ABSTRACT
Extracytoplasmic function (ECF) σ factors are the largest and the most diverse group of alternative σ factors, but their mechanisms of transcription are poorly studied. This subfamily is considered to exhibit a rigid promoter structure and an absence of mixing and matching; both −35 and −10 elements are considered necessary for initiating transcription. This paradigm, however, is based on very limited data, which bias the analysis of diverse ECF σ subgroups. Here we investigate DNA and protein recognition motifs involved in ECF σ factor transcription by a computational analysis of canonical ECF subfamily members, much less studied ECF σ subgroups, and the group outliers, obtained from recently sequenced bacteriophages. The analysis identifies an extended −10 element in promoters for phage ECF σ factors; a comparison with bacterial σ factors points to a putative 6-amino-acid motif just C-terminal of domain σ2, which is responsible for the interaction with the identified extension of the −10 element. Interestingly, a similar protein motif is found C-terminal of domain σ2 in canonical ECF σ factors, at a position where it is expected to interact with a conserved motif further upstream of the −10 element. Moreover, the phiEco32 ECF σ factor lacks a recognizable −35 element and σ4 domain, which we identify in a homologous phage, 7-11, indicating that the extended −10 element can compensate for the lack of −35 element interactions. Overall, the results reveal greater flexibility in promoter recognition by ECF σ factors than previously recognized and raise the possibility that mixing and matching also apply to this group, a notion that remains to be biochemically tested.
IMPORTANCE ECF σ factors are the most numerous group of alternative σ factors but have been little studied. Their promoter recognition mechanisms are obscured by the large diversity within the ECF σ factor group and the limited similarity with the well-studied housekeeping σ factors. Here we extensively compare bacterial and bacteriophage ECF σ factors and their promoters in order to infer DNA and protein recognition motifs involved in transcription initiation. We predict a more flexible promoter structure than is recognized by the current paradigm, which assumes rigidness, and propose that ECF σ promoter elements may complement (mix and match with) each other's strengths. These results warrant the refocusing of research efforts from the well-studied housekeeping σ factors toward the physiologically highly important, but insufficiently understood, alternative σ factors.
INTRODUCTION
Transcription initiation is a major regulatory checkpoint, which in prokaryotes is exhibited, in part, through interactions of RNA polymerase holoenzyme (RNAP) σ factor with DNA promoter elements (1). σ70 is a major σ factor family, encompassing most of the known σ factors (2), which share the same general promoter structure (1, 3). This structure is characterized by two canonical promoter elements, the −35 and −10 elements (where the coordinates correspond to the transcription start site). The binding of a σ factor in the context of RNAP leads to promoter melting, i.e., the transcription bubble is initiated within the −10 element (1, 4, 5); consequently, the upstream part of the −10 element (often called the extended −10 element) and the −35 element interact with the σ factor in the double-stranded DNA (dsDNA) form, while the downstream part of the −10 element (the short −10 element) is involved in single-stranded DNA (ssDNA) interactions (1).
Despite the general similarity of promoter structures, σ70 family members show significant differences in the complexity of their protein structures, and the family is accordingly divided into four different subfamilies (groups I to IV). Group I is the most complex and abundant of these subfamilies. Its members direct the expression of the majority of cellular genes (the housekeeping genes). Consequently, this group is essential for cell survival under normal conditions (1, 2). Group II is closely related to group I in terms of structural organization but is not strictly required for cell survival (1, 2). The remaining two groups respond to specific developmental/external stimuli and direct focused responses. Group III is implicated in the biosynthesis of flagella, sporulation, and the heat shock response (1, 2, 6).
Group IV σ factors, also named extracytoplasmic function (ECF) σ factors, respond to stimuli from outside the cell, often transcribe their own genes, and are implicated mainly in coping with stress and the uptake of nutrients. This group is the most abundant, and the most heterogeneous, among alternative σ factors, and is accordingly classified into more than 40 different subgroups (7, 8). Members of group IV are characterized by two domains (σ2 and σ4), which recognize −10 and −35 promoter elements, respectively, in contrast to other σ factors, which have three or four domains (1). Consequently, ECF σs are the most highly divergent members of the σ70 family, with only limited similarity to other σ70 factors.
Despite the established abundance and diversity within the σ70 protein family at the structural and functional levels, the mechanism of interaction with promoter sequences is well studied only for members of group I (the housekeeping σ factors) (1, 3, 5). This group recognizes variable promoter elements, which exhibit the mix-and-match mode of action (9). That is, it has been noticed that different promoter elements that interact with σ factors in the dsDNA form may complement each other to achieve a sufficient level of dsDNA binding strength. As an extreme example of this mechanism, it has been found that in group I, the extended −10 element can compensate for the absence of the −35 element (9–12).
While flexible promoter structure and mixing and matching are characteristic of interactions with housekeeping σ factors, an opposite paradigm for promoter recognition is presumed for ECF σ factors. It is thought that this group has a requirement for a rigid promoter structure, with highly conserved promoter elements, which do not mix and match (1). The extent to which these assumptions are valid is unclear, however, since only a small subset of ECF σ factors have experimentally established promoter recognition specificity. This limited data set may lead to a convergence toward possibly unjustified conclusions, particularly regarding the requirement of ECF σs for a rigid promoter structure. In fact, even for canonical ECF σ members (Escherichia coli σE and Bacillus subtilis σW), for which a reasonably large number of promoters are available, the possibility of mixing and matching has not been investigated, in contrast to the detailed qualitative and quantitative studies carried out for E. coli RpoD (9, 13, 14).
Furthermore, from Fig. 1, one can see that the promoters recognized by the σ70 family share the same general organization, in particular with respect to interactions with ssDNA and dsDNA. This is likely due to the same basic biophysical mechanism of transcription initiation within the family. In particular, the σ factors of the family should first bind to dsDNA, through interactions with the −35 element and the upstream part (and extension) of the −10 element, and subsequently open the two DNA strands, through interactions with the ssDNA of the downstream part of the −10 element (1, 4). This might indicate that promoters for ECF σ factors can adopt a less rigid structure than currently assumed. That is, different promoter elements involved in dsDNA and ssDNA interactions may complement—and, in more extreme cases, substitute for—each other, to achieve sufficiently strong kinetic parameters (i.e., binding affinity and transcription activity), which is the basic idea behind the mix-and-match model of promoter recognition (9, 13). Investigating the possibility of a less rigid promoter structure for the ECF σ group is the main motivation behind this work.
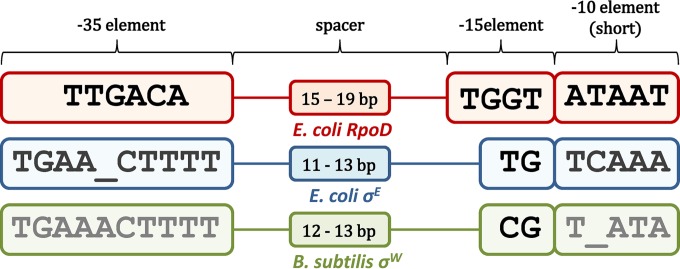
FIG 1.
Structure of promoters recognized by σ70 factors. The general structure of promoters recognized by the three well-studied representatives of the σ70 protein family—E. coli RpoD, E. coli σE, and B. subtilis σW—is shown. The indicated promoter elements correspond to the following consensus sequences: the −35 element and −15 element (9), which interact with the σ factor in the dsDNA form, and the short −10 element, which is involved in ssDNA interactions. The −15 element consensus is commonly shown as −15TGnT−12 in E. coli RpoD but is here shown as −15TGGT−12, since in reference 13, it was demonstrated that −13G is significantly conserved (more than −15T).
