Abstract
Squash was first domesticated in Mexico and is now found throughout North America (NA) along with Peponapis pruinosa, a pollen specialist bee species of the squash genus Cucurbita. The origin and spread of squash cultivation is well-studied archaeologically and phylogenetically; however, no study has documented how cultivation of this or any other crop has influenced species in mutualistic interactions. We used molecular markers to reconstruct the demographic range expansion and colonization routes of P. pruinosa from its native range into temperate NA. Populations east of the Rocky Mountains expanded from the wild host plant's range in Mexico and were established by a series of founder events. Eastern North America was most likely colonized from squash bee populations in the present-day continental Midwest USA and not from routes that followed the Gulf and Atlantic coasts from Mexico. Populations of P. pruinosa west of the Rockies spread north from the warm deserts much more recently, showing two genetically differentiated populations with no admixture: one in California and the other one in eastern Great Basin. These bees have repeatedly endured severe bottlenecks as they colonized NA, following human spread of their Cucurbita pollen hosts during the Holocene.
Keywords: Holocene, Cucurbita spp., approximate Bayesian computation, demographic inference
1. Introduction
Among the many interesting bee–plant relationships peculiar to the Americas are those that exist between two genera of solitary bees (Peponapis and Xenoglossa) and the genus Cucurbita (squashes, gourds and pumpkins). To these bees, commonly known as squash and gourd bees, it is a relationship on which their survival depends. It also seems to be the chief parameter of their evolution.
Hurd et al. [1, pp. 218–234].
Most populations and species distributions repeatedly expand and contract over evolutionary time [2,3], but how these changes affect population structure is species-specific. For example, responses to the sea-level changes and glacial extent in the last glacial maximum resulted in larger population sizes and broader distributions of cold-tolerant species such as pikas [4], and simultaneously reduced population size to refugia in many other species [5,6]. How species have responded to the spread of agriculture has also been complex. Pests of crops that are grown worldwide have ranges far beyond their ancestral distributions, in contrast to other species dependent on non-domesticated plants that occur in agriculturally valuable habitats that have been extirpated or diminished in population size. While domestication invariably reduces genetic variation despite great increases in population size, the population structure of the pests, pathogens and mutualists associated with agriculturally important species do not change predictably. Reduced genetic variation found in populations of potato late blight (Phytophora infestans) in France was attributed to recent colonization from the British Isles [7], and in the grape pest, Plasmospara viticola, from a founder event in the 1870s [8]. By contrast, the soft-skinned fruit pest, Drosophila suzukii, was first reported in the continental USA in 2008, but retains comparable nucleotide variation to that observed from populations near its ancestral range [9]. Common to each of the three examples above, however, is that the ancestral range is disjunct from that of the invading population, and each ‘pest’ species negatively impacts the fitness of their host. Here, we report on the genetic consequences to a species in a mutualistic interaction that has undergone a population range expansion into a continental region contiguous with the ancestral range.
Squashes of the species Cucurbita pepo were domesticated in Central and Southern Mexico during the early-Mid Holocene (5–10 kya) near geographical centres of domestication of other important crops, including maize (Zea mays), peppers (Capsicum annuum), common beans (Phaseolus vulgaris) and cotton (Gossypium hirsutum) [10]. Early New World hunter–gatherers used wild Cucurbita because the relatively large and conspicuous fruits could be dried to serve as storage vessels and floats for fish nets [11]. Furthermore, the oily seeds of cucurbits are edible and nutritious, unlike the bitter and usually unpalatable fruit [12].
Our understanding of the history of C. pepo cultivation is unusually well detailed from archaeological evidence of fossil seeds [13,14] as well as molecular data, including chloroplast restriction fragment polymorphisms [15], nuclear internal transcribed spacer sequences [16], allozymes [17] and chloroplast and mitochondrial sequence data [18–20]. Domesticated cultivars of C. pepo appear to be derived from two independent domestication events, one in south central Mexico ca. 10 000 years ago that gave rise to C. pepo var. pepo (pumpkins, zucchinis and marrows) [21], and a second ca. 5000 years ago in midwestern North America (NA; present-day western Missouri, USA) that gave rise to C. pepo var. ovifera (acorn, scallop and crookneck squashes) [22]. All species of Cucurbita are monocious, self-incompatible and bee-pollinated [1]. Therefore, the cultivation and spread of C. pepo by native American societies is intimately interwined with native pollinators. The honey bee (Apis mellifera) is the most common managed pollinator of cucurbits in NA but they were introduced to the New World by European colonists centuries later in the 1600s [23,24].
Domesticated and wild C. pepo are visited by many bee species, most of which are pollen generalists species that collect pollen from a range of host plant species [25]. Among the common visitors are bee species of two genera, Peponapis (N = 15 species) and Xenoglossa (N = 7 species), which are strict pollen specialists of Cucurbita. Other than Peponapis pruinosa, the focal species of this study, all species in these two genera have modern-day distributions limited to Mexico, Central and South America [1]. Peponapis pruinosa, occurs across much of continental NA from Central Mexico to the province of Ontario, Canada and from California to the eastern seaboard (figure 1) far beyond the distribution of its wild floral host (Cucurbita foetidissima), which is restricted to the warm deserts of Mexico and the USA. Where C. foetidissima does not occur, P. pruinosa relies on domesticated host plants (mostly C. pepo but also C. moschata and C. maxima) for pollen [1,26]. The current geographical distributions of P. pruinosa and its host plants clearly imply that this bee followed the pre-European cultivation of domesticated Cucurbita spp. and has considerably expanded its range beyond the ancestral distribution that presumably was defined by the occurrence of its native pollen host, C. foetidissima (figure 1). This association with a cultivated crop has allowed P. pruinosa to attain one of the largest geographical ranges of any native bee species in NA.
Figure 1.
