Abstract
Nrf2 (nuclear factor erythroid 2-related factor 2) is an antioxidant transcription factor. AMP-activated protein kinase (AMPK) functions as a central regulator of cell survival in response to stressful stimuli. Nrf2 should be coordinated with the cell survival pathway controlled by AMPK, but so far the mechanistic connections remain undefined. This study investigated the role of AMPK in Nrf2 trafficking and its activity regulation. A subnetwork integrating neighbor molecules suggested direct interaction between AMPK and Nrf2. In cells, AMPK activation caused nuclear accumulation of Nrf2. In the in vitro kinase and peptide competition assays, AMPK phosphorylated Nrf2 at the Ser558 residue (Ser550 in mouse) located in the canonical nuclear export signal. Nrf2 with an S550A mutation failed to be accumulated in the nucleus after AMPK activation. Leptomycin B, a nuclear export inhibitor, did not enhance nuclear accumulation of wild-type Nrf2 (WT-Nrf2) activated by AMPK or a phospho-Ser550-mimetic Nrf2 mutant, corroborating the finding that AMPK facilitated nuclear accumulation of Nrf2, probably by inhibiting nuclear export. Activated glycogen synthase kinase 3β (GSK3β) diminished the basal nuclear level of Myc-S550A-Nrf2. Taking the data collectively, AMPK phosphorylates Nrf2 at the Ser550 residue, which, in conjunction with AMPK-mediated GSK3β inhibition, promotes nuclear accumulation of Nrf2 for antioxidant response element (ARE)-driven gene transactivation.
INTRODUCTION
Redox homeostasis in the cell may be disturbed by a deficiency in energy production as well as by reactive oxygen species (ROS) generated from xenobiotic biotransformation or the process of fuel oxidation (1–3). Nrf2 (nuclear factor erythroid 2-related factor 2), a key antioxidant transcription factor, participates in maintaining redox homeostasis in the cell (4). Various physiological or pathological circumstances accompanying free radical generation activate Nrf2 as a consequence of an adaptive response to oxidative stress (5). However, the regulation of Nrf2 has been minimally studied in the context of cellular energy deficiency (e.g., starvation, hypoxia, and exercise).
The response of Nrf2 to oxidative challenge stress rapidly occurs mainly through phosphorylation at Ser40; protein kinase Cδ (PKCδ) induces activation of phosphorylation of Nrf2 at the residue, and the phosphorylated form is then released from Keap-1 and is stabilized for antioxidant response element (ARE)-driven gene expression (6, 7). An active form of Nrf2 has been shown to localize in the nucleus and binds to the ARE(s) present in the promoter regions of target genes (8). In this process, phosphoinositide 3-kinase controls nuclear translocation of Nrf2 (9). In contrast, glycogen synthase kinase 3β (GSK3β) catalyzes the inhibitory phosphorylation of Nrf2 for the tight activity control (10, 11). Other kinases, including mitogen-activated protein kinases and Fyn, may also affect Nrf2 activity (12, 13). Despite the identification of oxidative stress-associated Nrf2 kinases, the characteristics of regulation of Nrf2 in response to the change in energy status and the responsible kinase(s) remain largely unknown.
AMP-activated protein kinase (AMPK), a heterotrimeric complex composed of α-, β-, and γ-subunits, is an energy-sensing serine/threonine kinase ubiquitously expressed across different tissues (14). The AMPK α-subunit (α1 and α2 isoforms) contains the catalytic domain and is phosphorylated at the Thr172 residue upon activation (14). In mammalian cells, AMPK is activated by an increase in the AMP-to-ATP ratio, which occurs under a variety of stress conditions such as nutrient deprivation, ischemia, and hypoxia (e.g., prolonged exercise); AMPK activation allows the cell to recover energy homeostasis by enhancing fuel oxidation (14–16). In addition, AMPK induces manganese superoxide dismutase (MnSOD) and HO-1 (17–19), enabling the cell to recover its antioxidant capacity. Moreover, it has the ability to increase cell survival of harmful stresses (20–24). Nevertheless, information as to how AMPK induces the expression of cytoprotective or antioxidant enzymes is unavailable.
AMPK phosphorylates hepatocyte nuclear factor 4α (HNF4α), ChREBP, p300, and TORC2 (25). However, whether AMPK phosphorylates Nrf2 is unknown. In view of the lack of understanding of the link between AMPK and Nrf2, this study investigated whether AMPK phosphorylates Nrf2 and, if so, what the underlying basis is. Our findings revealed that AMPK phosphorylates Nrf2 at the Ser550 residue located in the canonical nuclear export signal (NES) sequence. Moreover, we found that AMPK causes accumulation of Nrf2 in the nucleus through phosphorylation in addition to GSK3β inhibition (10, 11). Our findings identifying AMPK as a novel kinase that activates Nrf2 through nuclear accumulation provide information on the distinct role of the energy-sensing enzyme in increasing antioxidant capacity and cell survival.
(A part of the data included in this paper was presented at the Annual Meeting of Experimental Biology [2016].)
MATERIALS AND METHODS
Materials.
Antibodies directed against Nrf2 and c-Myc were supplied by Santa Cruz Biotechnology (Santa Cruz, CA). The antibodies specifically recognizing phospho-AMPK, AMPK, phospho-glycogen synthase kinase 3β (phospho-GSK3β), GSK3β, and lamin A/C were obtained from Cell Signaling (Beverly, MA). Horseradish peroxidase-conjugated goat anti-rabbit and goat anti-mouse IgGs were obtained from Zymed Laboratories (San Francisco, CA). [32P]ATP (3,000 mCi/mmol) was supplied by PerkinElmer Life and Analytical Sciences (Waltham, MA). Anti-β-actin antibody and other reagents were obtained from Sigma-Aldrich (St. Louis, MO).
Analysis of protein-protein interaction network.
The protein-protein interaction network was mapped using the Mentha open database (http://mentha.uniroma2.it/). In the analysis of data from the microarray GEO database (accession number GSE24504), coefficients of correlation between median values of all gene pairs were calculated using R software (Bioconductor, pvclust) and Pearson's method. P values for each gene pair were calculated from the values corresponding to the distribution of the degree of correlation between all gene pairs. The human gene network obtained from HumanNet (http://www.functionalnet.org/humannet/) was converted to a mouse gene network using NCBI Human Genome (build 36.1). All networks were visualized using Cytoscape 3.0.0 software.
Cell culture.
HepG2 and HEK293 cells were supplied by the American Type Culture Collection (Manassas, VA). The cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS), 50 units/ml penicillin, and 50 μg/ml streptomycin at 37°C in a humidified atmosphere with 5% CO2. Cells with fewer than 20 passages were used.
Immunoblot analysis.
Cell lysates and subcellular fractions were prepared according to previously published methods (9). SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblot analyses were performed as previously described (26). Immunoreactive proteins were visualized using an ECL chemiluminescence detection kit (Amersham Biosciences, Buckinghamshire, United Kingdom).
Immunofluorescence.
Standard immunocytochemical methods were carried out for the immunostaining of Nrf2 as previously described (9). For immunocytochemistry, HepG2 cells were fixed in 4% formaldehyde for 10 min. After a blocking in 5% bovine serum albumin–PBS for 1 h, the cells were incubated with anti-Nrf2 antibody for 1 h and were then reacted with Alexa Fluor 546-conjugated rabbit anti-goat IgG antibody (Invitrogen, Carlsbad, CA). Stained cells were examined using an Eclipse Ti-U microscope (Nikon Inc., Japan).
Plasmids.
Specific base substitutions were made by oligonucleotide-mediated mutagenesis according to the instructions of the manufacturer (Stratagene, Santa Clara, CA). The Ser550 residue of Nrf2 (mouse form) was mutated to alanine or glutamic acid using a mutagenic primer (5′-CTGAAAAGGCGGCTCGCCACCTTGTATCTTG-3′ or 5′-CTGAAAAGGCGGCTCGAGACCTTGTATCTTG-3′). The authenticity of the DNA sequences was verified using an automatic DNA sequence analyzer. The construct encoding DN-AMPKα1 was a gift from J. Ha (Kyung Hee University, South Korea).
