Abstract
Although gene duplication is seen as the main path to evolution of new functions, molecular mechanisms by which selection favours the gain versus loss of newly duplicated genes and minimizes the fixation of pseudo-genes are not well understood. Here, we investigate in detail a duplicate honeybee gene obp11 belonging to a fast evolving insect gene family encoding odorant binding proteins (OBPs). We report that obp11 is expressed only in female bees in rare antennal sensilla basiconica in contrast to its tandem partner obp10 that is expressed in the brain in both females and males (drones). Unlike all other obp genes in the honeybee, obp11 is methylated suggesting that functional diversification of obp11 and obp10 may have been driven by an epigenetic mechanism. We also show that increased methylation in drones near one donor splice site that correlates with higher abundance of a transcript variant encoding a truncated OBP11 protein is one way of controlling its contrasting expression. Our data suggest that like in mammals and plants, DNA methylation in insects may contribute to functional diversification of proteins produced from duplicated genes, in particular to their subfunctionalization by generating complementary patterns of expression.
Keywords: epialleles, insect olfaction, genome evolution, insect epigenomics, Apis mellifera
1. Introduction
Several models have been proposed to explain how a newly arisen gene duplicate can be preserved in the genome and ultimately provide a novel function for an organism. Rodin & Riggs [1] consider epigenetic silencing by DNA methylation as one mechanism shifting the rate of duplicate genes conversion into pseudo-genes towards functional diversification. In their model, tissue- or stage-specific epigenetic silencing generates complementary roles for twin genes and consequently prevents them from becoming pseudo-genes, a process referred to as subfunctionalization. Indeed, in mammals and plants, whose genomes are heavily methylated, the occurrence of a functional new duplicate is much higher than in organisms that lost DNA methylation enzymology in particular flies and some nematodes [1,2].
In honeybees and other insects, genomic methylation is sparse and is predominantly associated with conserved genes encoding proteins involved in essential cellular functions, including metabolism, energy flux and nucleic acids binding [3–9]. In contrast, virtually all recently evolved, and in particular, lineage-specific genes have been found to be not methylated [4–6]. Unexpectedly, our analyses of the honeybee genome-wide methylation profiles have uncovered that one of the genes belonging to a recently evolved odorant binding protein (OBP) family, designated obp11, is methylated. This finding prompted us to investigate in more detail the impact of this epigenomic modification on obp11 expression and functional diversification. This gene encodes a putative small water-soluble molecule, which like all insect OBPs is predicted to facilitate delivering of hydrophobic compounds, such as odorants, to specialized receptors [10–13]. The high disparity of obps sequences implies a rapid rate of evolution of these important genes that have been implicated in various aspects of insect behaviour and development. Most insect genomes encode a large number of duplicated OBPs, for example, there are 57 OBPs known in the mosquito Anopheles gambiae and 51 in Drosophila melanogaster [14]. In the honeybee genome, a somewhat smaller number of 21 OBPs have been found with nine of them restricted to olfactory organs and others showing either more ubiquitous pattern of expression or specific confinement to non-sensory tissues [13]. Like in flies and mosquitoes, most of the obps in the honeybee are organized in clusters in the genome, reflecting the relatively recent expansions of genes belonging to this family. The biggest cluster contains nine obps, tandemly arranged in the same orientation within a 40 kb region on chromosome 15. Only three genes, obp1, obp9 and obp12 are encoded by single loci that have no other obps in close proximity. Although several studies have demonstrated selective binding of odorants or pheromones to different OBPs [15–18], the majority of insect OBPs remain in the orphan category awaiting functional characterization. Some of these genes are expressed in olfactory and gustatory sensilla, but many are found in other non-sensory tissues [10].
In this study, we investigate the impact of DNA methylation on obp11 expression and discuss the potential role of this epigenomic modification in its functional diversification.
