Abstract
Atrial natriuretic peptide (ANP) has a central role in regulating blood pressure in humans. Recently, microRNA 425 (miR-425) was found to regulate ANP production by binding to the mRNA of NPPA, the gene encoding ANP. mRNAs typically contain multiple predicted microRNA (miRNA)-binding sites, and binding of different miRNAs may independently or coordinately regulate the expression of any given mRNA. We used a multifaceted screening strategy that integrates bioinformatics, next-generation sequencing data, human genetic association data, and cellular models to identify additional functional NPPA-targeting miRNAs. Two novel miRNAs, miR-155 and miR-105, were found to modulate ANP production in human cardiomyocytes and target genetic variants whose minor alleles are associated with higher human plasma ANP levels. Both miR-155 and miR-105 repressed NPPA mRNA in an allele-specific manner, with the minor allele of each respective variant conferring resistance to the miRNA either by disruption of miRNA base pairing or by creation of wobble base pairing. Moreover, miR-155 enhanced the repressive effects of miR-425 on ANP production in human cardiomyocytes. Our study combines computational, genomic, and cellular tools to identify novel miRNA regulators of ANP production that could be targeted to raise ANP levels, which may have applications for the treatment of hypertension or heart failure.
INTRODUCTION
Atrial natriuretic peptide (ANP), which is encoded by the NPPA gene, is a cardiac hormone that exhibits diuretic, natriuretic, and vasorelaxant activities (1). In the PARADIGM-HF clinical trial, inhibition of neprilysin (an enzyme that degrades natriuretic peptides) in combination with renin-angiotensin-aldosterone system blockade reduced the number of hospitalizations for congestive heart failure and deaths from cardiovascular causes (2). The PARADIGM-HF trial has rekindled interest in the natriuretic peptide system as a therapeutic target and highlights the therapeutic importance of identifying factors that may increase endogenous natriuretic peptide levels.
Intravenous administration of ANP lowers blood pressure and induces natriuresis in animal models and in patients with heart failure (3). Transgenic mice overexpressing ANP are hypotensive (4), while ANP-deficient mice are hypertensive (5). However, the role of ANP in blood pressure regulation in the general population has remained uncertain until recent population genetic studies revealed that the minor allele of a common genetic variant (rs5068) in NPPA is associated with increased plasma ANP levels, lower blood pressure, and reduced risk of hypertension (6). These findings support a direct role of ANP in blood pressure regulation in humans and illustrate that modulating ANP levels could be a clinically relevant approach to treating hypertension or heart failure. The rs5068 single nucleotide polymorphism (SNP) is located in the 3′ untranslated region (3′ UTR) of NPPA, where it disrupts the target binding site of a microRNA (miRNA), miR-425 (7).
Binding of more than one miRNA can independently or coordinately modulate the expression of any given mRNA (8, 9). However, identifying bona fide miRNAs that regulate an mRNA of interest has been challenging. Although computational prediction algorithms are helpful in generating lists of potential miRNA candidates, the lists often contain a substantial number of false-positive miRNAs, which do not actually regulate the target mRNA (10). Due to the poor predictive accuracy of these algorithms, it is difficult to discern which of the candidate miRNAs warrant further experimental validation.
In the current study, we sought to identify additional miRNAs that target the NPPA 3′ UTR. We used a sequential screening strategy that involved (i) in silico prediction of miRNA candidates, (ii) prioritization of candidates based on (a) miRNA expression data in human atrial tissues and/or (b) the presence of common genetic variants within the predicted miRNA binding site that are associated with circulating ANP levels in human population genetics studies, (iii) experimental validation of the predicted interaction with NPPA mRNA using luciferase reporter assays, and (iv) confirmation of the effect of the miRNA on NPPA mRNA levels and ANP protein levels in human cardiomyocytes.
MATERIALS AND METHODS
In silico analysis.
The microRNA Data Integration Portal (mirDIP) (11) was used to generate a list of miRNAs that are predicted in silico to interact with the NPPA 3′ UTR (NM_006172). The mirDIP (version 1.1.2) was run with the default settings.
Cell culture.
COS-7 cells (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 10% fetal calf serum, 2 mM l-glutamine, 200 U/ml penicillin, and 200 μg/ml streptomycin (Fisher Scientific). Human cardiomyocytes were differentiated from human embryonic stem cells (hESCs) as follows: hESCs from the WA07 (H7) cell line were cultured on Matrigel-coated tissue culture polystyrene plates and maintained in mTeSR1 medium (Stem Cell Technologies). hESC medium was refreshed every 24 h, and hESCs were passaged using dispase (Sigma-Aldrich) at confluence. Cardiac differentiation of hESCs was induced using small molecules as previously described (12). Briefly, when hESCs maintained on Matrigel plates achieved confluence, cells were treated with CHIR99021 (Stemgent) in RPMI medium (Life Technologies) supplemented with Gem21 NeuroPlex without insulin (Gemini Bio Products) for 24 h (from day 0 to day 1). The medium was replaced with RPMI medium-Gem21–insulin at day 1. The cells were then treated with IWP4 (Stemgent) in RPMI medium-Gem21–insulin at day 3, and the medium was refreshed on day 5 with RPMI medium-Gem21–insulin. Cells were maintained in RPMI supplemented with Gem21 NeuroPlex (Gemini Bio Products) starting from day 7, with the medium changed every 3 days. Beating clusters were seen starting from day 10 of differentiation. Cardiomyocytes were harvested 5 to 10 days after the onset of beating, typically from day 15 to 20 of differentiation. For dissociation, cardiomyocytes were treated with collagenase A and B (Roche) in RPMI medium-Gem21 for 15 min. Afterward, the collagenase solution was removed from the cardiomyocytes and replaced with 0.05% trypsin-EDTA (Life Technologies) for a 3-min treatment to obtain single cells. Trypsin was neutralized with RPMI medium-Gem21, and cardiomyocytes were centrifuged and resuspended in RPMI medium-Gem21 for plating. Expression of cardiac troponin T was confirmed in all experiments. Cardiomyocytes were confirmed by sequencing to be homozygous for the rs5068, rs61764044, and rs5067 major alleles.
miRNA mimics and inhibitors.
Chemically modified, double-stranded RNAs designed to mimic endogenous mature miRNAs, as well as a scrambled negative-control miRNA mimic, were purchased from Life Technologies. Throughout this work, miRNA is used to indicate miRNA mimic when exogenously administered. Chemically modified single-stranded RNA designed to inhibit endogenous mature miRNAs (anti-miRs) and a single-stranded scrambled negative-control anti-miRNA were also purchased from Life Technologies.
Generation of luciferase-NPPA 3′ UTR reporter constructs.
The NPPA 3′ UTR sequence (299 bp) including the major alleles of rs5068, rs5067, and rs61764044 was PCR amplified from a deidentified human HapMap genomic DNA sample obtained from Coriell Repositories (Coriell Institute for Medical Research). The PCR product was cloned into the pIS0 vector (Addgene) 3′ of the sequence encoding firefly luciferase to generate the wild-type luciferase construct (WT-Luc). In addition, mutated NPPA 3′ UTR luciferase constructs were created by site-directed mutagenesis using the QuikChange II XL site-directed mutagenesis kit (Stratagene) and synthetic oligonucleotides (Life Technologies) according to the manufacturers' protocols. The synthetic oligonucleotides used to generate the 155 Mut-Luc construct were 5′-CCTCGCCTCTCCCACCCCATCGTGGCAATTTTAAGGTAGAACCTC-3′ and 5′-GAGGTTCTACCTTAAAATTGCCACGATGGGGTGGGAGAGGCGAGG-3′. The synthetic oligonucleotides used to generate the rs61764044 Minor-Luc construct were 5′-GCCTCTCCCACCCCACGCATTAAATTTTAAG-3′ and 5′-CTTAAAATTTAATGCGTGGGGTGGGAGAGGC-3′. The synthetic oligonucleotides used to generate the rs5067 Minor-Luc construct were 5′-GGTCTCTGCTGCATTCGTGTCATCTTGTTGC-3′ and 5′-GCAACAAGATGACACGAATGCAGCAGAGACC-3′. The synthetic oligonucleotides used to generate the 103/107 Mut-Luc construct were 5′-CTCCTGTCCCCTGGGGTCTCAACGCGATTTGTGTCATCTTGTTGC-3′ and 5′-GCAACAAGATGACACAAATCGCGTTGAGACCCCAGGGGACAGGAG-3′.
Transient transfection of luciferase constructs and miRNAs or anti-miRs into heterologous cells.
