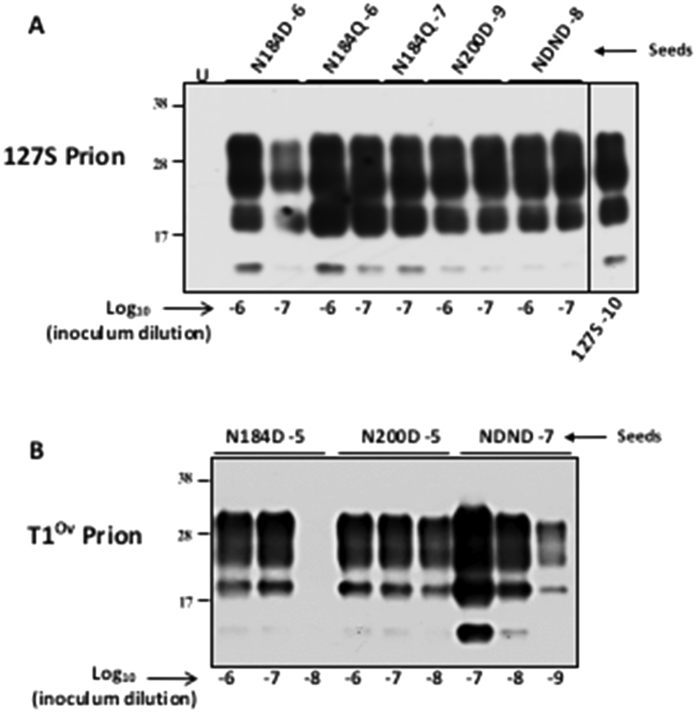
To further ascertain that PMCA-generated lowly glycosylated amplicons were good convertors of wild-type ovine PrPVRQ, 127S and T1Ov amplicons were submitted to mb-PMCA using tg338 mouse brain as substrate. The seeding activity of 127S and T1Ov amplicons was observed up to the 10−7 and 10−9 dilution (Fig. 7A,B), as with 127S amplicons generated from wild-type cells (Fig. 2) or with T1Ov prions from brains of terminally sick tg338 mice45, respectively.
Figure 7. Converting activity of PMCA-generated lowly glycosylated prions.
Serial dilutions of lowly-glycosylated 127S (A) and T1Ov (B) prions generated by cell-mb-PMCA (at the indicated Log10 dilution) were submitted to mb-PMCA using tg338 mouse brain as substrate. All the samples were PK-treated before western blot analyses. Amplification of 127S prions is shown for comparison (A). U: unseeded control.
One of the 127S PMCA products generated with unglycosylated PrPres seed (10−8) was inoculated to reporter tg338 mice to further confirm efficient conversion and maintaining of strain properties. The survival time of the mice, PrPres electrophoretic pattern in brain and spleen tissue, and PrPres/vacuolar deposition patterns in the brain were all consistent with the generation of (highly) infectious 127S prions (Table 2, Figs 6 and S3).
Collectively, these data indicate that 127S prion strain properties and T1Ov seeding capacity were essentially conserved despite intermediate replication on lowly glycosylated PrPC species. This lends support to the view that glycans do not play a major role in prion replication dynamics and strain biological properties.
Discussion
Following our simplification of the PMCA method, we now report that cell lysate expressing PrPC can conveniently replace brain substrate from PrP transgenic mice to achieve efficient amplification of prions from different species. Highly concentrated cell lysate may permit amplification at ‘maximal’ levels without the need to supplement the reaction mixture with PrP knockout brain substrate. Applying the cell-PMCA technique to a panel of cells expressing PrPC glycosylation mutants and two prion strains demonstrates that unglycosylated and monoglycosylated mutants are intrinsically convertible and that PrPC and/or PrPSc glycoforms stoichiometry does appear to alter neither PrPSc formation rate in vitro nor the biological properties of the formed prion (for at least the strain tested in vivo). PrP glycosylation may thus be dispensable to perpetuate prion strain information.
