Abstract
Clinical research is making toiling efforts for promotion and wellbeing of the health status of the people. There is a rapid increase in number and severity of diseases like cancer, hepatitis, HIV etc, resulting in high morbidity and mortality. Clinical research involves drug discovery and development whereas clinical trials are performed to establish safety and efficacy of drugs. Drug discovery is a long process starting with the target identification, validation and lead optimization. This is followed by the preclinical trials, intensive clinical trials and eventually post marketing vigilance for drug safety. Softwares and the bioinformatics tools play a great role not only in the drug discovery but also in drug development. It involves the use of informatics in the development of new knowledge pertaining to health and disease, data management during clinical trials and to use clinical data for secondary research. In addition, new technology likes molecular docking, molecular dynamics simulation, proteomics and quantitative structure activity relationship in clinical research results in faster and easier drug discovery process. During the preclinical trials, the software is used for randomization to remove bias and to plan study design. In clinical trials software like electronic data capture, Remote data capture and electronic case report form (eCRF) is used to store the data. eClinical, Oracle clinical are software used for clinical data management and for statistical analysis of the data. After the drug is marketed the safety of a drug could be monitored by drug safety software like Oracle Argus or ARISg. Therefore, softwares are used from the very early stages of drug designing, to drug development, clinical trials and during pharmacovigilance. This review describes different aspects related to application of computers and bioinformatics in drug designing, discovery and development, formulation designing and clinical research.
Key words: Argus, ARISg, bioinformatics, clinical research, eClinical
INTRODUCTION
The current drug discovery processes within pharmaceutical companies remain conservative, typically focused on discovery of a novel therapeutic target and new potential compound that modulates the activity of the identified target. Subsequently, preclinical and clinical investigations conducted are often slow, expensive, and risky process and take about 15 years and about 800 million USD to 1 billion USD to introduce a compound on the shelf of the market. Hence, an effective and innovative approach is required to predict the drug efficacy, thereby strengthening the success of drug development process. Bioinformatics is the applications of computer science in biology that can improve drug discovery with efficient statistical algorithms, rationale approaches for target identification, validation, and optimization. Computers and software tools greatly help creating databases, predict the function of proteins, model the structure of proteins, determine the coding regions of nucleic acid sequences, find suitable drug compounds from a large pool, perform data mining, analyzing, and interpret data faster thereby reducing time of drug discovery and eventually the cost involved in it.[1]
Clinical research is a branch of science that ensures the safety and effectiveness of medications, devices, diagnostic products, and treatment regimens for human use. Various software can predict the possible interactions, toxicities, and indications thereby, accurately defining the success of a novel compound or the repositioning for new uses. Clinical research varies from clinical practice as in the former established treatments as well as to establish a new treatment [Figure 1]. This review highlights about the emergence of clinical research along with speculated advantages with the application of bioinformatics tools and software. Information has been loaded on single platform that can be of use to researchers in different fields of medicine.
Figure 1.
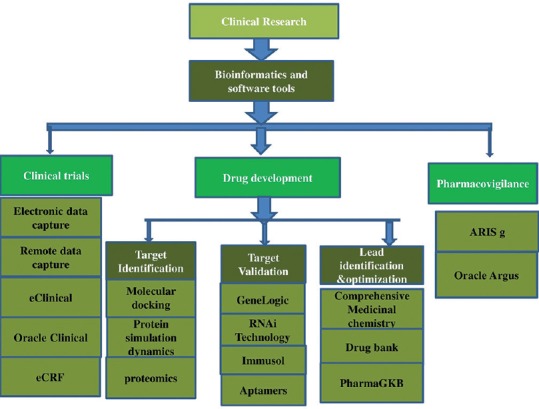
Illustrating the role of bioinformatics and software tools in clinical research
ROLE IN DRUG DEVELOPMENT
Major pharmaceutical companies have established high throughput screening facilities and have invested in automation to screen large compound libraries. Advances in bioinformatics have enabled genome-wide analysis for a broad range of research fields. Bioinformatics technology allows researchers to analyze the tetra bytes of data produced by the Human Genome Project. Gene sequence databases, gene expression databases, protein sequence databases, and related analysis tools all help to determine whether and how a particular molecule is directly involved in a disease process and in turn helps find new and better drug targets.[2] A successful and reliable drug design process could reduce the time and cost of developing useful pharmacological agents. Computational methods are used for the prediction of drug-likeness which means the identification and elimination of candidate molecules that are unlikely to survive the later stages of discovery and development. Drug-likeness could be predicted by genetic algorithm and neural network-based approaches.[3] The classical progression of the pharmaceutical discovery process goes from drug target to lead compound and finally to drug [Figure 2]. The ability to discover novel therapeutic targets for further research is the first critical step in this process. It is reported that approximately 483 drug targets account for nearly all drugs currently on the market (45% receptors, 28% enzymes, 5% ion channels, and 2% nuclear receptors).[4]
Figure 2.