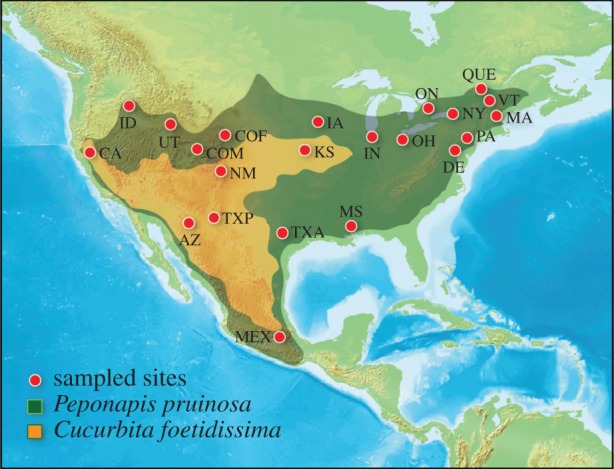
Map indicating sampling locations for Peponapis pruinosa. Dark green area indicates the approximate current distribution of P. pruinosa. Orange area bounds the range of the wild host plant, Cucurbita foetidissima, delimited using herbarium specimens. The stable range of C. foetidissima is smaller, because plants taken from the northern and eastern peripheries (NM and KS) often represent transient dispersal events and not persistent populations. See the electronic supplementary material, table S3 for population codes, geographical coordinates and sample size.
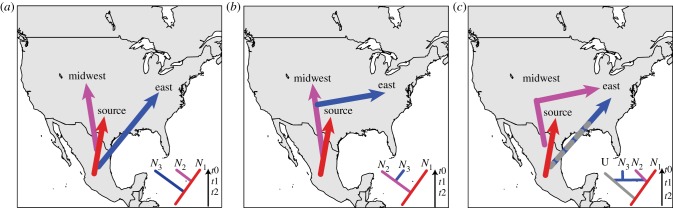
In this study, we use molecular markers to infer the demographic history and details of geographical range expansion of the squash bee, P. pruinosa, across NA. Specifically, we investigated: (i) signatures of range expansion in populations sampled from across the current distribution of the squash bee; (ii) centre(s) of origin of the expansion, and (iii) possible routes of colonization into eastern NA, where this squash bee is an abundant and important pollinator of Cucurbita crops. During range expansions, repeated founder effects generate a pattern of genetic diversity that steadily decreases along the expansion axis [3]. We thus expected a pattern wherein the centre of origin of the expansion maintains the highest genetic diversity among populations, and genetic diversity decreases in populations at greater distance from the centre of origin. To test this hypothesis, we used microsatellite markers and coalescent simulations to estimate levels of genetic diversity and demographic parameters (e.g. changes in population size). Using information from the archaeological record of C. pepo, we tested three alternative scenarios for the routes of colonization of P. pruinosa into the northeast of the USA: (i) a range expansion that initiated 10 000 years ago after the C. pepo v. pepo domestication event in Mexico, and accompanied the spread of cultivated cucurbits along the east coast by Native Americans [27]; (ii) a range expansion that initiated after the widespread cultivation of the second domesticated lineage of C. pepo v. ovifera in the Midwest ca. 5000 years ago [22]; and (iii) a scenario where the colonization of the northeast of the USA occurred from bee populations expanding through both the east coast and the Midwest (figure 2).
Figure 2.
Hypothesized scenarios of colonization of eastern North America tested by approximate Bayesian computation (ABC). Maps show routes of colonization and the tree topologies of hypothesized demographic scenarios. (a) Peponapis pruinosa colonized the east through the Atlantic coast of NA. (b) The colonization of the east resulted from the range expansion of populations from the Midwest region. (c) The population in the northeast is the product of admixture from an Atlantic and Midwest colonization. Grey on the phylogeny represents the unsampled (U) population that dispersed through the Atlantic coast. ‘Source’ indicates the population from Mexico; lineage ‘Midwest’ groups Arizona, Texas, Kansas, New Mexico, Iowa and Indiana; and lineage ‘East’ groups the remaining sampled populations.
These data are also used to investigate if the geographical range expansion of P. pruinosa has led to populations with large effective sizes and panmixia or small and isolated populations during northward migration. Founder effects are expected with colonization and range expansion [28], but there are few empirical data on how genetic bottlenecks affect bee populations [29]. Theory predicts that the haplodiploid sex determination system of bees increases their vulnerability to inbreeding [30]. Thus, we also measured the frequency of diploid males in populations to estimate risk of inbreeding depression throughout the range of P. pruinosa and assess the demographic stability of this important crop pollinator.
Our results support the view that (i) the geographical range expansion of P. pruinosa originated in Mexico, (ii) eastern NA was colonized through the continental Midwest, most likely after the second squash domestication event, and (iii) P. pruinosa is capable of thriving despite the greatly impoverished genetic diversity that has accompanied its rapid population expansion.
2. Material and methods
(a). Sampling
We collected 942 individuals of P. pruniosa (438 males and 504 females) from 22 populations in Mexico, the USA and eastern Canada (figure 1). All specimens were collected from flowers of cultivated Cucurbita plants except samples from Douglas, AZ that were collected from C. foetidissima flowers. Individuals were stored in 95% ethanol to preserve DNA for molecular analyses.
(b). Microsatellite development and variability
Microsatellites are hypervariable markers that are widespread across the genome, making them highly informative for studies of recent population demography [31]. Species that show limited genetic variability with allozymes and DNA sequence data often reveal more genetic variability with microsatellites [32]. We built genomic libraries enriched for microsatellites and used two different methods for microsatellite discovery: cloning and pyrosequencing (see methods in [33]). We designed primers for 24 DNA sequences, six of these primer pairs did not produce detectable PCR products and 12 were monomorphic. The remaining six variable microsatellite loci were used in this study (electronic supplementary material, table S1).
(c). Genetic diversity
We assessed population genetic diversity estimates as allele richness (Ar), expected heterozygosity (He) and Shannon diversity index (H′), standardizing for unequal sample sizes using MSA v. 4.05 [34]. We analysed both males and females in the same dataset treating haploid males as inbred genotypes. We visualized geographical patterns of genetic diversity by spatially interpolating Ar, He and H’ using a thin plate spline as implemented in the R package ‘fields’ [35]. To test for a linear relationship between genetic diversity and geographical distance, we regressed population genetic diversity estimates onto the linear geographical distance of all populations from the areas with highest genetic diversity using the R function ‘lm’. Euclidean distances between sampling locations were calculated according to the Earth's surface model implemented in the R package ‘fields’ [35]. Because the presence of diploid males indicates inbreeding and low levels of genetic variation in haplodiploid species [30], the frequency of diploid males (ϕ) was calculated for populations where males were sampled [36].