To approach with cell lysates the sensitivity obtained in one round of PMCA with the ad hoc transgenic mouse brain as substrate22, it was beneficial to use concentrated cell lysate with respect to total protein concentration, and to supplement it with PrP0/0 mouse brain lysate and 1% DSS. By using the 127S prions/ovine PrPC combination, we further showed that the PMCA amplification threshold obtained with brain material could be reached by using highly concentrated cell lysate alone, at least with 127S prions. The respective contributions of DSS, PrP0/0 mouse brain and concentrated cell lysate to efficient prion conversion remain to be determined. Non-PrPC cellular factors such as brain lipids or polyanionic scaffold molecules like sulphated glycans and RNA, which are known to improve PMCA37,39,40,43,48,49,50, may have been concentrated. The conditions used may also create a macromolecular crowded environment51 favouring highly efficient prion conversion. In ethical and practical terms, sensitive PMCA can thus be performed without requiring animal models.
Applying the cell-PMCA technique to a panel of cells expressing PrPC glycosylation mutants demonstrates that unglycosylated and monoglycosylated PrPC were intrinsically convertible by 127S prions, despite non convertibility in cultured Rov cells, even after apparent proper expression at the cell surface during biosynthesis14 or exposure to homologous prions (this study). The reasons for such discrepancies with regards to glycosylation requirements between cell-free and in-cell systems remain to be determined. Subtle alterations in the subcellular localisation/trafficking of the PrPC mutants or different turnover could explain their non-conversion in the cell models. Folding and/or stability and/or resistance to clearance of the nascent PrPSc assemblies in Rov cells may necessitate incorporation of a certain threshold of di-glycosylated species.
The molecular basis for prion strain-specific glycopattern and its perpetuation over serial passage is poorly understood. Host PrPC glycosylation has been reported to contribute to prion replication and to prion strain phenotype (reviews refs 52, 53, 54, 55). Both the infecting prions and the convertible PrPC isoforms in the recipient host or tissue determine the glycopattern of each strain. Use of biochemically deglycosylated native PrPC in PMCA reaction suggested that the stoichiometry of PrPC glycoforms regulated prion formation in a strain-specific manner56. For example, formation of PrPSc on seeding with mouse RML or hamster Sc237 prions necessitates presence or absence of unglycosylated PrPC, respectively. Oppositely, the failure of PrPC glycoform-specific antibodies57 to exert similar selectively towards PrPSc glycoforms58 lend to the proposal that the proportion of each PrPC glycoform incorporated into nascent PrPSc assemblies was controlled by the defined glycoforms stoichiometry in the starting infectious seeds52,58. Indirectly supporting this hypothesis is the observation that the PrPSc glycoform ratio (for a given strain) is conserved whatever PrPSc aggregation size32,34. What information does bring our PMCA modeling with cell expressing PrP glycosylation mutants? First, the high conversion rate of the mono- and un-glycosylated PrPC mutants relative to wild-type PrPC, despite expression at lowered levels in the cell lysates, would sustain the view that highly glycosylated PrPC species interfere with prion conversion or that presence of N-linked glycans on the two sites in PrPC cause steric hindrance for PrPSc formation or through stabilisation of the PrPC native state. The latter point would be consistent with the observation that the structural sequence important for PrP oligomerization lies between the two N-glycosylation sites59 or just upstream28,60. Diglycosylated PrPC species may thus have a dual role during the formation of PrPSc assemblies. Second, 127S prion seeds, which exhibit in tg338 mouse brain a determined PrPres glycotype (45% diglycosylated species, 35% monoglycosylated, 20% unglycosylated22), convert indifferently unglycosylated and monoglycosylated PrPC species alone or in combination. Because conversion is not monitored in real-time during PMCA reactions, a glycotypic preference may exist during the initial converting events but be trailed off within a 48 h round. It could be argued that in face of mono or unglycosylated PrPC species, mono and unglycosylated PrPSc may have been preferentially amplified. However, when the opposite experiment was done, that is when PMCA-generated unglycosylated or monoglycosylated PrPSc seeds were submitted to PMCA in the presence of wild-type PrPC, the initial 127S PrPSc glycotype was fully restored, thus suggesting no preferential compatibility between PrPC and PrPSc with regard to the occupancy of the glycosylation sites. The same observations were made with a Creutzfeldt-Jakob disease derived prion strain designated T1Ov, thus indicating that the non-requirement of PrP glycosylation for prion conversion is not limited to one peculiar strain.
