ABSTRACT
Multiple cellular pathways are regulated by small ubiquitin-like modifier (SUMO) modification, including ubiquitin-mediated proteolysis, signal transduction, innate immunity, and antiviral defense. In the study described in this report, we investigated the effects of SUMO on the replication of two members of the Rhabdoviridae family, vesicular stomatitis virus (VSV) and rabies virus (RABV). We show that stable expression of SUMO in human cells confers resistance to VSV infection in an interferon-independent manner. We demonstrate that SUMO expression did not alter VSV entry but blocked primary mRNA synthesis, leading to a reduction of viral protein synthesis and viral production, thus protecting cells from VSV-induced cell lysis. MxA is known to inhibit VSV primary transcription. Interestingly, we found that the MxA protein was highly stabilized in SUMO-expressing cells. Furthermore, extracts from cells stably expressing SUMO exhibited an increase in MxA oligomers, suggesting that SUMO plays a role in protecting MxA from degradation, thus providing a stable intracellular pool of MxA available to combat invading viruses. Importantly, MxA depletion in SUMO-expressing cells abrogated the anti-VSV effect of SUMO. Furthermore, SUMO expression resulted in interferon-regulatory factor 3 (IRF3) SUMOylation, subsequently decreasing RABV-induced IRF3 phosphorylation and interferon synthesis. As expected, this rendered SUMO-expressing cells more sensitive to RABV infection, even though MxA was stabilized in SUMO-expressing cells, since its expression did not confer resistance to RABV. Our findings demonstrate opposing effects of SUMO expression on two viruses of the same family, intrinsically inhibiting VSV infection through MxA stabilization while enhancing RABV infection by decreasing IFN induction.
IMPORTANCE We report that SUMO expression reduces interferon synthesis upon RABV or VSV infection. Therefore, SUMO renders cells more sensitive to RABV but unexpectedly renders cells resistant to VSV by blocking primary mRNA synthesis. Unlike the interferon-mediated innate immune response, intrinsic antiviral resistance is mediated by constitutively expressed restriction factors. Among the various anti-VSV restriction factors, only MxA is known to inhibit VSV primary transcription, and we show here that its expression does not alter RABV infection. Interestingly, MxA depletion abolished the inhibition of VSV by SUMO, demonstrating that MxA mediates SUMO-induced intrinsic VSV resistance. Furthermore, MxA oligomerization is known to be critical for its protein stability, and we show that higher levels of oligomers were formed in cells expressing SUMO than in wild-type cells, suggesting that SUMO may play a role in protecting MxA from degradation, providing a stable intracellular pool of MxA able to protect cells from viral infection.
INTRODUCTION
In addition to ubiquitin, several ubiquitin-like (UBL) proteins have been reported to function as protein modifiers that regulate various cellular functions (1). The best-characterized member of the UBL protein family is the small ubiquitin-like modifier (SUMO) family (2). SUMOylation is a posttranslational modification where a reversible covalent bond is formed between the SUMO molecule and the target protein. In humans, the SUMO protein family consists of SUMO1 and two highly homologous proteins, SUMO2 and SUMO3 (collectively known as SUMO2/3), which share only 18% homology with ubiquitin. SUMO2 and SUMO3, which share 97% sequence identity, cannot be distinguished by currently available antibodies and are expressed at significantly higher levels than SUMO1, with which they share approximately 50% sequence identity (3). SUMO2 and SUMO3 contain a lysine residue at position 11 (K11) that can be used for self-conjugation or conjugation with SUMO1 and that is usually the site of poly-SUMOylation chains. In contrast, SUMO1 does not contain K11 and therefore does not form chains. However, SUMO1 can be attached to lysine residues within SUMO2/3 chains, leading to chain termination.