Transient-transfection and luciferase reporter assays.
Transient transfection was done with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA). Briefly, cells were replated 24 h before transfection at a density of 7 × 105 cells in six-well plates. The cells were incubated with the plasmid of interest and Lipofectamine reagent for 3 h for transfection and were continuously incubated for additional 24 h in minimal essential medium containing 1% FBS. Control cells were transfected with an equal amount of the respective empty plasmid (i.e., mock transfection). For luciferase assays, the cells were transiently transfected with pGL-NQO1-ARE construct, which contained three copies of ARE derived from the promoter of NQO1. Luciferase activity was measured by adding Luciferase assay reagent (Promega, Madison, WI).
In vitro kinase assay.
Recombinant glutathione S-transferase (GST)-tagged human Nrf2 and human AMPK were purchased from Abnova (Taipei City, Taiwan) and Upstate Biotechnology (Lake Placid, NY), respectively. For in vitro kinase assay, recombinant AMPK, Nrf2, and [32P]ATP were mixed in a final volume of 20 μl of a kinase buffer containing Tris-HCl (25 mM; pH 7.4), 10 mM MgCl2, 25 mM β-glycerophosphate, 1 mM Na3VO4, 2 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, and 200 μM ATP, and the reaction mixture was incubated for 30 min at 30°C. Protein phosphorylation was visualized by autoradiography after SDS-PAGE analysis.
Statistical analysis.
The differences between groups were analyzed using one-way analysis of variance (ANOVA) with Bonferroni correction or the two-tailed Student t test. The criterion for statistical significance was set at a P value of <0.05 or a P value of <0.01.
RESULTS
The coexpression network between AMPK and Nrf2.
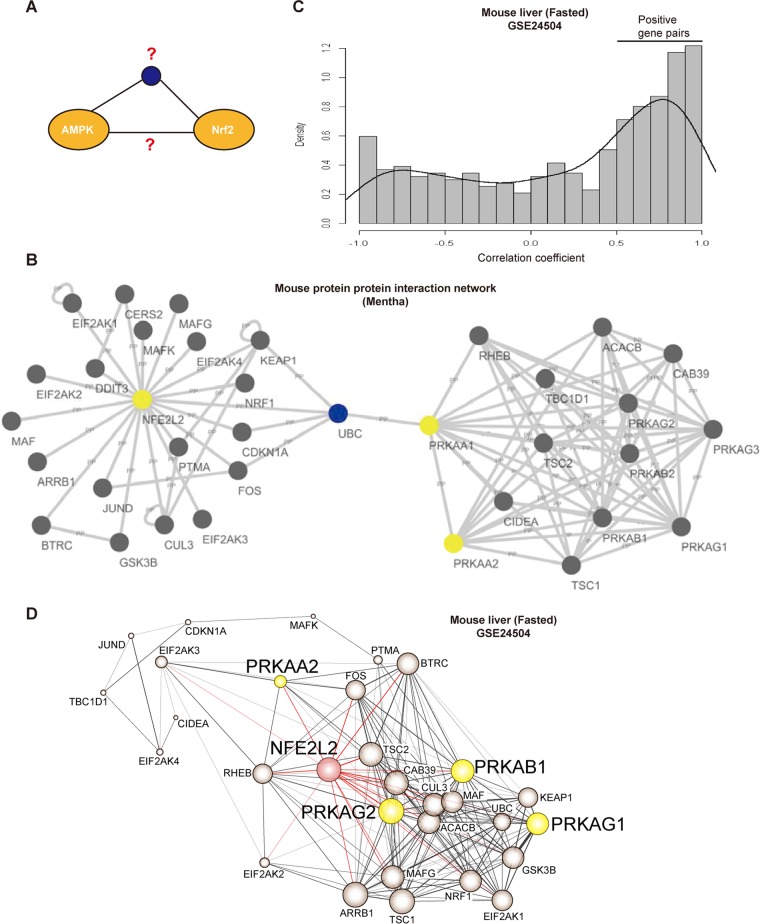
To explore the molecular interactions (Fig. 1A), we first obtained a subnetwork integrating neighbor molecules of AMPK and Nrf2 using the open database of protein-protein interactions and Mentha software (http://mentha.uniroma2.it/) and merged them to acquire the integrated protein interactions (Fig. 1B). The subnetwork included Prkaa1 (AMPKα1), Prkaa2 (AMPKα2), and Nfe2l2 (Nrf2) as first-neighbor molecules, and there was no direct interaction between AMPK and Nrf2. As an effort to find the first-neighbor interactions in a physiologically relevant situation, we next chose a fasting animal model and used the cDNA array GEO database (GSE24504) derived from the liver of mice fasted for 24 to 72 h; global gene coexpression analysis emerged as a powerful approach to identify the tissues and the conditions under which important interactions occur (27, 28). We then calculated the coefficient of correlation between all gene pairs comprised in the subnetwork and plotted the density distribution of each gene pair interaction versus correlation coefficient (Fig. 1C). The density distribution was negatively skewed under the fasting condition. In this analysis, the P values were tested from the distribution of the degree of correlation between each gene pair of all array-embedded genes; 13,894 genes made 151,547 gene pairs. Of 436 gene pairs among 30 genes in the subnetwork, data from 110 gene pairs were statistically significant (P < 0.01). In particular, the P value of the correlation between AMPK and Nrf2 was 0.009 (see Data File S1 in the supplemental material). The network redrawn using the correlation coefficient of gene pairs displayed a direct association between Nrf2 (Nfe2l2) and each subunit of AMPK (i.e., Prkaa2 [AMPKα2], Prkab1 [AMPKβ1], Prkag1 [AMPKγ1], and Prkag2 [AMPKγ2]) (Fig. 1D).
FIG 1.
The coexpression network between AMPK and Nrf2. (A) Schematic diagram of hypothesis. AMPK and Nrf2 may directly or indirectly interact with each other. (B) A subnetwork of protein-protein interactions between PRKAA1 (AMPKα1), PRKAA2 (AMPKα2), and NFE2L2 (Nrf2). (C) Density distribution of correlation coefficients between all gene pairs comprised in the subnetwork shown in panel B. Correlation coefficients were calculated using public cDNA microarray data (GEO database accession no. GSE24504). (D) Gene interaction network between AMPK and Nrf2. Gene interactions are based on the correlation profile. Filled colors indicate Nrf2 subunits (red) or AMPK subunits (yellow). The node size is proportional to the number of neighboring genes. The line thickness reflects the correlation coefficient between nodes.
Nuclear accumulation of Nrf2 by AMPK activation.
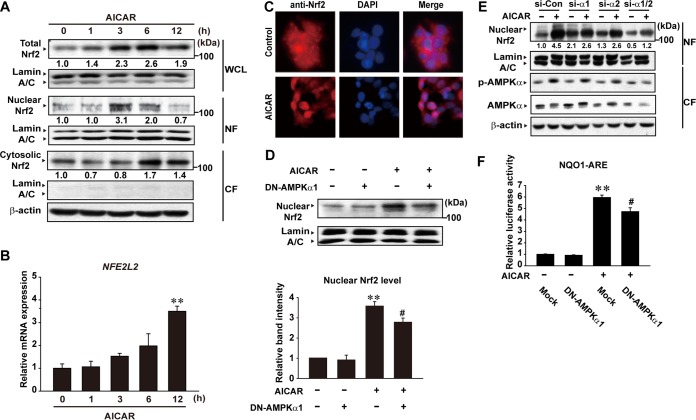
To understand the role of AMPK in regulating Nrf2 activity in greater depth, we first examined the subcellular localization of Nrf2 after treatment of HepG2 cells with 5-aminoimidazole-4-carboxamide-1-β-d-ribofuranoside (AICAR), an AMP analog that activates AMPK. AICAR treatment increased Nrf2 levels in whole-cell lysate or in nuclear fractions from 3 h to 12 h (Fig. 2A). However, Nrf2 levels in the cytoplasmic fractions were slightly increased only at later times, which might have been due to Nrf2-dependent transcriptional autoinduction (29). This idea is supported by an increase in the mRNA level (Fig. 2B). In the immunocytochemical analysis, Nrf2 was predominantly located in the cytoplasm of control cells. AICAR treatment promoted nuclear localization of Nrf2 (Fig. 2C), confirming the functional role of AMPK in its nuclear accumulation. Immunoblotting assay results showed that transfection with dominant-negative (DN)-AMPKα1 significantly attenuated AICAR-inducible accumulation of Nrf2 in the nucleus (Fig. 2D). Consistently, knockdown of AMPKα1/2 diminished nuclear accumulation of Nrf2 by AICAR (Fig. 2E).