We finally show that monoglycosylated and unglycosylated 127S amplicons share similar strain properties as normally glycosylated 127S prions in tg338 mice, including the PrPres glycotype in the brain of the mice. Collectively, we can conclude that a defined stoichiometry of PrPSc glycoforms and of PrPC glycoforms is not necessary for efficient conversion by PMCA, and to dictate strain-specific properties, at least for 127S prions. Prion strain properties, including the glycotype stoichiometry of PrPSc, may thus be solely enciphered within PrPSc structural backbone or within the way PrPSc molecules do assemble.
Methods
Ethics Statement
All animal experiments were carried out in accordance with the European Union directive 2010/63 and were approved by COMETHEA, the local ethics committee of the authors’ institution (permit number 12/034).
Transgenic mice and Prion strains
The transgenic lines (tg338, tg7, tga20 and tg650 lines) and prions (127S, T1Ov, 139A, 263K and vCJD) have been previously described22,33,34,45. Pools of prion-sick mouse brains were prepared as 20% (wt/vol) homogenate in 5% glucose by use of tissue homogenizer (Precellys 24 Ribolyzer, Ozyme, Bertin technologies, France). The homogenate was diluted half to 10% in PMCA buffer (see below) to obtain the 10−1 dilution of the inoculum and stored at −80 °C. The Zürich I mouse line on an Sv129 mouse background was used as PrP0/0 line61.
Cell culture
The Rabbit kidney epithelial RK13 cell line was used to establish cells expressing sheep (Rov9, P2FJ6 and glycosylation mutants), hamster (HaRK13), human (HuRK13) and mouse PrP (MoRK13). Rov9, P2FJ6 clones, cells expressing glycosylation mutants and MoRK13 cells have been described previously30,32,34,44. The open reading frame of hamster and human PrPC was PCR amplified from Syrian hamster and human (Met 129 allele) genomic DNA, cloned into pBluescript plasmid, before subcloning in the pTRE and pCDNA plasmids (Clontech), respectively. After sequencing, each plasmid was introduced into RK13 cells as described previously30, and puromycin-resistant cell clones were selected for doxycycline-inducible and constitutive expression of PrPC, respectively. Cells were cultivated at 37 °C in 5% CO2 in Opti-MEM (Gibco) supplemented with 10% foetal calf serum and 0.1% penicillin and streptomycin. Cells were passaged once a week at a ¼ dilution. For production of large amount of concentrated cell lysates, cells were cultured in 2 or 4 layers of multilayer cell culture flasks (Thermo-Scientific Nunc).
Preparation of cell lysate for PMCA
Cultured cell lines in either T175 cm2 or in multilayer culture flasks were rinsed three times with sterile Ca++ and Mg++ free PBS. The cells were dissociated by incubation with trypsin-free dissociation media (Sigma) for 10 min at 37 °C. They were flushed with PBS, recovered in a falcon tubes and harvested by 5 min centrifugation at 1000 g at 4 °C. The pellet was then resupended in a given volume of cold and 0.2 μm filtered PMCA buffer (Tris-HCl 50 mM pH 7.4, EDTA 5 mM, NaCl 300 mM, 1% Triton-X-100). The lysed cells were incubated at 4 °C during 15–30 min with gentle vortexing. The lysates were centrifuged at 2000 g during 6 min to pellet the insoluble and chromatin materials. Supernatants were collected, aliquoted and stored at −80 °C until use as substrate in Cell mb-PMCA reactions. Protein content of cell lysates was measured by Bradford protein concentration determination kit (BCA kit, Pierce) using BSA as standard.
Cell-miniaturized beads-Protein Misfolding Cyclic Amplification (Cell-mb-PMCA)
The standard mb-PMCA, using brain lysate as source of PrPC substrate was realized as described22, by using 96-well PCR microplates and one 2.384 mm teflon beads. The Cell-mb-PMCA was set up with either 100% cell lysate or a mix with 10% mouse PrP0/0 brain lysate (ratio 1:1) in the presence or absence of 1% of Dextran Sulfate Sodium (DSS > 500 kDa Sigma Aldrich, Saint Quentin Fallavier, France) as indicated. Practically, a 4 μl aliquot of the analyte inoculum (10−n dilution) was suspended in 36 μl of PMCA substrate (brain or cell lysates) to obtain the 10−n+1 dilution. A series of 10-fold dilution was made by diluting 4 μL of the previous inoculum dilution to the next 36 μL containing well. Microplates were subjected to 96 cycles of 30 sec sonication at 200–220 Watt power (36–40% amplitude of the Q700 sonicators, Misonix, Farmingdale USA; or Delta Labo, Colombelles France) followed by 29.5 min of incubation at 37 °C. When needed, a second round of PMCA was realized with 1/10 diluted aliquot of the first round in fresh lysates. At the end of the PMCA, aliquots from each sample were analysed for PrPres content by Western blotting.