SUMO modification occurs through the formation of an isopeptide bond between the amino group of a lysine residue on the substrate and the carboxyl terminus group of SUMO. SUMOylation involves a three-enzyme cascade: a single SUMO activation enzyme (E1) that exists as a dimer (SAE1/SAE2), an E2-conjugating enzyme (Ubc9), and multiple substrate-specific E3 SUMO ligases (PIAS1, PIAS3, PIASxα, PIASxβ, PIASy, RanBP2, and Pc2) (4, 5). SUMOylation is a highly dynamic process whereby SUMOylation patterns are frequently altered in response to different cell stimuli. Other key players in this process are the SUMO-specific proteases (SENPs), which are responsible for cleaving the isopeptide bond on specific SUMO substrates.
SUMOylation has been involved in several cellular processes, such as transcriptional regulation, promyelocytic leukemia (PML) nuclear body formation, protein stability, subcellular localization, signal transduction, and innate immunity (4–9).We recently reported that the expression of different SUMO paralogs reduces STAT1 phosphorylation in response to alpha interferon (IFN-α) and IFN-γ, resulting in a decrease in the IFN-γ-induced transcriptional response without affecting the IFN-α-induced transcriptional response (10). Another study showed that the SUMOylation of IFN-regulatory factor 3 (IRF3) or IRF7 negatively regulates type I IFN production upon viral infection (11, 12). Nonetheless, besides its physiological relevance, the role of SUMO in intrinsic antiviral activity still remains to be determined.
In the study described in this report, we investigated the effects of SUMOylation on the replication of two members of the Rhabdoviridae family (Mononegavirales order), vesicular stomatitis virus (VSV) and rabies virus (RABV). Both viruses contain a single-stranded negative-sense RNA genome (∼12 kb) which encodes five proteins, nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), and the polymerase (L), in the order 3′-N-P-M-G-L-5′ (13). During transcription, a positive-stranded leader RNA and five capped and polyadenylated mRNAs are synthesized. The replication process yields nucleocapsids containing full-length antigenome sense RNA, and these, in turn, serve as the templates for the synthesis of genome sense RNA. The genomes then associate with viral proteins to form new viruses budding from the cell membranes.
We report here that the stable expression of SUMO in human cell lines inhibited RABV- and VSV-induced IFN production. As expected, cells were more sensitive to RABV, but, remarkably, cells were resistant to VSV in an IFN- and STAT1-independent manner. Furthermore, we demonstrate that SUMO expression did not alter VSV entry but inhibited primary mRNA synthesis, resulting in the reduction of viral protein synthesis and subsequent viral production and thus protecting cells from VSV-induced cell lysis. Human MxA is a member of the large dynamin-like GTPases, a long-established IFN-stimulated gene (ISG) restriction factor of a broad range of RNA and DNA viruses (14) which targets VSV at the level of the level of primary transcription (15, 16). Therefore, we investigated MxA and demonstrate that MxA is highly stabilized through its oligomerization in SUMO-expressing cells and that its depletion blocks SUMO-induced VSV resistance.
MATERIALS AND METHODS
Materials.
Human recombinant IFN-α2 was sourced from Schering (USA), and rabbit anti-STAT1 (sc-346), rabbit anti-SUMO1 (Sc-9060), rabbit anti-IRF3 (Sc-9082), and rabbit polyclonal anti-actin clone C-11 (Sc-1615) antibodies were from Santa Cruz Biotechnology. Rabbit anti-SUMO2/3 antibodies were from Invitrogen, rabbit anti-phospho-IRF3 (Ser396) antibodies were from Cell Signaling, anti-Ubc9 monoclonal antibody (MAb) was from Abgent, and monoclonal anti-6×His antibody was from Clontech. Rabbit anti-MxA antibodies were from Proteintech or were a gift from S. R. Valentini (UNESP, Araraquara, Brazil) (17), and mouse anti-MxA monoclonal antibody M143 was provided by G. Kochs (Institute of Virology, Freiburg, Germany). The 64G12 monoclonal antibody against the human IFN-α/β receptor (anti-IFNAR1 antibody) was a gift from P. Eid (INSERM UMR1014). Monoclonal anti-RABV P antibody and rabbit anti-VSV polyclonal antibodies were produced as described previously (18, 19). Small interfering RNA (siRNA) targeting Ubc9-1 was purchased from Dharmacon. Ubc9-2 and STAT1 expression was silenced using ON-TARGETplus siRNA SMARTpool riRNA reagents purchased from GE Healthcare; the different siRNAs were transfected into cells using the HiPerFect transfection reagent (Qiagen).