Consequently, we investigated DNA and protein recognition motifs involved in transcription by ECF σ factors, through a bioinformatics analysis of bacterial and bacteriophage ECF σ factors and their promoters. To avoid inferring conclusions from a limited data set, we looked to a substantially different system, an opportunity provided by bacteriophages. Transcription of bacteriophage late genes is often exhibited by the phage-encoded σ factors; bacteriophage phiEco32, whose gene expression strategy was recently studied in detail, provides an example of a bacteriophage-encoded σ factor belonging to an ECF subfamily (15). Moreover, the recently sequenced bacteriophage 7-11 (16), a close relative of phiEco32, is also of great significance. The 7-11 ECF σ factor exhibits homology with the phiEco32 ECF σ factor but is more closely related to bacterial ECF σ factors; consequently, its specificity can be compared with those of both phiEco32 and bacterial ECF σ factors.
Therefore, we investigated the specificities (at the levels of both DNA and protein sequence) of the characterized bacteriophage σ factors. We then compared how these σ factors relate to the classified bacterial ECF σ subgroups. This helped to identify the σ factor–DNA interactions associated with these subgroups, which may point to greater flexibility in promoter recognition by ECF σ factors. Overall, the work provides previously unavailable information about the similarities among ECF σ factors and their promoter elements. This may, in turn, provide a starting point for experiments testing for unifying mechanisms of promoter recognition within the σ70 family, e.g., investigating whether mixing and matching is exhibited in the entire family.
MATERIALS AND METHODS
Sequence data sets. (i) DNA sequences.
The DNA sequence data sets used in the analysis were composed of promoters recognized by the following σ factors: E. coli σE, B. subtilis σW, the E. coli RpoD σ factor, bacteriophage-encoded σ factors from 7-11 and phiEco32, and bacterial ECF σ factors from subgroups 28 and 32.
The σE promoter data set is composed of 60 experimentally verified promoters and contains aligned −35 and −10 elements (17); information on the promoters that are active (and inactive) under in vitro conditions is also given in reference 17.
σW promoters were retrieved from DBTBS, a database of Bacillus subtilis promoters and σ factors (18). This data set is composed of 34 experimentally determined promoters. Of these, one promoter sequence (upstream of ywbLMN) was not used in further analysis, due to difficulty in aligning its −35 element (at least five mismatches from the consensus).
RpoD promoters contain 322 sequences with experimentally determined transcription start sites that were aligned de novo (13) with the Gibbs Motif Sampler.
Three phage 7-11 promoters have been inferred through genome sequence analysis (19). Six phage phiEco32 promoters have been determined experimentally (15). These data sets with phage promoters contain only the aligned −10 elements.
For the ECF28 subgroup, we analyzed two DNA sequence data sets, which do not contain already aligned promoters. The first data set was used for an unsupervised search of ECF28 promoters and is composed of 50 bp upstream of the genes encoding selected representatives (with protein sequences that differ significantly from each other) of the ECF28 subgroup (all GenInfo Identifier [GI] numbers for the sequences in this section are provided in Table S1 in the supplemental material). The second data set was used for a supervised search of ECF28 promoters, based on the weight matrices (20), for which we extended the previous data set to include the entire length of the upstream intergenic regions.
For the ECF32 subgroup, we analyzed those promoters recognized by the subgroup members with the conserved protein motif, for which the corresponding promoter sequences can be inferred. These promoter sequences were either obtained from reference 7 or identified by us through MLSA (multiple local sequence alignment), as described below.
(ii) Protein sequences.
We analyzed a protein sequence data set containing bacterial ECF σ factors classified into 43 different subgroups (7). Among these, we analyzed the following subgroups in detail: ECF01, which contains σW; ECF02 (σE); ECF28, which is similar to the phage 7-11 ECF σ factor; and ECF32.
We also analyzed the protein sequences of the σ factors encoded by bacteriophages 7-11 and phiEco32, which were retrieved from GenBank. The protein sequences classified as RpoD σ factors, which consist of nearly 44,000 σ factors, were also retrieved from GenBank.
DNA motif alignment. (i) Multiple sequence alignments.
For multiple local DNA sequence alignments, we used a Gibbs search (Gibbs Motif Sampler) (21), as a standard approach for identifying short motifs that are conserved in a set of DNA sequences. The Gibbs Motif Sampler was used in both the Site Sampler and Motif Sampler modes.
In the Site Sampler mode, the algorithm finds exactly one motif in each query sequence; this provides an estimate of the presence of the conserved motif in the entire set of sequences analyzed. On the other hand, the Motif Sampler mode enables identification of the motif, which is not contained in all query sequences; note that in the last cycle of the search, the algorithm adds/subtracts motifs from the sequences analyzed based on their impact on the informational content of the alignment (21). Consequently, the total number of motifs detected per sequence in the final alignment can be either larger or smaller than 1.
In every Gibbs Motif Sampler analysis performed, only the direct strand was searched, the number of motifs was set to 1, and the estimated total number of sites was set to the number of query sequences. The motif length was set to several different values (to check for the robustness of the motif detected) in the analysis of the same query set, with the remaining parameters at their default values.
(ii) Sequence logos.
We used a MATLAB function (MathWorks) and enoLOGOS (22) to generate DNA sequence logos. enoLOGOS and MATLAB were used with their default values, with GC content set to that of the species to which the sequences analyzed corresponded.
The sequence logos were generated through enoLOGOS for phage (7-11 and phiEco32), ECF σE and σW, and RpoD −10 elements, which were defined in the data sets described above. The alignments in the data sets for σE and σW promoters were first checked through the Gibbs Motif Sampler (whose output matched the alignments in the original data sets), and then these alignments were used for generating −10 element sequence logos, with the additional 3 bp upstream of the −10 element. Sequence logos were also generated for the −35 elements found upstream of two 7-11 promoters. The −35 element found in the third sequence was omitted, since it is notably degenerated and therefore significantly obscures the alignment. In the analysis of noncanonical σ factor–DNA interactions, the sequence logos were generated by MATLAB, which includes σE promoters that are inactive in vitro, σW promoters, and selected ECF32 σ factor promoters. For σW and selected σE promoters, we aligned the respective −10 elements, along with the spacer sequences extending to the downstream edge of the −35 element, while for ECF32, the sequence logos correspond to the entire promoter sequences.
(iii) PhiEco32 upstream promoter sequence conservation.
To check whether the sequences upstream of all the phiEco32 promoters (as defined in our data set) share a conserved motif, the analysis was performed using Gibbs Motif Sampler in the Site Sampler mode, as described above (see “Mulitiple sequence alignments”). The motif length was set to 8, 7 or 6 bp to check the robustness of the detected motif.
(iv) ECF28 promoter identification.
An unsupervised search was our first approach for identifying ECF28 promoters; the DNA data set, described above, was analyzed by both the Site Sampler and the Motif Sampler (see above), with motif lengths set to 7, 6 or 5 bp—again, to verify the robustness of predicted motifs. As an alternative unsupervised strategy, we aligned the same set of DNA sequences pairwise through BLAST (Blast2seq) (23): each of the query sequences was aligned against the remaining query sequences, and the motif length was set to the lowest value (7 bp).
Alternatively, ECF28 promoters were searched by a supervised strategy (24), which is based on using the σW −35 element weight matrix. The weight matrix elements are calculated as reported previously (24, 25). We used this weight matrix for searching the sequence data set (see “DNA sequences” above); the highest-scoring motifs were identified in all the query sequences, and 35-bp segments downstream of these motifs were extracted. The extracted segments were then analyzed with the Gibbs Motif Sampler (see above) to search for ECF28 promoter elements.