Steps in new drug development
TARGET IDENTIFICATION
Before any potential new therapeutic is discovered, the disease under consideration needs to be understood, to unravel the causative underlying condition. The disease mechanism defines the possible cause or causes of a particular disorder, as well as the path or phenotype of a disease. The identification of new and clinically relevant molecular targets for drug intervention is of outstanding importance. Drug targets could be receptors, proteins, enzymes, deoxyribonucleic acid, or ribonucleic acid (RNA) that are pivotally involved in disease processes. These drug molecules physically attach to the drug target, triggering a cascade of intracellular biochemical reactions, and eventually a cellular reaction. The classical method of target identification included the use of animal and human cell lines that identifies key avenues involved: The enzyme that metabolizes the molecule (drug) and protein that act as receptors. Thus, ideal drug targets are specific and have a strong effect on the targeted biological pathway without disturbing the other pathways.[5]
Deploying genomics and proteomics, potential drug targets are identified by elucidating the interaction at the molecular level of a disease. These molecular interactions are assessed by a thorough analysis of sequences of a gene or a protein, three-dimensional (3D) protein structure and its interaction in various expression/metabolic pathways. With the aid of bioinformatics and computational tools, a network is constructed on the basis of endogenous metabolic, regulatory, and signaling networks with which the potential drug target interacts. The network generated can reveal the interaction relationship of a drug target under consideration. Based on their interaction relationship, the selection of the drug target could be narrowed to the most competent drug target to a great extent.
Target identification can be done by computational methods
Molecular docking
This technique predicts the structure of intermolecular complex found between two molecules and to find the best orientation of ligand which would form a complex with overall minimum energy. The 3D pose of the bound ligand can be visualized using different visualizing tools such as pymol, rasmol which could help in inference of the best fit of ligand. It gives the result as a score based on the docking algorithms generated due to various possible structural combinations.[6] During signal transduction, various biomolecules such as nucleic acids, proteins, and lipids play a critical role. Docking technique could predict the affinity between these biomolecules or receptors (drug target) and potential drug candidate. The foremost requirement of molecular docking is the structure of the protein or receptor of interest. The structure is determined by X-ray crystallography or nuclear magnetic resonance (NMR) spectroscopy. The scoring function generates the scores based on which drug candidate best fit to the target is identified.
Molecular dynamics simulation
This computer simulation method calculates the time-dependent behavior of a molecular system and provides the information about the structure or the microscopic interaction between the molecules. It is routinely used in determination of ligand docking, structure of lipid bilayer, and prediction of protein structure from polypeptide chain from the data generated by X-ray crystallography and NMR spectroscopy.[7] This method developed a vast platform for the modern computational protein folding.
Proteomics
Proteomics are studies on the structure and function of proteins. In proteomics, a large-scale comprehensive study is performed on modification/variation of protein abundance, their respective interacting partners and networks with an aim to elucidate cellular processes. Using bioinformatics tools for proteomics, the large data can be stored appropriately, and analysis can be performed rapidly. Bioinformatics tools and proteomic information are now also available for biomarker discovery, integrating biofluids, and tissue information. This new approach takes advantage of functional synergy between certain biofluids and tissues with the potential for clinically significant findings (for e.g., proteomics is highly useful in identification of candidate biomarkers (proteins in body fluids that are of value for diagnosis), identification of the bacterial antigens that are targeted by the immune response, and identification of possible immunohistochemistry markers of infectious or neoplastic diseases).
Target validation
With the technological advancement in drug discovery, the availability of potential target is not rate limiting rather the problem is in the selection of the most potent drug target. This imposes a challenge to the bioinformaticians to develop tools which could select a short number of genes or the drug targets having the strongest association with the disease out of a huge data.