Cell cultures.
Human glioblastoma astrocytoma U373MG cells, cervical cancer HeLa cells, and BSR cells (cloned from BHK-21 [baby hamster kidney] cells) were purchased from ATCC (http://www.lgcstandards-atcc.org) and grown at 37°C in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. HeLa cells stably expressing His-tagged SUMO1 (His-SUMO1) or His-SUMO3, U373MG cells stably expressing His-SUMO2 or His-SUMO3 (10), and NIH 3T3 cells harboring the empty vector or the plasmid carrying the gene for MxA (kindly provided by J. Pavlovic [15]) were kept in medium supplemented with 0.5 mg/ml of neomycin.
Viral stocks.
VSV (Mudd-Summer strain, Indiana serotype) and RABV (CVS strain) were grown in BSR cells, and the virus titers were determined by standard plaque assays on BSR cells. Murine leukemia virus (MLV)-derived vectors carrying green fluorescent protein (GFP) pseudotyped with the VSV glycoprotein (VSV-G), provided by F. L. Cosset, ENS Lyon, had a titer of 107 IU/ml and were generated as previously described (20).
Knockdown of MxA.
We purchased short hairpin RNA (shRNA) DNA clones containing a hygromycin selection marker that target MxA (shMxA; CTCACCAGAGAATAACAGA) and its nontargeting control from GeneCopoeia (Rockville, MD). The shRNA DNA clones were transfected into HEK293 cells together with a plasmid carrying DNA encoding HIV-1 Gag-Pol and pVSV-G (carrying the glycoprotein of VSV). The virus particles subsequently made were used to transduce wild-type (wt) HeLa cells (HeLa-wt cells), HeLa cells stably expressing His-SUMO1 (HeLa-SUMO1 cells), and HeLa cells stably expressing His-SUMO3 (HeLa-SUMO3) cells to knock down MxA. Stably transduced cells were selected for 4 weeks using hygromycin (final concentration, 0.5 mg/ml).
MxA oligomerization.
In order to assess the endogenous oligomerization state of MxA, whole extracts were collected from HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells and lysed, using a needle, in 0.5% Triton X-100 in phosphate-buffered saline (PBS) containing protease inhibitor tablets and 10 mM iodoacetamide. The extracts were centrifuged for 10 min at 1,500 × g, 100 μg/ml of disuccinimidyl suberate (DSS; a covalent cross-linking agent) was added, and the samples were incubated at room temperature for 1 h. The reaction was quenched using 50 mM Tris-HCl for 15 min at room temperature. The samples were then immunoprecipitated with MxA antibodies and protein IgG beads. The eluates were subsequently run on a 4 to 12% gradient gel and revealed using anti-MxA, anti-SUMO1, anti-SUMO2/3, or anti-Ubc9 antibodies. Extracts from wt NIH 3T3 cells (NIH 3T3-wt cells) and NIH 3T3 stably expressing MxA (NIH 3T3-MxA cells) were used as controls.
Northern blot analysis.
Cells were infected with VSV at a multiplicity of infection (MOI) of 3 in the absence or the presence of cycloheximide (100 μg/ml). After 1 h of adsorption, the cells were washed and fresh medium with or without cycloheximide (100 μg/ml) was added. After 4 h of infection, total RNA was isolated from the cells with an RNA Now kit (Ozyme). Total RNA was separated on a 1.5% agarose gel under denaturing conditions and blotted onto nylon membranes (Roche Molecular Biochemicals). Hybridizations were performed with digoxigenin (DIG)-labeled oligonucleotides recognizing the VSV N-gene sequence and by incubation with anti-DIG antibody conjugated to alkaline phosphatase followed by incubation with CDP Star chemiluminescent substrate (Roche).