FIG 2.
AMPK activation of Nrf2. (A) Immunoblottings for Nrf2. Nrf2 was immunoblotted on whole-cell lysates (WCL) and on the nuclear fraction (NF) and cytoplasmic fraction (CF) of HepG2 cells treated with AICAR (500 μM) for the indicated times. (B) Quantitative real-time PCR assays. NFE2L2 mRNA levels were measured in HepG2 cells treated with AICAR (500 μM). Data represent means ± standard errors of the means (SEM) of results from three different cell preparations (one-way ANOVA with Bonferroni correction; results significantly different from time zero control results [**, P < 0.01]). (C) Immunocytochemistry for Nrf2. HepG2 cells were treated with vehicle (Control) or AICAR for 3 h and were immunochemically stained for Nrf2 (red). (D) The effect of DN-AMPKα1 on Nrf2 nuclear accumulation by AICAR. Nrf2 was immunoblotted on the nuclear fractions of HepG2 cells treated with vehicle or AICAR for 5 h following DN-AMPKα1 transfection. Data represent means ± SEM of results from three independent experiments (significantly different from mock control results [**, P < 0.01] and from AICAR treatment results [#, P < 0.05]). (E) The effect of small interfering AMPKα1 (siAMPKα1) and/or siAMPKα2 on Nrf2 nuclear accumulation by AICAR. Nrf2 was immunoblotted on the nuclear fractions of HepG2 cells treated with vehicle or AICAR for 5 h following siAMPKα1/2 transfection. (F) Effect of DN-AMPKα1 on the NQO1-ARE reporter activity. Luciferase expression from NQO1-ARE was measured in HepG2 cells treated with vehicle or AICAR for 18 h following DN-AMPKα1 transfection. Data represent means ± SEM of results from three different cell preparations (significantly different from mock control results [**, P < 0.01] and from AICAR treatment results [#, P < 0.05]).
AICAR has the ability to induce luciferase expression from the NAD(P)H quinone oxidoreductase 1 (NQO1)-ARE reporter that contains three copies of the antioxidant response element (ARE) derived from the promoter region of the NQO1 gene. Overexpression of DN-AMPKα1 reduced AICAR-inducible ARE reporter activity (i.e., 30% compared to mock transfection) (Fig. 2F). These results indicate that AMPK may enhance the activity of Nrf2 through nuclear localization.
Direct phosphorylation of Nrf2 by AMPK.
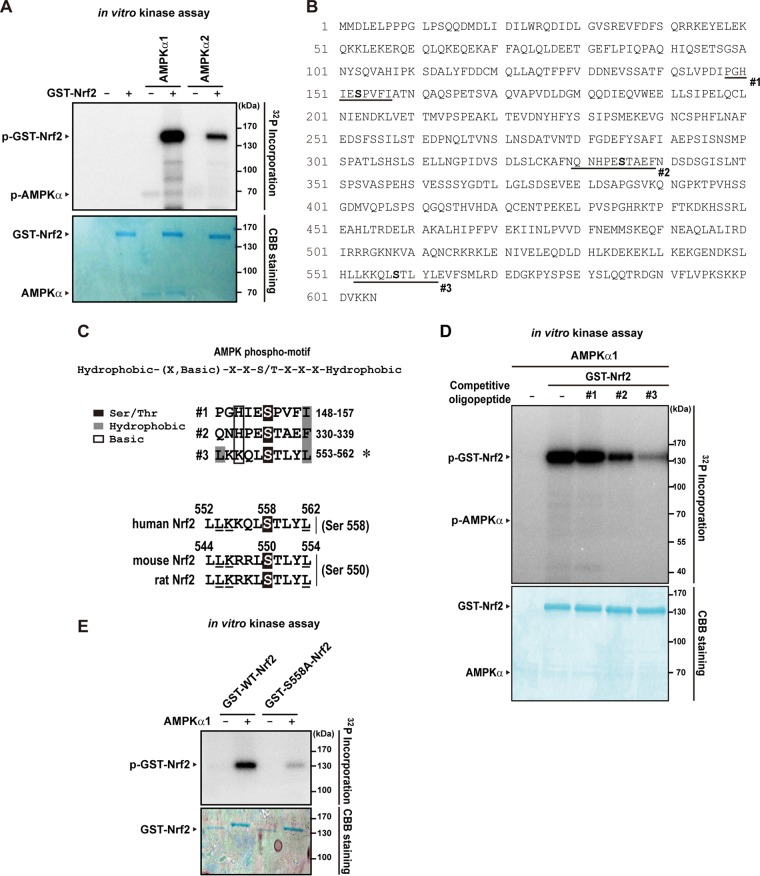
To investigate the molecular basis of AMPK's activation of Nrf2, we then determined whether AMPK phosphorylates Nrf2. In the in vitro kinase assay, AMPK directly phosphorylated Nrf2 (Fig. 3A). Biochemical and bioinformatics studies have identified the optimal consensus recognition motif for AMPK phosphorylation (30, 31). We found three putative AMPK recognition motifs within Nrf2 (Fig. 3B). Of them, the third one (having Ser558 in the human form and Ser550 in the mouse and rat homologous form) was especially worthy of notice because it is conserved across the species, including human, mouse, and rat, and has a perfect match with the known recognition motif (Fig. 3C). To find the functional phosphorylation site(s) in Nrf2 corresponding to AMPK, three different 10-residue oligopeptides mimicking the putative target sites (site 1, residues 148 to 157 comprising Ser153; site 2, residues 330 to 339 comprising Ser335; site 3, residues 553 to 562 comprising Ser558) were synthesized and used as competitors in the in vitro kinase assay (they were readily soluble in the final solution for the kinase reaction performed at 0.2 μg/μl). Of note, the ability of AMPK to phosphorylate Nrf2 was greatly diminished only by the presence of oligopeptide 3 (Fig. 3D). Next, a site-directed mutagenesis assay was done to corroborate the outcome of the peptidomimetic experiment. Given the inhibitory effect of oligopeptide 3 on AMPK-mediated Nrf2 phosphorylation (Fig. 3D), we generated S558A-Nrf2 mutant and compared the in vitro phosphorylation with wild-type Nrf2 (WT-Nrf2) results. The single amino acid substitution (Ser→Ala at 558) completely abrogated Nrf2 phosphorylation by AMPK (Fig. 3E), strengthening the authenticity of Ser558 phosphorylation. Our findings provide strong evidence that Ser558 in Nrf2 (Ser550 in mouse Nrf2) is an authentic site of phosphorylation by AMPK.
FIG 3.