(v) ECF32 promoter identification.
For ECF32 subgroup members that have the conserved protein motif, but for which promoter sequences are not available in reference 7, we identified promoters by MLSA. For MLSA, we used Gibbs Motif Sampler in the Motif Sampler mode, with the motif lengths set to 4, 5, and 6 bp; segments comprising 100 bp upstream of the translation start sites of ECF σ factor genes were aligned.
Protein sequence alignments.
We used ClustalW (26) for multiple alignments of protein sequences, BLAST for pairwise alignments, and CD-Search (27) for domain identification. ClustalW was used with default parameters; BLAST was used with the blastp version and the option for aligning 2 or more sequences (the remaining parameters were at default values); and CD-Search was also used with default parameters, except for the E value threshold, which was lowered in a stepwise fashion (to the final value of 10), with the purpose of predicting the σ4 domain in the phiEco32 σ factor protein sequence.
For the assessment of similarities between phage (7-11 and phiEco32)-encoded ECF σ factors, bacterial ECF members (σE and σW), and RpoD σ factors, we analyzed their sequences with CD-Search and aligned the predicted σ2 and σ4 domains pairwise with BLAST; for phiEco32 σ, the entire ECF σ domain was used in the analysis.
Further, for the assessment of similarities between the phage 7-11 σ factor and all the representatives of ECF and RpoD subfamilies (note the data sets defined above), we aligned them pairwise by BLAST, as described above. When the phage 7-11 σ factor was compared with ECF σ factors, the analysis was conducted separately against all 43 different ECF subgroups.
In addition to pairwise sequence alignment, the phage 7-11 σ factor was compared with selected members of ECF subgroups ECF01 and ECF28 through multiple-sequence alignment by ClustalW. For comparison with ECF01, the data set was narrowed to the members most closely related to the phage 7-11 σ factor (i.e., those with the best pairwise alignment E values). On the other hand, for comparison with ECF28, the data set was narrowed so as to contain one member from each bacterial genus that appears in the ECF28 subgroup. Multiple-sequence alignment was also carried out for the entire ECF28 subgroup; further, sequence logos were generated (through enoLOGOS) from segments of ECF28 σ factors that span the region from the C terminus of the σ2 domain to the appearance of the first gap in the multiple alignment (which almost coincides with the beginning of the σ4 domain). Along the same lines (by ClustalW), we analyzed the selected ECF02 members and ECF32 σ subgroups, and for the sequences containing the conserved protein motif in both subgroups, sequence logos were generated through enoLOGOS; flanking sequences were added to these motifs in order to observe their conservation.
RESULTS
Comparing binding specificities.
Until now, all the available information on how σ factors from ECF subfamily operate was obtained through the analysis of bacterial ECF members. Therefore, bacteriophages 7-11 and phiEco32, which encode highly similar ECF σ factors (15, 19), are suitable candidates for ECF σ outliers. The specificity of phiEco32 σ was experimentally established, while the specificity for its relative (phage 7-11) was determined by genome sequence analysis and was confirmed through homology with the phiEco32 promoters (at both the sequence and promoter layout levels) (15, 19). Therefore, comparison of the promoters recognized by the outlier phage ECF σ factors with those for the well-characterized ECF members (E. coli σE, B. subtilis σW) and a canonical group I member (E. coli RpoD) would allow one to (i) notice differences within the ECF σ factor subfamily and possibly modify the current paradigm of promoter conservation and rigidness and (ii) examine whether promoters recognized by σ70 members from distinct subfamilies are built on similar scaffolds, which could point to similarities in their mechanisms of promoter recognition (28).
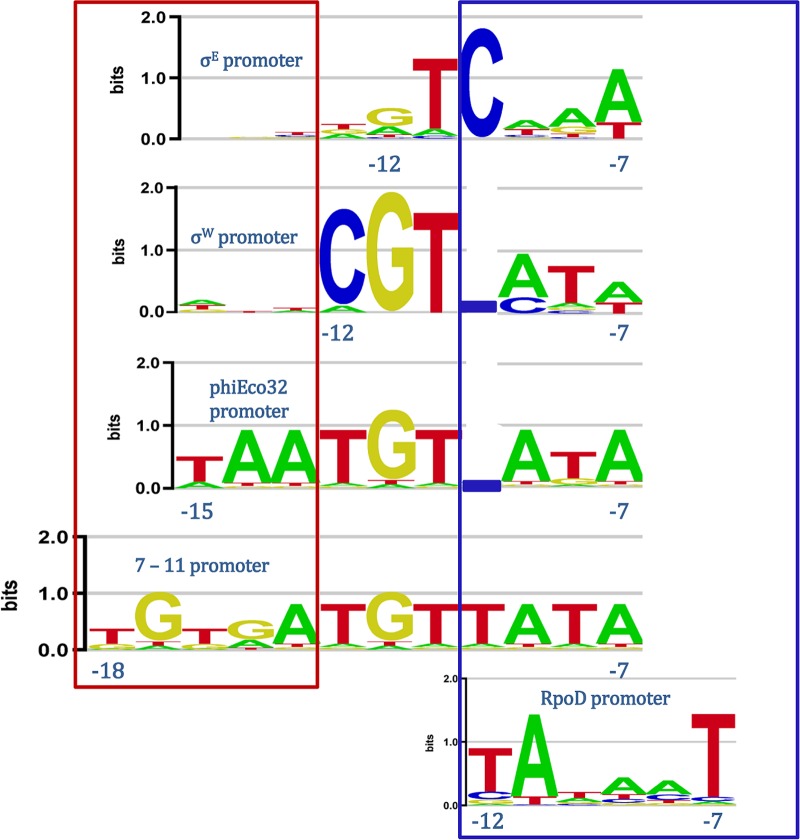
As can be seen from the alignment shown in Fig. 2, there are notable differences between the −10 elements of promoters recognized by bacterial and phage ECF σ factors. Specifically, there is a notable similarity among the downstream segments of all the aligned promoters; compare, e.g., the sequence logo of the σW promoter with those of the two phage promoters. On the other hand, the upstream parts of the −10 elements of the two phage promoters are substantially longer than those of the σW and σE promoters. Note that Fig. 2 clearly shows an absence of conservation in the sequences upstream of the −10 elements for σW and σE, in contrast to the upstream parts of the −10 elements for the phage σ factors (sequences boxed in red).
FIG 2.
Alignment of −10 elements in promoters recognized by σ70 factors. Shown is an alignment of the sequence logos for −10 elements recognized by bacterial ECF (σE and σW), phage ECF (phiEco32 and 7-11), and group I (E. coli RpoD) σ factors. For promoters recognized by σE and σW, we extended the −10 elements 3 bp upstream in order to observe an absence of conservation in this extension. The TATA box of group I (RpoD) promoters and the analogous sequences in ECF σ promoters are boxed in blue; the sequences in the upstream −10 element extension, found in phage promoters, and the equivalent positions in σE and σW promoters are boxed in red. The coordinates of the upstream and the downstream edges of the promoter elements are indicated relative to the transcription start site.
Furthermore, the extension of the phage −10 element also suggests an equivalence with RpoD (group I σ) promoter function, where such extension (the so-called −15 element [9, 13]) may compensate for the absence of σ factor interactions with a −35 element, this being a hallmark of the mixing and matching of promoter elements (9). As can be observed in Fig. 2, this notion is further supported by the presence of a TA-rich segment in the downstream part of the phage promoters, which is a well-known feature of RpoD −10 elements (5).