COS-7 cells were transfected with wild-type or mutated luciferase-NPPA 3′ UTR constructs using X-tremeGENE HP DNA transfection reagent (Roche) according to the manufacturer's protocol. Twenty-four hours after transfection of the constructs, mirVana miRNA mimics (5 nM), scrambled negative-control miRNA mimic (5 nM), anti-miRNA inhibitors (100 nM), or scrambled negative-control anti-miR (100 nM) were transfected using Lipofectamine RNAiMax (Life Technologies). (For miRNA transfection using two miRNA mimics, the total amount of transfected miRNA was held constant under all experimental conditions. For the experimental conditions labeled with “miR-425” or “miR-155,” the total concentration of transfected miRNA was held constant by the addition of the scrambled negative-control miRNA mimic. For anti-miRNA cotransfection, the total amount of transfected anti-miR was held constant under all experimental conditions. For the experimental conditions labeled with “anti-miR-425” or “anti-miR-155,” the total concentration of transfected miRNA was held constant by the addition of the scrambled negative-control anti-miRNA.) After an additional 48 h, firefly and renilla luciferase activities in cell extracts were measured using the Dual-Luciferase reporter assay system (Promega).
Transient transfection of miRNAs or anti-miRNAs into hESC-CMs.
hESC-derived cardiomyocytes (hESC-CMs) were transfected with either miRNA mimic (50 nM), scrambled negative-control miRNA mimic (50 nM), anti-miRNA (100 nM), or scrambled negative-control anti-miRNA (100 nM) using Lipofectamine RNAiMax. (For miRNA cotransfection, the total amount of transfected miRNA was held constant under all experimental conditions. For the experimental conditions labeled with “miR-425” or “miR-155,” the total concentration of transfected miRNA was held constant by the addition of the scrambled negative-control miRNA mimic.) After 24 h, cells were washed and incubated in serum-free medium for an additional 24 h. NPPA gene expression was measured using quantitative reverse transcription-PCR (qRT-PCR), and ANP protein production and secretion were assessed using an enzyme-linked immunosorbent assay (ELISA) (proANP 1-98; Biomedica Medizinprodukte GmbH & Co KG) to detect N-terminal-proANP (Nt-proANP) levels in culture medium.
Transient transfection of NPPA cDNA expression plasmid and miR-105 or anti-miR-105 into heterologous cells.
A full-length human NPPA cDNA expression plasmid containing the rs5067 major allele (OriGene; catalogue no. SC122740) and a plasmid specifying renilla luciferase (as a transfection efficiency control) were transfected into COS-7 cells using X-tremeGENE HP DNA transfection reagent. Twenty-four hours after transfection of the constructs, miR-105 mimic (5 nM), scrambled negative-control miRNA mimic (5 nM), anti-miR-105 (100 nM), or scrambled negative-control anti-miRNA (100 nM) was transfected using Lipofectamine RNAiMax. Cells and culture medium were collected 24 h after miRNA or anti-miRNA transfection for measurement of renilla luciferase activity and secreted Nt-proANP levels by ELISA, respectively.
Measurement of mRNA and miRNA levels.
Total RNA was extracted from cultured cells using TRIzol (Life Technologies), and cDNA was synthesized using the high-capacity cDNA reverse transcription kit (Applied Biosystems) according to the manufacturer's protocol. NPPA and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels were measured by qRT-PCR using TaqMan assays (Applied Biosystems) in a 7500 Fast real-time PCR system (Life Technologies) according to the manufacturer's protocol. Relative changes in NPPA mRNA levels normalized to GAPDH mRNA levels were determined using the relative cycle threshold (CT) method. TaqMan miRNA reverse transcription and real-time assay kits (Applied Biosystems) were used to detect mature miRNAs and U6 small nuclear RNA (U6 snRNA). Relative changes in miRNA levels normalized to U6 snRNA levels were determined using the relative CT method.
Human population genetic analysis.
The midregional fragment of proANP (MR-proANP) was measured in plasma samples from the Malmö Diet and Cancer Study using an immunoluminometric assay (BRAHMS), as previously described (13). Genotypes were generated using a genome-wide SNP microarray (Illumina Omni Express Exome; Illumina). MR-proANP showed right-skewed distributions and underwent natural logarithmic transformation (lnMR-proANP) before analysis. Genetic association was tested using linear regression with an additive genetic model adjusted for age and sex.
Statistical analysis.
For all figures, data are presented as means ± standard errors of the means (SEMs) from 6 biological replicates (n = 6), and statistical significance was assessed by two-tailed independent t test and one-way analysis of variance (ANOVA) with Bonferroni post hoc testing, as appropriate. In all cases, a P value of <0.05 was considered statistically significant. t tests are reported without adjustments for multiple comparisons. Each experiment was repeated at least three times, with consistent results. Representative figures are presented in this work.
RESULTS
miRNAs predicted to target the NPPA 3′ UTR.
The mirDIP (11) was used to generate a list of miRNAs predicted in silico to interact with the NPPA 3′ UTR. The 7 algorithms in mirDIP use a number of criteria to identify potential miRNA-mRNA interactions, including base pairing with the seed sequence (i.e., nucleotides 2 to 8 at the miRNA 5′ end), evolutionary conservation of target sites, thermal stability of the miRNA-mRNA duplex, and secondary structure surrounding the miRNA binding site (14). Four hundred ninety-four miRNAs were predicted to target the NPPA 3′ UTR by at least one of the 7 mirDIP algorithms, and 37 miRNAs were predicted to interact with the NPPA 3′ UTR by 3 or more of the algorithms (Fig. 1). Thirty of the 37 miRNAs were expressed in human atrial tissues (15). We were interested in the miRNAs that were expressed in the atria because ANP is synthesized predominantly in the atria (1). As expected, miR-425 was among the 30 miRNAs predicted to target NPPA. We focused our attention on 12 of the remaining 29 miRNAs because they were expressed at high levels in human atrial tissues (15). Seed binding sites of 3 of these 12 miRNAs were predicted to include common genetic variants, either SNPs (miR-155 and miR-769-5p) or an insertion-deletion (miR-497-3p), in the NPPA 3′ UTR (see Table S1 in the supplemental material).
FIG 1.
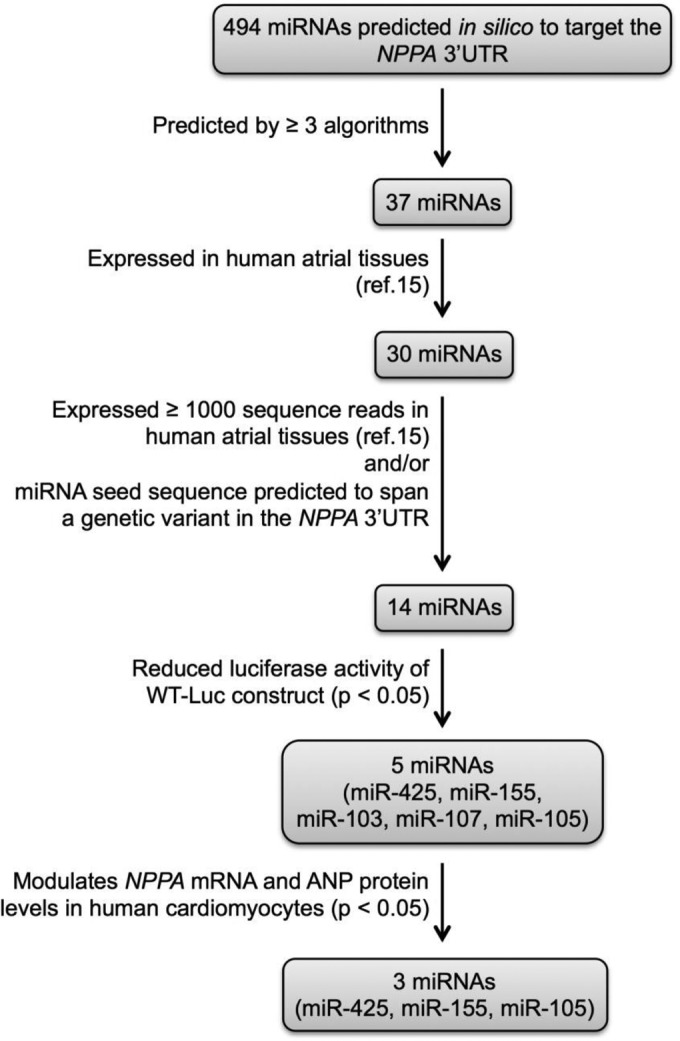
Schematic workflow for identification of NPPA-targeting miRNAs. In silico miRNA predictions were based on mirDIP. Evidence of expression in human atrial tissues was based on the work of Hsu et al. (15).