(d). Population structure
Population differentiation was estimated using Nei's GST in the software MSA v. 4.05 [34]. To identify genetic clusters in our data, we used the discriminant analysis of principal components (DAPC) implemented in the package ADEGENET v. 1.3–9.2 for R [37]. We performed the DAPC analysis using the number of sampled populations as the prior representing the maximum number of possible clusters. DAPC is a multivariate approach that identifies clusters of genetically related organisms by partitioning genetic variability into clusters that maximize between-group and minimize within-group differentiation. This multivariate approach does not assume Hardy–Weinberg equilibrium, making it an ideal clustering algorithm for datasets where this assumption is violated. Because individual membership probability changes with the number of PCA axes retained, we used the alpha score function to choose the optimal number of principal components for the analysis of our dataset [37].
(e). Demographic parameter estimation
We used the coalescent-based approach incorporated in MSVAR to estimate demographic parameters of change in effective population sizes for populations of P. pruinosa across NA. Wide uniform priors were chosen for all parameters (electronic supplementary material, table S2) to allow comparisons of parameter estimates from each run among different populations. We assumed a linear change in population size and a stepwise mutation model for microsatellite evolution. We independently analysed each population where we sampled more than 30 haploid chromosome sets using the same priors to compare relative values of the estimated demographic parameters. For each dataset, we ran four independent chains of 108 generations, sampling parameter values every 103 generations. For several populations, we ran longer chains of 109 generations to reach convergence. Sampling parameter values were recorded every 2500 generations. The first 10% of the generations of all chains were discarded as burn-in. We analysed MSVAR outputs using the R packages ‘locfit’, ‘coda’ and ‘runjags’ [38–40]. For parameter estimation, we combined all chains that reached convergence based on the ‘Gelman & Rubin diagnostic’ to obtain the mode and 90% highest probability density (90%HPD) limits for each parameter.
(f). Reconstruction of colonization history
We tested different hypotheses about the colonization of P. pruinosa from Mexico across NA using an approximate Bayesian computation (ABC) framework in the software DIYABC v. 2.1.0 [41,42]. The DAPC analysis (see Results) indicated populations from California, Idaho and Utah were highly divergent from populations east of the Rocky Mountains. We therefore excluded these populations from the ABC analysis so we could specifically test three demographic scenarios for how P. pruinosa reached eastern NA from northern Mexico: (i) southerly range extension along the Gulf coastal plains and/or Piedmont from Mexico to the eastern seaboard (figure 2a); (ii) passage through the Great Plains (Midwest) then eastward to the Atlantic seaboard (Midwest hypothesis), (figure 2b) or (iii) the joint invasion from the eastern seaboard and the Great Plains (figure 2c). We assumed a generalized stepwise mutation model to simulate mutations at microsatellite loci [43]. The mean mutation rate (μ) was drawn from a broad, uniform prior distribution ranging from 10−5 to 10−3. To differentiate between the three possible scenarios of colonization to the eastern part of NA, priors for the demographic parameter time since the population started diverging (ta) were defined based on information from the archaeological evidence of domestication of C. pepo and results from the MSVAR analysis. For each scenario, we simulated 3 × 106 datasets. Within populations, we compared the summary statistics: mean number of alleles per locus (NA), and mean expected heterozygosity (HE). Between populations, we compared NA, HE, FST and shared allele distance (DSA). The posterior probability of each competing scenario was estimated using a logistic regression on 104 simulated datasets. To assess the effect of single loci on the reconstruction of the colonization history P. pruinosa, we reran the ABC analysis removing one locus at the time. We chose the best scenario based on the highest significant probability values with non-overlapping 95% confidence intervals. We tested the performance of the best demographic scenario by reproducing the observed data with 3 × 105 pseudo-replications and using the model checking procedure implemented in DIYABC v. 2.1.0 [41].
3. Results
(a). Genetic diversity summary statistics
Electronic supplementary material, table S1 summarizes information on the six nuclear microsatellite loci analysed. Mean expected heterozygosity (He) was 0.374 (min = 0.049; max = 0.584) and mean number of alleles (Na) was 9.67 (min = 7; max = 14). Out of the 138 possible population-locus combinations, 19 Hardy–Weinberg tests could not be calculated because of the presence of one fixed allele. Forty-nine of the remaining tests showed GIS negative values significantly different from zero, indicating deviations from Hardy–Weinberg equilibrium due to heterozygote excess. In the population from Mexico, all loci were at Hardy–Weinberg equilibrium (p = 0.0001) and no diploid males were found. We did find diploid males in five populations: one each in Utah (ϕ = 0.026), Mississippi (ϕ = 0.037), New York (ϕ = 0.012), two in Vermont (ϕ = 0.091) and seven in California (ϕ = 0.053).
(b). Signatures of range expansion and centre of origin
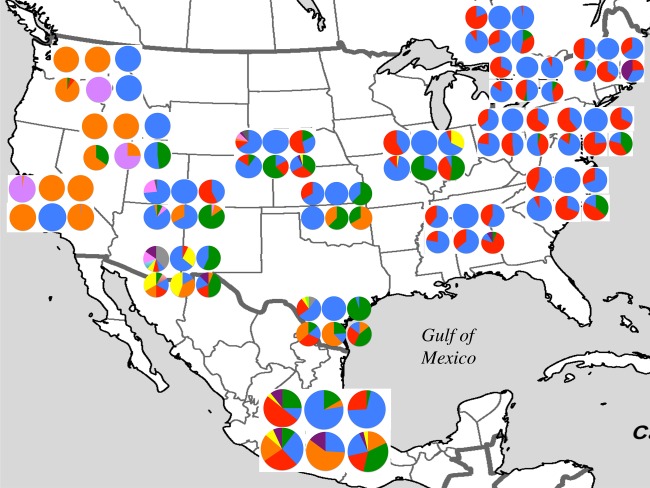
The distribution of genetic diversity across the geographical range of P. pruinosa supports a demographic scenario of spatial range expansion with a clear pattern of decreasing genetic diversity towards the species' northernmost present-day limits (electronic supplementary material, figure S1a,b). Squash bee populations that co-occur with wild C. foetidissima populations are more genetically diverse than populations on the Atlantic and Pacific coasts where Cucurbita is only represented by cultivated squashes and pumpkins (figure 3). Populations of P. pruinosa from warm and arid Mexico, Arizona and western Texas had the greatest genetic diversity of the sites we sampled (electronic supplementary material, table S3). There was a significant negative correlation between geographical distance and genetic diversity, using the population sampled from Mexico as the nearest to the centre of origin of the range expansion (Ar: r2 = 0.345, p < 0.005; He: r2 = 0.309, p < 0.05; H’: r2 = 0.385, p < 0.005). No significant correlation emerged when populations from Arizona and western Texas (El Paso) were considered the centres of origin (electronic supplementary material, figure S2). The population from California was least diverse genetically (Ar = 1.26; He = 0.084; H′ = 0.15), being characterized by a single-dominant allele at each locus with frequencies ranging between 0.7 and 1.