Consequently, we investigated the following questions. What is the organization of ECF phage promoters (particularly with respect to their −35 elements) with this peculiar −10 element structure? What protein sequence features of the bacteriophage σ factor enable the recognition of such −10 element extensions, and how do these features relate to the remaining ECF σ subgroups? Through this investigation, we inquired (i) if the classical qualitative trademark of mixing and matching in the housekeeping (group I) σ factors—i.e., compensation for the absence of a −35 element by the extension of the −10 element (9)—also appears for ECF σ factors and (ii) by what mechanism, at the level of protein-DNA interactions, this −10 element extension is exhibited. Moreover, we also explored whether additional interactions outside of the canonical ECF −35 and −10 elements are exhibited.
Comparing protein sequences.
We next investigated whether the established similarity between the −10 promoter elements shown in Fig. 2 is also observed in their respective protein sequences. We therefore started by examining relationships between the phage (7-11 and phiEco32) σ factor sequences and those of the well-studied σ70 family members (σW, σE, and RpoD). Since we are interested in the DNA sequence specificity of the σ factors, we first identified domains σ2 and σ4 in each of the protein sequences and then used these domains for comparison of the protein sequences of the σ factors for which −10 promoter elements are shown in Fig. 2 (see Materials and Methods, “Protein sequence alignments”). Note that domain σ4 of phiEco32 was not detected even when the search threshold was substantially lowered, so the entire phiEco32 ECF domain was used in the analysis. The absence of domain σ4 in phiEco32 in the context of its interactions with the −35 element is analyzed further below.
The results of the comparisons are summarized in Tables 1 and 2, where we see that they are consistent overall for the two domains. It is clear that the phage σ factors are indeed outliers, highly divergent from the E. coli RpoD σ factor and also distant from σE, which is a member of the ECF σ group. Only the alignment of σW with phage σ factors is statistically significant, in agreement with the higher level of similarity between the −10 promoter elements for phage (7-11 and phiEco32) σ factors and σW. Note that the 7-11 σ factor is more similar to σW than is its relative, phiEco32, a finding also supported by the absence of domain σ4 in the phiEco32 σ factor. One may consequently expect that the phage phiEco32 σ factor, as an extreme outlier, could provide an example of a qualitatively different regulatory paradigm of ECF σ factor functioning. Furthermore, the 7-11 σ factor, with its higher similarity to σW, can be compared with both the phiEco32 σ factor and bacterial ECF σ factors.
TABLE 1.
Domain 2 similaritiesa
| Domain 2 | E-value for similarity to domain 2 of: |
|||
|---|---|---|---|---|
| E. coli σ70 | Phage 7-11 σ | Phage phiEco32 σ | E. coli σE | |
| Phage 7-11 σ | 0.046 | |||
| Phage phiEco32 σ | No alignment | 2e−7 | ||
| E. coli σE | 4e−6 | 0.86 | 0.61 | |
| B. subtilis σW | 6e−4 | 1e−6 | 3e−5 | 2e−15 |
Pairwise alignment of σ2 domains predicted in protein sequences of phage ECF (7-11 and phiEco32), bacterial ECF (σE and σW), and group I (E. coli RpoD) σ factors. For the phiEco32 σ factor, the entire ECF domain was used in the analysis.
TABLE 2.
Domain 4 similaritiesa
| Domain 4 | E-value for similarity to domain 4 of: |
||
|---|---|---|---|
| E. coli σE | Phage 7-11 σ | E. coli σ70 | |
| B. subtilis σW | 2e−15 | 2e−4 | No alignment |
| E. coli σE | No alignment | 0.42 | |
| Phage 7-11 σ | No alignment | ||
Pairwise alignment of σ4 domains predicted in protein sequences of phage ECF (7-11 and phiEco32), bacterial ECF (σE and σW), and group I (E. coli RpoD) σ factors. For the phiEco32 σ factor, the entire ECF domain was used in the analysis.
Accordingly, we further investigated, by pairwise alignment of the 7-11 σ factor protein sequence with the representatives of the 43 ECF subgroups, which of these subgroups are most closely related to the phage σ factors (see Materials and Methods, “Protein sequence alignments”). We generally obtained statistically significant (sometimes highly statistically significant) alignments, where ECF28 emerged as the subgroup most closely related to the phage σ factors. For the well-studied ECF subgroups, the closest similarity was found between 7-11 σ and ECF01 (with B. subtilis σW). Furthermore, we also aligned the phage σ factors with the members of the RpoD (group I) subfamily (∼44,000 protein sequences annotated in GenBank) and obtained statistically insignificant alignments for a large majority of the sequences analyzed, in agreement with the results obtained by aligning the domains.
Therefore, there are often substantial differences at the protein sequence level between the σ factors, as exemplified by the clearly unrelated phage ECF and RpoD σ factors. On the other hand, the structures of the promoters recognized by σ70 factors exhibit the same general scaffold; note, in particular, the similarities in −10 element organization between the promoters for phage σ factors and RpoD, established in Fig. 2. Consequently, the similarities established at the level of promoter structure could indeed be due to a common mechanism of promoter recognition (such as mixing and matching) rather than to similarities in σ factor protein sequences.
Analyzing −35 promoter elements.
As discussed above, the promoters for 7-11 and phiEco32 σ factors have a notable extension of the upstream segment of the −10 element, which, in RpoD σ factors, is associated with complementing the strength of the −35 element (9). Therefore, our next goal was to analyze the sequences upstream of the phage −10 elements and to search for putative −35 elements (see Materials and Methods, “DNA motif alignment,” “Multiple sequence alignments,” and “phiEco32 upstream promoter sequence conservation”). We started with the sequences upstream of the predicted promoters for phage 7-11; note that the 7-11 σ factor protein sequence has a significant similarity to that of σW (see above), which has an established promoter specificity. In accordance with this notion, we identified the −35 elements of the promoters recognized by phage 7-11 σ factors, which display extensive similarity to those recognized by σW. This is illustrated in Fig. 3, where we show the alignment of the predicted 7-11 −35 element with the established σW −35 element.
FIG 3.
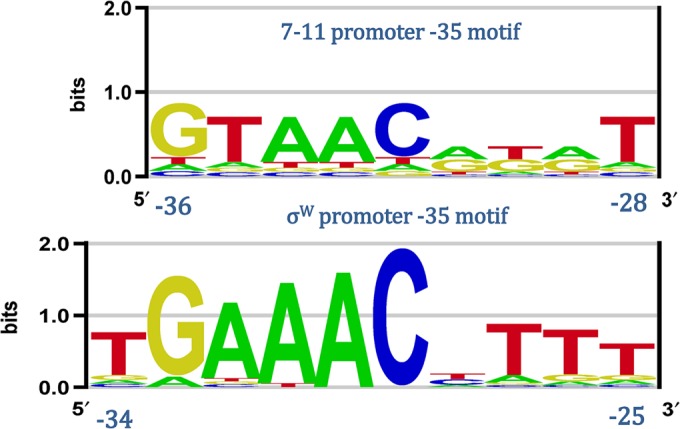
Alignment of the phage 7-11 and σW −35 elements. The upper sequence logo was generated from the motifs localized upstream of the phage 7-11 −10 elements; the lower sequence logo was generated from the aligned −35 promoter elements recognized by σW. The coordinates of the promoter elements relative to the transcription start site are indicated.