Despite lower expression of miR-105 in human atrial tissue (15), this miRNA was included as a 13th potential candidate NPPA-modulating miRNA because the seed sequence of miR-105 spans a genetic variant in the NPPA 3′ UTR (see Fig. S1A and D in the supplemental material) that is associated with plasma Nt-proANP levels in humans (see below).
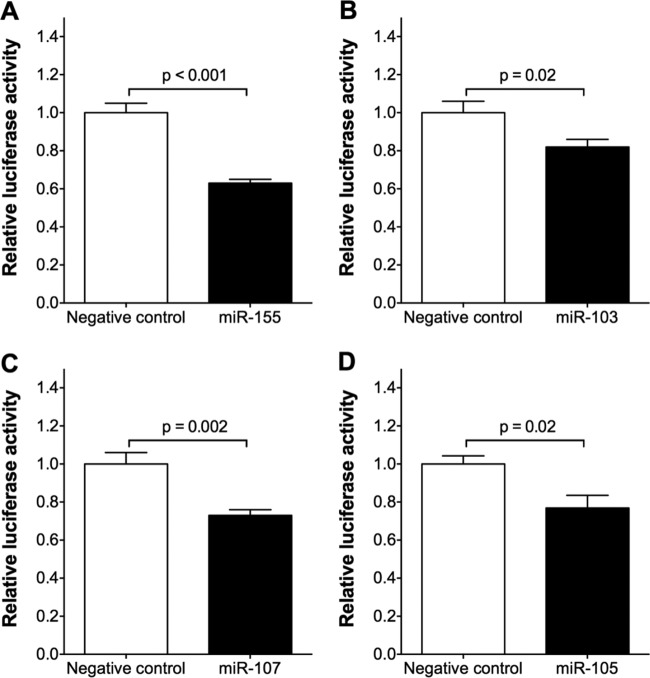
To test the predicted interaction of each of the 13 candidate miRNAs with the NPPA 3′ UTR, we generated a reporter construct in which the firefly luciferase gene was ligated to the wild-type NPPA 3′ UTR (WT-Luc). The WT-Luc construct and a second plasmid directing constitutive expression of renilla luciferase to control for transfection efficiency were cotransfected with each of the candidate miRNAs into COS-7 cells. Of the 13 candidate miRNAs, miR-155, miR-103, miR-107, and miR-105 each reduced the luciferase activity in cells containing the WT-Luc construct (Fig. 2), suggesting that these 4 miRNAs could interact with the NPPA 3′ UTR. Transfection of the other 9 candidate miRNAs failed to alter luciferase activity (see Table S1 in the supplemental material). Therefore, we chose to further investigate the potential roles of miR-155, miR-103, miR-107, and miR-105 in the regulation of NPPA expression.
FIG 2.
Four miRNAs predicted in silico to target the NPPA 3′ UTR reduce the luciferase activity produced by the firefly luciferase-NPPA 3′ UTR (WT-Luc) construct. The ratio of firefly luciferase activity to renilla luciferase activity was normalized to the ratio in COS-7 cells transfected with constructs encoding luciferase without the NPPA 3′ UTR and renilla. The effect of each miRNA was compared to that of a scrambled negative-control miRNA mimic (relative luciferase activity). miR-155 (A), miR-103 (B), miR-107 (C), and miR-105 (D) reduced the activity of the WT-Luc construct. Data are means ± SEMs (n = 6 biological replicates; two-tailed independent t test).
Validation of the predicted binding of miR-155 with the NPPA 3′ UTR.
To confirm that the predicted seed binding sequence for miR-155 was required for the interaction between miR-155 and the NPPA 3′ UTR, a reporter construct (155 Mut-Luc) in which mutations were introduced in 6 of the NPPA 3′ UTR seed binding nucleotides was generated. Transfection of miR-155 reduced the luciferase activity in COS-7 cells containing the wild-type but not the mutant construct (Fig. 3A). We confirmed that COS-7 cells express miR-155 (Table 1). To further establish that miR-155 interacts with the NPPA 3′ UTR, we transfected an anti-miRNA directed against miR-155 (anti-miR-155) together with the wild-type or mutant construct into COS-7 cells. Inhibition of endogenous miR-155 by anti-miR-155 increased the luciferase activity in cells containing the wild-type but not the mutant construct (Fig. 3B). These results suggest that the NPPA 3′ UTR is a direct target of miR-155 and that the effect is mediated by the sequence complementary to the seed sequence.
FIG 3.
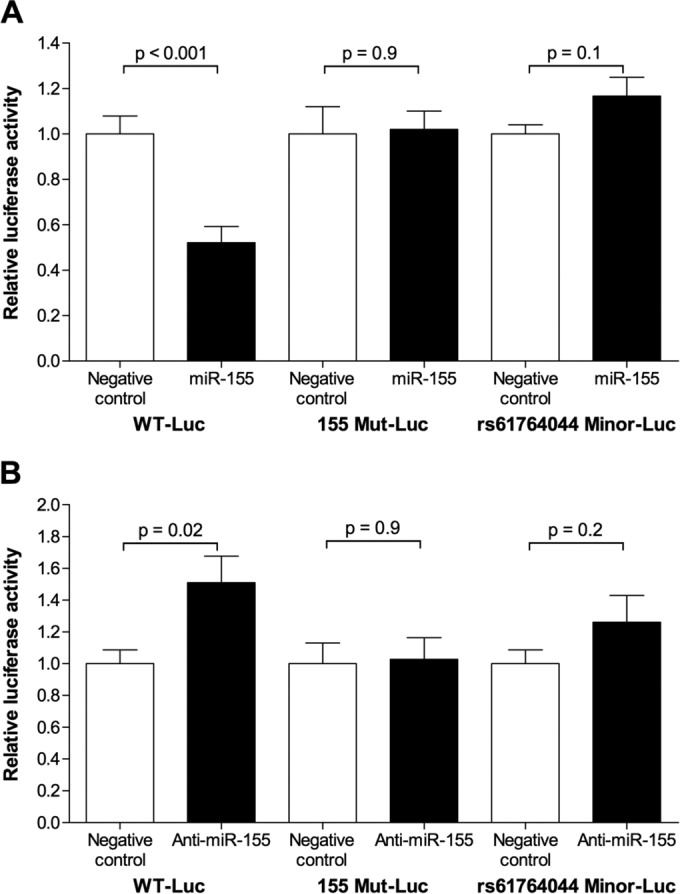
miR-155 interacts with the NPPA 3′ UTR in an rs61764044 allele-specific manner. (A) miR-155 reduced the activity of the WT-Luc construct but not the 155 Mut-Luc construct (containing a 6-nucleotide mutation at the predicted miR-155 seed binding site) or the rs61764044 Minor-Luc construct. The ratio of firefly luciferase activity to renilla luciferase activity was normalized to the ratio in COS-7 cells transfected with constructs encoding luciferase without the NPPA 3′ UTR and renilla. For each construct (WT-Luc, 155 Mut-Luc, or rs61764044 Minor-Luc), the effect of the miRNA was compared to that of a scrambled negative-control miRNA mimic (relative luciferase activity). (B) Anti-miR-155 increased the activity of the WT-Luc construct but not the 155 Mut-Luc construct or the rs61764044 Minor-Luc construct. The ratio of firefly luciferase activity to renilla luciferase activity was normalized to the ratio in COS-7 cells transfected with constructs encoding luciferase without the NPPA 3′ UTR and renilla. For each construct (WT-Luc, 155 Mut-Luc, or rs61764044 Minor-Luc), the effect of the anti-miR was compared to that of a scrambled negative-control anti-miRNA (relative luciferase activity). The rs61764044 Minor-Luc construct contains the minor alleles of both rs61764044 and rs5068. Data are means ± SEMs (n = 6 biological replicates; two-tailed independent t test).
TABLE 1.