However, approximately 324 drug targets have been identified for possessing clinical importance.[8] This indicates that the current pharmaceutical research and development are dependent and relies on a small pool of drug targets, despite the availability of numerous genome data on human as well as the pathogen.[9] A significant number of drugs fail at early preclinical stages due to the wrong target identification. Recently, a drug target prediction method, i.e., support vector machine has been developed[10] that is based on the physiochemical properties of protein sequences rather than homology annotation and protein 3D structure. This method can distinguish known drug targets from presumptive nondrug target with an accuracy of 84% in 10-fold cross-validation test.
Databases and computational tools used for target validation are:
Gene logic
Gene logic is a leading integrated genomics company providing comprehensive genomic reference databases and life science laboratory information management solutions. It makes use of its large database of tumor and normal gene expression to generate a short list of targets that are preferentially expressed in disease situations. The targets in this list can then be screened for functional target validation. The use of the database provides the investigator with a differential expression pattern of the target as well as its expression levels in different tissue and disease types. The database also allows the researcher to investigate the expression levels of other proteins known to be involved in certain pathways.
Ribonucleic interference technology
Intradigm (Rockville, MD, USA) makes use of RNA interference (RNAi) technology in which small interfering RNA (siRNA) oligos are used as gene inhibitors to degrade homologous mRNA with high specificity and efficacy. Intradigm focuses on an angiogenesis pathway shown to be important in cancer, inflammation, autoimmune, and other diseases. The potential to execute efficacious in vivo target validation with clinically viable siRNA delivery provides high-value information to understand the role of a particular gene or protein in the disease process, multiple genes of the same pathway, as well as the role of the pathway in the disease. This information is not only critical to the drug discovery process but also important for potential therapeutic siRNA development.
Immusol
Immusol (San Diego) recently launched a proprietary technology that allows fast and efficient in vivo target validation for efficacy and safety in multiple disease models using siRNA vectors. Immusol has inducible RNAi vectors that can be stably introduced into cultured tumor cells or cell lines that result in the expression of RNAi and can be used for target validation. The inducible vector can also be studied in a mouse xenograft tumor model.
Aptamers
Nascacell works with aptamers, the synthetic nucleic acid ligands for target validation as well as screening. Aptamers bind the active binding site to which the small molecule drug binds and inactivates the specific functional epitope on protein without disturbing the rest of the molecule. Hence, aptamers mimic the effect of a small drug molecule. In addition, aptamers can differentiate between various posttranslational modifications by inactivating the stable protein with physiological turnover rate.
Lead identification/optimization
Lead identification process starts with screening of compound libraries. Compounds which interact with target protein and modulate its activity are identified. Lead optimization is a complex process of drug discovery where the chemical structure of a confirmed hit is extensively optimized to produce a preclinical drug candidate.[11] The optimization of the promising lead candidate is done by the modifying the primary and secondary structure of the compound. This complex step can be enhanced by the recent innovation and progress in computation which scrutinize related compound to give a lead candidate. Another major hurdle is the accurate prediction of drug toxicity.
Software used for lead identification/optimization is:
Comprehensive medicinal chemistry
This database provides valuable information about the biochemical properties such as drug class, pKa, and Log P data of over 8,400 pharmaceutical molecules.
Drug bank
It is a database which associates the chemical and the pharmacological data with various drug targets and provides comprehensive information about the sequence, structure, and pathway information. It combines detailed drug (i.e., chemical, pharmacological, and pharmaceutical) data with comprehensive drug target (i.e., sequence, structure, and pathway) information. Its extensive drug and drug target data have enabled the discovery of a number of existing drugs to treat rare and newly identified illnesses.
PharmaGKB
It is a computational tool which predicts the response of a drug with respect to the variation in the human genetics. The PharmGKB is a pharmacogenomics knowledge resource that encompasses clinical information including dosing guidelines and drug labels, potentially clinically actionable gene-drug associations and genotype-phenotype relationships. PharmGKB collects, curates, and disseminates knowledge about the impact of human genetic variation on drug responses.
Quantitative structure activity relationship
Quantitative structure activity relationship is a technique employed to predict the properties and activities of a naïve of untested chemical compound which have structural homology and their physical and chemical properties are well established. The various software and databases used during drug discovery are summarized in Table 1.