Figure 3.
Geographical distribution of allele frequencies for 17 populations of Peponapis pruinosa. Pie charts and colours represent microsatellite marker and allele frequencies, respectively. Populations from Arizona, Delaware, Iowa, Massachusetts, and Vermont, are not shown in the figure for clarity.
(c). Population structure and clustering
We found significant overall population structure across all populations (GST = 0.366). Multivariate genetic analyses show that populations from west of the Rocky Mountains (California, Idaho and Utah) are genetically distinct from all other populations (figure 4). Unlike populations east of the Rocky Mountains, the high membership probabilities for individuals from California and Idaho + Utah, suggest that each of these populations has a distinct genetic composition, the colonization of these two areas is recent, and there is little or no admixture among them and populations east of the Rocky Mountains (figure 4). Individuals from the geographical area where the wild host plant and P. pruinosa are co-distributed show a distinct genetic composition, but the proportion of admixture increased with increasing latitude (from Mexico to Colorado; electronic supplementary material, figure S3).
Figure 4.
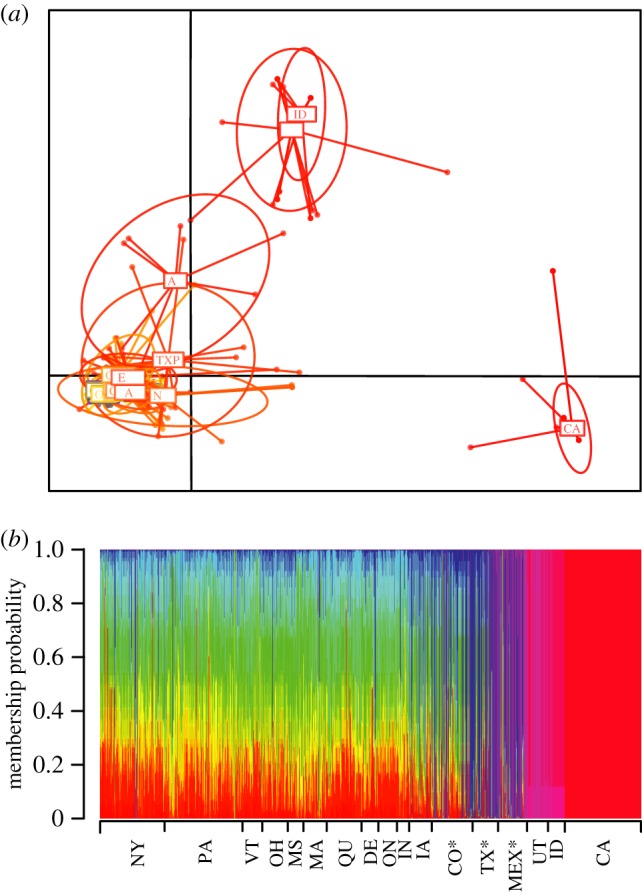
Clustering analysis of individuals. (a) Scatterplot of the discriminant analysis of principal components (DAPC). Each ellipse represents a population that groups individuals based on the first principal component. (b) Membership probabilities based on DAPC. Vertical bars represent individuals coloured by sampling location. Partitioned vertical lines represent individuals with admixed membership.
(d). Demographic parameters and routes of colonization
We used a coalescent approach to investigate the magnitude of the major and most recent demographic change in P. pruinosa populations by estimating the relative difference in effective population size between the current (N0) and ancestral populations (N1). All coalescent simulations included in our parameter estimation converged, as indicated by the Gelman–Rubin convergence statistic (less than 1.1). Furthermore, all independent chains provided consistent marginal posterior probability distribution (electronic supplementary material, figure S4). Comparisons between current and ancestral population sizes show reductions in population size that varied between two- and eightfold on a log scale (table 1). Estimated times of the drastic reductions in population sizes were highly dependent on the priors we set for each analysis and are not reported.
Table 1.
Highest posterior probability values and 90% highest posterior density (HPD) intervals for current (N0) and ancestral (N1) population sizes estimated with MSVAR.
| population | current Ne (N0) | N0 HPD 95% | ancestral Ne (N1) | N1 HPD 95% |
|---|---|---|---|---|
| Mexico | 0.067 | [0.031–230] | 119 564 | [31 019–511 964] |
| Colorado | 10.5 | [0.088–176] | 58 321 | [8075–511 964] |
| Utah | 0.252 | [0–424] | 91 770 | [231–45 167 390] |
| Quebec | 0.131 | [0–5] | 440 | [28–18 581] |
| Midwest + East | 10 564 | [25–2 976 870] | 547 237 688 | [4 916 918–67 435 720 193] |
We compared the three models of colonization route followed by P. pruinosa to the northern part of the Atlantic coast, one along the Gulf coast, the second one through the Midwest and the third scenario assuming an admixed migration from the Atlantic coast and the Midwest (figure 2). The ABC analysis strongly supported the hypothesis that P. pruinosa colonized the east coast of NA after the second domestication event of C. pepo that approximately occurred 5000 ya in the present-day midwestwern USA (0.643, CI (0.607–0.678); table 2). The Midwest hypothesis was consistently supported by all analyses after removing a single locus from the dataset (electronic supplementary material, table S4). Evaluation of the performance of each model agrees with these results. We simulated 500 random pseudo-replicates under each scenario. Twenty per cent failed to display the higher posterior probability of better-supported scenario (type I error); statistical power was on average 81% (1 − β [type II error]). This evaluation of model choice shows that, given the polymorphism of the markers and the sample sizes of our dataset, this procedure was consistent and powerful in differentiating between the three competing colonization route hypotheses that we tested.
Table 2.