Phosphorylation of Nrf2 at the Ser558 residue by AMPK. (A) In vitro phosphorylation of Nrf2 by AMPK. Recombinant GST-tagged Nrf2 was incubated with recombinant AMPK and [32P]ATP for 20 min at 30°C, and proteins in the mixture were resolved by SDS-PAGE. The band was visualized by autoradiography of 32P-labeled protein. Loading of proteins was confirmed by Coomassie brilliant blue (CBB) staining. (B) Amino acid sequence of human Nrf2. Putative AMPK phosphorylation sites are shown in bold. Synthetic peptides used to mimic the sites are underlined. (C) The consensus motif of AMPK phosphorylation. Data represent predicted AMPK phosphorylation motifs in Nrf2. The region comprising Ser558 is evolutionally conserved. (D) Inhibition by oligopeptide 3 of AMPK's phosphorylation of Nrf2. In vitro kinase assays were performed using the reaction mixture (20 μl) containing 0.4 μg of purified GST-tagged Nrf2, 0.15 μg of AMPK, and 1 μCi of [32P]ATP in the presence of 4.3 μg of the indicated oligopeptide for 15 min at 30°C, and proteins in the mixture were resolved by SDS-PAGE. The band was visualized by autoradiography of 32P-labeled protein. Loading of proteins was confirmed by CBB staining. (E) In vitro kinase assays using WT-Nrf2 or S558A-Nrf2 mutant.
Accumulation of S550-phosphorylated Nrf2 in the nucleus.
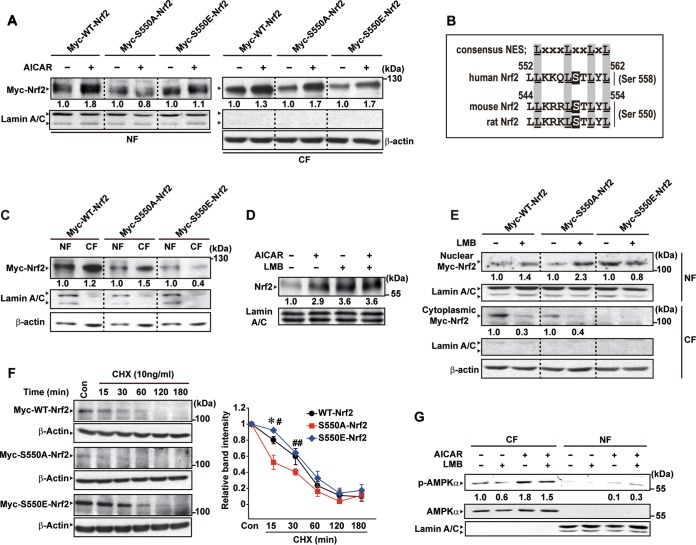
To link nuclear Nrf2 accumulation and Ser550 phosphorylation by AICAR, we next generated plasmids encoding Myc-tagged mutants of Nrf2 by site-directed mutagenesis. AICAR-inducible nuclear localization of Nrf2 was abolished by the mutation of Ser550 to alanine (Myc-S550A-Nrf2, a mutant that mimics dephosphorylated Nrf2) (Fig. 4A). Nevertheless, Myc-S550A-Nrf2 existed basally in the nuclear fraction to a certain degree. Another mutant encoding Myc-S550E-Nrf2 with glutamate substituted for Ser550 mimics phosphorylated Nrf2. AICAR treatment had no effect on the nuclear level of Myc-S550E-Nrf2, presumably because of its predominant basal localization in the nucleus.
FIG 4.
The effect of Ser550 phosphorylation of Nrf2 on nuclear localization. (A) Nuclear fraction (NF) or cytoplasmic fraction (CF) Nrf2 levels. HEK293 cells were transfected with the plasmid encoding Myc-tagged WT-Nrf2 or the site-directed mutant (S550A-Nrf2 or S550E-Nrf2) and were treated with AICAR for 6 h. (B) Alignment of the NES motif in Nrf2 with the consensus sequence. (C) Immunoblottings for Nrf2 in the nuclear fraction (NF) or cytoplasmic fraction (CF) of HEK293 cells transfected with Myc-tagged WT-Nrf2, S550A-Nrf2, or S550E-Nrf2 for 24 h. (D) The effect of leptomycin B (LMB) on the activation of Nrf2 by AICAR. HepG2 cells were treated with AICAR and/or leptomycin B (10 nM) for 3 h. (E) Immunoblottings for Nrf2 in HEK293 cells treated with leptomycin B for 6 h after transfection with Myc-tagged S550A-Nrf2 or S550E-Nrf2. (F) Protein stability experiment using cycloheximide (CHX). HEK293 cells were treated with 10 μg/ml CHX for the indicated times after transfection with Myc-tagged WT-Nrf2, S550A-Nrf2, or S550E-Nrf2. Data represent means ± SEM of results from three independent experiments (significantly different at the respective times from WT-Nrf2 results [*, P < 0.05] and from S550A-Nrf2 results [#, P < 0.05; ##, P < 0.05]). Con, control. (G) Activation and subcellular localization of AMPKα. Each protein was measured in the subcellular fractions of HepG2 cells treated with AICAR and/or leptomycin B for 30 min. For panels A, C, and E, relative band intensities were compared to those of the respective controls of each construct.
Nrf2 has the motifs of nuclear export signal (NES) and nuclear localization signal (NLS) (32). The canonical (leucine-rich) NES motif in Nrf2 (544LLKRRLSTLYL554), a consensus sequence conserved across different species (human, mouse, and rat), contains the Ser550 residue (Fig. 4B). In subcellular fractionation assays, the nucleus-to-cytoplasm ratio of Myc-S550E-Nrf2 was greater than that of Myc-WT-Nrf2 or Myc-S550A-Nrf2 (Fig. 4C). In a continuing effort to verify the role of AMPK in Nrf2 trafficking and its activity regulation, we next adopted leptomycin B as an inhibitor of nuclear export; leptomycin B binds covalently to CRM1, an export receptor for leucine-rich NES, inhibiting its association with cargo protein (33). As expected, leptomycin B treatment enhanced nuclear accumulation of Nrf2, as did AICAR treatment (Fig. 4D). Combined treatment with leptomycin B and AICAR resulted in a comparable effect, showing no enhancement. Moreover, leptomycin B treatment caused nuclear accumulation of Myc-S550A-Nrf2 at a level similar to that seen with Myc-WT-Nrf2, supporting the idea that it may inhibit the export of Myc-S550A-Nrf2 already translocated into the nucleus irrespective of AMPK catalysis (Fig. 4E). Leptomycin B treatment did not further increase the nuclear level of Myc-S550E-Nrf2, probably because the phosphomimetic mutant had been localized in the nucleus. These results support the concept that the phosphorylation of Nrf2 at the Ser550 residue prevents nuclear export of Nrf2, probably by impeding CRM1 binding to Nrf2.
Nrf2 shows delayed degradation when accumulated in the nucleus due to hindering nuclear export signal (32). Next, we assessed whether Ser550 phosphorylation alters the Nrf2 degradation rate. In the experiment using cycloheximide (CHX), the degradation of Myc-S550E-Nrf2 was retarded compared to the degradation of either Myc-WT-Nrf2 or Myc-S550A-Nrf2 (Fig. 4F), being consistent with the previous observation with respect to NES-lacking Nrf2 (32). Our results and those reported by others support the conclusion that the phosphorylation of Nrf2 at the Ser550 residue may hinder CRM1-mediated nuclear export.
AMPK is also modulated by CRM1-mediated export via its C-terminal NES (34). AMPK catalyzes the phosphorylation of several transcription factors (e.g., FoxO3 and PGC-1) (35, 36). We wondered whether nuclear accumulation of Nrf2 by leptomycin B accompanied AMPK activation. The phosphorylated form (Thr172) or total form of AMPK was found predominantly in the cytoplasmic fraction but minimally in the nuclear fraction (Fig. 4G). As expected, AICAR treatment increased the level of phospho-AMPK (Fig. 4G). Leptomycin B treatment did not enhance the basal or inducible phosphorylation of AMPK, raising the notion that AMPK phosphorylates Nrf2 in the cytoplasm. Together, these results support the conclusion that Nrf2 may be translocated into the nucleus after AMPK-mediated phosphorylation at Ser550 in the cytoplasm and that the Ser550-phosphorylated form stays in the nucleus.
Differential roles of PKCδ and AMPK for the activation of Nrf2.