Real-time PCR.
Total RNAs were extracted using an RNeasy minikit (Qiagen) following the manufacturer's instructions. RNA samples were converted to cDNA using a RevertAid H Minus first-strand cDNA synthesis kit (Thermo Scientific). Real-time PCRs were performed in duplicate using 5 μl of cDNA diluted 10 times in water and Takyon carboxy-X-rhodamine SYBR MasterMix blue dTTP (Eurogentec). The following program was used on a 7900HT Fast real-time PCR system (Applied Biosystems): 3 min at 95°C, followed by 35 cycles of 15 s at 95°C, 25 s at 60°C, and 25 s at 72°C. Values for each transcript were normalized to the expression levels of RPL13A (60S ribosomal protein L13a) using the 2−ΔΔCT threshold cycle (CT) method. The primers used for quantification of the transcripts by real-time quantitative PCR are shown in Table 1.
TABLE 1.
Primers used for quantification of transcripts by qRT-PCR
| Primer | Sequence (5′-3′) |
|
|---|---|---|
| Forward | Reverse | |
| RABV-P | ACCTTGGTGAGATGGTTAGGG | AGTTGACCGTGACATAGGATAC |
| RPL13A | CCTGGAGGAGAAGAGGAAAGAGA | TTGAGGACCTCTGTGTATTTGTCAA |
| IFN-α1 | CCAGTTCCAGAAGGCTCCAG | TCCTCCTGCATCACACAGGC |
| IFN-β | TGCATTACCTGAAGGCCAAGG | AGCAATTGTCCAGTCCCAGTG |
| IFN-λ1 | GGACGCCTTGGAAGAGTCAC | GAGGAGGCGGAAGAGGTTGA |
| IFN-λ2/3 | GGGCCTGTATCCAGCCTCAG | CTGGTCTAGGACGTCCTCCA |
| IFN-γ | GGCAGCCAACCTAAGCAAGAT | CAGGGTCACCTGACACATTCA |
Immunofluorescence analysis.
Cells grown on glass coverslips were fixed for 20 min with 4% paraformaldehyde (PFA) in PBS and permeabilized for 5 min in 0.1% Triton X-100 in PBS. The cells were then prepared for immunofluorescence staining using the appropriate primary antibody and the corresponding secondary antibody conjugated to Alexa Fluor (Molecular Probes). Cells were mounted onto glass slides by using Immu-Mount (Shandon) containing DAPI (4′,6-diamidino-2-phenylindole). Confocal laser microscopy was performed on a Zeiss LSM 710 microscope.
Purification of His6-tagged SUMO conjugates.
HeLa-wt and HeLa-SUMO3 cells (107) that were noninfected or that had been infected with RABV for 20 h were lysed in denaturing buffer A (6 M guanidinium-HCl, 0.1 M NaH2PO4, 0.01 M Tris-HCl, pH 8.0, 5 mM imidazole, 10 mM β-mercaptoethanol). After sonication, the lysates were mixed with 50 μl of Ni-nitrilotriacetic acid (NTA)-agarose beads (Qiagen) for 3 h at room temperature. The beads were successively washed with buffer B (0.1% Triton X-100, 8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-HCl, pH 6.3, 10 mM β-mercaptoethanol) and subsequently eluted with 200 mM imidazole in 0.15 M Tris-HCl, pH 6.7, 30% glycerol, and 0.72 M β-mercaptoethanol. The eluates were then analyzed by Western blotting.
RESULTS
Protective effect of SUMO in VSV-infected cells.