Model choice for colonization scenarios of Peponapis pruinosa into eastern North America based on the approximate Bayesian computation (ABC) analysis. Scenario 1 assumes a southerly range extension from Mexico through the east coast. Scenario 2 assumes the expansion passaged through the Great Plains (Midwest), then eastward to the Atlantic coast. Scenario 3 assumes that migrants from the east coast and the Midwest colonized northeastern North America. See figure 2 for details.
| Scenario 1 | Scenario 2 | Scenario 3 | |
|---|---|---|---|
| posterior probabilitya | 0.043 [0.017–0.048] | 0.643 [0.607–0.678] | 0.325 [0.290–0.360] |
| confidence in scenario choiceb | |||
| type I error | 0.201 | ||
| type II error | 0.105 | 0.283 | |
| number of outlying statisticsc | |||
| p < 0.05 | 4 | 3 | 6 |
| p < 0.01 | 1 | 0 | 0 |
| p < 0.001 | 0 | 0 | 0 |
aThe median and 95% confidence intervals of the posterior probability indicate the revised probability distribution of each scenario after taking into consideration the prior information of the model.
bModel performance of best scenario. Type I error indicates the probability with which the best model is rejected. Type II error indicates the probability of deciding for best scenario when it is not true.
cThe number of summary statistics significantly different than the observed data. These statistics were used to discriminate between competing scenarios.
4. Discussion
Our results strongly support the hypothesis that the current distribution of the squash bee, P. pruinosa, is the result of a massive spatial range expansion from Mesoamerica into the temperate regions of NA. To the best of our knowledge, this is the first study to infer an effect of plant domestication and cultivation by early human societies on the demographic history of a pollinating species. Specialist insect pests have invaded NA from Mesoamerica following the spread of cultivated plants (e.g. boll weevil and cotton) [44], and many others have spread along with their host plant and silviculture [45]. Remarkably, our study represents the only known case of a specialist pollinator expanding along with the spread of a domesticated plant after first cultivation. Other animal-pollinated North American plants that have been domesticated either are still grown largely in their native range (low blueberries) or have been spread widely for cultivation in recent time, but not accompanied by the specialist component of their pollinator guild [46,47]. All other examples of pollinators moving outside their native ranges represent either intentional human introductions [48,49], inadvertent transoceanic transport [29,50,51] or response to recent climate change [52]. By contrast, our study shows that this specialist pollinator naturally followed the spread of its cultivated host plant.
The DAPC analysis revealed three genetic clusters of P. pruinosa: one in California, one grouping populations from Utah + Idaho (Intermountain West) and a more genetically heterogeneous group comprising all populations from the Rockies east to the Atlantic coast. The lack of genetic diversity in California and Utah + Idaho populations implies that these geographical areas were independently and recently colonized by P. pruinosa after a severe bottleneck. Furthermore, the lack of evidence for admixture indicates that these three genetic clusters remain isolated and migration between these areas does not occur or is exceedingly rare. Unlike the colonized areas east of the Rocky Mountains, dispersal into western NA was through corridors among tall mountain ranges. In the Middle Holocene, during a climatic period marked by warmer, wetter summers, Fremont peoples grew squash, beans and corn north into central Utah (eastern Great Basin) and the northern Colorado Plateau [53]. Their agriculture disappeared from the Intermountain West around 1350. Five centuries later, squash cultivation resumed in the Intermountain West, this time grown by European settlers [54]. Cultivated squash in homestead gardens likely provided the string of floral ‘stepping-stones’ that facilitated northward dispersal of P. pruinosa from its native range shared with C. foetidissima, probably the Four Corners Region in southeastern Utah. In California, there is no archaeological evidence of squash cultivation. However, both P. pruinosa and another squash bee, Xenoglossa angustior, as well as their shared wild host C. foetidissima, are found today in California's Central Valley [55]. This implies that the occurrence of P. pruinosa in California is the result of a long-distance dispersal event either in the recent past after cultivation became widespread, or after C. foetidissima became established. The shortest distance from source populations of P. pruinosa in the Mojave Desert to the Central Valley would be through Tehachapi Pass. A chance long-distance dispersal event is consistent with the remarkable genetic uniformity of P. pruinosa in California, and we predict a similar pattern of low genetic diversity will be found in sympatric populations of X. angustior.
A surprising finding is the low genetic diversity and small current effective population sizes of P. pruinosa outside the range of its wild Cucurbita host plant. This pattern suggests that the spatial expansion of P. pruinosa has not been followed by demographic expansions, as is usually observed among pest and pathogenic species of crops that spread with cultivation [7,8]. Demographic parameters estimated with coalescent analyses detected smaller present than ancestral effective populations sizes across the distribution of P. pruinosa. Preliminary results with mitochondrial DNA sequence data and genome-wide SNP markers corroborate these findings (MM López-Uribe 2013, unpublished data). Thus, effective population sizes are unexpectedly small, a counterintuitive finding based on frequent estimates of hundreds to thousands of P. pruinosa in local populations surveyed in squashes and pumpkins across NA (JH Cane 2004–2015, unpublished data). Low levels of genetic variability and unbalanced allele frequency spectra at the periphery of a species' geographical range are expected under a demographic model of consecutive bottlenecks after a range expansion [56]. However, we detected these signatures in populations from the ancestral range and periphery of the present-day distribution of P. pruinosa, suggesting that other factors may be driving the apparent low effective population sizes in this wild pollinator species. We hypothesize that this is a result of this specialist bees' reliance on cultivated Cucurbita throughout most of its current distribution, and that P. pruinosa populations are subject to sometimes frequent disturbance, such as deep tillage (which disrupts nest sites), widely spaced crop rotation, misapplied insecticides and local gardening decisions [25]. Therefore, the demographic instability of P. pruinosa populations in NA may result from ongoing extinction–colonization dynamics driven by recent farming and gardening practices. An alternative explanation is that our findings are the result of an artefact due to violations to the assumptions of the model used in MSVAR (e.g. microsatellite mutation model or complex demographic scenarios). However, simulation studies have demonstrated that MSVAR is based on a robust coalescent approach that efficiently detects both signatures of expansion and decline using microsatellite markers [57].
Despite severe reductions in genetic variability, diploid males were rare (seven individuals in five populations; ϕ = 0.012–0.091), which raises the possibility that P. pruinosa may possess mechanisms to avoid or reduce high frequencies of diploid males despite genetic impoverishment (e.g. strong balancing selection). In haplodiploid insects, sterile diploid males are produced when fertilized eggs are homozygous at the single complementary sex determination (csd) locus [30]. Low genetic variability and small effective population sizes increase homozygosity at the population level, leading to increased production of diploid males in insects with csd and inbreeding depression [58]. Further work on this problem is needed. For now, mechanisms by which P. pruinosa and other bee species avoid diploid male production remain speculative. Lasioglossum leucozonium, an invasive bee species from Europe, is currently widespread in NA and also experienced a severe bottleneck upon colonization in NA [29]. Populations of L. leucozonium and P. pruinosa in NA suggest that solitary bees can be effective colonizers of new areas despite severe founder events. However, we cannot assert the ubiquity of this pattern based on just two species. Levels of genetic variability in other successful exotic bee species (e.g. Anthidium manicatum [59], Anthophora plumipes [60], Megachile sculpturalis [61], Megachile rotundata [62] and Osmia cornifrons [63]) should be investigated.