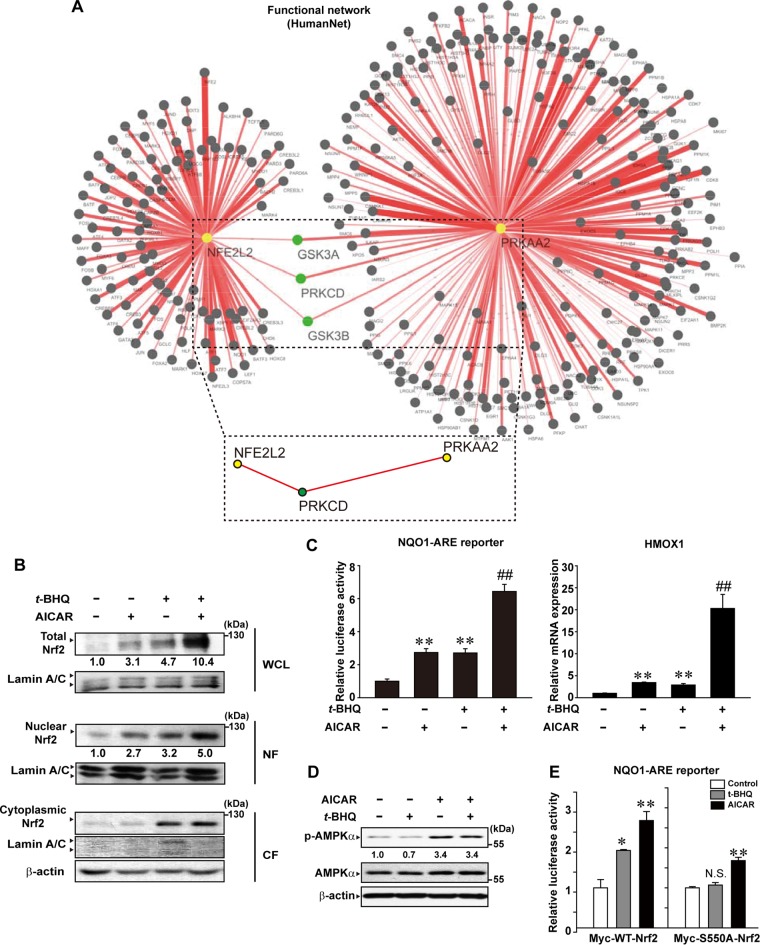
We further investigated the relationship between AMPK and Nrf2 using the functional network program (HumanNet). In this approach, a mouse subnetwork was obtained by mapping human genes to mouse orthologs. The network contained 304 first neighbors of Prkaa2 and Nfe2l2 as well as 305 interactions with the two gene products (Fig. 5A); Prkaa2 and Nfe2l2 commonly interacted with Prkcd, Gsk3a, and Ggk3b as first neighbors, indicative of the association of PKCδ and GSK3 with AMPK and Nrf2.
FIG 5.
The effects of AICAR and t-BHQ on the activation of Nrf2. (A) Functional network between AMPK and Nrf2. The network was obtained from an open database (HumanNet). A dotted box captures the association of PKCδ and AMPK or Nrf2. (B) Immunoblottings for Nrf2 in HepG2 cells treated with AICAR and/or 30 μM t-BHQ for 3 h. WCL, whole-cell lysates; NF, nuclear fraction; CF, cytoplasmic fraction. (C) NQO1-ARE reporter assays and qRT-PCR assays for HMOX1 in HepG2 cells treated with AICAR and/or t-BHQ for 18 h. (D) Immunoblottings for p-AMPKα in HepG2 cells treated with AICAR or AICAR plus t-BHQ for 30 min. (E) NQO1-ARE reporter assays. HEK293 cells were treated with either t-BHQ or AICAR for 6 h after cotransfection with Myc-tagged WT-Nrf2 or S550A-Nrf2 for 24 h. For panels C and E, data represent means ± SEM of the results from three different cell preparations (significantly different from vehicle control results [**, P < 0.01; *, P < 0.05] and from AICAR-treated control results [##, P < 0.01]; N.S., not significant).
PKCδ, a protein product encoded by Prkcd, phosphorylates Nrf2 at the Ser40 residue, leading to nuclear translocation and activation of Nrf2 (7). In the next experiment, we focused on the role of PKCδ in the activation of Nrf2 by AMPK. Here, t-BHQ (tert-butylhydroquinone) was used as the agent that activates Nrf2 through PKCδ (7). Treatment of HepG2 cells with t-BHQ synergistically enhanced the ability of AICAR to induce Nrf2 nuclear accumulation (Fig. 5B) or NQO1-ARE gene transactivation (Fig. 5C, left). This effect was verified by quantitative real-time PCR (qRT-PCR) assays for HMOX1 (Fig. 5C, right). t-BHQ treatment did not increase the basal or AICAR-inducible AMPK phosphorylation levels (i.e., p-AMPKα levels) (Fig. 5D). Our result showed that PKCδ may have a complementary role in the activation of Nrf2 by AMPK.
To assess the impact of Nrf2 phosphorylation at Ser550 by AMPK in the transactivation of ARE reporter gene by t-BHQ, we did more experiments using Myc-WT-Nrf2 and Myc-S550A-Nrf2. Treatment with either t-BHQ or AICAR promoted NQO1-ARE luciferase activity in the cells expressing Myc-WT-Nrf2 (Fig. 5E, left). Of note, t-BHQ treatment failed to increase the gene transactivation in S550A-Nrf2-transfected cells, whereas AICAR treatment showed a marginal effect (Fig. 5E, right). The results indicate that AMPK and PKCδ play distinct roles in the cellular trafficking of Nrf2, showing mutual dependency for full activation.
Effect of CA-GSK3β on the basal nuclear level of S550A-Nrf2.
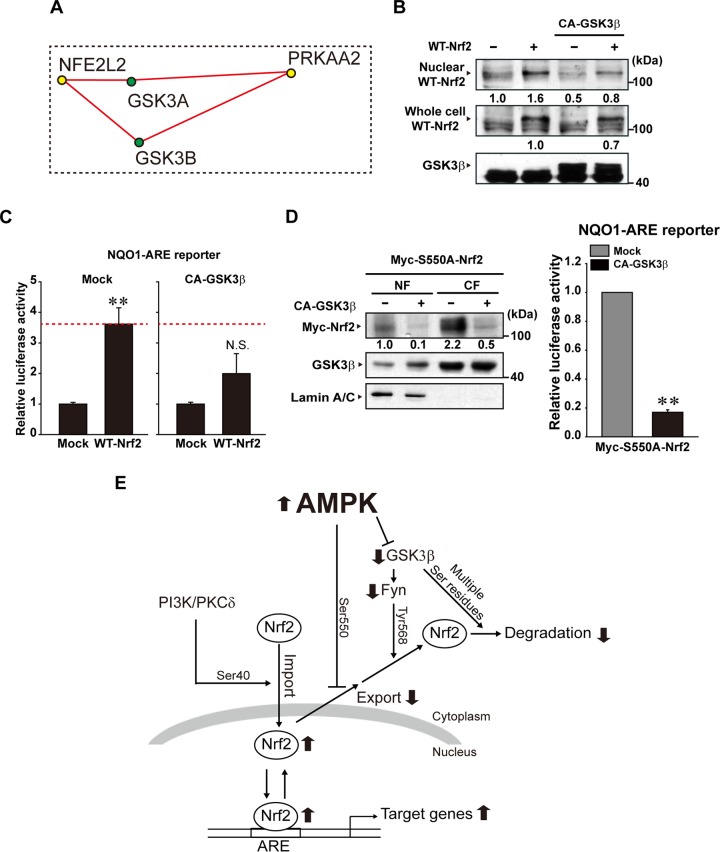
AMPK inhibits GSK3β, a constitutively active Ser/Thr kinase encoded by GSK3B (37, 38). GSK3β increases Fyn-mediated Nrf2 phosphorylation at Tyr568, enhancing export of Nrf2 from the nucleus (10, 11). The HumanNet network enabled us to find GSK3β and GSK3α as the network motif components engaged in the AMPK-Nrf2 pathway (Fig. 6A). To verify GSK3β association with the identified signaling pathway, we examined the effect of the constitutively active mutant of GSK3β (CA-GSK3β, a mutant with Ser9-to-Ala substitution [39, 40]) transfection on Nrf2 regulation. Because GSK3β and GSK3α have 98% identity in their catalytic domains and functional redundancy (41, 42), we used GSK3β for this experiment. CA-GSK3β overexpression attenuated both nuclear accumulation of Myc-WT-Nrf2 and Myc-WT-Nrf2-inducible ARE activity (Fig. 6B and C). Since Myc-S550A-Nrf2 was barely present in the nuclear fraction (Fig. 4A), we also determined the effect of CA-GSK3β on the basal nuclear level of Myc-S550A-Nrf2 in an effort to understand the coherent ON-OFF switch. As expected, CA-GSK3β transfection markedly diminished the nuclear and cytoplasmic levels of Myc-S550A-Nrf2 (Fig. 6D, left). Consistently, CA-GSK3β overexpression similarly reduced ARE reporter activity in HEK293 cells transfected with Myc-S550A-Nrf2 (Fig. 6D, right). Overall, our results show that AMPK directly phosphorylates Nrf2 at Ser550, which facilitates nuclear accumulation of Nrf2 in conjunction with AMPK's inhibition of GSK3β-mediated export (Fig. 6E).