Protease digestion of PMCA products
To analyse the production of Proteinase K (PK)-resistant PrPSc species during PMCA, 10 μL of each sample were supplemented with SDS (up to 0.6% final concentration) and treated with PK (125 μg/mL final concentration) at 37 °C for 1 hour. The PK digestion was stopped by adding an equal volume of 2x Laemmli denaturation sample buffer and heating at 100 °C for 5 min. The samples were then stored at −20 °C. The levels of thermolysin-resistant PrP species in the PMCA amplicons were determined, as previously described22.
SDS-PAGE and western blotting
PMCA samples were run on Criterion XT 12% Bis-Tris precast gels (Biorad, Hercules, CA, USA), electrotransferred onto nitrocellulose membranes with the semi-dry electrotransfer system (Biorad) and probed with biotinylated Sha31 anti-PrP monoclonal antibody62, as described above. PrPC content of the cell lysates was determined by western blotting with SAF3462 anti octarepeat region of PrP. Quantification was determined with the GeneTools software after acquisition of the signals with a GeneGnome digital imager.
Endpoint-titration of PMCA products in tg338 mice
Standard protocol based on the use of disposable equipment and preparation of all inocula in a class II microbiological safety cabinet was followed. Serial ten-fold dilutions of PMCA products were prepared in sterile 5% glucose containing 5% bovine serum albumin. Individually identified 6- to 10-week-old tg338 recipient mice (n = 5 mice per dilution) were inoculated intracerebrally with 20 μL of each sample. The inoculated animals were observed daily for the appearance of prion disease symptoms. Animals at terminal stage of disease were euthanized. The survival time was defined as the number of days from inoculation to euthanasia. Their brain and spleen were removed for PrPres analysis by western blotting and histoblotting as previously described22,33. For histoblotting procedure, brains were rapidly removed from euthanized mice and frozen on dry ice. Cryosections were cut at 8–10 μm, transferred onto Superfrost slides and kept at −20 °C until use. Histoblot analyses were performed on 3 brains per dilution per amplicon, using the 12F1063 anti-PrP antibody.
To quantify vacuolar degeneration, brains were fixed in neutral-buffered 10% formalin (4% formaldehyde) before paraffin embedding. After deparaffinization, 2-μm-thick sections were stained with hematoxylin-eosin. Vacuolation profiles were established according to the standard method described by Fraser and Dickinson64, using three brains per experiment.
Additional Information
How to cite this article: Moudjou, M. et al. Glycoform-independent prion conversion by highly efficient, cell-based, protein misfolding cyclic amplification. Sci. Rep. 6, 29116; doi: 10.1038/srep29116 (2016).
Supplementary Material
Acknowledgments
We thank the staff of Animalerie Rongeurs (INRA, Jouy-en-Josas, France) for animal care, and Christel Michel (INRA, Jouy-en-Josas) for help with western blots. This work was funded by the Fondation pour la Recherche Médicale (Equipe FRM DEQ20150331689), by Région Ile de France (DIM MALINF) and by Agence Nationale pour la Sécurité du Médicament et des produits de Santé (ANSM, HAP-2014-051).
Footnotes
Author Contributions M. Moudjou, J.C., P.S., H.L., D.V., H.R., M.D. and V.B. conceived and designed the experiments. M. Moudjou, J.C., M. Mekrouti, F.R., L.H., P.S. and V.B. performed the experiments. M. Moudjou, J.C., M. Mekrouti, F.R., L.H., H.R., M.D. and V.B. analysed the data. M. Moudjou and V.B. wrote the manuscript. All authors reviewed the manuscript.