Summary of data relating to miR-425, miR-155, miR-105, miR-103, and miR-107a
| miRNA | Endogenous miRNA expression level in cell type: |
WT-Luc luciferase activity (COS-7) |
NPPA mRNA and Nt-proANP protein level (hESC-CMs) |
Human population genetics studies: minor allele associated with higher plasma Nt-proANP level | ||||
|---|---|---|---|---|---|---|---|---|
| Human atrial tissue | COS-7 | hESC-CMs | Effect of miRNA | Effect of anti-miRNA | Effect of miRNA | Effect of anti-miRNA | ||
| miR-425 | ↑↑↑ | ↑↑ | ↑↑ | ↓ | ↑ | ↓ | ↑ | rs5068 |
| miR-155 | ↑↑ | ↑↑↑ | ↑↑ | ↓ | ↑ | ↓ | ↑ | rs61764044 |
| miR-105 | ↑ | ↑ | ↑ | ↓ | ↑ | — | ↑ | rs5067 |
| miR-103 | ↑↑↑ | ↑↑↑ | ↑↑↑ | ↓ | ↑ | — | — | — |
| miR-107 | ↑↑ | ↑↑ | ↑ | ↓ | ↑ | — | — | — |
Expression levels in human atrial tissues are based on RNA sequencing data in the work of Hsu et al. (15). Expression levels in COS-7 cells and hESC-CMs were determined by qRT-PCR (↑, low; ↑↑, intermediate; ↑↑↑, high). For effect of miRNA/anti-miRNA in luciferase assays, ↑ indicates an increase and ↓ indicates a decrease in luciferase activity in cells containing the WT-Luc construct. For effect of miRNA/anti-miRNA in hESC-CMs, ↑ indicates an increase and ↓ indicates a decrease in NPPA mRNA and secreted Nt-proANP protein levels. —, none.
miR-155 regulates endogenous NPPA mRNA and ANP protein levels in human cardiomyocytes.
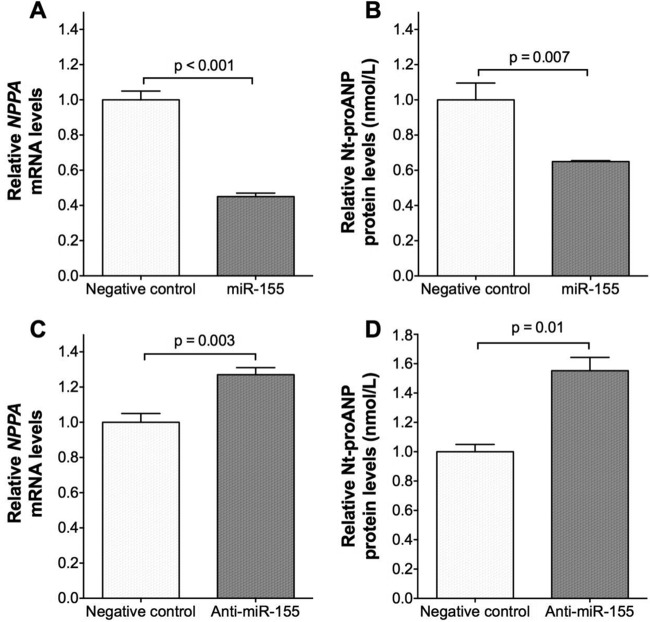
To determine whether miR-155 regulates endogenous NPPA mRNA expression and ANP protein production in cardiomyocytes, we transfected miR-155 into human embryonic stem cell-derived cardiomyocytes (hESC-CMs) and evaluated its effect on NPPA mRNA levels by qRT-PCR and production of the secreted Nt-proANP protein by ELISA. Transfection of miR-155 into hESC-CMs reduced endogenous NPPA mRNA (Fig. 4A), as well as secreted Nt-proANP protein levels (Fig. 4B), while the opposite effects were obtained with the transfection of anti-miR-155 (Fig. 4C and D). These findings provide support for the biological relevance of miR-155 in regulating NPPA mRNA and ANP protein levels in human cardiomyocytes.
FIG 4.
miR-155 reduces and anti-miR-155 increases NPPA mRNA and secreted Nt-proANP protein levels in human cardiomyocytes. hESC-CMs (∼1 × 105 per well) were transfected with miR-155, a scrambled negative-control miRNA mimic, anti-miR-155, or a scrambled negative-control anti-miRNA. Twenty-four hours later, cells were washed and incubated in 1 ml of medium. After an additional 24 h, cells and media were harvested. (A) NPPA mRNA levels in hESC-CMs transfected with miR-155 relative to cells transfected with negative-control miRNA mimic. (B) Nt-proANP protein levels (nanomoles/liter) in the medium of hESC-CMs transfected with miR-155 relative to negative-control miRNA mimic. (C) NPPA mRNA levels in hESC-CMs transfected with anti-miR-155 relative to negative-control anti-miRNA. (D) Nt-proANP protein levels in the medium of hESC-CMs transfected with anti-miR-155 relative to negative-control anti-miRNA. Data are means ± SEMs (n = 6 biological replicates; two-tailed independent t test).
Genetic variant rs61764044 is associated with plasma ANP levels and blood pressure in humans and disrupts miR-155 binding of NPPA mRNA.
During the course of studies to identify additional variants in the NPPA locus, we performed deep sequencing analyses on 268 samples from individuals of European descent and identified SNP rs61764044, which lies in the miR-155 binding site of the NPPA 3′ UTR (see Fig. S1C in the supplemental material); this SNP has also been reported in the 1000 Genomes Project database. This variant is perfectly correlated with rs5068, which lies 123 nucleotides upstream in the 3′ UTR (see Fig. S1A), indicating that the minor alleles of rs61764044 and rs5068 are always inherited together (perfect linkage disequilibrium, r2 = 1). Previous studies showed that the rs5068 minor allele disrupts miR-425 binding and is strongly associated with higher plasma ANP levels and lower blood pressure (6, 7). Because of perfect correlation of the rs61764044 and rs5068 minor alleles, the rs61764044 minor allele is associated with increased plasma ANP levels, lower blood pressure, and reduced risk of hypertension in humans.
The miR-155 seed sequence spans the major allele of rs61764044, and the minor allele of rs61764044 is predicted to introduce G-U wobble base pairing within the miR-155 seed binding region (see Fig. S1C in the supplemental material). G-U wobbles in the miRNA seed binding region impair the ability of an miRNA to repress the mRNA target (16–18). We generated a reporter construct (rs61764044 Minor-Luc) in which the firefly luciferase gene was ligated to the NPPA 3′ UTR sequence containing the minor alleles of both rs61764044 and rs5068. Transfection of miR-155 reduced the luciferase activity in COS-7 cells containing the major allele (WT-Luc) construct but not the minor allele construct (Fig. 3A). Anti-miR-155 increased the luciferase activity in COS-7 cells containing the major but not minor allele construct (Fig. 3B). Taken together, these results indicate that the NPPA 3′ UTR containing the rs61764044 major allele is a direct target of miR-155 and that the presence of the rs61764044 minor allele confers resistance to miR-155-mediated repression.
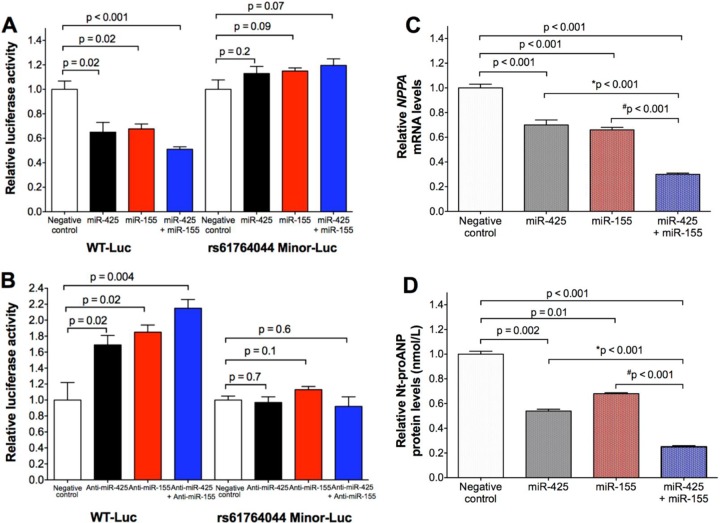
miR-155 augments the miR-425-mediated decreases in endogenous NPPA mRNA and ANP protein levels in human cardiomyocytes.
Given the fact that rs61764044 is perfectly correlated with rs5068 and previous studies demonstrated that miR-425 targets the NPPA 3′ UTR with the rs5068 major allele and inhibits ANP levels in human cardiomyocytes (7), we sought to determine whether miR-155 can enhance the repressive effects of miR-425 on the NPPA mRNA. Transfection of either miRNA alone reduced the luciferase activity in the presence of the WT-Luc construct containing the major alleles of rs61764044 and rs5068 but not the rs61764044 Minor-Luc construct containing the minor alleles of rs61764044 and rs5068 (Fig. 5A). Cotransfection of miR-425 and miR-155 decreased luciferase activity in the presence of the WT-Luc construct containing the major alleles but not the rs61764044 Minor-Luc construct containing the minor alleles of rs61764044 and rs5068 (Fig. 5A). Cotransfection of anti-miR-425 and anti-miR-155 increased the luciferase activity in the presence of the WT-Luc construct containing the major alleles but not the rs61764044 Minor-Luc construct containing the minor alleles of rs61764044 and rs5068 (Fig. 5B). Moreover, in hESC-CMs, cotransfection of miR-425 and miR-155 decreased endogenous NPPA mRNA levels and secreted Nt-proANP protein levels to a greater extent than did either miRNA alone (Fig. 5C and D). Taken together, these findings indicate that miR-155 regulates NPPA expression and ANP protein production through its interaction with the rs61764044 major allele and can act together with miR-425 to further suppress ANP levels. Thus, association of the rs5068 and rs61764044 minor alleles with higher plasma ANP and Nt-proANP levels, as well as lower blood pressure and reduced risk of hypertension in humans, is likely due to coinheritance of resistance to two independent miRNAs (6, 7).