FIG 6.
The effect of CA-GSK3β on WT-Nrf2 or S550A-Nrf2 localization. (A) Interactions of GSK3α and GSK3β with NFE2L2 (Nrf2) or PRKAA2 (AMPKα2). The interactions were captured from the functional network shown in Fig. 5A. (B) The effect of CA-GSK3β overexpression on nuclear WT-Nrf2. HEK293 cells were cotransfected with CA-GSK3β and WT-Nrf2 for 24 h. (C) The effect of CA-GSK3β on WT-Nrf2-inducible ARE reporter activity (24 h). (D) Left, immunoblotting for Myc-S550A-Nrf2 of HEK293 cells transfected with CA-GSK3β (or mock transfected) and Myc-S550A-Nrf2. Immunoblotting for lamin A/C was done to confirm the purity of nuclear fractions. Right, data represent NQO1-ARE reporter activity in the cells cotransfected with CA-GSK3β (or mock transfected [Mock]) and S550A-Nrf2. NF, nuclear fraction; CF, cytoplasmic fraction. (E) A schematic diagram illustrating the proposed signaling pathway by which AMPK regulates Nrf2 trafficking. For panels C and D, data represent means ± SEM of results from three different cell preparations (significantly different from mock control results [**, P < 0.01; *, P < 0.05]; N.S., not significant).
DISCUSSION
Oxidative stress in the cell occurs in the physiological situations subjected to oxygen resupply following energy deficiency and/or hypoxic preconditioning (e.g., hypoxia and reperfusion injury) (43). In this process, Nrf2 is activated in response to oxidative stress, playing a key role in the ARE-mediated induction of a battery of defensive genes. However, whether an increase in the AMP/ATP ratio (frequently occurring prior to oxidative stress) regulates Nrf2 had been unknown. A bioinformatics approach using the open database of protein-protein interactions, Mentha program, and global gene coexpression analysis in fasted animals (i.e., energy deficiency) enabled us to extract AMPK as a potential Nrf2 kinase. Indeed, AMPK plays a role in the sensing and modulation of energy homeostasis by monitoring energy status in eukaryotic cells (i.e., the AMP/ATP ratio) (14, 44). Our results shown here demonstrate that AMPK directly phosphorylates Nrf2 for its accumulation in the nucleus. Our finding showing the novel mechanism of Nrf2 trafficking (independent of oxidative stress regulation) explains the basis of preparatory adaptation of cells for homeostatic redox recalibration.
Several physiological substrates of AMPK have been identified (25). In all cases, AMPK phosphorylated serine residues in the consensus motif, a sequence representing Φ-(β,X)-X-X-Ser/Thr-X-X-X-Φ. In the consensus sequence, Φ represents bulky or hydrophobic amino acids such as valine (V), leucine (L), methionine (M), and isoleucine (I), whereas β represents basic amino acids such as lysine (K), arginine (R), and histidine (H). Bulky hydrophobic residues are found at the positions of P−5 and P+4, and at least 1 basic residue is located at either P−4 or P−3. The outcome showing the ability of AMPK to phosphorylate Nrf2 matches the enzyme specificity for other substrates. Moreover, the results of experiments performed using the mutants of Nrf2 strengthen the notion that AMPK serves as an Nrf2 kinase, as corroborated by the in vitro kinase data.
Great progress has been made in elucidating the mechanisms underlying nucleocytoplasmic transportation. Nrf2 has a canonical leucine-rich NES sequence (LKRRLSTLYL) at the C terminus between amino acids 545 and 554 (32). In a Ran GTPase-dependent fashion, nuclear importin and exportin recognize the NLSs and NESs on cargo proteins, facilitating their transport (45). Our finding that AMPK phosphorylated Nrf2 at Ser550 matches its effect on Nrf2 nuclear accumulation, since the serine residue is located within the canonical NES motif. The results obtained from experiments using leptomycin B and CHX also strengthen this concept. The fact that the NES motif of Nrf2 overlaps ZIP motifs supports the notion that the NES motif may be masked when Nrf2 forms heterodimers via its leucine zipper with bZIP proteins such as small MafG/K (32), which would sustain the nuclear presence of activated Nrf2. Hence, AMPK modification of Nrf2-NES and simultaneous heterodimerization of Nrf2 with other molecules would prevent its nuclear export. In addition, Nrf2-NES appears to be redox insensitive (46). Thus, the signaling pathway regulating Nrf2-NES seems to be independent of the redox-sensitive Keap1-regulatory pathway. Our results showing Nrf2 modification within the NES may help decipher a mechanism governing nuclear localization of Nrf2.
AMPKα may move out of the nucleus using the CRM1 carrier, shuttling between the nucleus and the cytoplasm. It has also been shown that AMPKα1 exists predominantly in the cytoplasm, whereas the AMPKα2 subunit showed main localization in the nucleus (47, 48). Various stresses such as heat shock, energy depletion, and oxidative stress affect the phosphorylation state of AMPKα1 and AMPKα2, sometimes causing AMPK's nuclear accumulation (49). Our findings indicate that Thr172-phosphorylated AMPKα mainly existed in the cytoplasm. Despite the activation of Nrf2, leptomycin B had no effect on basal or inducible p-AMPKα levels or its subcellular distribution. Hence, it is highly likely that Nrf2 phosphorylation by AMPK occurs in the cytoplasm and that the Ser550-phosphorylated form of Nrf2 is located in the nucleus.
Nrf2 released from its Keap1 inhibitor through modification of cysteine residues of Keap1 translocates to the nucleus (50). In this event, PKCδ phosphorylates Nrf2 at Ser40 (7). Both Cys151 modification of Keap1 and PKCδ-mediated Nrf2 phosphorylation are required for the release of Nrf2 from Keap1, leading to the nuclear localization of Nrf2 (7). Hence, it is plausible that the nuclear accumulation of Nrf2 elicited by AMPK may prolong the half-life of Nrf2, as shown in Fig. 4F, due to a decrease in the Nrf2 level in the Keap1-binding pool. Our finding that Nrf2 activation elicited by t-BHQ was independent of AMPK and was augmented by AICAR treatment raises the notion that the AMPK pathway is distinct from the PKCδ pathway in the activation of Nrf2. Since we cannot exclude the possibility of the existence of other pleiotropic effects of t-BHQ, studies using genetic modulation of PKCδ may be necessary. Moreover, leptomycin B had an additional effect on t-BHQ's effect of Nrf2, as did AICAR (32). So, an additional redox-insensitive AMPK pathway may be necessary for full ARE-dependent gene transcription. Phytochemicals may have the ability to activate Nrf2. However, not all of them can rescue cells from injury, implying the existence of another signal(s) required for cell protection. Some phytochemicals increase the activity of Nrf2 without AMPK activation. Thus, the agents that activate both PKCδ and AMPK may possess a higher antioxidant capacity.