Our study reveals previously unknown details about the geographical expansion of P. pruinosa in NA, a specialist bee of an economically important and widespread crop [64,65]. These results strongly support the hypothesis that P. pruinosa colonized eastern NA after C. pepo was domesticated the second time [22], a finding first proposed based on wing morphometrics [66]. We also show that some bee species can be resilient to the negative effects of low genetic variability. Peponapis pruinosa has successfully undergone a massive range expansion in spite of severe and repeated bottlenecks [67]. The extent of this pattern in bees deserves further attention, as does understanding the mechanisms by which they avoid inbreeding depression. Such information has important management implications, including reintroducing native bee populations where habitat loss and intense agricultural systems have extinguished elements of the native bee community.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank all the people involved in the sampling of Peponapis pruinosa across Mexico, the USA and Canada: Derek Artz, Ta'i Roulston, Karen Stickler, Beatriz Moisset, Leif Richardson, Carlos Vergara, Jason Gibbs, John Wenzel, Blair Simpson, Elizabeth Evans, Michael Veit, Andre Payette, Heather Harmon, Bob Hammon, Scott Prajzner, Katharina Ullmann, Karen Goodell, John Purdy, Sheena Shidu, Christopher Mayack, Jack Neff, Liz Maynard, Michael Welker, Randy Ritland, Jennifer Thomas and Victor Gonzalez. We also thank Steve Bogdanowicz for help with microsatellite development, Christine Santiago for assistance with microsatellite genotyping, Neil McCoy for figure improvements, and Shannon Hedtke and Rayna Bell for comments on earlier versions of the manuscript.
Ethics
Squash bees are not endangered or protected species. Sampled sites in this study included private agricultural property where landowners granted collection permits.
Data accessibility
Raw genotypic data and geographic information of individuals: Dryad http://dx.doi.org/10.5061/dryad.5j354.
Authors' contributions
M.M.L.U., R.L.M. and B.N.D. designed the study; M.M.L.U. performed the molecular laboratory work and data analysis; M.M.L.U., J.H.C., R.L.M. and B.N.D. drafted the manuscript. All authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
Funding for this study was provided by grants from the Grace Griswold Endowment (Cornell University to M.M.L.U.), and by awards from the National Science Foundation (DEB-0814544 and DEB-0742998 to B.N.D.).
References
- 1.Hurd PD, Linsley EG, Whitaker TW. 1971. Squash and gourd bees (Peponapis, Xenoglossa) and the origin of the cultivated Cucurbita. Evolution 25, 218–234. ( 10.2307/2406514) [DOI] [PubMed] [Google Scholar]
- 2.Hewitt G. 2004. Genetic consequences of climatic oscillations in the Quaternary. Phil. Trans. R. Soc. Lond. B 359, 183–195. ( 10.1098/rstb.2003.1388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li JZ, et al. 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319, 1100–1104. ( 10.1126/science.1153717) [DOI] [PubMed] [Google Scholar]
- 4.Brown JL, Knowles LL. 2012. Spatially explicit models of dynamic histories: examination of the genetic consequences of Pleistocene glaciation and recent climate change on the American Pika. Mol. Ecol. 21, 3757–3775. ( 10.1111/j.1365-294X.2012.05640.x) [DOI] [PubMed] [Google Scholar]
- 5.Provan J, Bennett K. 2008. Phylogeographic insights into cryptic glacial refugia. Trends Ecol. Evol. 23, 564–571. ( 10.1016/j.tree.2008.06.010) [DOI] [PubMed] [Google Scholar]
- 6.Soltis DE, Morris AB, McLachlan JS, Manos PS, Soltis PS. 2006. Comparative phylogeography of unglaciated eastern North America. Mol. Ecol. 15, 4261–4293. ( 10.1111/j.1365-294X.2006.03061.x) [DOI] [PubMed] [Google Scholar]
- 7.Montarry J, Andrivon D, Glais I, Corbiere R, Mialdea G, Delmotte F. 2010. Microsatellite markers reveal two admixed genetic groups and an ongoing displacement within the French population of the invasive plant pathogen Phytophthora infestans. Mol. Ecol. 19, 1965–1977. ( 10.1111/j.1365-294X.2010.04619.x) [DOI] [PubMed] [Google Scholar]
- 8.Fontaine MC, Austerlitz F, Giraud T, Labbé F, Papura D, Richard-Cervera S, Delmotte F. 2013. Genetic signature of a range expansion and leap-frog event after the recent invasion of Europe by the grapevine downy mildew pathogen Plasmopara viticola. Mol. Ecol. 22, 2771–2786. ( 10.1111/mec.12293) [DOI] [PubMed] [Google Scholar]
- 9.Adrion JR, et al. 2014. Drosophila suzukii: the genetic footprint of a recent, worldwide invasion. Mol. Biol. Evol. 31, 3148–3163. ( 10.1093/molbev/msu246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lentz DL, Pohl MD, Alvarado JL, Tarighat S, Bye R. 2008. Sunflower (Helianthus annuus L.) as a pre-Columbian domesticate in Mexico. Proc. Natl Acad. Sci. USA 105, 6232–6237. ( 10.1073/pnas.0711760105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hart JP, Daniels RA, Sheviak CJ. 2004. Do Cucurbita pepo gourds float fishnets? Am. Antiq. 69, 141–148. ( 10.2307/4128352) [DOI] [Google Scholar]
- 12.Smith BD. 1992. Rivers of change: essays on early agriculture in eastern North America. Tuscaloosa, AL: University of Alabama Press. [Google Scholar]
- 13.Whitaker TW. 1981. Archeological cucurbits. Econ. Bot. 35, 460–466. ( 10.1007/BF02858596) [DOI] [Google Scholar]