References
- Falsone A. & Falsone S. F. Legal but lethal: functional protein aggregation at the verge of toxicity. Front Cell Neurosci 9, 45, 10.3389/fncel.2015.00045 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S. B. Biology and genetics of prions causing neurodegeneration. Annu Rev Genet 47, 601–623, 10.1146/annurev-genet-110711-155524 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C. Transmissible proteins: expanding the prion heresy. Cell 149, 968–977, 10.1016/j.cell.2012.05.007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringue V., Vilotte J. L. & Laude H. Prion agent diversity and species barrier. Veterinary research 39, 47, 10.1051/vetres:2008024 (2008). [DOI] [PubMed] [Google Scholar]
- Bruce M. E. TSE strain variation. Br Med Bull 66, 99–108 (2003). [DOI] [PubMed] [Google Scholar]
- Collinge J. & Clarke A. R. A general model of prion strains and their pathogenicity. Science 318, 930–936, 10.1126/science.1138718 (2007). [DOI] [PubMed] [Google Scholar]
- Weissmann C., Li J., Mahal S. P. & Browning S. Prions on the move. EMBO Rep 12, 1109–1117, 10.1038/embor.2011.192 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge J., Sidle K. C., Meads J., Ironside J. & Hill A. F. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature 383, 685–690 (1996). [DOI] [PubMed] [Google Scholar]
- Somerville R. A. & Ritchie L. A. Differential glycosylation of the protein (PrP) forming scrapie-associated fibrils. J Gen Virol 71 (Pt 4), 833–839 (1990). [DOI] [PubMed] [Google Scholar]
- DeArmond S. J. et al. Selective neuronal targeting in prion disease. Neuron 19, 1337–1348, S0896-6273 (1997). [DOI] [PubMed] [Google Scholar]
- Tuzi N. L. et al. Host PrP glycosylation: a major factor determining the outcome of prion infection. PLoS Biol 6, e100, 10.1371/journal.pbio.0060100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman F. K. et al. The glycosylation status of PrPC is a key factor in determining transmissible spongiform encephalopathy transmission between species. J Virol 89, 4738–4747, 10.1128/JVI.02296-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancellotti E. et al. Post-translational changes to PrP alter transmissible spongiform encephalopathy strain properties. EMBO J 32, 756–769, 10.1038/emboj.2013.6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamat M. K., Dron M., Chapuis J., Langevin C. & Laude H. Prion propagation in cells expressing PrP glycosylation mutants. J Virol 85, 3077–3085, 10.1128/JVI.02257-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saborio G. P., Permanne B. & Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 411, 810–813, 10.1038/35081095 (2001). [DOI] [PubMed] [Google Scholar]
- Bannach O. et al. Detection of prion protein particles in blood plasma of scrapie infected sheep. PLoS One 7, e36620, 10.1371/journal.pone.0036620 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilla J., Saa P. & Soto C. Detection of prions in blood. Nat Med 11, 982–985, 10.1038/nm1286 (2005). [DOI] [PubMed] [Google Scholar]
- Lacroux C. et al. Preclinical detection of variant CJD and BSE prions in blood. PLoS Pathog 10, e1004202, 10.1371/journal.ppat.1004202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroux C. et al. Prionemia and leukocyte-platelet-associated infectivity in sheep transmissible spongiform encephalopathy models. J Virol 86, 2056–2066, 10.1128/JVI.06532-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safar J. G. et al. Transmission and detection of prions in feces. J Infect Dis 198, 81–89, 10.1086/588193 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilla J. et al. Crossing the species barrier by PrP(Sc) replication in vitro generates unique infectious prions. Cell 134, 757–768, 10.1016/j.cell.2008.07.030 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudjou M. et al. Highly infectious prions generated by a single round of microplate-based protein misfolding cyclic amplification. MBio 5, e00829–00813, 10.1128/mBio.00829-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikiya R. A. & Bartz J. C. In vitro generation of high-titer prions. J Virol 85, 13439–13442, 10.1128/JVI.06134-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P. et al. Cell-free formation of misfolded prion protein with authentic prion infectivity. Proc Natl Acad Sci USA 103, 15818–15823, 10.1073/pnas.0605608103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saborio G. P. et al. Cell-lysate conversion of prion protein into its protease-resistant isoform suggests the participation of a cellular chaperone. Biochem Biophys Res Commun 258, 470–475 (1999). [DOI] [PubMed] [Google Scholar]