GSK3β inhibits Nrf2 activity through phosphorylation and nuclear exclusion (11). In another study, GSK3β phosphorylated Nrf2 at several serine residues (51). Moreover, GSK3β induces nuclear localization of Fyn through threonine phosphorylation. Tyr568 phosphorylation of Nrf2 by Fyn facilitates Nrf2 export from the nucleus for its binding to Keap1 and degradation (10). In the present study, we confirmed the inhibitory effect of GSK3β on Nrf2. Our finding that CA-GSK3β reduced the basal nuclear level of Myc-S550A-Nrf2 and ARE activity indicates that AMPK-mediated nuclear accumulation of Nrf2 may be coupled to the inhibition of GSK3β. The phosphatidylinositol 3-kinase (PI3K)/Akt pathway is necessary for the activation of Nrf2 (52). Since the PI3K/Akt pathway causes the inhibitory phosphorylation of GSK3β, PI3K-dependent activation of Nrf2 may be explained in part by AMPK activity.
In conclusion, our results demonstrate that AMPK directly phosphorylates Nrf2 at Ser550, promoting nuclear accumulation of Nrf2 for ARE-mediated gene transcription, which occurs in addition to AMPK's inhibition of GSK3β, a kinase catalyzing the nuclear exclusion of Nrf2. The findings provide novel information on the coherent ON-OFF switch signaling loop for Nrf2 and help expand our understanding of the regulatory mechanism of the important mammalian antioxidant transcription factor.
Supplementary Material
ACKNOWLEDGMENTS
The overall study was conceived and designed by M. S. Joo, W. D. Kim, and S. G. Kim; M. S. Joo and W. D. Kim performed the experiments; M. S. Joo, W. D. Kim, and J. H. Koo analyzed data; K. Y. Lee, J. H. Kim, and J. H. Koo analyzed bioinformatic data; M. S. Joo and S. G. Kim wrote the paper.
We declare that we have no competing interests.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00118-16.
REFERENCES
- 1.Sorensen M, Sanz A, Gomez J, Pamplona R, Portero-Otin M, Gredilla R, Barja G. 2006. Effects of fasting on oxidative stress in rat liver mitochondria. Free Radic Res 40:339–347. doi: 10.1080/10715760500250182. [DOI] [PubMed] [Google Scholar]
- 2.Morel Y, Barouki R. 1999. Repression of gene expression by oxidative stress. Biochem J 342(Pt 3):481–496. [PMC free article] [PubMed] [Google Scholar]
- 3.Lu Y, Cederbaum AI. 2008. CYP2E1 and oxidative liver injury by alcohol. Free Radic Biol Med 44:723–738. doi: 10.1016/j.freeradbiomed.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 5.Cullinan SB, Diehl JA. 2004. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J Biol Chem 279:20108–20117. doi: 10.1074/jbc.M314219200. [DOI] [PubMed] [Google Scholar]
- 6.Huang HC, Nguyen T, Pickett CB. 2002. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem 277:42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 7.Niture SK, Jain AK, Jaiswal AK. 2009. Antioxidant-induced modification of INrf2 cysteine 151 and PKC-delta-mediated phosphorylation of Nrf2 serine 40 are both required for stabilization and nuclear translocation of Nrf2 and increased drug resistance. J Cell Sci 122:4452–4464. doi: 10.1242/jcs.058537. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Kensler TW, Wakabayashi N, Biswal S. 2007. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 9.Kang KW, Lee SJ, Park JW, Kim SG. 2002. Phosphatidylinositol 3-kinase regulates nuclear translocation of NF-E2-related factor 2 through actin rearrangement in response to oxidative stress. Mol Pharmacol 62:1001–1010. doi: 10.1124/mol.62.5.1001. [DOI] [PubMed] [Google Scholar]
- 10.Jain AK, Jaiswal AK. 2007. GSK-3beta acts upstream of Fyn kinase in regulation of nuclear export and degradation of NF-E2 related factor 2. J Biol Chem 282:16502–16510. doi: 10.1074/jbc.M611336200. [DOI] [PubMed] [Google Scholar]
- 11.Salazar M, Rojo AI, Velasco D, de Sagarra RM, Cuadrado A. 2006. Glycogen synthase kinase-3beta inhibits the xenobiotic and antioxidant cell response by direct phosphorylation and nuclear exclusion of the transcription factor Nrf2. J Biol Chem 281:14841–14851. doi: 10.1074/jbc.M513737200. [DOI] [PubMed] [Google Scholar]
- 12.Sun Z, Huang Z, Zhang DD. 2009. Phosphorylation of Nrf2 at multiple sites by MAP kinases has a limited contribution in modulating the Nrf2-dependent antioxidant response. PLoS One 4:e6588. doi: 10.1371/journal.pone.0006588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain AK, Jaiswal AK. 2006. Phosphorylation of tyrosine 568 controls nuclear export of Nrf2. J Biol Chem 281:12132–12142. doi: 10.1074/jbc.M511198200. [DOI] [PubMed] [Google Scholar]
- 14.Hardie DG, Ross FA, Hawley SA. 2012. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lage R, Dieguez C, Vidal-Puig A, Lopez M. 2008. AMPK: a metabolic gauge regulating whole-body energy homeostasis. Trends Mol Med 14:539–549. doi: 10.1016/j.molmed.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Zmijewski JW, Banerjee S, Bae H, Friggeri A, Lazarowski ER, Abraham E. 2010. Exposure to hydrogen peroxide induces oxidation and activation of AMP-activated protein kinase. J Biol Chem 285:33154–33164. doi: 10.1074/jbc.M110.143685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kukidome D, Nishikawa T, Sonoda K, Imoto K, Fujisawa K, Yano M, Motoshima H, Taguchi T, Matsumura T, Araki E. 2006. Activation of AMP-activated protein kinase reduces hyperglycemia-induced mitochondrial reactive oxygen species production and promotes mitochondrial biogenesis in human umbilical vein endothelial cells. Diabetes 55:120–127. doi: 10.2337/diabetes.55.01.06.db05-0943. [DOI] [PubMed] [Google Scholar]
- 18.Park CE, Yun H, Lee EB, Min BI, Bae H, Choe W, Kang I, Kim SS, Ha J. 2010. The antioxidant effects of genistein are associated with AMP-activated protein kinase activation and PTEN induction in prostate cancer cells. J Med Food 13:815–820. doi: 10.1089/jmf.2009.1359. [DOI] [PubMed] [Google Scholar]
- 19.Liu XM, Peyton KJ, Shebib AR, Wang H, Korthuis RJ, Durante W. 2011. Activation of AMPK stimulates heme oxygenase-1 gene expression and human endothelial cell survival. Am J Physiol Heart Circ Physiol 300:H84–H93. doi: 10.1152/ajpheart.00749.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peralta C, Bartrons R, Serafin A, Blazquez C, Guzman M, Prats N, Xaus C, Cutillas B, Gelpi E, Rosello-Catafau J. 2001. Adenosine monophosphate-activated protein kinase mediates the protective effects of ischemic preconditioning on hepatic ischemia-reperfusion injury in the rat. Hepatology 34:1164–1173. doi: 10.1053/jhep.2001.29197. [DOI] [PubMed] [Google Scholar]
- 21.Ido Y, Carling D, Ruderman N. 2002. Hyperglycemia-induced apoptosis in human umbilical vein endothelial cells: inhibition by the AMP-activated protein kinase activation. Diabetes 51:159–167. doi: 10.2337/diabetes.51.1.159. [DOI] [PubMed] [Google Scholar]
- 22.Shin SM, Cho IJ, Kim SG. 2009. Resveratrol protects mitochondria against oxidative stress through AMP-activated protein kinase-mediated glycogen synthase kinase-3beta inhibition downstream of poly(ADP-ribose)polymerase-LKB1 pathway. Mol Pharmacol 76:884–895. doi: 10.1124/mol.109.058479. [DOI] [PubMed] [Google Scholar]