2. Results
(a). Genomic locus encoding OBP11
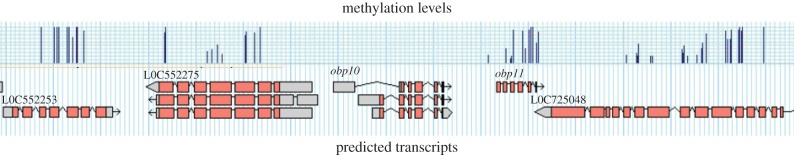
obp11 (GB50937) maps to the linkage group 10 in a region that is highly methylated (figure 1). It codes for six exons spanning approximately 580 bp and is arranged in a head to tail tandem, approximately 1 kb apart, with another member of this family obp10 (GB50936). Interestingly, these two closely linked loci show contrasting methylation patterns, obp10 follows the expected non-methylated pattern, whereas obp11 is methylated resembling other genes in this genomic region (in all examined methylomes). There are no predicted transcript variants for obp11 in the official genome assembly, but at least three alternatively spliced transcripts of obp10 have been compiled. It should be noted, however, that rare obp11 splice variants may not have been detected in the available transcriptomes, because there are no good antennal RNAseq datasets, in contrast to multiple brain RNAseq libraries in which obp10 is abundant. Indeed, our study indicates that an alternatively spliced variant is produced from the obp11 locus (see below).
Figure 1.
A snapshot of a genomic landscape shows the tandem arrangement of obp10 and obp11. The level of individual CpGs methylation for each gene is shown at the top, and the transcript variants deduced from the available transcriptomes at the bottom. Extracted from the honeybee genome browser (BeeBase, dna.anu.edu.au). (Online version in colour.)
(b). OBP11 is expressed in sensilla basiconica in the antennae of female honeybees
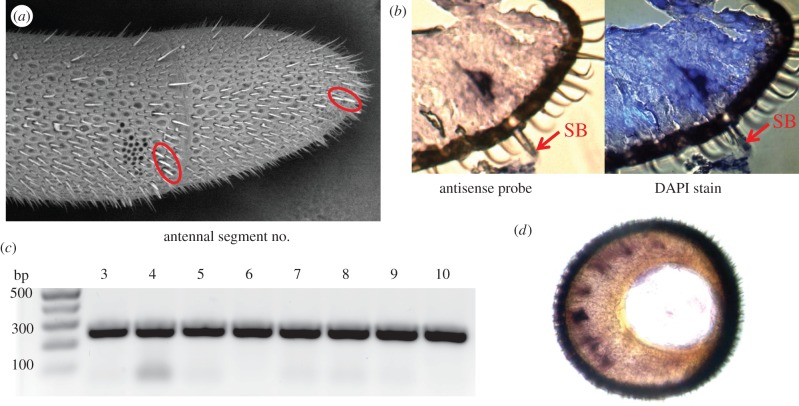
Using in situ hybridization and qPCR, we have examined the expression of obp11 in various tissues in all three castes, queens, workers and drones. As shown in figure 2, mRNAs encoding OBP11 are found in antennal sensilla basiconica also known as pegs that are found only in female bees, queens and workers [19]. In comparison with other highly abundant sensilla such as pore plates (placodea), basiconica are very rare. There are only small populations of approximately 150 basiconica on each antenna localized in clusters of 15–20 near the top of segments 3–10 (figure 2b). However, the expression of obp11 is very high suggesting that OBP11 protein is required at elevated levels. OBP11 is essentially female-specific with only marginal or no expression present in drones, confirming the restricted localization of this protein in sensilla basiconica. Both workers and queens have very high levels of OBP11, but foraging bees and older nurses have slightly higher levels than queens (electronic supplementary material, figure S1b). There is no expression of note in brains in all types of female bees (electronic supplementary material, figure S1a) and in ovaries and testes (not shown). By comparison, obp10 is expressed in adult brains of all bees with low expression detectable at larval stages. A markedly higher expression of obp10 is found in optic lobes than in central brains of adult bees (electronic supplementary material, figure S1b).
Figure 2.
Analysis of obp11 expression in the worker honeybee antenna. (a) Scanning electron microscopy image shows the localization of sensilla basiconica on the antennal tip (segments 3–10). (b) In situ analysis on a longitudinal section of the antenna showing the expression in the nuclei of basiconica. (c) PCR detection of a 250 bp obp11 amplicon in antennal segments 3–10. (d) In situ hybridization on a cross section of the antenna with the same antisense probe as in panel (b). (Online version in colour.)