Table 1.
Software and bioinformatics tools used in clinical research (drug discovery)
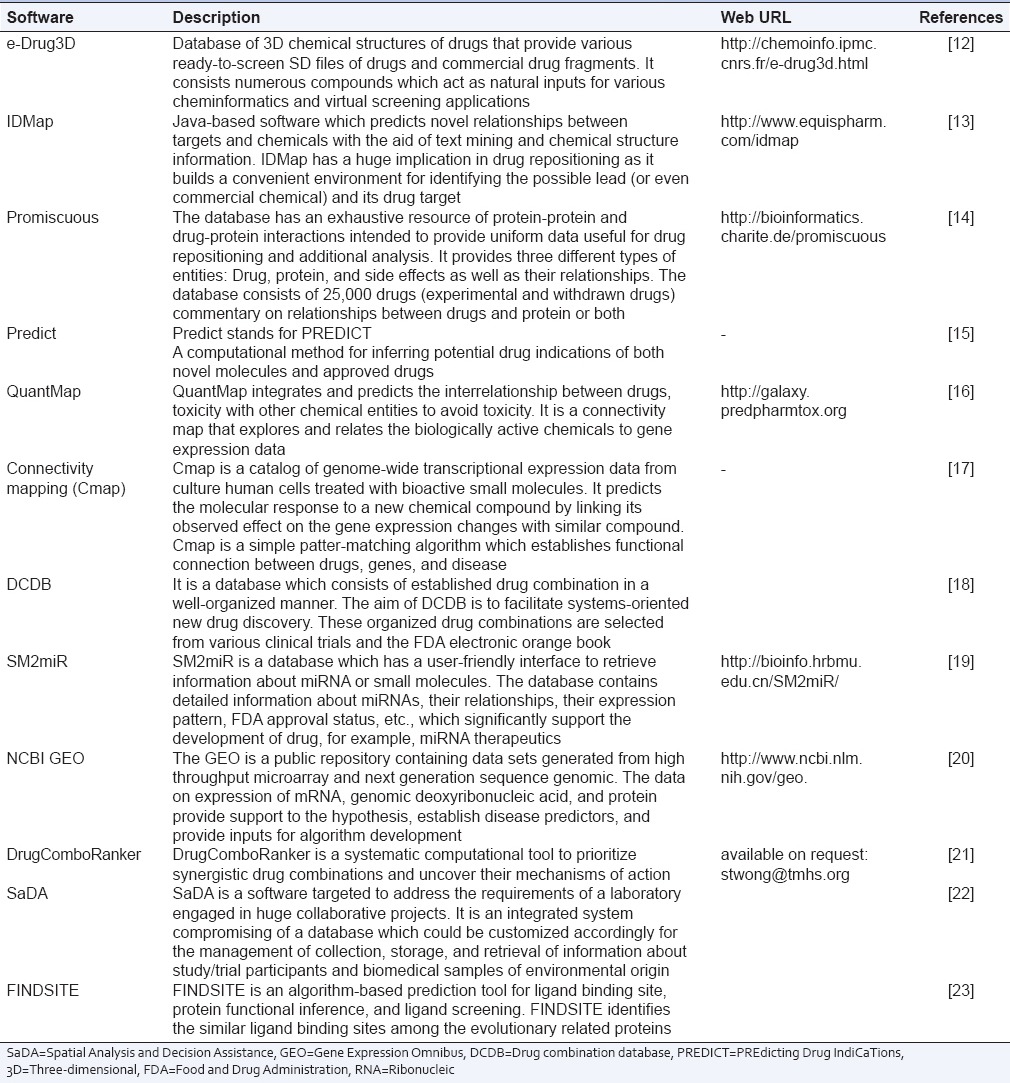
Software and bioinformatics tools in clinical trials
Besides, expanding exponentially in volume, clinical trials are becoming more complex too. In addition, the regulatory authorities demand more safety monitoring data that must be collected, compiled, and analyzed. The increasing accessibility of computers and advancement in information technology has significantly improved the clinical trial practices. Hence, there is a huge demand of computational tools in the clinical trials. In addition, technological developments have to address the need of Big Data Management (BDM) on the aspects of acquisition, storage, distribution, and analysis of data. BDM technologies are used to solve the current data storage issues and support specialized tools and facilities that can be used for normalizing and harmonizing data, used with advanced analytics. With BDM, the medical records and follow-up data can be more efficiently stored and extracted. The enormous variety of data may be structured, unstructured, and semi-structured is a dimension that makes health-care data both interesting and challenging. The unstructured data are the office medical records, handwritten notes of nurse and doctor, hospital admission and discharge records, paper prescriptions, radiograph films, magnetic resonance imaging, computed tomography, and other images. Structured data are data which can be easily stored, queried, recalled, analyzed, and manipulated by machine (although humans may not so easily read or interpret them). The structured data in electronic medical records and electronic health records include familiar input record fields such as patient name, date of birth, address, physician's name, hospital name and address, treatment reimbursement codes, and other information easily coded into and handled by automated databases. The software companies are investing more in developing tools for drug discovery. Now, various clinical data management tools are available in the market. Besides, electronic data capture (EDC), eCRF, etc., various clinical data management tools and software available in the market are clinical conductor CTMS (by Bio-Optronics), Clindex, Ascend (by Biopharm). These clinical trials are referred as electronic clinical trial when the planning, data collections, data accession, exchange, and archival for the execution, management, analysis, and reporting of the trial is performed electronically. Following are the software available for the BDM and address the aspects of acquisition, storage, distribution, and analysis.
Predixion Software uses cloud-based predictive analytic software to explain patterns in hospital datasets
Health fidelity is using natural language processing to turn unstructured data (e.g., narrative medical records) into structured data suitable for computer management, to address needs in revenue cycle management, compliance, and analytics
Informatica Corporation (NASDAQ: INfA) is the world's leading independent provider of data integration software. The Informatica Platform provides all the capabilities; the pharmaceutical industry needs to ensure that it can integrate and manage ever-growing volumes of data while using that data to innovate faster and achieve optimal results