- 23.Shin SM, Kim SG. 2009. Inhibition of arachidonic acid and iron-induced mitochondrial dysfunction and apoptosis by oltipraz and novel 1,2-dithiole-3-thione congeners. Mol Pharmacol 75:242–253. doi: 10.1124/mol.108.051128. [DOI] [PubMed] [Google Scholar]
- 24.Stefanelli C, Stanic I, Bonavita F, Flamigni F, Pignatti C, Guarnieri C, Caldarera CM. 1998. Inhibition of glucocorticoid-induced apoptosis with 5-aminoimidazole-4-carboxamide ribonucleoside, a cell-permeable activator of AMP-activated protein kinase. Biochem Biophys Res Commun 243:821–826. doi: 10.1006/bbrc.1998.8154. [DOI] [PubMed] [Google Scholar]
- 25.Towler MC, Hardie DG. 2007. AMP-activated protein kinase in metabolic control and insulin signaling. Circ Res 100:328–341. doi: 10.1161/01.RES.0000256090.42690.05. [DOI] [PubMed] [Google Scholar]
- 26.Ki SH, Cho IJ, Choi DW, Kim SG. 2005. Glucocorticoid receptor (GR)-associated SMRT binding to C/EBPbeta TAD and Nrf2 Neh4/5: role of SMRT recruited to GR in GSTA2 gene repression. Mol Cell Biol 25:4150–4165. doi: 10.1128/MCB.25.10.4150-4165.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bossi A, Lehner B. 2009. Tissue specificity and the human protein interaction network. Mol Syst Biol 5:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piya S, Shrestha SK, Binder B, Stewart CN Jr, Hewezi T. 2014. Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front Plant Sci 5:744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwak MK, Itoh K, Yamamoto M, Kensler TW. 2002. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol Cell Biol 22:2883–2892. doi: 10.1128/MCB.22.9.2883-2892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardie DG. 2011. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev 25:1895–1908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jain AK, Bloom DA, Jaiswal AK. 2005. Nuclear import and export signals in control of Nrf2. J Biol Chem 280:29158–29168. doi: 10.1074/jbc.M502083200. [DOI] [PubMed] [Google Scholar]
- 33.Fornerod M, Ohno M, Yoshida M, Mattaj IW. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051–1060. doi: 10.1016/S0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 34.Kazgan N, Williams T, Forsberg LJ, Brenman JE. 2010. Identification of a nuclear export signal in the catalytic subunit of AMP-activated protein kinase. Mol Biol Cell 21:3433–3442. doi: 10.1091/mbc.E10-04-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A. 2007. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem 282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 36.Chaturvedi RK, Adhihetty P, Shukla S, Hennessy T, Calingasan N, Yang L, Starkov A, Kiaei M, Cannella M, Sassone J, Ciammola A, Squitieri F, Beal MF. 2009. Impaired PGC-1alpha function in muscle in Huntington's disease. Hum Mol Genet 18:3048–3065. doi: 10.1093/hmg/ddp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horike N, Sakoda H, Kushiyama A, Ono H, Fujishiro M, Kamata H, Nishiyama K, Uchijima Y, Kurihara Y, Kurihara H, Asano T. 2008. AMP-activated protein kinase activation increases phosphorylation of glycogen synthase kinase 3beta and thereby reduces cAMP-responsive element transcriptional activity and phosphoenolpyruvate carboxykinase C gene expression in the liver. J Biol Chem 283:33902–33910. doi: 10.1074/jbc.M802537200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi SH, Kim YW, Kim SG. 2010. AMPK-mediated GSK3beta inhibition by isoliquiritigenin contributes to protecting mitochondria against iron-catalyzed oxidative stress. Biochem Pharmacol 79:1352–1362. doi: 10.1016/j.bcp.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 39.Stambolic V, Woodgett JR. 1994. Mitogen inactivation of glycogen synthase kinase-3 beta in intact cells via serine 9 phosphorylation. Biochem J 303(Pt 3):701–704. doi: 10.1042/bj3030701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacAulay K, Blair AS, Hajduch E, Terashima T, Baba O, Sutherland C, Hundal HS. 2005. Constitutive activation of GSK3 down-regulates glycogen synthase abundance and glycogen deposition in rat skeletal muscle cells. J Biol Chem 280:9509–9518. doi: 10.1074/jbc.M411648200. [DOI] [PubMed] [Google Scholar]
- 41.Doble BW, Patel S, Wood GA, Kockeritz LK, Woodgett JR. 2007. Functional redundancy of GSK-3alpha and GSK-3beta in Wnt/beta-catenin signaling shown by using an allelic series of embryonic stem cell lines. Dev Cell 12:957–971. doi: 10.1016/j.devcel.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plyte SE, Hughes K, Nikolakaki E, Pulverer BJ, Woodgett JR. 1992. Glycogen synthase kinase-3: functions in oncogenesis and development. Biochim Biophys Acta 1114:147–162. [DOI] [PubMed] [Google Scholar]
- 43.Ferrari R, Alfieri O, Curello S, Ceconi C, Cargnoni A, Marzollo P, Pardini A, Caradonna E, Visioli O. 1990. Occurrence of oxidative stress during reperfusion of the human heart. Circulation 81:201–211. doi: 10.1161/01.CIR.81.1.201. [DOI] [PubMed] [Google Scholar]
- 44.Hardie DG, Scott JW, Pan DA, Hudson ER. 2003. Management of cellular energy by the AMP-activated protein kinase system. FEBS Lett 546:113–120. doi: 10.1016/S0014-5793(03)00560-X. [DOI] [PubMed] [Google Scholar]
- 45.Ullman KS, Powers MA, Forbes DJ. 1997. Nuclear export receptors: from importin to exportin. Cell 90:967–970. doi: 10.1016/S0092-8674(00)80361-X. [DOI] [PubMed] [Google Scholar]
- 46.Li W, Jain MR, Chen C, Yue X, Hebbar V, Zhou R, Kong AN. 2005. Nrf2 Possesses a redox-insensitive nuclear export signal overlapping with the leucine zipper motif. J Biol Chem 280:28430–28438. doi: 10.1074/jbc.M410601200. [DOI] [PubMed] [Google Scholar]
- 47.Salt I, Celler JW, Hawley SA, Prescott A, Woods A, Carling D, Hardie DG. 1998. AMP-activated protein kinase: greater AMP dependence, and preferential nuclear localization, of complexes containing the alpha2 isoform. Biochem J 334(Pt 1):177–187. doi: 10.1042/bj3340177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turnley AM, Stapleton D, Mann RJ, Witters LA, Kemp BE, Bartlett PF. 1999. Cellular distribution and developmental expression of AMP-activated protein kinase isoforms in mouse central nervous system. J Neurochem 72:1707–1716. [DOI] [PubMed] [Google Scholar]
- 49.Kodiha M, Rassi JG, Brown CM, Stochaj U. 2007. Localization of AMP kinase is regulated by stress, cell density, and signaling through the MEK–>ERK1/2 pathway. Am J Physiol Cell Physiol 293:C1427–1436. doi: 10.1152/ajpcell.00176.2007. [DOI] [PubMed] [Google Scholar]
- 50.Kobayashi M, Yamamoto M. 2005. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid Redox Signal 7:385–394. doi: 10.1089/ars.2005.7.385. [DOI] [PubMed] [Google Scholar]
- 51.Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, Cuadrado A. 2011. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol 31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L, Chen Y, Sternberg P, Cai J. 2008. Essential roles of the PI3 kinase/Akt pathway in regulating Nrf2-dependent antioxidant functions in the RPE. Invest Ophthalmol Vis Sci 49:1671–1678. doi: 10.1167/iovs.07-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.