(c). Methylation and alternative splicing of obp11
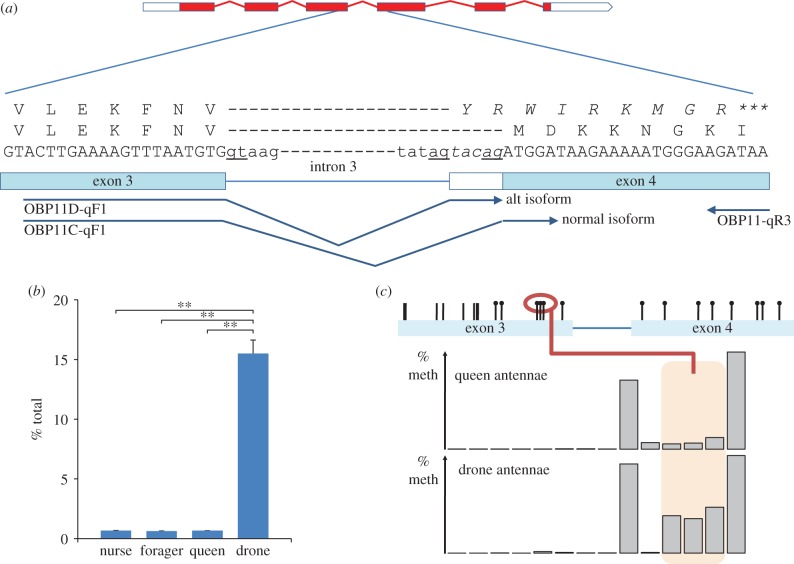
Our analysis of RNAseq datasets that recently became available has revealed the existence of an alternative splice acceptor site tatag instead of tatagtacag in intron 3 generating transcripts with a premature stop codon (figure 3a). Using a set of variant-specific primers, the expression of both the normal and variant transcript was examined in antennae of drones, foragers, young nurses and queens. As shown in figure 3b, the spliced variant is more abundant in drones than in females indicating that essentially only non-functional OBP11 peptides are produced in drones. Next, we conducted ultra-deep obp11 amplicon sequencing to compare methylation patterns in male versus female antennae. This approach generates hundreds of thousands of reads per amplicon and has a resolving power to detect methylation signatures in all cell types in a given tissue [20,21]. Figure 3c shows an interesting difference in the methylation of three CpGs in exon 3 between female and male antennae with much higher methylation found in males. This significantly increased methylation in exon 3 occurs close to the donor splice site in intron 3 and is predicted to have an effect on obp11 transcription by influencing the splicing process. In many organisms, including the honeybee, methylation has been found to be more frequent near or in splice sites [4,6,7,22], and several studies have provided experimental evidence that these epigenomic modifications can directly influence the splicing process [22,23]. We conclude that higher methylation in the 3′ end of exon 3 has the capacity to enforce the preferential transcription of a variant encoding truncated OBP11.
Figure 3.
Effect of methylation on obp11 alternative splicing. (a) Alternative splice acceptor site in intron 3 generates transcripts with a premature stop codon resulting in truncated peptide. Primers used to examine the expression of full-length (FL) and truncated (T) transcripts are shown below the exon/intron map. (b) Expression of obp11 alternatively spliced variant shown as percentage of total obp11 expression. In drone antennae, the spliced variant with the premature stop codon constitutes a much higher proportion of obp11 transcripts. This result is based on five replicates. (c) Increased methylation of three CpG sites in exon 3 correlates with higher abundance of the spliced variant encoding the truncated obp11. Based on three libraries. **p < 0.01, Student's t-test. (Online version in colour.)
3. Discussion
The unique methylation status of obp11 among the 21 members of the honeybee OBP family suggests that this gene's functional specialization conceivably involved an epigenetic mechanism. In terms of evolutionary invention, one scenario is that obp11 and its tandem partner obp10 may have been translocated to a methylated genomic region where one of them became a target of DNA methyltransferases. Alternatively, both genes in this tandem were initially methylated after translocation, but further rearrangements or sequence changes eliminated the signatures in obp10 required for the DNA methylation toolkit to recognize this gene as a potential target. In the case of obp11, successive epigenomic modifications affecting its sequence, such as cytosine loss, may have changed its regulation, for example by recruiting a set of DNA-binding proteins or histone modifiers creating a novel expression pattern. It is well established that epigenetic effectors, directly or indirectly, converge on chromatin, orchestrating changes in chromatin structure and consequently gene expression [24].