- Jones M. et al. In vitro amplification and detection of variant Creutzfeldt-Jakob disease PrPSc. J Pathol 213, 21–26, 10.1002/path.2204 (2007). [DOI] [PubMed] [Google Scholar]
- Mays C. E. et al. In vitro amplification of misfolded prion protein using lysate of cultured cells. PLoS One 6, e18047, 10.1371/journal.pone.0018047 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt T. D., Jiang L., Bett C., Eisenberg D. & Sigurdson C. J. A proposed mechanism for the promotion of prion conversion involving a strictly conserved tyrosine residue in the beta2-alpha2 loop of PrPC. J Biol Chem 289, 10660–10667, 10.1074/jbc.M114.549030 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilette D. Cell models of prion infection. Veterinary research 39, 10, 10.1051/vetres:2007049 (2008). [DOI] [PubMed] [Google Scholar]
- Vilette D. et al. Ex vivo propagation of infectious sheep scrapie agent in heterologous epithelial cells expressing ovine prion protein. Proc Natl Acad Sci USA 98, 4055–4059, 10.1073/pnas.061337998 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuendorf E. et al. Glycosylation deficiency at either one of the two glycan attachment sites of cellular prion protein preserves susceptibility to bovine spongiform encephalopathy and scrapie infections. J Biol Chem 279, 53306–53316, 10.1074/jbc.M410796200 (2004). [DOI] [PubMed] [Google Scholar]
- Laferriere F. et al. Quaternary structure of pathological prion protein as a determining factor of strain-specific prion replication dynamics. PLoS Pathog 9, e1003702, 10.1371/journal.ppat.1003702 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin C., Andreoletti O., Le Dur A., Laude H. & Beringue V. Marked influence of the route of infection on prion strain apparent phenotype in a scrapie transgenic mouse model. Neurobiol Dis 41, 219–225, 10.1016/j.nbd.2010.09.010 (2011). [DOI] [PubMed] [Google Scholar]
- Tixador P. et al. The physical relationship between infectivity and prion protein aggregates is strain-dependent. PLoS Pathog 6, e1000859, 10.1371/journal.ppat.1000859 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dron M. et al. Endogenous proteolytic cleavage of disease-associated prion protein to produce C2 fragments is strongly cell- and tissue-dependent. J Biol Chem 285, 10252–10264, 10.1074/jbc.M109.083857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarradin P. et al. Transgenic Rabbits Expressing Ovine PrP Are Susceptible to Scrapie. PLoS Pathog 11, e1005077, 10.1371/journal.ppat.1005077 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault N. R. et al. Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions. Proc Natl Acad Sci USA 109, E1938–E1946, 10.1073/pnas.1206999109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mays C. E., Titlow W., Seward T., Telling G. C. & Ryou C. Enhancement of protein misfolding cyclic amplification by using concentrated cellular prion protein source. Biochem Biophys Res Commun 388, 306–310, 10.1016/j.bbrc.2009.07.163 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama Y. et al. Sulfated dextrans enhance in vitro amplification of bovine spongiform encephalopathy PrP(Sc) and enable ultrasensitive detection of bovine PrP(Sc). PLoS One 5, 10.1371/journal.pone.0013152 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saa P. et al. Strain-specific role of RNAs in prion replication. J Virol 86, 10494–10504, 10.1128/JVI.01286-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne L. & Terry L. A. In vitro amplification of PrPSc derived from the brain and blood of sheep infected with scrapie. J Gen Virol 89, 3177–3184, 10.1099/vir.0.2008/004226-0 (2008). [DOI] [PubMed] [Google Scholar]
- Wong C. et al. Sulfated glycans and elevated temperature stimulate PrP(Sc)-dependent cell-free formation of protease-resistant prion protein. EMBO J 20, 377–386, 10.1093/emboj/20.3.377 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama T. et al. Heparin enhances the cell-protein misfolding cyclic amplification efficiency of variant Creutzfeldt-Jakob disease. Neurosci Lett 498, 119–123, 10.1016/j.neulet.2011.04.072 (2011). [DOI] [PubMed] [Google Scholar]