On the other hand, analysis of the phiEco32 upstream sequences failed to identify −35 elements. This finding is consistent with the absence of a recognizable domain σ4 (implicated in −35 element interactions) in the phiEco32 σ protein sequence. However, we identified a conserved motif (AAGACCT) in a minority of the upstream sequences—i.e., in two of the six sequences, with a total of three repeats – corresponding to the phage late promoters. We argue that the predicted motifs are putative binding sites of a transcription factor (possibly phage encoded), which may be responsible, in part, for the different regulatory patterns for phiEco32 middle and late genes. The implications of this finding for the ECF σ regulatory paradigm are further assessed in the Discussion.
The protein sequence features recognizing the −10 element extension.
We next investigated what protein sequence elements are responsible for recognizing the −10 element extension in the phage promoters. We reasoned that these elements should appear C-terminal of the predicted domain σ2, since this domain is implicated in −10 element recognition. Note that for the 7-11 σ factor, the domain σ2 predictions are based on the domain boundaries of σE and σW (the canonical ECF members). Since these factors do not recognize −10 elements with upstream extensions, the segment that is implicated in recognizing the −10 element extension is expected to be found C-terminal of the predicted domain σ2 boundary. Furthermore, if functional, this segment should be conserved in at least some bacterial ECF σ group members. Finally, these sequence elements should not be conserved in ECF σ factors that are closely related to σW (ECF01 subgroup), which have protein sequences similar to those of phage σ factors but do not have extended −10 regions. We then compared the sequences C-terminal of domain σ2 of phage 7-11 σ (i) with those of bacterial ECF σ factors, in particular with those of the ECF28 subgroup, which are, among bacterial ECF σ factors (see above) the closest to the bacteriophage 7-11 σ, and (ii) with those of ECF01 subgroup representatives, whose canonical member is σW (see Materials and Methods, “Protein sequence alignments”).
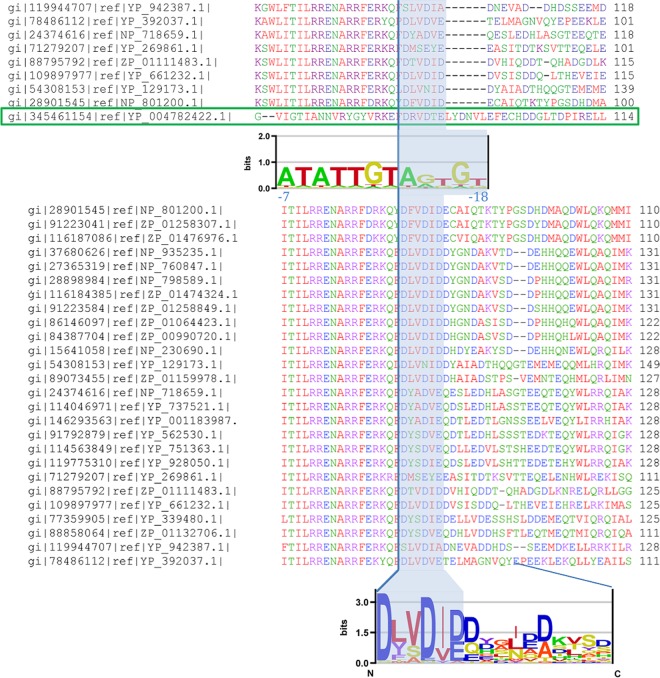
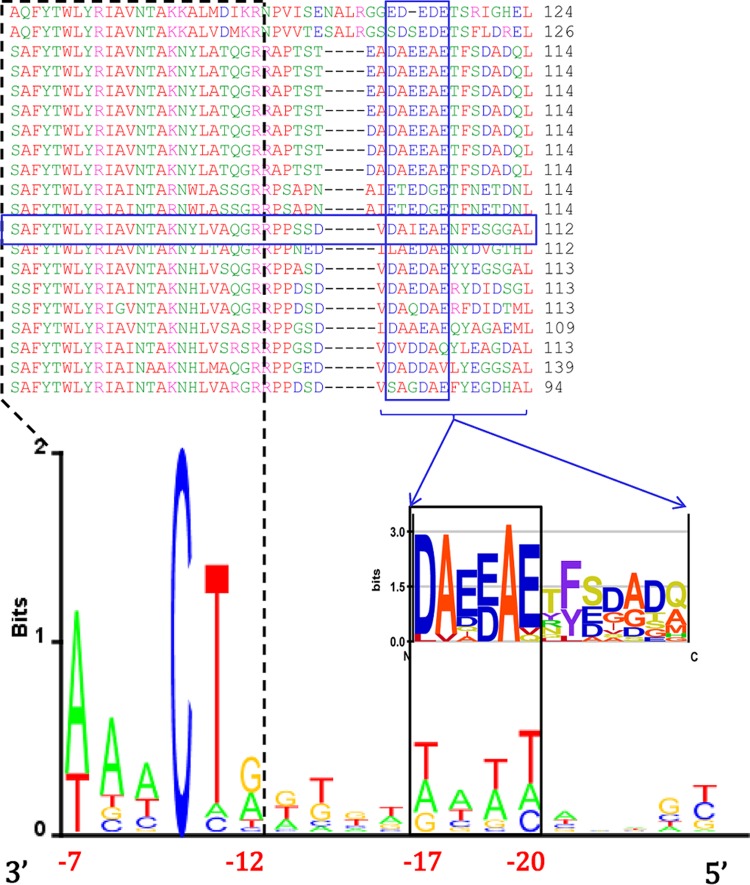
Hence, we started by aligning the 7-11 σ factor with selected representatives of the ECF28 subgroup (Fig. 4, top). One can observe that the segment just C-terminal of the 7-11 σ2 domain boundary (indicated by a vertical line in Fig. 4) is well aligned with the corresponding segments in the selected ECF28 representatives. The segment is composed of 6 amino acids (aa), about the same length as the sequence responsible for interacting with the extended −10 element in group I σ factors (RpoD) (3). Furthermore, in order to observe whether this feature is present in all the ECF28 members, and to establish the extent of the sequence conservation, we performed a multiple alignment of the entire ECF28 group and used it to construct a logo of the sequences C-terminal of the σ2 domain (Fig. 4, bottom). As Fig. 4 indicates, the relevant segment is present in all ECF28 members; more importantly, as can be seen from the sequence logo, the protein sequence C-terminal of this segment—extending to the N terminus of the σ4 domain—is notably less conserved.
FIG 4.
Phage 7-11–ECF28 subgroup σ factor alignment. (Top) Multiple alignment of the phage 7-11 σ factor (boxed in green) and selected representatives of the ECF28 subgroup. The segment of the alignment that is shown corresponds to the C-terminal part of the σ2 domain (with the vertical line indicating the C terminus) and the sequences C-terminal of the indicated σ2 domain boundary. The shaded area in the alignment corresponds to the segment implicated in interacting with the −10 element extension in the promoter recognized by phage 7-11 σ factor. Below the alignment is the sequence logo of that promoter, with the −10 element extension shaded and the coordinates shown relative to the transcription start site. (Bottom) Multiple alignment of the entire ECF28 subgroup. The conserved segment interacting with the −10 element extension is shaded. Below the alignment is the sequence logo of the shaded segment, together with the C-terminal flanking sequences that reach down to the first gap in the multiple alignment.
Next, in Fig. 5 we compared the phage 7-11 σ factor protein sequence with the multiple alignment of the ECF01 σ subgroup (whose representative is σW). One may clearly observe the absence of the conserved sequence element (noted in the ECF28 subgroup) in the ECF01 subgroup; instead, gaps and poorly aligned sequences appear in the multiple alignment. This finding is consistent with the expectation stated above, i.e., that the sequence element of interest is not present in the ECF01 subgroup.