- 14.Simon ML. 2011. Evidence for variability among squash seeds from the Hoxie site (11CK4), Illinois. J. Archaeol. Sci. 38, 2079–2093. ( 10.1016/j.jas.2010.12.006) [DOI] [Google Scholar]
- 15.Wilson HD, Doebley J, Duvall M. 1992. Chloroplast DNA diversity among wild and cultivated members of Cucurbita (Cucurbitaceae). Theor. Appl. Genet. 84, 859–865. ( 10.1007/bf00227397) [DOI] [PubMed] [Google Scholar]
- 16.Jobst J, King K, Hemleben V. 1998. Molecular evolution of the internal transcribed spacers (ITS1 and ITS2) and phylogenetic relationships among species of the family Cucurbitaceae. Mol. Phylogenet. Evol. 9, 204–219. ( 10.1006/mpev.1997.0465) [DOI] [PubMed] [Google Scholar]
- 17.Decker-Walters DS, Staub JE, Chung S-M, Nakata E, Quemada HD. 2002. Diversity in free-living populations of Cucurbita pepo (Cucurbitaceae) as assessed by random amplified polymorphic DNA. Syst. Bot. 27, 19–28. [Google Scholar]
- 18.Sanjur OI, Piperno DR, Andres TC, Wessel-Beaver L. 2002. Phylogenetic relationships among domesticated and wild species of Cucurbita (Cucurbitaceae) inferred from a mitochondrial gene: implications for crop plant evolution and areas of origin. Proc. Natl Acad. Sci. USA 99, 535–540. ( 10.1073/pnas.012577299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng YH, Alverson AJ, Wang QF, Palmer JD. 2013. Chloroplast phylogeny of Cucurbita: evolution of the domesticated and wild species. J. Syst. Evol. 51, 326–334. ( 10.1111/jse.12006) [DOI] [Google Scholar]
- 20.Kistler L, Newsom LA, Ryan TM, Clarke AC, Smith BD, Perry GH. 2015. Gourds and squashes (Cucurbita spp.) adapted to megafaunal extinction and ecological anachronism through domestication. Proc. Natl Acad. Sci. USA 112, 15 107–15 112. ( 10.1073/pnas.1516109112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith BD. 1997. The initial domestication of Cucurbita pepo in the Americas 10 000 years ago. Science 276, 932–934. ( 10.1126/science.276.5314.932) [DOI] [Google Scholar]
- 22.Smith BD. 2006. Eastern North America as an independent center of plant domestication. Proc. Natl Acad. Sci. USA 103, 12 223–12 228. ( 10.1073/pnas.0604335103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheppard W. 1989. A history of the introduction of honey bee races into the United States. I. Am. Bee J. 129, 617–619. [Google Scholar]
- 24.Sheppard W. 1989. A history of the introduction of honey bee races into the United States. II. Am. Bee J. 129, 664–667. [Google Scholar]
- 25.Shuler RE, Roulston TH, Farris GE. 2005. Farming practices influence wild pollinator populations on squash and pumpkin. J. Econ. Entomol. 98, 790–795. ( 10.1603/0022-0493-98.3.790) [DOI] [PubMed] [Google Scholar]
- 26.Hurd PD, Linsley EG. 1964. The squash and gourd bees—genera Peponapis Robertson and Xenoglossa Smith—Inhabiting America north of Mexico: (Hymenoptera: Apoidea). Berkeley, CA: University of California. [Google Scholar]
- 27.Whitaker TW, Bemis W. 1964. Evolution in the genus Cucurbita. Evolution 18, 553–559. ( 10.2307/2406209) [DOI] [Google Scholar]
- 28.Prugnolle F, Manica A, Balloux F. 2005. Geography predicts neutral genetic diversity of human populations. Curr. Biol. 15, R159–R160. ( 10.1016/j.cub.2005.02.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zayed A, Constantin ŞA, Packer L. 2007. Successful biological invasion despite a severe genetic load. PLoS ONE 2, e868 ( 10.1371/journal.pone.0000868) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zayed A, Packer L. 2005. Complementary sex determination substantially increases extinction proneness of haplodiploid populations. Proc. Natl Acad. Sci. USA 102, 10 742–10 746. ( 10.1073/pnas.0502271102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Putman AI, Carbone I. 2014. Challenges in analysis and interpretation of microsatellite data for population genetic studies. Ecol. Evol. 4, 4399–4428. ( 10.1002/ece3.1305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boys J, Cherry M, Dayanandan S. 2005. Microsatellite analysis reveals genetically distinct populations of red pine (Pinus resinosa, Pinaceae). Am. J. Bot. 92, 833–841. ( 10.3732/ajb.92.5.833) [DOI] [PubMed] [Google Scholar]
- 33.López-Uribe MM, Santiago CK, Bogdanowicz SM, Danforth BN. 2013. Discovery and characterization of microsatellites for the solitary bee Colletes inaequalis using Sanger and 454 pyrosequencing. Apidologie 44, 163–172. ( 10.1007/s13592-012-0168-3) [DOI] [Google Scholar]
- 34.Dieringer D, Schlötterer C. 2003. Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol. Ecol. Notes. 3, 167–169. ( 10.1046/j.1471-8286.2003.00351.x) [DOI] [Google Scholar]
- 35.Furrer R, Nychka D, Sain S. 2012. Fields: tools for spatial data, version 6. R package version 6.
- 36.Owen RE, Packer L. 1994. Estimation of the proportion of diploid males in populations of Hymenoptera. Heredity 72, 219 ( 10.1038/hdy.1994.31) [DOI] [Google Scholar]
- 37.Jombart T, Devillard S, Balloux F. 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11, 94 ( 10.1186/1471-2156-11-94) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loader C. 2007. Locfit: Local regression, likelihood and density estimation. R package version, 1.5–4.
- 39.Denwood M. 2013. runjags: Interface utilities for MCMC models in Just Another Gibbs Sampler (JAGS) using parallel and distributed computing methods. See http://cran.r-project.org/web/packages/runjags.
- 40.Plummer M, Best N, Cowles K, Vines K. 2006. CODA: Convergence diagnosis and output analysis for MCMC. R package v. 6, 7–11.