- Courageot M. P. et al. A cell line infectible by prion strains from different species. J Gen Virol 89, 341–347, 10.1099/vir.0.83344-0 (2008). [DOI] [PubMed] [Google Scholar]
- Chapuis J. et al. Emergence of two prion subtypes in ovine PrP transgenic mice infected with human MM2-cortical Creutzfeldt-Jakob disease prions. Acta Neuropathol Commun 4, 10, 10.1186/s40478-016-0284-9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreoletti O. et al. Atypical/Nor98 scrapie infectivity in sheep peripheral tissues. PLoS Pathog 7, e1001285, 10.1371/journal.ppat.1001285 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalife M. et al. Mutated but Not Deleted Ovine PrP(C) N-Terminal Polybasic Region Strongly Interferes with Prion Propagation in Transgenic Mice. J Virol 90, 1638–1646, 10.1128/JVI.02805-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Wang X., Yuan C. G. & Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science 327, 1132–1135, 10.1126/science.1183748 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault N. R. et al. Protease-resistant prion protein amplification reconstituted with partially purified substrates and synthetic polyanions. J Biol Chem 280, 26873–26879, 10.1074/jbc.M503973200 (2005). [DOI] [PubMed] [Google Scholar]
- Deleault N. R., Lucassen R. W. & Supattapone S. RNA molecules stimulate prion protein conversion. Nature 425, 717–720, 10.1038/nature01979 (2003). [DOI] [PubMed] [Google Scholar]
- Zhou Z. et al. Crowded cell-like environment accelerates the nucleation step of amyloidogenic protein misfolding. J Biol Chem 284, 30148–30158, 10.1074/jbc.M109.002832 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguzzi A., Heikenwalder M. & Polymenidou M. Insights into prion strains and neurotoxicity. Nat Rev Mol Cell Biol 8, 552–561, 10.1038/nrm2204 (2007). [DOI] [PubMed] [Google Scholar]
- Lawson V. A., Collins S. J., Masters C. L. & Hill A. F. Prion protein glycosylation. J Neurochem 93, 793–801, 10.1111/j.1471-4159.2005.03104.x (2005). [DOI] [PubMed] [Google Scholar]
- Poggiolini I., Saverioni D. & Parchi P. Prion protein misfolding, strains, and neurotoxicity: an update from studies on Mammalian prions. Int J Cell Biol 2013, 910314, 10.1155/2013/910314 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancellotti E. et al. The role of host PrP in Transmissible Spongiform Encephalopathies. Biochim Biophys Acta 1772, 673–680, 10.1016/j.bbadis.2006.10.013 (2007). [DOI] [PubMed] [Google Scholar]
- Nishina K. A. et al. The stoichiometry of host PrPC glycoforms modulates the efficiency of PrPSc formation in vitro. Biochemistry 45, 14129–14139, 10.1021/bi061526k (2006). [DOI] [PubMed] [Google Scholar]
- Beringue V. et al. Regional heterogeneity of cellular prion protein isoforms in the mouse brain. Brain 126, 2065–2073, 10.1093/brain/awg205 (2003). [DOI] [PubMed] [Google Scholar]
- Khalili-Shirazi A. et al. PrP glycoforms are associated in a strain-specific ratio in native PrPSc. J Gen Virol 86, 2635–2644, 10.1099/vir.0.80375-0 (2005). [DOI] [PubMed] [Google Scholar]
- Chakroun N. et al. The oligomerization properties of prion protein are restricted to the H2H3 domain. FASEB J 24, 3222–3231, 10.1096/fj.09-153924 (2010). [DOI] [PubMed] [Google Scholar]
- Kurt T. D. et al. Prion transmission prevented by modifying the beta2-alpha2 loop structure of host PrPC. J Neurosci 34, 1022–1027, 10.1523/JNEUROSCI.4636-13.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueler H. et al. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature 356, 577–582, 10.1038/356577a0 (1992). [DOI] [PubMed] [Google Scholar]
- Feraudet C. et al. Screening of 145 anti-PrP monoclonal antibodies for their capacity to inhibit PrPSc replication in infected cells. J Biol Chem 280, 11247–11258, 10.1074/jbc.M407006200 (2005). [DOI] [PubMed] [Google Scholar]
- Krasemann S., Groschup M. H., Harmeyer S., Hunsmann G. & Bodemer W. Generation of monoclonal antibodies against human prion proteins in PrP0/0 mice. Mol Med 2, 725–734 (1996). [PMC free article] [PubMed] [Google Scholar]
- Fraser H. & Dickinson A. G. The sequential development of the brain lesion of scrapie in three strains of mice. J Comp Pathol 78, 301–311 (1968). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.