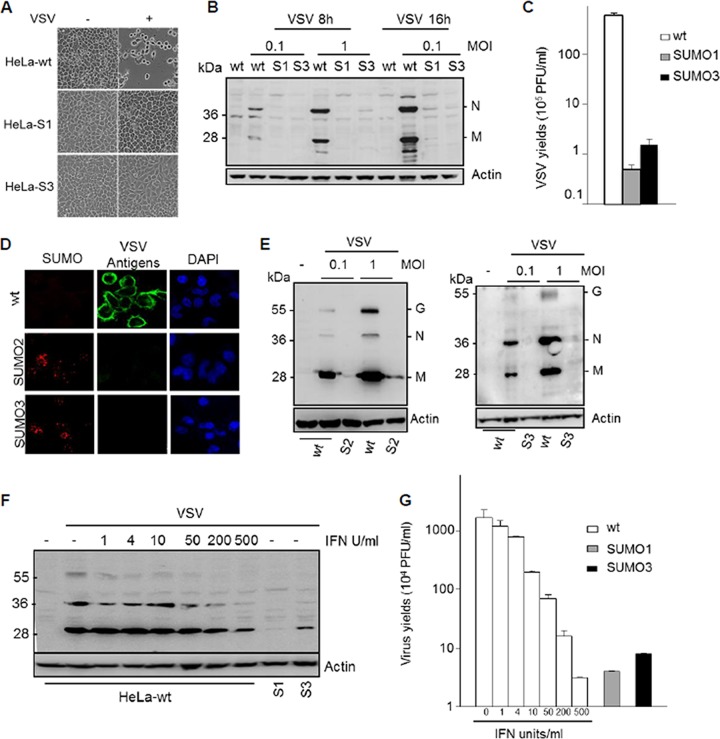
HeLa and U373MG cells stably expressing His-SUMO1 (HeLa-SUMO1 cells), U373MG cells stably expressing His-SUMO2 (U373MG-SUMO2 cells), and HeLa and U373MG cells stably expressing His-SUMO3 (HeLa-SUMO3 and U373MG-SUMO3 cells, respectively) were used throughout the course of this study and have been previously characterized using immunofluorescence and Western blot analysis (10). To determine the impact of the different SUMO paralogs on VSV replication, wild-type HeLa cells (HeLa-wt cells), HeLa-SUMO1 cells, and HeLa-SUMO3 cells were infected and the effect of SUMO expression on VSV-induced cell lysis, VSV protein synthesis, and viral production was subsequently determined. Compared to the effect on infected HeLa-wt cells, which lysed, HeLa-SUMO1 and HeLa-SUMO3 cells were protected against cell lysis during VSV infection (Fig. 1A). Western blot analysis using extracts from cells infected for 8 h and 16 h at different MOIs revealed that the synthesis of VSV proteins was inhibited in HeLa cells expressing SUMO1 or SUMO3 (Fig. 1B). The supernatants of HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells infected for 16 h at an MOI of 0.1 were used for the determination of virus yields. As seen in Fig. 1C, the VSV titers in infected HeLa-SUMO1 and HeLa-SUMO3 cells were reduced by 2 log units compared to those in infected HeLa-wt cells. Resistance to VSV infection was also demonstrated in U373MG cells stably expressing SUMO2 or SUMO3 (Fig. 1D and E). Dual immunofluorescence staining for SUMO2 or SUMO3 and the VSV antigens (Fig. 1D), as well as Western blot analysis using extracts from cells infected at an MOI of 0.1 or 1, showed that the expression of SUMO2 or SUMO3 in U373MG cells inhibited viral protein expression (Fig. 1E), reduced VSV multiplication, and protected cells from VSV-induced cell lysis (data not shown).
FIG 1.