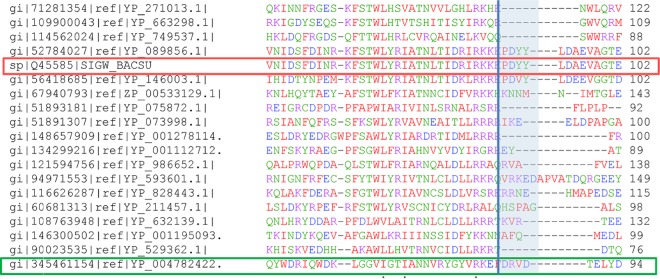
FIG 5.
Phage 7-11–ECF01 subgroup σ factor alignment. Shown is a multiple alignment of selected ECF01 representatives, including σW (boxed in red)—comprising the group recognizing promoters with no −10 element extension—and the phage 7-11 σ factor (boxed in green). The segment of the alignment shown corresponds to the C-terminal part of the σ2 domain (with the C terminus indicated by the vertical line) and the sequences C-terminal of the boundary. The shaded area corresponds to the segment implicated in interacting with the −10 element extension in the promoter recognized by phage 7-11.
Overall, the results presented above indicate that the segment appearing just C-terminal of domain σ2 in some ECF σ subfamily members is involved in recognizing the extension of the −10 element. Moreover, we compared multiple alignments of ECF28 and 7-11 sequences with the sequences of representatives of the other ECF σ subgroups. The results of that comparison indicate that the conserved segment located just C-terminal of the σ2 domain is absent in the other ECF σ subgroups. Consequently, the appearance of this feature is rather specific, as would be expected for recognition of the extended −10 element, since it is evidently not a widespread property of ECF σ factors (being absent from σE and σW). This makes ECF28 a candidate for a bacterial ECF subgroup with a putatively distinct regulatory paradigm, in contrast to the current viewpoint on bacterial ECF σ factor functioning.
Therefore, we next analyzed the promoter specificity of ECF28 σ factors. Note that this subgroup is composed of entirely unexamined ECF σ factors. Here we started from the paradigm of ECF σ factor autoregulation, so that we searched the intergenic sequences upstream of the ECF σ factor genes, by using both supervised and unsupervised methods for regulatory element detection, as described in Materials and Methods. This analysis did not identify ECF28 promoters, for which there are two possible reasons: either the subgroup members are not autoregulated, so that the promoters are not contained in the intergenic regions analyzed, or their promoters do not contain a recognizable −35 element. Both of these possibilities indicate a departure from the current paradigm of ECF σ promoter recognition, whose implications are further analyzed in the Discussion.
Analyzing the promoter specificity of ECF subgroups.
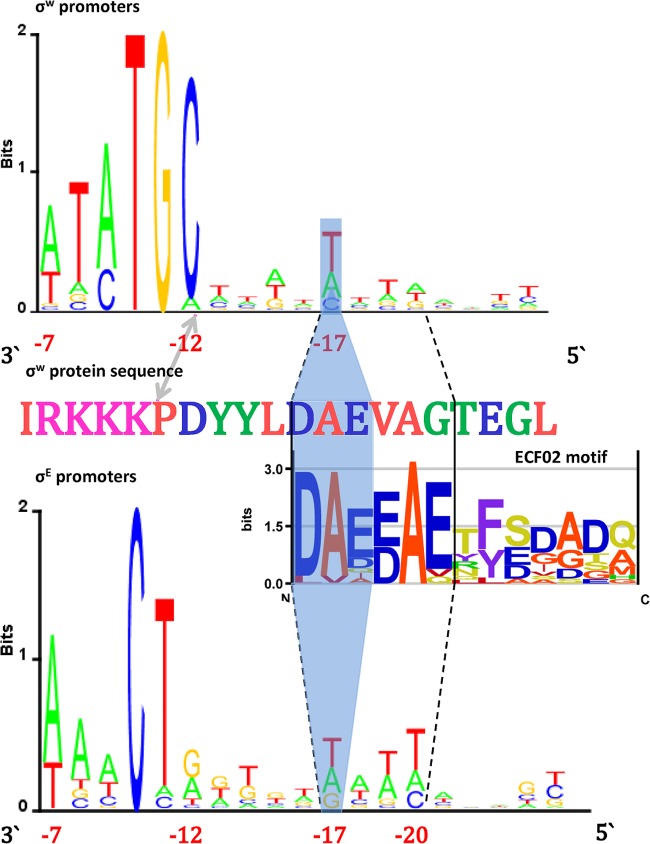
We further analyzed the canonical bacterial ECF members σE and σW, for which a large number of experimentally verified promoters are available, and whose interactions outside of canonical −35 and −10 elements can provide additional examples of flexibility within the ECF σ subfamily. We started by aligning the available promoters for σE (see Materials and Methods, “Sequence logos”), where we unexpectedly observed conservation in the spacer between the −10 and −35 elements, which is prominent for promoter sequences that are inactive in vitro (Fig. 6, bottom sequence logo). Note that this conservation is located near the upstream edge of the −10 element (positions −17 to −20 in the sequence logo) but not as its direct extension. Furthermore, note that this conserved promoter element is not prominent in the sequences that are active in vitro, a point further elaborated in the Discussion.
FIG 6.
Putative σE protein-DNA spacer interactions. (Top) Multiple alignment of the protein sequence of σE (boxed in blue) and those of selected ECF02 members (the corresponding GI numbers are provided in Table S1 in the supplemental material), showing the C-terminal part of the σ2 domain and the flanking sequence C-terminal of domain σ2. The conserved protein motif identified is boxed in blue. Below the alignment is a sequence logo presenting the conserved protein motif and the (unconserved) flanking sequence C-terminal of that motif. (Bottom) The −10 element of the promoter, together with the spacer sequence, is presented in the form of a 3′-to-5′-oriented sequence logo (coordinates are indicated relative to the transcription start site). The putative interaction between the conserved protein motif and the conserved promoter spacer motif is indicated by outlining (solid black lines), as is the −10 element–σ2 domain interaction (dashed black lines).
Next, to identify the motif at the protein sequence level that putatively interacts with this conserved spacer element, we compared the σE protein sequence with those of selected representatives of the ECF02 subgroup and searched for a conserved motif further C-terminal of the σ2 domain (see Materials and Methods, “Protein sequence alignments”). This search identified a conserved 6-aa motif in the ECF02 subgroup, which was found in close proximity to the σ2 domain C terminus but not as a direct extension of this domain (Fig. 6). Therefore, the location of this protein motif puts it in an optimal position for interacting with the conserved spacer element in the promoters recognized by σE.
Next, by comparing the σE with the σW sequence, we observed the upstream half of the motif identified in σE (DAE) C-terminal of σW domain σ2 (Fig. 7). Interestingly, at the DNA level, this is complemented by the conserved T nucleotide at position −17 in the σW promoter sequence, which corresponds to the most downstream base of the σE spacer motif. Clearly, in σW, we find that the partial conservation of the σE protein motif leads to a corresponding partial conservation of the protein-DNA interaction. The absence of full conservation of this interaction is consistent with the fact that multiple alignment of ECF01 members shows no conservation of the protein motif across this subgroup.
FIG 7.
Putative σE–promoter spacer versus σW–promoter spacer interactions. (Top) σW promoter sequence logo (in the 3′-to-5′ orientation) showing the −10 element and the upstream spacer flanking sequence. (Center) Segment of the σW protein sequence including the C-terminal part of domain σ2 and the flanking sequence C-terminal of domain σ2. The edge of domain σ2 and the edge of the −10 element in the promoter sequence are marked by the arrow. Below the sequence is the logo of the conserved ECF02 subgroup protein motif. (Bottom) Sequence logo of promoters recognized by σE, showing the −10 element and the upstream spacer flanking sequence. The conserved protein/DNA spacer segments found in σE are outlined with dashed lines. Shading indicates the part of this conserved domain that also appears in σW, as well as the corresponding protein-DNA interactions. The coordinates of the promoter elements relative to the transcription start site are indicated.