One important advantage of subfunctionalization of obp10 and obp11 is the female-specific expression of obp11 in sensilla basiconica that are missing in males, with obp10 retaining its male and female brain expression. According to Lynch et al. [25], close proximity of two genes could influence the probability of duplicate preservation, increasing the probability of subfunctionalization but decreasing the probability of neofunctionalization. Gene duplicates are frequently preserved by subfunctionalization, whereby both members of a pair experience degenerative mutations that reduce their joint levels and patterns of activity to that of the single ancestral gene [2]. Several authors have argued that DNA methylation has been the driving mechanism facilitating functional divergence by epigenetic silencing in one situation, most likely by promoter methylation [1,2]. Additionally, analyses of paralogous gene pairs in Arabidopsis and rice suggest that the changes in gene body methylation could provide a way for duplicate genes to develop differential patterns of expression [26]. This line of reasoning is consistent with a proposed divergence of duplicate genes in mammals whereby high levels of methylation of two adjacent duplicate genes not only gradually decreases, but also becomes more divergent with evolutionary age [27]. It is conceptually easy to perceive how such a strategy operating via an epigenetic mechanism would be beneficial for a fast evolving honeybee gene that arose from multiple duplications of an ancestral gene. Unfortunately, it is difficult or even impossible to prove that obp10 also was initially methylated. While this is a likely possibility, the evolutionary history of obp genes is too recent to leave a detectable mark on their sequences resulting from the loss of cytosines that, in some cases, can be theoretically estimated by computational analyses using the observed-to-expected (o/e) CpG ratios. This ratio is greater than one for all obps, which in bioinformatics terms classifies them as non-methylated genes. However, it is important to note that computational analyses are not always good predictors of methylation status in a given genomic region and even the best algorithms have no more than 86% prediction accuracy [28].
At the molecular level, methylation of obp11, which appears to be more prevalent in drones than in female bees might be part of a mechanism that keeps in check its expression in male tissues. The very high level of obp11 transcripts in sensilla basiconica of female antennae suggests that this gene is driven by a strong promoter whose regulation in males may require an additional level of control to prevent undesirable transcription in a caste that does not even have these sensilla. Because there is no promoter methylation in insects, such a mechanism would have to operate at the gene body level in accord with an established hypothesis that gene body methylation reduces transcriptional noise associated with spurious transcription of genes [29–31]. Indeed, in recent times, more evidence supporting the idea that gene body methylation may lead to either increase or decrease of transcript levels and to changes of alternative splicing patterns to generate different protein isoforms has become available [21,31–33].
Our data suggest that one mechanism by which methylation can reduce the level of functional OBP11 protein is alternative splicing, in particular the choice of alternative acceptor sites in intron 3 that generate two messages, one encoding a full-length OBP11 and the other a truncated non-functional peptide. This notion is in line with some evidence that DNA methylation is part of a control system regulating splicing in insects [3,4,6,34]. In the case of obp11, DNA methylation can enforce preferential selection of a transcript variant with a premature stop codon that cannot produce a full-length functional protein. A similar situation has been reported for another honeybee gene encoding a highly conserved transmembrane protein with the Ynnh domain (lysoplasmalogenase-like protein TMEM86A) [4].
Although the specific ligand of obp11 is not known, its expression in females and the recent evidence in two ant species that sensilla basiconica are involved in sensing cuticular hydrocarbon (CH) pheromones [35] suggests a similar function in honeybees. These sensilla enable discrimination of CHs detected by the antenna with remarkable sensitivity even for enantiomers of a queen pheromone and are primary sensors of both nest-mates and non-nest-mates. In this context, the lack of sensilla basiconica and suppressed obp11 expression in drones is less surprising as this caste is not involved in social interactions within the colony, and kin recognition may not be beneficial for mating flights, during which drones find unrelated queens.
To fully understand how these and other duplicated obp genes in the honeybee are maintained under natural conditions, the properties of their ligands need to be characterized. If obp10 and obp11 show different binding properties, then their evolution involved not only subfunctionalization, but also neofunctionalization. In Drosophila, a combination of subfunctionalization and neofunctionalization after gene duplication was the evolutionary driving force for two OBPs that arose by tandem gene duplication [36]. In vitro binding studies have shown that compared with the ancestral protein, one of them, OBP57d, is more specialized to tridecanoic acid, whereas the other, OBP57e, is generalized to a wide range of fatty acids. Although mutations or knockouts in OBP genes in Drosophila and mosquitoes are not lethal, they nonetheless result in serious behavioural defects. For example, the Drosophila OBP mutant, lush, is defective in pheromone-driven social aggregation behaviour [37].