FIG 5.
miR-155 interacts with the rs61764044 major allele and acts together with miR-425 to further suppress NPPA mRNA and ANP protein levels in human cardiomyocytes. (A and B) Allele-specific effects of miR-425 plus miR-155 (A) and anti-miR-425 plus anti-miR-155 (B) on the luciferase activity produced by the WT-Luc and rs61764044 Minor-Luc constructs in COS-7 cells. The rs61764044 Minor-Luc construct contains the minor alleles of rs61764044 and rs5068. (C) NPPA mRNA levels in hESC-CMs transfected with the indicated miRNAs relative to scrambled negative-control miRNA mimic. (D) Nt-proANP protein levels in the medium of hESC-CMs transfected with the indicated miRNAs relative to scrambled negative-control miRNA mimic. For miR-425 plus miR-155 cotransfection, the total amount of transfected miRNA was held constant under all experimental conditions (10 nM total concentration in COS-7 cells, 100 nM total concentration in hESC-CMs). For the conditions labeled with “miR-425” or “miR-155,” the total concentration of transfected miRNA was held constant by the addition of the scrambled negative-control miRNA mimic. For anti-miR-425 plus anti-miR-155 cotransfection, the total amount of transfected anti-miRNA was held constant under all experimental conditions (200 nM total concentration in COS-7 cells). For the experimental conditions labeled with “anti-miR-425” or “anti-miR-155,” the total concentration of transfected miRNA was held constant by the addition of the scrambled negative-control anti-miRNA. Data are means ± SEMs (n = 6 biological replicates). P values are from a two-tailed independent t test. *, P < 0.001 versus miR-425, and #, P < 0.001 versus miR-155, by one-way ANOVA with Bonferroni post hoc testing.
miR-103 and miR-107 do not appear to regulate NPPA mRNA and ANP protein levels in human cardiomyocytes.
Of the 13 candidate miRNAs, miR-103 and miR-107 share the same seed sequence and thus are predicted to target the same complementary sequence in the NPPA 3′ UTR (see Fig. S1A and E in the supplemental material). Exogenous administration of either miRNA reduced the luciferase activity in COS-7 cells containing the wild-type construct but not a construct (103/107 Mut-Luc) with a mutated seed binding site (see Fig. S2A and S3A in the supplemental material), confirming that the seed binding site is required for interaction of miR-103 and miR-107 with the NPPA 3′ UTR. Inhibition of endogenous miR-103 or miR-107 with their respective anti-miRNAs increased the luciferase activity in COS-7 cells containing the wild-type but not mutant construct (see Fig. S2B and S3B). These findings indicate that the NPPA 3′ UTR is a direct target of miR-103 and miR-107.
Human ESC-CMs express both miR-103 and miR-107 (Table 1). The effects of overexpressing miR-103 and miR-107, as well as each of their anti-miRNAs, on NPPA mRNA and ANP protein levels were examined. Transfection of the miRNAs or their anti-miRNAs had no effect on NPPA mRNA or secreted Nt-proANP protein levels in hESC-CMs (data not shown). These findings suggest that these miRNAs may not have a role in regulating endogenous ANP levels in human cardiomyocytes.
Genetic variant rs5067 is associated with plasma Nt-proANP levels in humans and disrupts miR-105 binding of NPPA mRNA.
MicroRNA-105 was considered a candidate NPPA-targeting miRNA for further investigation because a common polymorphism (rs5067) lies in the miR-105 binding site (see Fig. S1A and D in the supplemental material). The miR-105 seed sequence binds to the rs5067 major allele, but the rs5067 minor allele is predicted to disrupt base pairing of miR-105 with the NPPA 3′ UTR. In a study of 5,453 Swedish individuals in the Malmö Diet and Cancer Study, the rs5067 minor allele was found to be associated with higher plasma Nt-proANP levels (0.079 standard deviations of lnMR-proANP per G-allele of rs5067, 95% confidence intervals = 0.024 to 0.135, P = 0.004). The mean and standard deviation of lnMR-proANP are 4.20 and 0.416. The correlation of rs5067 with rs5068 is low (r2 = 0.012, HapMap CEU), although with high linkage disequilibrium (D′ = 1) due to the fact that the haplotypes containing the minor alleles of the two variants are mutually exclusive (i.e., the minor alleles are never coinherited). The variants rs5067 and rs5068 are thus independent genetic association signals with plasma Nt-proANP levels. We hypothesized that disruption of the binding of miR-105 to the NPPA 3′ UTR by the rs5067 minor allele could represent a mechanism by which rs5067 is associated with plasma Nt-proANP levels in humans.
To test whether the presence of the rs5067 minor allele alters interaction of miR-105 with the NPPA 3′ UTR, we performed luciferase reporter assays using constructs including either the major or the minor rs5067 allele in COS-7 cells. Transfection of miR-105 reduced the luciferase activity in the presence of the WT-Luc construct containing the rs5067 major allele but not the construct containing the rs5067 minor allele (Fig. 6A). We confirmed that COS-7 cells express miR-105 (Table 1). Anti-miR-105 increased the luciferase activity in the presence of the WT-Luc construct containing the rs5067 major allele but not the construct containing the rs5067 minor allele (Fig. 6B). Taken together, these results suggest that the NPPA 3′ UTR containing the rs5067 major allele is a direct target of miR-105. The presence of the rs5067 minor allele confers resistance to miR-105-mediated repression, which could be a mechanism by which rs5067 alters ANP levels in humans.
FIG 6.
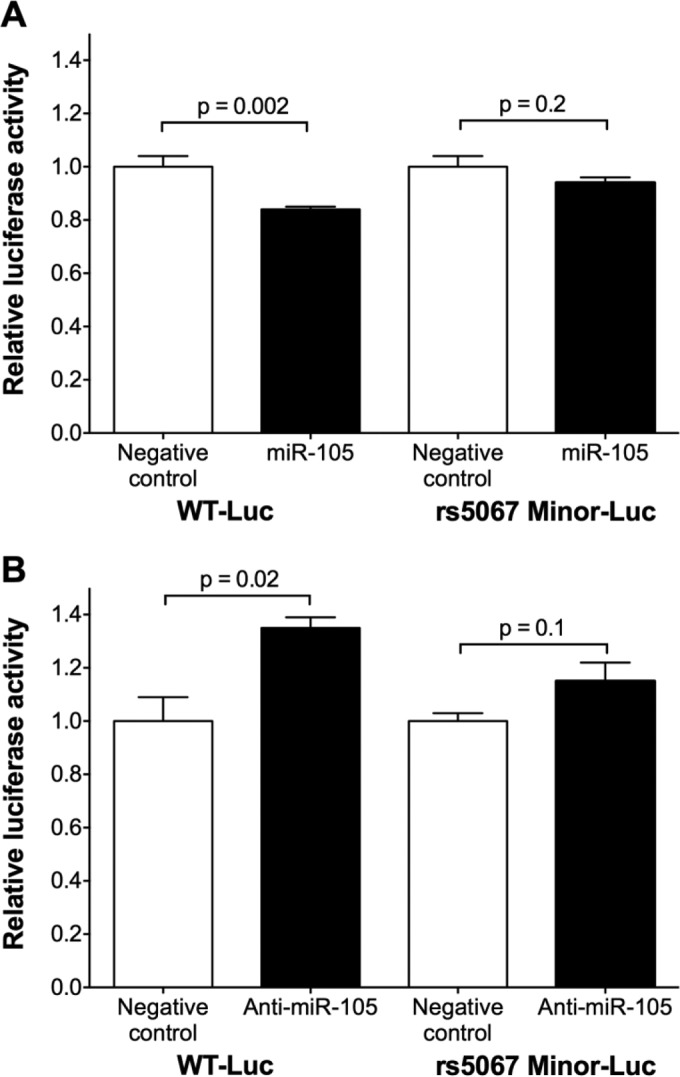
miR-105 interacts with the NPPA 3′ UTR in an rs61764044 allele-specific manner. (A) miR-105 reduced the activity of the WT-Luc construct but not the rs5067 Minor-Luc construct. The ratio of firefly luciferase activity to renilla luciferase activity was normalized to the ratio in COS-7 cells transfected with constructs encoding luciferase without the NPPA 3′ UTR and renilla. For each construct (WT-Luc or rs5067 Minor-Luc), the effect of the miRNA was compared to that of a scrambled negative-control miRNA mimic (relative luciferase activity). (B) Anti-miR-105 increased the activity of the WT-Luc construct but not rs5067 Minor-Luc construct. The ratio of firefly luciferase activity to renilla luciferase activity was normalized to the ratio in COS-7 cells transfected with constructs encoding luciferase without the NPPA 3′ UTR and renilla. For each construct (WT-Luc or rs5067 Minor-Luc), the effect of the anti-miRNA was compared to that of a scrambled negative-control anti-miRNA (relative luciferase activity). Data are means ± SEMs (n = 6 biological replicates; two-tailed independent t test).