MIM cloud: It makes data collection and distribution easy. Data are stored securely and can be accessed from anywhere on the internet.
Data management in clinical research
Clinical Data Management (CDM) is an integral part of clinical research as it efficiently gathers the data of trial subjects at the investigator site and ensures the validity, quality, and integrity of the same. Thus, the data stored in the database is reliable, of high quality, and statistically sound. Implementation of CDM to the clinical trial process significantly makes the drug development process cost-efficient and remarkably reduces the time required from the laboratory to the market. Various procedures including database design, data entry, data validation, and the other crucial procedures including the CRF designing and data locking are evaluated for quality at regular intervals during the trials. At present, there is a huge demand of CDMS to make the clinical trial efficient, for compliance with regulatory requirement and to lead ahead in the pharmaceutical competitive market.
Few of the currently available off-the-shelf software for clinical trials are:
e-Clinical: Innovative e-Clinical technologies are now becoming essential to make clinical data acquisition, aggregation, analysis, and decision-making for the new product. Two companies e-Clinical solutions and e-Clinical works are leader in clinical solutions. The eClinical application (e.g. elluminate by e-Clinical solutions) provides a cost cutting, broad applicability with functionally rich solution to manage clinical trial, support EDC all integrated into one system. The workflows developed in e-Clinical enables clinical operations to effectively plan each stage of a trial
Oracle clinical and oracle remote data capture: It is also known as relational database management system. It is used for managing database design and data acquisition for clinical study. This allows objects to be reused for multiple studies, saves time in study setup, and ensures that there are standardization and consistency of data collection and reporting. Oracle clinical can be customized to contain views that allow the data to be browsed. System generated error messages are programed to conduct data validation. The Oracle clinical application allows electronic data to be created, modified, maintained, and transmitted without compromising the authenticity, integrity, and confidentiality of data
Electronic case report form: Electronic case report form is an electronic tool replica of paper CRF where the clinical trial subject data are captured in an electronic format. The clinical and nonclinical data of the subject (including medical procedures) are gathered directly into the interface of central clinical database, thereby accelerating the data transmission to the sponsor. In multicentric trial, the importance of eCRF greatly increases as the data managers have continuous insight of data collection at one point. Thus, making data collection more efficient which significantly contribute to the efficacy of the whole clinical trial.