Our findings imply that an evolutionarily conserved epigenetic strategy can be used in honeybees and possibly other insects to preserve duplicate genes by generating complementary patterns of expression, or by combining subfunctionalization with neofunctionalization. In a broader context, this study improves our understanding of the effects of gene body methylation on gene expression in purely mechanistic terms.
4. Material and methods
(a). Biological material
All honeybee specimens came from our Canberra colonies. Queens were purchased from local beekeepers. Larvae were harvested from brood frames taken from the hive and incubated at 35°C, 80% humidity and snap frozen in liquid nitrogen if required. Tissue dissections were carried out in a standard bee Ringer solution. A video showing our method of adult brain extraction can be access at: db.tt/wSj9BBxL. Antennae were separated from heads and processed for in situ hybridization as described previously [38]. Two sets of specimens were collected in 2013 and 2016 using hives founded by unrelated queens. Age-matched bees were used for expressional profiling. Pollen foragers were 17 days old, nurses were 11 days old, drones 14–15 days old and egg-laying queens 14 days old.
(b). DNA and RNA extraction
DNA and RNA isolation were performed as described elsewhere [20,39]. Briefly, DNA was purified using the MasterPure DNA purification kit from Epicentre Biotechnologies. The RNase digestion step was not performed. For expression analysis, RNA was extracted using TRIzol® (Invitrogen), and where DNA was required from the same starting material DNA was extracted from the Trizol interface using a standard phenol–chloroform method. For RNA alone, a combined Trizol/column procedure was used as per Zymogen RNA Clean & Concentrator-5 kit, catalogue no. R1015. Two individuals per replicate, i.e. two brains or four antennae equivalent to 2.5 µg total RNA from brain samples and 0.5 µg total RNA from purified antennal samples were used. To assess RNA quality, 1–2 µl of each RNA sample was denatured with formamide, containing 0.01% SybrGreen and evaluated by agarose gel electrophoresis (1.5% agarose; 20 V cm−1).
(c). DNA bisulfite conversion and amplicon preparation
Genomic DNA of 1.5 µg was bisulfite converted using the QIAGEN Epitect® bisulfite kit, as per the manufacturer's protocol. The converted DNA was amplified via a nested PCR with AmLAM-specific primers, see electronic supplementary material, table S1 for BS-seq-specific primers. The PCR products were purified using Agencourt® AMPure® XP PCR purification system (Beckman–Coulter).
(d). Next-generation sequencing library preparation for methylation analysis
Three libraries for each condition were prepared from 500 to 600 ng of every amplicon using the NEBNext® DNA Library Prep Master Mix for Illumina®, and NEBNext® Multiplex Oligos for Illumina® Index Primers Set 1 (New England Biolabs). Size selection of adaptor ligated DNA was performed using Agencourt AMPure XP beads (Beckman Coulter), with the bead : DNA ratio of the first bead selection 0.9X, followed by a second bead selection with bead : DNA ratio at 0.2X. Each library was eluted in 30 µl of 0.1X TE, library size confirmed via agarose gel electrophoresis, and diluted to a final concentration of 4 nM. A detailed description of our protocol has been published elsewhere [21].
(e). Next-generation sequencing MiSeq sequencing
Next-generation sequencing was performed on Illumina MiSeq instrument using MiSeq reagent kit v3 (Illumina) and 600 cycles. PhiX spike was added at 5% concentration as recommended by Illumina for low-diversity libraries [20,21].
(f). Analysis of bs-seq results
We have used a previously described method that combines custom Python scripts and open-source software. Methylation density was calculated as the percentage of methylated CpG motifs found across the OBP11 amplicons [20,21].
(g). Expression analysis
In situ hybridization was performed on antennal sections using in vitro synthesized DIG-labelled riboprobes. The primers for generating obp11 amplicon used for cloning and subsequent transcription as in electronic supplementary material, table S1. Other experimental details can be found in [38–40].
OBP11 transcripts levels were quantitated via RT-PCR (see electronic supplementary material, table S1 for quantitative PCR primers). cDNA was synthesized from 2.5 µg of RNA using either Maxima (Thermo Scientific), or SuprtScript (Invitrogen) reverse transcriptase as per the manufacturer's protocols and amplified using a SYBR® green I-based assay. All RT-PCR experiments were performed using the Applied Biosystems® StepOnePlus™ real-time PCR system. Gene expression was normalized against both calmodulin and utg, and relative expression calculated using the 2−ΔΔCT method, as previously described [6,39]. OBP10 transcript levels were evaluated using the available RNAseq datasets [39]. For more details, see [39] and electronic supplementary material, figure S1 legend.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Sylvain Foret for helpful discussions and Paul Helliwell for providing biological material.