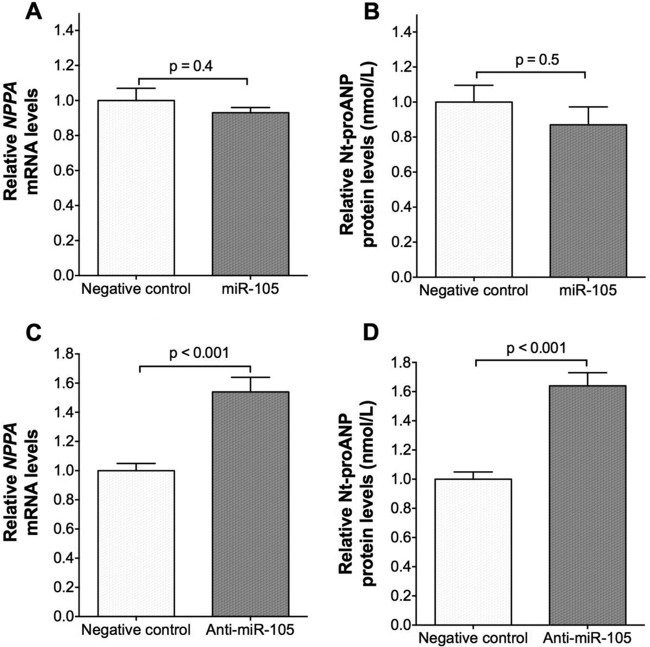
miR-105 regulates endogenous NPPA mRNA and ANP protein levels in human cardiomyocytes.
Human ESC-CMs, homozygous for the rs5067 major allele, were transfected with miR-105 to determine whether miR-105 can regulate endogenous NPPA mRNA expression and ANP production. Transfection of miR-105 had no effect on NPPA mRNA (Fig. 7A) or secreted Nt-proANP protein levels (Fig. 7B) in hESC-CMs. As with human atrial tissues (15), hESC-CMs express miR-105 (Table 1). Inhibition of endogenous miR-105 with anti-miR-105 did increase NPPA mRNA (Fig. 7C), as well as secreted Nt-proANP protein levels (Fig. 7D), in hESC-CMs. Although overexpression of miR-105 did not have an effect on NPPA mRNA or Nt-proANP protein levels in hESC-CMs, inhibition of endogenous miR-105 with anti-miR-105 increased NPPA mRNA and secreted Nt-proANP protein levels, suggesting that endogenous levels of miR-105 could modulate NPPA mRNA and ANP protein levels in human cardiomyocytes.
FIG 7.
Anti-miR-105 increases NPPA mRNA and secreted Nt-proANP protein levels in human cardiomyocytes. hESC-CMs (∼1 × 105 per well) were transfected with miR-105, a scrambled negative-control miRNA mimic, anti-miR-105, or a scrambled negative-control anti-miRNA. Twenty-four hours later, cells were washed and incubated in 1 ml of medium. After an additional 24 h, cells and medium were harvested. (A) NPPA mRNA levels in hESC-CMs transfected with miR-105 relative to negative-control miRNA mimic. (B) Nt-proANP protein levels in the medium of hESC-CMs transfected with miR-105 relative to negative-control miRNA mimic. (C) NPPA mRNA levels in hESC-CMs transfected with anti-miR-105 relative to negative-control anti-miRNA. (D) Nt-proANP protein levels in the medium of hESC-CMs transfected with anti-miR-105 relative to negative-control anti-miRNA. Data are means ± SEMs (n = 6 biological replicates; two-tailed independent t test).
DISCUSSION
In this study, we used a multifaceted screening strategy that combined in silico miRNA target prediction, miRNA expression data, human genetic association data, and cellular models to identify four miRNAs, in addition to miR-425, that target the NPPA 3′ UTR. Two of the four miRNAs (miR-155 and miR-105) were shown to have a functional role in modulating endogenous NPPA mRNA levels and secreted Nt-proANP levels in human cardiomyocytes (Table 1; see also Fig. S4 in the supplemental material). We observed that a genetic variant (rs61764044), which disrupts the miR-155 binding site, contributes to the ANP-raising and blood pressure-lowering effects of the previously published rs5068 variant (6, 7) by virtue of perfect linkage disequilibrium with rs5068. Similarly to miR-425 and miR-155, miR-105 interacts with a genetic variant (rs5067), specifically binding to the rs5067 major but not minor allele, consistent with the finding that the rs5067 minor allele is associated with increased plasma Nt-proANP levels in humans. Thus, by integrating computational, genomic, and cellular approaches, we have identified novel regulatory mechanisms controlling ANP levels and highlighted the potential of applying a similar integrated screening strategy to uncover miRNA regulators of other genes of interest.
In silico analysis revealed that as many as 494 miRNAs are predicted to target the NPPA 3′ UTR. Because miRNA-mRNA interactions predicted by multiple algorithms appear to provide greater specificity without excessive loss of sensitivity (14, 19), miRNAs predicted to interact with the NPPA 3′ UTR by at least 3 of the algorithms were given priority. To further narrow down the list of miRNAs for subsequent experimental validation, we focused on miRNAs that are expressed at abundant levels in human atrial tissues (15), where the target gene NPPA is highly expressed. This strategy identified miR-155 as a novel miRNA that interacted with the NPPA 3′ UTR, as confirmed by luciferase reporter assays, and decreased NPPA mRNA and secreted Nt-proANP protein levels in hESC-CMs. Although miR-103 and miR-107 were also identified as miRNAs that can target the NPPA 3′ UTR based on luciferase reporter assays, neither the miRNAs nor their anti-miRNAs altered NPPA expression in hESC-CMs, the cell type of interest (Table 1). Examination of the levels of miR-103 and miR-107 in COS-7 cells and hESC-CMs transfected with the respective miRNA mimics showed that compared to the endogenous miRNA levels in cells transfected with the negative control, there were similar levels of overexpression of the transfected miRNAs in COS-7 cells (see Fig. S5C and D in the supplemental material) and in hESC-CMs (see Fig. S6C and D in the supplemental material). (COS-7 cells and hESC-CMs transfected with miR-425, miR-155, and miR-105 also showed similar levels of overexpression of each of the miRNAs compared to cells transfected with the negative control [see Fig. S5 and S6].) As such, differences in the responses obtained for miR-103 and miR-107 in the COS-7 cells and hESC-CMs are most likely not due to differences in the levels of overexpression of these transfected miRNAs in the two cell types. A role for miR-103 and miR-107 in regulating endogenous NPPA expression in the heart therefore remains unproven and requires further investigation. Others have also reported instances in which an miRNA was shown to target a 3′ UTR of interest in luciferase reporter assays but had no effects on endogenous target mRNA/protein levels in the cell type of interest (20). It has been proposed that target secondary structure (21, 22) and the presence of RNA-binding proteins (23, 24), the levels of which may vary between different cell types, can influence the ability of miRNAs to regulate target mRNAs and interfere with the ability of miRNAs to directly interact with the 3′ UTR of target mRNAs.