SUMO confers resistance to VSV. (A) SUMO1 and SUMO3 protected cells from VSV-induced cell lysis. HeLa-wt, HeLa-SUMO1 (HeLa-S1), and HeLa-SUMO3 (HeLa-S3) cells were not infected (−) or infected (+) with VSV at an MOI of 0.1 for 16 h. The phase-contrast picture was acquired using a Nikon Eclipse TS100 microscope and Coolpix lens camera. Magnifications, ×10. (B) HeLa-wt, HeLa-SUMO1 (S1), and HeLa-SUMO3 (S3) cells were infected with VSV for 8 h or 16 h. Equal amounts of cell extracts were analyzed by Western blotting for the expression of VSV antigens and actin. (C) The supernatants of cells infected at an MOI of 0.1 for 16 h were used for the determination of virus yields. Mean values and standard deviations from three independent experiments are shown. (D) U373MG-wt, U373MG-SUMO2, and U373MG-SUMO3 cells were infected for 8 h at an MOI of 0.1 or 1. Double immunofluorescence was performed with cells infected at an MOI of 1 with anti-SUMO2/3 (red) and anti-VSV (green) antibodies. The nucleus was stained with DAPI. (E) Extracts from cells infected at different MOIs were analyzed by Western blotting for the expression of VSV antigens and actin. S2, U373MG-SUMO2 cells; S3, U373MG-SUMO3 cells. (F, G) Comparative effects of IFN and SUMO on VSV inhibition. HeLa-wt cells were treated for 24 h with 1, 4, 10, 50, 200, and 500 units/ml of IFN-α. Then, IFN-α-treated HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells were infected with VSV at an MOI of 1 for 8 h. (F) Extracts of infected cells were analyzed by Western blotting for VSV proteins and actin. (G) The supernatants were used to determine the virus yields. Mean values and standard deviations from three independent experiments are shown.
To compare the inhibition of VSV by SUMO1 or SUMO3 with that induced by IFN, HeLa-wt cells were treated for 24 h with 1, 4, 10, 50, 200, and 500 units/ml of IFN-α. Subsequently, IFN-α-treated HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells were infected with VSV at an MOI of 1 for 8 h. Extracts of the infected cells were analyzed by Western blotting for VSV proteins, and the supernatants were used to determine viral yields. Both the inhibition of viral protein synthesis (Fig. 1F) and the inhibition of VSV multiplication (Fig. 1G) in HeLa-SUMO1 and HeLa-SUMO3 cells (not treated with IFN-α) were comparable to those in HeLa-wt cells treated with 500 units/ml of IFN-α.
SUMO does not alter VSV entry but inhibits primary viral transcription.
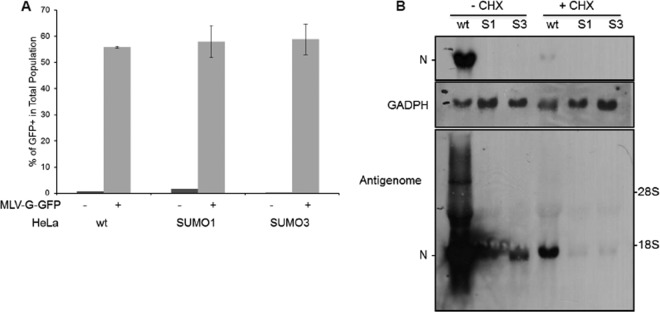
To investigate which viral step is targeted by SUMO, we first studied whether VSV entry was altered. To do this, we used murine leukemia virus (MLV)-derived vectors encoding GFP pseudotyped with the receptor-binding VSV G protein (VSV-G) (MLV-G-GFP) (20). This pseudovirus can undergo VSV-G-mediated entry but cannot produce its own VSV-G envelope and, hence, is capable of undergoing only a single round of infection. MLV-G-GFP was used to transduce wild-type U373MG (U373MG-wt), U373MG-SUMO3, HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells (Fig. 2A and data not shown). The results revealed that HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells had similar levels of GFP staining, with the levels reaching 55.8%, 58%, and 58.8% GFP-positive cells, respectively (Fig. 2A). The same results were obtained with U373MG cells expressing SUMO3 (data not shown). Taken together, the data demonstrate that VSV entry was not affected in SUMO-expressing cells.
FIG 2.