Competing interests
We declare we have no competing interests.
Funding
This research was supported by the Australian Research Council grant DP160103053 awarded to R.M.
References
- 1.Rodin SN, Riggs AD. 2003. Epigenetic silencing may aid evolution by gene duplication. J. Mol. Evol. 56, 718–729. ( 10.1007/s00239-002-2446-6) [DOI] [PubMed] [Google Scholar]
- 2.Lynch M, Conery JS. 2000. The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. ( 10.1126/science.290.5494.1151) [DOI] [PubMed] [Google Scholar]
- 3.Flores K, Wolschin F, Corneveaux JJ, Allen AN, Huentelman MJ, Amdam GV. 2012. Genome-wide association between DNA methylation and alternative splicing in an invertebrate. BMC Genomics 13, 480 ( 10.1186/1471-2164-13-480) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. 2010. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 8, e1000506 ( 10.1371/journal.pbio.1000506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foret S, Kucharski R, Pittelkow Y, Lockett GA, Maleszka R. 2009. Epigenetic regulation of the honey bee transcriptome: unravelling the nature of methylated genes. BMC Genomics 10, 472 ( 10.1186/1471-2164-10-472) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foret S, et al. 2012. DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. Proc. Natl Acad. Sci. USA 109, 4968–4973. ( 10.1073/pnas.1202392109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonasio R, et al. 2012. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr. Biol. 22, 1755–1764. ( 10.1016/j.cub.2012.07.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maleszka R. 2014. The social honey bee in biomedical research: realities and expectations. Drug Discov. Today Dis. Models 12, 7–13. ( 10.1016/j.ddmod.2014.06.001) [DOI] [Google Scholar]
- 9.Maleszka R. 2016. Epigenetic code and insect behavioural plasticity. Curr. Opin. Insect Sci. 15, 45–52. ( 10.1016/j.cois.2016.03.003) [DOI] [PubMed] [Google Scholar]
- 10.Pelosi P, Calvello M, Ban L. 2005. Diversity of odorant-binding proteins and chemosensory proteins in insects. Chem. Senses 30(Suppl 1), 291–292. ( 10.1093/chemse/bjh229) [DOI] [PubMed] [Google Scholar]
- 11.Krieger J, Breer H. 1999. Olfactory reception in invertebrates. Science 286, 720–723. ( 10.1126/science.286.5440.720) [DOI] [PubMed] [Google Scholar]
- 12.Deyu Z, Leal WS. 2002. Conformational isomers of insect odorant-binding proteins. Arch. Biochem. Biophys. 397, 99–105. ( 10.1006/abbi.2001.2660) [DOI] [PubMed] [Google Scholar]
- 13.Foret S, Maleszka R. 2006. Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera). Genome Res. 16, 1404–1413. ( 10.1101/gr.5075706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong DP, Zhang HJ, Zhao P, Xia QY, Xiang ZH. 2009. The odorant binding protein gene family from the genome of silkworm, Bombyx mori. BMC Genomics 10, 332 ( 10.1186/1471-2164-10-332) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danty E, et al. 1999. Cloning and expression of a queen pheromone-binding protein in the honeybee: an olfactory-specific, developmentally regulated protein. J. Neurosci. 19, 7468–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plettner E, Lazar J, Prestwich EG, Prestwich GD. 2000. Discrimination of pheromone enantiomers by two pheromone binding proteins from the gypsy moth Lymantria dispar. Biochemistry 39, 8953–8962. ( 10.1021/bi000461x) [DOI] [PubMed] [Google Scholar]
- 17.Pophof B. 2002. Moth pheromone binding proteins contribute to the excitation of olfactory receptor cells. Naturwissenschaften 89, 515–518. ( 10.1007/s00114-002-0364-5) [DOI] [PubMed] [Google Scholar]
- 18.Zhou JJ, et al. 2004. Revisiting the odorant-binding protein LUSH of Drosophila melanogaster: evidence for odour recognition and discrimination. FEBS Lett. 558, 23–26. ( 10.1016/S0014-5793(03)01521-7) [DOI] [PubMed] [Google Scholar]
- 19.Esslen J, Kaissling KE. 1976. Number and distribution of sensilla on antennal flagellum of honeybee (Apis mellifera L). Zoomorphologie 83, 227–251. ( 10.1007/BF00993511) [DOI] [Google Scholar]