Importance of database in clinical research
Databases are created in clinical research for recording and propagating scientific information. The investigators are exposed to eCRF which is the desired format in which is investigator captures the data.[24] Database manages information and is more than repositories as it allows:
Efficient data collection at one point
Rapid data output in desired format
Easy but restricted accessibility
Audit trail function
Ensure data completion, integrity, and quality
Easy archival.
Software and bioinformatics tools in pharmacovigilance
Drug safety software used by pharmaceutical companies
ARISg
It is leading platform for both pharmacovigilance and clinical safety system. This application provides an efficient and end-to-end requirement for managing adverse event reporting in compliance with regulatory requirements and Food and Drug Administration 21CFR part 11. ARISg provides an integrated system for pharmacovigilance and risk management, thereby enabling pharmaceutical companies to monitor and evaluate their products for safety risk.
Argus
Oracle Argus is software for pharmacovigilance application which enables pharmaceutical companies to infer fast and better safety decisions, optimize global compliance, and with integrate risk management system. Argus provides customizable end-to-end safety process with automated case processing, periodic reporting, E2B intake and submission, detailed analytics, and safety operations integrated into a single system.
CONCLUSION
Clinical trial is the key link between advances in medical research technology and improved health care. It is an integral part of medical health research dedicated to elucidate human disease, its prevention, treatment, and promoting health. Clinical trial of a novel drug candidate is increasingly highly complex, time–consuming, and very expensive process. The pharmaceutical marketplace is very competitive, and the demand for rapid access to approval of new drug is high. Hence, pharmaceutical companies face huge pressure to increase the efficiency and efficacy of the drug discovery and development. The technology initiative is considered as the only way to achieve this goal.
With the advent of electronic clinical trials and computer-aided drug design research, there was a revolution in the drug discovery and development processes. The software companies developed various tools for analysis of genomic, sequence analysis, genetic algorithms, phylogenetic inference, genome database organization and mining, optical computation and holographic memory, pattern recognition, and image analysis. In addition, various stages of clinical trials such as target identification, target validation, randomization, data collection, data integration; trial management and pharmacovigilance became streamlined, efficient, and manageable. The adoption of the technologies by the pharmaceutical companies and regulators not only improved the efficiency but also the evaluation of clinical data (i.e., the transforming trial data in useable knowledge) and data security significantly improved while the overall cost of clinical trial reduced. Speculated advantages of bioinformatics in clinical research are e stablishing and using huge data strategy for clinical trials, leveraging the new techniques to provide the patient-centric approach in trials, bringing evolution in existing processes and systems with new techniques, provide help in case studies from existing data sources for advanced trials, easier data sharing, and would combat data privacy issues [Figure 3]. Thus, the drive to innovate has prompted researchers and the regulators to explore novel and more complicated ways to investigate promising new products with the aid of bioinformatics and software tools yielding trial design that are faster, more flexible, and more targeted. Looking forward, we can only anticipate that the future will bring more rapid technological changes for the new level of drug discovery that would have never been achieved through traditional review of the raw data.
Figure 3.
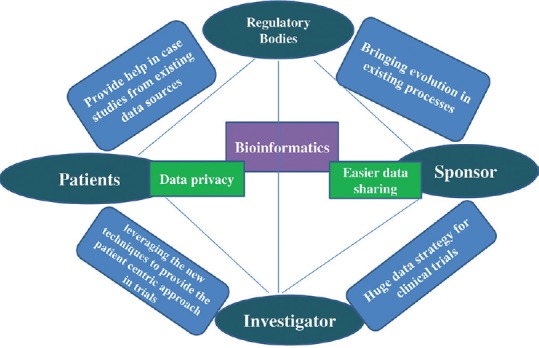
Speculated advantages of bioinformatics in clinical research
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Zerhouni EA. Clinical research at a crossroads: The NIH roadmap. J Investig Med. 2006;54:171–3. doi: 10.2310/6650.2006.X0016. [DOI] [PubMed] [Google Scholar]