Since the results obtained for σE and σW indicate a notable flexibility of promoter specificity within the ECF subfamily, in particular with respect to interactions with conserved motifs outside of the canonical promoter elements, we also explored the presence of these interactions across the other subgroups. However, this effort was complicated by a very restricted set of available ECF promoters, which made it hardly feasible to conduct a detailed analysis, as in the case of canonical ECF members. We consequently limited our investigation to assessing conservation between domains σ2 and σ4, and a corresponding conservation in the spacer sequences—i.e., outside of the −10 and −35 elements—of the interacting promoters, comparable to what was observed for σE and σW (see Materials and Methods, “Sequence logos,” “ECF32 promoter identification,” and “Protein sequence alignments”). This investigation identified ECF32 as another ECF σ subgroup, for which we found interactions that were not restricted to extensions of the σ2 domain and −10 element. In particular, it is interesting that in ECF32, the conserved sequence element is located at the opposite end of the spacer sequence, i.e., near the downstream edge of the −35 element, a finding consistent with the position of the conserved protein sequence, which is located N-terminal of the σ4 domain (see Fig. S1 in the supplemental material). Moreover, note that mutating a base within this ECF32 conserved element (the conserved G at position −25) has been shown experimentally to decrease promoter transcriptional activity significantly (29), suggesting that the conserved motif is indeed functional.
DISCUSSION
We provided here a comprehensive analysis of DNA and protein motifs involved in promoter recognition by ECF σ factors. The analysis was motivated by the limited data available on the mechanisms of promoter recognition within this large and heterogeneous, but poorly studied, σ factor group. This limited information on ECF σ specificity has been applied to searching for promoters for other ECF σ subgroups, based on the assumption of autoregulation, i.e., that the promoters are located upstream of ECF σ factor genes (7). To break this cycle of limited data inducing similar data, we here computationally investigated the binding specificity of phage ECF σ factors, which represent the outliers of the ECF family, examined their relationships to the classified bacterial ECF σ factor subgroups, and provided an extensive analysis of promoter elements for ECF σ factors.
First, we showed that the promoters for the bacteriophage ECF σ factors examined can accommodate a significant −10 element extension, which was previously unnoticed in the ECF σ subfamily (1). In particular, promoters for σW and σE have 2-bp motifs involved in dsDNA interactions. This should be compared to the 4-bp −15 element in RpoD σ factors, which is the upstream part of the −10 element involved in dsDNA interactions (9). In fact, the −10 element for bacteriophage ECF σ factors not only is notably longer than those for σE and σW but also is longer than those for RpoD σ factors, since the part interacting in dsDNA form (equivalent to the −15 element) is 5 to 7 bp long.
For the bacteriophage phiEco32 σ factor, this −10 element extension is accompanied by an absence of the −35 element, together with the loss of the σ4 domain. We predict, however, that such loss may be complemented by phiEco32 ECF σ factor interactions with a transcription factor, since we find conserved motifs upstream of −10 elements in a subset of the genes transcribed by phiEco32 σ. Such complementation of the σ4 domain with a transcription factor would then be another (putative) example of variability in ECF σ functioning, particularly since examples of ECF promoter regulation by DNA binding transcription factors are rare (1, 30). Such complementation would also have a clear role in bacteriophage temporal gene regulation, i.e., in distinguishing between bacteriophage middle and late gene expression. Note that it is the bacteriophage late genes (15) that are associated with this upstream motif.
We also predicted the protein motif that is responsible for interactions with the −10 element extension in the bacteriophage σ factors and in ECF28, which is an entirely unexamined bacterial ECF σ factor subgroup. Furthermore, in ECF28, the sequence motif is located just C-terminal of the predicted domain σ2 boundary, so that this sequence is clearly an extension of domain σ2 as it appears in σE and σW. This domain σ2 extension is rather specific and, notably, does not appear in the subgroups represented by σE and σW, which are known not to contain a −10 element extension; interestingly, a similar motif appears C-terminal of domain σ2 in the σE subgroup, as we discuss further below. In fact, it is commonly assumed that ECF σ factors depend on the −35 element to accomplish transcription, since they do not contain domain σ3 (1). Note that in RpoD σ factors, the upstream part of the −10 element, which is involved in dsDNA interactions, is recognized by the C-terminal part of the σ2 domain and the N-terminal part of the σ3 domain (3). However, the results presented here indicate that in ECF σ factors, an even longer extension of the −10 element can be achieved through a C-terminal extension of the σ2 domain.
Consequently, the study of ECF28 subgroup members may notably extend our understanding of ECF functioning. We were unable to locate their promoters either by searching for shared motifs or by using a −35 element ECF σ weight matrix. This suggests that either the ECF28 subgroup is not self-regulated or its promoters do not contain a recognizable ECF −35 element. In fact, we hypothesize that the absence of self-regulation may be due to more-efficient transcription related to the protein motif that can recognize the −10 element extension. This possibility may make obsolete the enhancement in transcription due to the positive-feedback loop, i.e., such enhancement could be achieved by the −10 element extension.
Furthermore, in ECF02 (σE), ECF32, and partially in σW, we found additional examples of flexibility in ECF σ factor promoter recognition, as exemplified through cooccurrence of the conserved protein/DNA motifs, located C-terminal of the σ2 domain and further upstream of the −10 element; in the case of ECF32, some of these interactions were also confirmed experimentally. Such flexibility of ECF σ promoter recognition appears even more pronounced compared to promoter recognition by group I σ factors. In particular, note the presence of the putative spacer motif in ECF promoters, where such spacer sequence interactions do not appear for group I σ factors, i.e., they are limited to extensions of the −10 promoter element and σ2 domain. Moreover, the spacer motif for σE is more pronounced in the promoters with low measured in vitro transcription activity. This may suggest that the appearance of this motif is related to increasing the otherwise low strength of these promoters. If so, this may provide another example of complementation (mixing and matching) of ECF σ promoter elements.
For the ECF σ factor in phiEco32, we have seen that extension of the −10 element can compensate for interactions with the −35 element. This suggests that mixing and matching might be also exhibited for ECF σ factors, since the absence of a −35 element accompanied by a −10 element extension is an often-quoted example of mixing and matching for housekeeping σ factors. This may also be suggested by a bacteriophage ECF σ promoter structure that is closer to those of RpoD promoters than to those of promoters for σW and σE (note the characteristic TATA box). On the other hand, domain σ2 of the bacteriophage σ factors is more divergent from that of RpoD than from those of σW and σE. This may suggest that the similar promoter organization is due not to similar protein sequences in the σ70 family but rather to a possibility of a common general promoter recognition mechanism, such as mixing and matching.
Here, overall, we have predicted novel interactions of ECF σ factors with their promoters, which may point to a significantly greater flexibility than recognized previously. The results also suggest that mixing and matching may occur in ECF σ factor recognition, which should be biochemically tested. In particular, this can be exhibited by in vitro transcription analysis, starting with a specific promoter and changing one base pair or one element at a time (31–33). If corroborated, mixing and matching in ECF σ factors would be a significant finding, since it is believed that mixing and matching does not appear in this group, which is most divergent from RpoD σ factors. That is, demonstration of mixing and matching in ECF σ factors would strongly suggest that this is a common kinetic mechanism for promoter recognition in the entire σ70 family. Such a common mechanism could then provide a unifying framework for understanding promoter recognition within the diverse σ70 family.