In addition to prioritizing miRNAs that are highly expressed in human atria, we leveraged human population genetic data to investigate whether any of the candidate miRNAs were predicted to target variants in the NPPA 3′ UTR that are associated with plasma ANP levels in humans. This approach led to the identification of miR-105, which was validated to interact with rs5067 in an allele-dependent manner. The rs5067 minor allele disrupted base pairing between miR-105 and the NPPA 3′ UTR, which could underlie the observed association of the rs5067 minor allele with higher plasma Nt-proANP levels in humans. The cause of the failure of miR-105 to decrease NPPA mRNA and protein levels in hESC-CMs is unclear and warrants further investigation, but a potential explanation is that the levels of miR-105 in cardiomyocytes, albeit low (Table 1), may at endogenous levels already exert a maximal effect on the target NPPA mRNA. It has been reported that the threshold for saturation of miRNA activity varies for different miRNAs, independent of the expression levels of the miRNA (25, 26). In this model, introducing more miR-105 into the hESC-CMs would not have any further effect, but inhibiting endogenous miR-105 would have an effect on NPPA expression by relieving the repression mediated by endogenous miR-105. In COS-7 cells transfected with an NPPA cDNA expression plasmid containing the rs5067 major allele, we observed that cotransfection of miR-105 reduced secreted Nt-proANP protein levels (see Fig. S7A in the supplemental material), while the opposite effect was obtained with cotransfection of anti-miR-105 (see Fig. S7B). These results suggest that the repressive effect of overexpressing miR-105 on ANP production can be revealed when the target NPPA mRNA is also overexpressed, providing support for the possibility of saturation.
While miR-155 was predicted to target the NPPA 3′ UTR and was expressed in human atria, we discovered the rs61764044 variant in the miR-155 binding site only when additional sequencing identified the variant. The perfect correlation between rs5068 and rs61764044 was not known at the time of the prior studies of rs5068 (6, 7). The finding of an allele-specific negative regulatory effect of miR-155 in the current study indicates that rs61764044 contributes to the association between rs5068 and plasma ANP levels, as well as blood pressure. Furthermore, our results highlight the importance of having a complete catalogue of genetic variation. In this model, the presence of the coinherited minor alleles prevents both miR-425 and miR-155 from binding to the NPPA 3′ UTR, resulting in increased ANP production, lower systolic and diastolic blood pressures, and reduced risk of hypertension in humans. The rs61764044 minor allele creates a G-U wobble base pairing in the miR-155 seed binding site. Studies have shown that the presence of even a single G-U wobble in the seed binding region impairs miRNA-mediated repression of the mRNA target (16–18). In the case of the FGF20 3′ UTR, for example, a similar disruption of a seed binding site was reported for the T allele of rs12720208, which introduces a G-U wobble base pairing in the miR-433 seed binding site and disrupts the ability of miR-433 to repress FGF20 (27).
The observation that cotransfection of miR-425 and miR-155 resulted in greater NPPA gene repression in hESC-CMs than did transfection of either miRNA alone has implications for the therapeutic potential of using combinations of anti-miRNAs to increase ANP levels. In fact, coinheritance of the rs5068 and rs61764044 minor alleles in humans can be viewed as an experiment of nature that demonstrates the physiologic impact of these ANP-raising mechanisms on blood pressure and suggests therapeutic opportunities to increase ANP levels with potential benefits in hypertension or heart failure. Recent studies have implicated the therapeutic potential of miR-155 inhibition in suppressing cardiac hypertrophy and heart failure (28, 29). Moreover, miR-155 levels were also reported to be increased in human hearts with hypertrophy compared to nonhypertrophic controls and increased miR-155 levels were correlated with depressed cardiac function and increased wall thickness (29). Our study is the first to report that miR-155 can target the NPPA 3′ UTR to directly regulate ANP levels in human cardiomyocytes. While miR-155 is a multifunctional miRNA with multiple targets (30), its effects on human NPPA expression could contribute to an effect on cardiac hypertrophy and heart failure.
From our multitiered screening strategy, we observed that of the 14 candidate miRNAs (including miR-425) that we tested in our NPPA 3′ UTR luciferase reporter assay, only 5 of them (miR-425, miR-155, miR-105, miR-103, and miR-107) were able to reduce the luciferase activity of the WT NPPA 3′ UTR luciferase reporter construct. Examination of the predicted binding sites of these 5 miRNAs revealed that the binding sites of 4 of these miRNAs (miR-425, miR-105, miR-103, and miR-107) are located close to each other in the NPPA 3′ UTR (see Fig. S1A in the supplemental material). As mRNA secondary structure has been reported to affect the ability of miRNAs to target mRNAs (21, 22), it is reasonable to speculate that this region of the NPPA 3′ UTR may contain secondary structure elements that increase the accessibility of these miRNA binding sites, thereby facilitating the interaction between these miRNAs and the NPPA 3′ UTR. Additional work investigating the secondary structure of the NPPA mRNA will be required to confirm this possibility. Moreover, out of these 5 miRNAs that reduced the luciferase activity of the WT NPPA 3′ UTR luciferase reporter construct, we found that only 3 of them (miR-425, miR-155, and miR-105) were able to modulate ANP levels in hESC-CMs. A unique characteristic shared by these 3 miRNAs that is not seen for miR-103 and miR-107 is that miR-425, miR-155, and miR-105 all target sequences in the NPPA 3′ UTR that include genetic variants that are associated with plasma ANP levels in humans. Our findings suggest that in attempts to narrow down the list of candidate miRNAs obtained from computational prediction algorithms, it is worthwhile to consider prioritizing the miRNAs which target genetic variants that are associated with phenotypes of interest. Such an approach may help increase the likelihood that the miRNA will have biological relevance in regulating the endogenous levels of its target mRNA.
Several limitations of the present study merit consideration. We chose to experimentally validate a subset of predicted NPPA-targeting miRNAs based on their expression levels in human atrial tissues. It remains likely that there are other miRNAs, expressed at lower levels in human atria, that can interact with the NPPA 3′ UTR and regulate ANP levels. We have not examined primary human cardiomyocytes because of technical challenges, including difficulties associated with obtaining human cardiac tissue, low cardiomyocyte yields from the isolation process, and inability to maintain the viability of isolated cardiomyocytes for an extended period of time (31). However, hESC-CMs have been well characterized and exhibit structural and functional properties of native human cardiomyocytes (32, 33). Gene expression profiling studies have also shown that gene expression patterns of cardiac transcription factors and cardiac structural markers (including NPPA) in hESC-CMs are consistent with gene expression patterns in human cardiac tissues (34–36). Moreover, miRNA profiling has demonstrated that known cardiomyocyte-specific miRNAs are expressed in hESC-CMs and exhibit the expected expression pattern (34). These observations suggest that hESC-CMs are a biologically relevant model for studying the effect of miRNAs on NPPA mRNA levels and ANP production in the human heart.
In conclusion, we identified miR-155 and miR-105 as novel regulators of ANP production. These miRNAs target sequences including genetic variants that are associated with plasma ANP levels in humans. Our findings suggest the potential for miRNA-targeted therapies to increase ANP levels, which could supplement current therapy that reduces natriuretic peptide degradation. Further experiments in animals examining the in vivo effects of anti-miRNAs in derepressing ANP production (which lie beyond the scope of this study) would help to characterize phenotypic responses that will further expand our understanding of the clinical applications of such treatments.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by funds of the Department of Anesthesia, Critical Care and Pain Medicine, Massachusetts General Hospital; award number T32HL007208 from the National Heart, Lung, and Blood Institute (C.W.); the Howard Hughes Medical Institute (P.J.); grant support PF10/014 by KU Leuven (S.T. and S.P.J.); the Leducq Foundation (C.W., K.D.B., and D.B.B.); and grants from the NIH (R01HL098283, T.J.W. and C.N.-C.; R01HL113933, C.N.-C.; and R01HL124262, C.N.-C.).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
None of the authors report any significant relationships with industry or financial disclosures related to the manuscript. C. Wu, P. Arora, T. J. Wang, K. D. Bloch, and C. Newton-Cheh are named as coinventors on a patent application relating to the use of miRNA antagonists to increase ANP production to treat hypertension and heart failure.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01114-15.