- 41.Cornuet J-M, Santos F, Beaumont MA, Robert CP, Marin J-M, Balding DJ, Guillemaud T, Estoup A. 2008. Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics 24, 2713–2719. ( 10.1093/bioinformatics/btn514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cornuet J-M, Ravigné V, Estoup A. 2010. Inference on population history and model checking using DNA sequence and microsatellite data with the software DIYABC (v.1. 0). BMC Bioinform. 11, 401 ( 10.1186/1471-2105-11-401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Estoup A, Jarne P, Cornuet JM. 2002. Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol. Ecol. 11, 1591–1604. ( 10.1046/j.1365-294X.2002.01576.x) [DOI] [PubMed] [Google Scholar]
- 44.Kim KS, Sappington TW. 2006. Molecular genetic variation of boll weevil populations in North America estimated with microsatellites: implications for patterns of dispersal. Genetica 127, 143–161. ( 10.1007/s10709-005-2673-z) [DOI] [PubMed] [Google Scholar]
- 45.Mayer F, Piel FB, Cassel-Lundhagen A, Kirichenko N, Grumiau L, Økland B, Bertheau C, Grégoire JC, Mardulyn P. 2015. Comparative multilocus phylogeography of two Palaearctic spruce bark beetles: influence of contrasting ecological strategies on genetic variation. Mol. Ecol. 24, 1292–1310. ( 10.1111/mec.13104) [DOI] [PubMed] [Google Scholar]
- 46.Simpson B, Neff J. 1987. Pollination ecology in the arid southwest. Aliso 11, 417–440. [Google Scholar]
- 47.Vander Kloet SP. 1988. The genus Vaccinium in North America. Ottawa, ON: Agriculture Canada. [Google Scholar]
- 48.Batra S. 1979. Osmia cornifrons and Pithitis smaragdula, two Asian bees introduced into the United States for crop pollination. In Proc. of the IVth Int. Symp. on Pollination, 11–13 October, College Park, MD, Contrib No. 5591. pp. 307–312. College Park, MD: Maryland Agricultural Experimentation Station.
- 49.Whitfield CW, et al. 2006. Thrice out of Africa: ancient and recent expansions of the honey bee, Apis mellifera. Science 314, 642–645. ( 10.1126/science.1132772) [DOI] [PubMed] [Google Scholar]
- 50.Hinojosa-Díaz IA, Yáñez-Ordóñez O, Chen G, Peterson A, Engel M. 2005. The North American invasion of the giant resin bee (Hymenoptera: Megachilidae). J. Hymenopt. Res. 14, 69–77. [Google Scholar]
- 51.Strickler K, Cane JH. 2003. For non-native crops, whence pollinators of the future? Lanham, MD: Entomological Society of America. [Google Scholar]
- 52.Dellicour S, Mardulyn P, Hardy OJ, Hardy C, Roberts S, Vereecken N. 2014. Inferring the mode of colonization of the rapid range expansion of a solitary bee from multilocus DNA sequence variation. J. Evol. Biol. 27, 116–132. ( 10.1111/jeb.12280) [DOI] [PubMed] [Google Scholar]
- 53.Grayson DK. 1993. The desert's past: a natural prehistory of the Great Basin. Washington, DC: Smithsonian Institution Press. [Google Scholar]
- 54.Hartley WG. 1970. Mormons, crickets, and gulls: a new look at an old story. Utah Hist. Q. 38, 224–239. [Google Scholar]
- 55.Hurd PD, Linsley EG. 1967. Squash and gourd bees of the genus Xenoglossa (Hymenoptera: Apoidea). Ann. Entomol. Soc. Am. 60, 988–1007. ( 10.1093/aesa/60.5.988) [DOI] [Google Scholar]
- 56.Excoffier L, Foll M, Petit RJ. 2009. Genetic consequences of range expansions. Ann. Rev. Ecol. Evol. Sci. 40, 481–501. ( 10.1146/annurev.ecolsys.39.110707.173414) [DOI] [Google Scholar]
- 57.Girod C, Vitalis R, Leblois R, Fréville H. 2011. Inferring population decline and expansion from microsatellite data: a simulation-based evaluation of the Msvar method. Genetics 188, 165–179. ( 10.1534/genetics.110.121764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hedrick PW, Gadau J, Page RE. 2006. Genetic sex determination and extinction. Trends Ecol. Evol. 21, 55–57. ( 10.1016/j.tree.2005.11.014) [DOI] [PubMed] [Google Scholar]
- 59.Gibbs J, Sheffield CS. 2009. Rapid range expansion of the wool-carder bee, Anthidium manicatum (Linnaeus)(Hymenoptera: Megachilidae), in North America. J. Kans. Entomol. Soc. 82, 21–29. ( 10.2317/JKES805.27.1) [DOI] [Google Scholar]
- 60.Hinojosa-Díaz I. 2008. The giant resin bee making its way west: first record in Kansas (Hymenoptera: Megachilidae). ZooKeys 1, 67–71. ( 10.3897/zookeys.1.17) [DOI] [Google Scholar]
- 61.Mangum WA, Sumner S. 2003. A survey of the North American range of Megachile (Callomegachile) sculpturalis, an adventive species in North America. J. Kans. Entomol. Soc. 76, 658–662. [Google Scholar]
- 62.Goulson D. 2003. Effects of introduced bees on native ecosystems. Annu. Rev. Ecol. Evol. Syst. 34, 1–26. ( 10.1146/annurev.ecolsys.34.011802.132355) [DOI] [Google Scholar]
- 63.Batra S. 1978. Osmia cornifrons and Pithitis smaragdula, two Asian bees introduced into the United States for crop pollination. In Proc. IV Int. Symp. Pollination, 11–13 October, College Park, MD, Contrib No. 5591. College Park, MD: University of Maryland Agricultural Experiment Station Special Miscellaneous Publication I.
- 64.Cane JH, Sampson BJ, Miller SA. 2011. Pollination value of male bees: the specialist bee Peponapis pruinosa (Apidae) at summer squash (Cucurbita pepo). Environ. Entomol. 40, 614–620. ( 10.1603/EN10084) [DOI] [PubMed] [Google Scholar]
- 65.Hurd PD, Linsley EG, Michelbacher AE. 1974. Ecology of the squash and gourd bee, Peponapis pruinosa, on cultivated cucurbits in California (Hymenoptera: Apoidea). Washington, DC: Smithsonian Institution Press. [Google Scholar]
- 66.Bischoff I, Schröder S, Misof B. 2009. Differentiation and range expansion of North American squash bee, Peponapis pruinosa (Apidae: Apiformes) populations assessed by geometric wing morphometry. Ann. Entomol. Soc. Am. 102, 60–69. ( 10.1603/008.102.0106) [DOI] [Google Scholar]
- 67.Payette A, Payette M. 2003. Première mention de l'abeille Peponapis pruinosa (Say)(Hymenoptera: Apidae) pour le Québec. Fabreries 28, 37–47. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw genotypic data and geographic information of individuals: Dryad http://dx.doi.org/10.5061/dryad.5j354.