- 2.DiMasi JA, Hansen RW, Grabowski HG. The price of innovation: New estimates of drug development costs. J Health Econ. 2003;22:151–85. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- 3.Clark DE, Pickett SD. Computational methods for the prediction of drug-likeness. Drug Discov Today. 2000;5:49–58. doi: 10.1016/s1359-6446(99)01451-8. [DOI] [PubMed] [Google Scholar]
- 4.Drews J. Genomic sciences and the medicine of tomorrow. Nat Biotechnol. 1996;14:1516–8. doi: 10.1038/nbt1196-1516. [DOI] [PubMed] [Google Scholar]
- 5.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–55. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 6.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat Rev Drug Discov. 2004;3:935–49. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 7.Levitt M, Warshel A. Computer simulation of protein folding. Nature. 1975;253:694–8. doi: 10.1038/253694a0. [DOI] [PubMed] [Google Scholar]
- 8.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–6. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 9.Li Q, Lai L. Prediction of potential drug targets based on simple sequence properties. BMC Bioinformatics. 2007;8:353. doi: 10.1186/1471-2105-8-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang JT, Liu W, Tang H, Xie H. Screening drug target proteins based on sequence information. J Biomed Inform. 2014;49:269–74. doi: 10.1016/j.jbi.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Shaikh SA, Jain T, Sandhu G, Latha N, Jayaram B. From drug target to leads – Sketching a physicochemical pathway for lead molecule design in silico. Curr Pharm Des. 2007;13:3454–70. doi: 10.2174/138161207782794220. [DOI] [PubMed] [Google Scholar]
- 12.Pihan E, Colliandre L, Guichou JF, Douguet D. e-Drug3D: 3D structure collections dedicated to drug repurposing and fragment-based drug design. Struct Bioinform. 2012;28:1540–1. doi: 10.1093/bioinformatics/bts186. [DOI] [PubMed] [Google Scholar]
- 13.Ha S, Seo YJ, Kwon MS, Chang BH, Han CK, Yoon JH. ID Map: Facilitating the detection of potential leads with therapeutic targets. Bioinformatics. 2008;24:1413–5. doi: 10.1093/bioinformatics/btn138. [DOI] [PubMed] [Google Scholar]
- 14.von Eichborn J, Murgueitio MS, Dunkel M, Koerner S, Bourne PE, Preissner R. Promiscuous: A database for network-based drug-repositioning. Nucleic Acids Res. 2011;39:D1060–6. doi: 10.1093/nar/gkq1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottlieb A, Stein GY, Ruppin E, Sharan R. PREDICT: A method for inferring novel drug indications with application to personalized medicine. Mol Syst Biol. 2011;7:496. doi: 10.1038/msb.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaal W, Hammerling U, Gustafsson MG, Spjuth O. Automated quant map for rapid quantitative molecular network topology analysis. Syst Biol. 2013;29:2369–70. doi: 10.1093/bioinformatics/btt390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caiment F, Tsamou M, Jennen D, Kleinjans J. Assessing compound carcinogenicity in vitro using connectivity mapping. Carcinogenesis. 2014;35:201–7. doi: 10.1093/carcin/bgt278. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Hu B, Fu C, Chen X. DCDB: Drug combination database. Databases Ontol. 2010;26:587–8. doi: 10.1093/bioinformatics/btp697. [DOI] [PubMed] [Google Scholar]
- 19.Liu X, Wang S, Meng F, Wang J, Zhang Y, Dai E, et al. SM2miR: A database of the experimentally validated small molecules’ effects on microRNA expression. Databases Ontol. 2013;29:409–11. doi: 10.1093/bioinformatics/bts698. [DOI] [PubMed] [Google Scholar]
- 20.Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, et al. NCBI GEO: Mining millions of expression profiles – Database and tools. Nucleic Acids Res. 2005;33:D562–6. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang L, Li F, Sheng J, Xia X, Ma J, Zhan M, et al. Drug combo ranker: Drug combination discovery based on target network analysis. Bioinformatics. 2014;30:228–36. doi: 10.1093/bioinformatics/btu278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh KS, Thual D, Spurio R, Cannata N. SaDA: From sampling to data analysis-an extensible open source infrastructure for rapid, robust and automated management and analysis of modern ecological high-throughput microarray data. Int J Environ Res Public Health. 2015;12:6352–66. doi: 10.3390/ijerph120606352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brylinski M, Skolnick J. A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. Proc Natl Acad Sci U S A. 2008;105:129–34. doi: 10.1073/pnas.0707684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang J, Zhu X. Bioinformatics databases: Intellectual property protection strategy. J Intellect Property Rights. 2010;15:447–54. [Google Scholar]