REFERENCES
- 1.Volpe M, Rubattu S, Burnett J Jr. 2014. Natriuretic peptides in cardiovascular diseases: current use and perspectives. Eur Heart J 35:419–425. doi: 10.1093/eurheartj/eht466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McMurray JJ, Packer M, Desai AS, Gong J, Lefkowitz MP, Rizkala AR, Rouleau JL, Shi VC, Solomon SD, Swedberg K, Zile MR. 2014. Angiotensin-neprilysin inhibition versus enalapril in heart failure. N Engl J Med 371:993–1004. doi: 10.1056/NEJMoa1409077. [DOI] [PubMed] [Google Scholar]
- 3.Potter LR. 2011. Regulation and therapeutic targeting of peptide-activated receptor guanylyl cyclases. Pharmacol Ther 130:71–82. doi: 10.1016/j.pharmthera.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinhelper ME, Cochrane KL, Field LJ. 1990. Hypotension in transgenic mice expressing atrial natriuretic factor fusion genes. Hypertension 16:301–307. doi: 10.1161/01.HYP.16.3.301. [DOI] [PubMed] [Google Scholar]
- 5.John SW, Krege JH, Oliver PM, Hagaman JR, Hodgin JB, Pang SC, Flynn TG, Smithies O. 1995. Genetic decreases in atrial natriuretic peptide and salt-sensitive hypertension. Science 267:679–681. doi: 10.1126/science.7839143. [DOI] [PubMed] [Google Scholar]
- 6.Newton-Cheh C, Larson MG, Vasan RS, Levy D, Bloch KD, Surti A, Guiducci C, Kathiresan S, Benjamin EJ, Struck J, Morgenthaler NG, Bergmann A, Blankenberg S, Kee F, Nilsson P, Yin X, Peltonen L, Vartiainen E, Salomaa V, Hirschhorn JN, Melander O, Wang TJ. 2009. Association of common variants in NPPA and NPPB with circulating natriuretic peptides and blood pressure. Nat Genet 41:348–353. doi: 10.1038/ng.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arora P, Wu C, Khan AM, Bloch DB, Davis-Dusenbery BN, Ghorbani A, Spagnolli E, Martinez A, Ryan A, Tainsh LT, Kim S, Rong J, Huan T, Freedman JE, Levy D, Miller KK, Hata A, Del Monte F, Vandenwijngaert S, Swinnen M, Janssens S, Holmes TM, Buys ES, Bloch KD, Newton-Cheh C, Wang TJ. 2013. Atrial natriuretic peptide is negatively regulated by microRNA-425. J Clin Invest 123:3378–3382. doi: 10.1172/JCI67383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balaga O, Friedman Y, Linial M. 2012. Toward a combinatorial nature of microRNA regulation in human cells. Nucleic Acids Res 40:9404–9416. doi: 10.1093/nar/gks759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. 2005. Combinatorial microRNA target predictions. Nat Genet 37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 10.Akbari Moqadam F, Pieters R, den Boer ML. 2013. The hunting of targets: challenge in miRNA research. Leukemia 27:16–23. doi: 10.1038/leu.2012.179. [DOI] [PubMed] [Google Scholar]
- 11.Shirdel EA, Xie W, Mak TW, Jurisica I. 2011. NAViGaTing the micronome—using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS One 6:e17429. doi: 10.1371/journal.pone.0017429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP. 2012. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A 109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith JG, Newton-Cheh C, Almgren P, Struck J, Morgenthaler NG, Bergmann A, Platonov PG, Hedblad B, Engstrom G, Wang TJ, Melander O. 2010. Assessment of conventional cardiovascular risk factors and multiple biomarkers for the prediction of incident heart failure and atrial fibrillation. J Am Coll Cardiol 56:1712–1719. doi: 10.1016/j.jacc.2010.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Rooij E. 2011. The art of microRNA research. Circ Res 108:219–234. doi: 10.1161/CIRCRESAHA.110.227496. [DOI] [PubMed] [Google Scholar]
- 15.Hsu J, Hanna P, Van Wagoner DR, Barnard J, Serre D, Chung MK, Smith JD. 2012. Whole genome expression differences in human left and right atria ascertained by RNA sequencing. Circ Cardiovasc Genet 5:327–335. doi: 10.1161/CIRCGENETICS.111.961631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doench JG, Sharp PA. 2004. Specificity of microRNA target selection in translational repression. Genes Dev 18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yue D, Liu H, Huang Y. 2009. Survey of computational algorithms for microRNA target prediction. Curr Genomics 10:478–492. doi: 10.2174/138920209789208219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brennecke J, Stark A, Russell RB, Cohen SM. 2005. Principles of microRNA-target recognition. PLoS Biol 3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karbiener M, Glantschnig C, Scheideler M. 2014. Hunting the needle in the haystack: a guide to obtain biologically meaningful microRNA targets. Int J Mol Sci 15:20266–20289. doi: 10.3390/ijms151120266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Q, Feng MG, Mo YY. 2009. Systematic validation of predicted microRNAs for cyclin D1. BMC Cancer 9:194. doi: 10.1186/1471-2407-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. 2007. Potent effect of target structure on microRNA function. Nat Struct Mol Biol 14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 22.Li F, Zheng Q, Ryvkin P, Dragomir I, Desai Y, Aiyer S, Valladares O, Yang J, Bambina S, Sabin LR, Murray JI, Lamitina T, Raj A, Cherry S, Wang LS, Gregory BD. 2012. Global analysis of RNA secondary structure in two metazoans. Cell Rep 1:69–82. doi: 10.1016/j.celrep.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, Lund AH, Perrakis A, Raz E, Agami R. 2007. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 24.Beillard E, Ong SC, Giannakakis A, Guccione E, Vardy LA, Voorhoeve PM. 2012. miR-Sens—a retroviral dual-luciferase reporter to detect microRNA activity in primary cells. RNA 18:1091–1100. doi: 10.1261/rna.031831.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gentner B, Schira G, Giustacchini A, Amendola M, Brown BD, Ponzoni M, Naldini L. 2009. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods 6:63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- 26.Brown BD, Naldini L. 2009. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet 10:578–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- 27.Wang G, van der Walt JM, Mayhew G, Li YJ, Zuchner S, Scott WK, Martin ER, Vance JM. 2008. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet 82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seok HY, Chen J, Kataoka M, Huang ZP, Ding J, Yan J, Hu X, Wang DZ. 2014. Loss of microRNA-155 protects the heart from pathological cardiac hypertrophy. Circ Res 114:1585–1595. doi: 10.1161/CIRCRESAHA.114.303784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heymans S, Corsten MF, Verhesen W, Carai P, van Leeuwen RE, Custers K, Peters T, Hazebroek M, Stoger L, Wijnands E, Janssen BJ, Creemers EE, Pinto YM, Grimm D, Schurmann N, Vigorito E, Thum T, Stassen F, Yin X, Mayr M, de Windt LJ, Lutgens E, Wouters K, de Winther MP, Zacchigna S, Giacca M, van Bilsen M, Papageorgiou AP, Schroen B. 2013. Macrophage microRNA-155 promotes cardiac hypertrophy and failure. Circulation 128:1420–1432. doi: 10.1161/CIRCULATIONAHA.112.001357. [DOI] [PubMed] [Google Scholar]
- 30.Faraoni I, Antonetti FR, Cardone J, Bonmassar E. 2009. miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta 1792:497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Mitcheson JS, Hancox JC, Levi AJ. 1998. Cultured adult cardiac myocytes: future applications, culture methods, morphological and electrophysiological properties. Cardiovasc Res 39:280–300. doi: 10.1016/S0008-6363(98)00128-X. [DOI] [PubMed] [Google Scholar]
- 32.Harding SE, Ali NN, Brito-Martins M, Gorelik J. 2007. The human embryonic stem cell-derived cardiomyocyte as a pharmacological model. Pharmacol Ther 113:341–353. doi: 10.1016/j.pharmthera.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 33.Dick E, Rajamohan D, Ronksley J, Denning C. 2010. Evaluating the utility of cardiomyocytes from human pluripotent stem cells for drug screening. Biochem Soc Trans 38:1037–1045. doi: 10.1042/BST0381037. [DOI] [PubMed] [Google Scholar]
- 34.Synnergren J, Ameen C, Lindahl A, Olsson B, Sartipy P. 2011. Expression of microRNAs and their target mRNAs in human stem cell-derived cardiomyocyte clusters and in heart tissue. Physiol Genomics 43:581–594. doi: 10.1152/physiolgenomics.00074.2010. [DOI] [PubMed] [Google Scholar]
- 35.Puppala D, Collis LP, Sun SZ, Bonato V, Chen X, Anson B, Pletcher M, Fermini B, Engle SJ. 2013. Comparative gene expression profiling in human-induced pluripotent stem cell–derived cardiocytes and human and cynomolgus heart tissue. Toxicol Sci 131:292–301. doi: 10.1093/toxsci/kfs282. [DOI] [PubMed] [Google Scholar]
- 36.Babiarz JE, Ravon M, Sridhar S, Ravindran P, Swanson B, Bitter H, Weiser T, Chiao E, Certa U, Kolaja KL. 2012. Determination of the human cardiomyocyte mRNA and miRNA differentiation network by fine-scale profiling. Stem Cells Dev 21:1956–1965. doi: 10.1089/scd.2011.0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.