- 20.Kucharski R, Foret S, Maleszka R. 2015. EGFR gene methylation is not involved in Royalactin controlled phenotypic polymorphism in honey bees. Sci. Rep. UK 5, 14070 ( 10.1038/srep14070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wedd L, Kucharski R, Maleszka R. 2016. Differentially methylated obligatory epialleles modulate context-dependent LAM gene expression in the honey bee Apis mellifera. Epigenetics 11, 1–10. ( 10.1080/15592294.2015.1107695) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maunakea AK, Chepelev I, Cui K, Zhao K. 2013. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 23, 1256–1269. ( 10.1038/cr.2013.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shukla S, et al. 2011. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479, 74–79. ( 10.1038/nature10442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Badeaux AI, Shi Y. 2013. Emerging roles for chromatin as a signal integration and storage platform. Nat. Rev. Mol. Cell Biol. 14, 211–224. ( 10.1038/nrm3545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch M, O'Hely M, Walsh B, Force A. 2001. The probability of preservation of a newly arisen gene duplicate. Genetics 159, 1789–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Marowsky NC, Fan C. 2014. Divergence of gene body DNA methylation and evolution of plant duplicate genes. PLoS ONE 9, e110357 ( 10.1371/journal.pone.0110357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller TE, Yi SV. 2014. DNA methylation and evolution of duplicate genes. Proc. Natl Acad. Sci. USA 111, 5932–5937. ( 10.1073/pnas.1321420111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das R, et al. 2006. Computational prediction of methylation status in human genomic sequences. Proc. Natl Acad. Sci. USA 103, 10 713–10 716. ( 10.1073/pnas.0602949103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki MM, Kerr ARW, De Sousa D, Bird A. 2007. CpG methylation is targeted to transcription units in an invertebrate genome. Genome Res. 17, 625–631. ( 10.1101/gr.6163007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huh I, Zeng J, Park T, Yi SV. 2013. DNA methylation and transcriptional noise. Epigenet. Chromatin 6, 9 ( 10.1186/1756-8935-6-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lev Maor G, Yearim A, Ast G. 2015. The alternative role of DNA methylation in splicing regulation. Trends Genet. 31, 274–280. ( 10.1016/j.tig.2015.03.002) [DOI] [PubMed] [Google Scholar]
- 32.Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. 2014. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 6, 577–590. ( 10.1016/j.ccr.2014.07.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagner JR, Busche S, Ge B, Kwan T, Pastinen T, Blanchette M. 2014. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol. 15, R37 ( 10.1186/gb-2014-15-2-r37) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyko F, Maleszka R. 2011. Insects as innovative models for functional studies of DNA methylation. Trends Genet. 27, 127–131. ( 10.1016/j.tig.2011.01.003) [DOI] [PubMed] [Google Scholar]
- 35.Sharma KR, et al. 2015. Cuticular hydrocarbon pheromones for social behavior and their coding in the ant antenna. Cell Rep. 12, 1261–1271. ( 10.1016/j.celrep.2015.07.031) [DOI] [PubMed] [Google Scholar]
- 36.Harada E, et al. 2012. Functional evolution of duplicated odorant-binding protein genes, Obp57d and Obp57e, in Drosophila. PLos ONE 7, e29710 ( 10.1371/journal.pone.0029710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu P, Atkinson R, Jones DN, Smith DP. 2005. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron 45, 193–200. ( 10.1016/j.neuron.2004.12.031) [DOI] [PubMed] [Google Scholar]
- 38.Maleszka J, Foret S, Saint R, Maleszka R. 2007. RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera). Dev. Genes Evol. 217, 189–196. ( 10.1007/s00427-006-0127-y) [DOI] [PubMed] [Google Scholar]
- 39.Wojciechowski M, Rafalski D, Kucharski R, Misztal K, Maleszka J, Bochtler M et al. 2014. Insights into DNA hydroxymethylation in the honeybee from in-depth analyses of TET dioxygenase. Open Biol. 4, 140110 ( 10.1098/rsob.140110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ashby R, Foret S, Searle I, Maleszka R. 2016. MicroRNAs in honey bee caste determination. Sci Rep. 6, 18794 ( 10.1038/srep18794) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.