Supplementary Material
ACKNOWLEDGMENTS
We thank Konstantin Severinov and Evgeny Zdobnov for useful discussions.
Funding Statement
This work is supported by a Marie Curie International Reintegration Grant within the 7th European Community Framework Programme (PIRG08-GA-2010-276996), by the Ministry of Education and Science of the Republic of Serbia under project ON173052, and by the Swiss National Science Foundation under SCOPES project IZ73Z0_152297.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00244-16.
REFERENCES
- 1.Feklístov A, Sharon BD, Darst SA, Gross CA. 2014. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu Rev Microbiol 68:357–376. doi: 10.1146/annurev-micro-092412-155737. [DOI] [PubMed] [Google Scholar]
- 2.Paget M, Helmann J. 2003. The σ70 family of sigma factors. Genome Biol 4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borukhov S, Nudler E. 2003. RNA polymerase holoenzyme: structure, function and biological implications. Curr Opin Microbiol 6:93–100. doi: 10.1016/S1369-5274(03)00036-5. [DOI] [PubMed] [Google Scholar]
- 4.Djordjevic M, Bundschuh R. 2008. Formation of the open complex by bacterial RNA polymerase—a quantitative model. Biophys J 94:4233–4248. doi: 10.1529/biophysj.107.116970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murakami KS, Darst SA. 2003. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol 13:31–39. doi: 10.1016/S0959-440X(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 6.Koo B-M, Rhodius VA, Nonaka G, Gross CA. 2009. Reduced capacity of alternative σs to melt promoters ensures stringent promoter recognition. Genes Dev 23:2426–2436. doi: 10.1101/gad.1843709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staroń A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, Mascher T. 2009. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) σ factor protein family. Mol Microbiol 74:557–581. doi: 10.1111/j.1365-2958.2009.06870.x. [DOI] [PubMed] [Google Scholar]
- 8.Ulrich LE, Zhulin IB. 2010. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res 38(Suppl 1):D401–D407. doi: 10.1093/nar/gkp940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hook-Barnard IG, Hinton DM. 2007. Transcription initiation by mix and match elements: flexibility for polymerase binding to bacterial promoters. Gene Regul Syst Bio 1:275–293. [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar A, Malloch R, Fujita N, Smillie D, Ishihama A, Hayward R. 1993. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J Mol Biol 232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- 11.Minakhin L, Severinov K. 2003. On the role of the Escherichia coli RNA polymerase 70 region 4.2 and α-subunit C-terminal domains in promoter complex formation on the extended −10 galP1 promoter. J Biol Chem 278:29710–29718. doi: 10.1074/jbc.M304906200. [DOI] [PubMed] [Google Scholar]
- 12.Ponnambalam S, Webster C, Bingham A, Busby S. 1986. Transcription initiation at the Escherichia coli galactose operon promoters in the absence of the normal −35 region sequences. J Biol Chem 261:16043–16048. [PubMed] [Google Scholar]
- 13.Djordjevic M. 2011. Redefining Escherichia coli σ70 promoter elements: −15 motif as a complement of the −10 motif. J Bacteriol 193:6305–6314. doi: 10.1128/JB.05947-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchell JE, Zheng D, Busby SJW, Minchin SD. 2003. Identification and analysis of ‘extended −10′ promoters in Escherichia coli. Nucleic Acids Res 31:4689–4695. doi: 10.1093/nar/gkg694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pavlova O, Lavysh D, Klimuk E, Djordjevic M, Ravcheev DA, Gelfand MS, Severinov K, Akulenko N. 2012. Temporal regulation of gene expression of the Escherichia coli bacteriophage phiEco32. J Mol Biol 416:389–399. doi: 10.1016/j.jmb.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kropinski AM, Lingohr EJ, Ackermann H-W. 2011. The genome sequence of enterobacterial phage 7-11, which possesses an unusually elongated head. Arch Virol 156:149–151. doi: 10.1007/s00705-010-0835-5. [DOI] [PubMed] [Google Scholar]
- 17.Rhodius VA, Mutalik VK. 2010. Predicting strength and function for promoters of the Escherichia coli alternative sigma factor, σE. Proc Natl Acad Sci U S A 107:2854–2859. doi: 10.1073/pnas.0915066107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishii T, Yoshida K-I, Terai G, Fujita Y, Nakai K. 2001. DBTBS: a database of Bacillus subtilis promoters and transcription factors. Nucleic Acids Res 29:278–280. doi: 10.1093/nar/29.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzina J, Djordjevic M. 2015. Inferring bacteriophage infection strategies from genome sequence: analysis of bacteriophage 7-11 and related phages. BMC Evol Biol 15(Suppl 1):S1. doi: 10.1186/1471-2148-15-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stormo GD, Fields DS. 1998. Specificity, free energy and information content in protein-DNA interactions. Trends Biochem Sci 23:109–113. doi: 10.1016/S0968-0004(98)01187-6. [DOI] [PubMed] [Google Scholar]
- 21.Thompson W, Rouchka EC, Lawrence CE. 2003. Gibbs Recursive Sampler: finding transcription factor binding sites. Nucleic Acids Res 31:3580–3585. doi: 10.1093/nar/gkg608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Workman CT, Yin Y, Corcoran DL, Ideker T, Stormo GD, Benos PV. 2005. enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic Acids Res 33(Suppl 2):W389–W392. doi: 10.1093/nar/gki439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatusova TA, Madden TL. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett 174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 24.Stormo GD. 2000. DNA binding sites: representation and discovery. Bioinformatics 16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 25.Hertz GZ, Stormo GD. 1999. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 26.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 27.Marchler-Bauer A, Bryant SH. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32(Suppl 2):W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Djordjevic M. 2013. Efficient transcription initiation in bacteria: an interplay of protein–DNA interaction parameters. Integr Biol 5:796–806. doi: 10.1039/c3ib20221f. [DOI] [PubMed] [Google Scholar]
- 29.Nissan G, Manulis S, Weinthal DM, Sessa G, Barash I. 2005. Analysis of promoters recognized by HrpL, an alternative sigma-factor protein from Pantoea agglomerans pv. gypsophilae. Mol Plant Microbe Interact 18:634–643. doi: 10.1094/MPMI-18-0634. [DOI] [PubMed] [Google Scholar]
- 30.Abellón-Ruiz J, Bernal-Bernal D, Abellán M, Fontes M, Padmanabhan S, Murillo FJ, Elías-Arnanz M. 2014. The CarD/CarG regulatory complex is required for the action of several members of the large set of Myxococcus xanthus extracytoplasmic function σ factors. Environ Microbiol 16:2475–2490. doi: 10.1111/1462-2920.12386. [DOI] [PubMed] [Google Scholar]
- 31.Hook-Barnard I, Johnson XB, Hinton DM. 2006. Escherichia coli RNA polymerase recognition of a σ70-dependent promoter requiring a −35 DNA element and an extended −10 TGn motif. J Bacteriol 188:8352–8359. doi: 10.1128/JB.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burr T, Mitchell J, Kolb A, Minchin S, Busby S. 2000. DNA sequence elements located immediately upstream of the −10 hexamer in Escherichia coli promoters: a systematic study. Nucleic Acids Res 28:1864–1870. doi: 10.1093/nar/28.9.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miroslavova NS, Busby SJ. 2006. Investigations of the modular structure of bacterial promoters. Biochem Soc Symp 73:1–10. doi: 10.1042/bss0730001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.