SUMO does not alter VSV entry but inhibits viral mRNA transcription. (A) HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells were infected with MLV-G-GFP for 48 h. The transduction efficiency, expressed as the percentage of GFP-positive (GFP+) cells, was measured at 48 h postinfection using flow cytometry. Mean values and standard deviations from three independent experiments are shown. (B) HeLa-wt, HeLa-SUMO1 (S1), and HeLa-SUMO3 (S3) cells were infected by VSV at an MOI of 4 in the absence of cycloheximide (−CHX) or the presence of cycloheximide (+CHX). After 6 h of infection, samples were analyzed for the presence of VSV N mRNA, antigenome mRNA, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA, as described in Materials and Methods. A lower level of exposure of the membrane shows the presence of N mRNA exclusively in the wt cells (in the absence of cycloheximide [−CHX]) (top); a higher level of exposure allowed the detection of a small amount of antigenome and N mRNA (bottom).
To determine whether viral transcription was altered by SUMO expression, HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells were infected with VSV at an MOI of 4 for 6 h. Total RNA isolated from extracts of the infected cells was analyzed by Northern blotting for VSV N mRNA (Fig. 2B). The amount of N mRNA in HeLa-SUMO1 and HeLa-SUMO3 cells was significantly reduced compared to that in control infected cells (Fig. 2B, first three lanes). To determine whether the decrease in the amount of N mRNA was due to the inhibition of VSV primary transcription, cells were treated with the protein synthesis inhibitor cycloheximide (CHX), which restricts viral transcription to the synthesis of primary mRNA, as genome replication requires the ongoing synthesis of N protein (21). CHX was added to each culture 1 h before infection and was maintained throughout the experiment. The results revealed that VSV primary transcription was inhibited by SUMO1 or SUMO3 expression (Fig. 2B), since N mRNA was undetectable in extracts from infected HeLa-SUMO1 and HeLa-SUMO3 cells treated with CHX (Fig. 2B, bottom, three last lanes). This result suggests that SUMO1 or SUMO3 expression affects primary transcription or an upstream step between viral entry and primary transcription.
SUMO confers resistance to VSV in an IFN-independent manner.
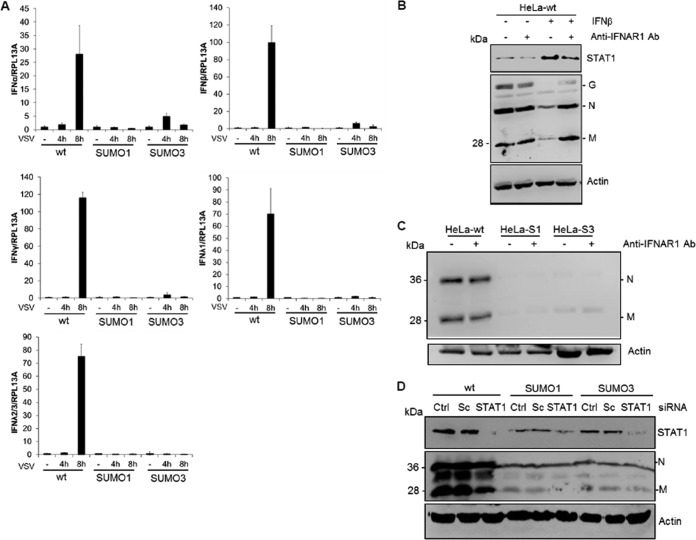
It has previously been shown that IFN inhibits VSV primary transcription (22). Furthermore, a key response of mammalian cells to virus infection is the production of IFN. As such, we tested whether SUMO expression alters IFN synthesis upon VSV infection. This was carried out by the quantification of IFN-α, IFN-β, IFN-λ1, IFN-λ2/λ3, and IFN-γ mRNAs by quantitative real-time PCR (qRT-PCR) in extracts of HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells infected at different times with VSV at an MOI of 1 (Fig. 3A). IFN-α, IFN-β, IFN-λ1, IFN-λ2/λ3, and IFN-γ mRNAs were induced in HeLa-wt cells at 8 h after VSV infection. In contrast, in SUMO1- or SUMO3-expressing cells, there was no significant increase in the expression of the mRNAs at any time after VSV infection (Fig. 3A). These results show that VSV-induced IFN production was highly decreased in SUMO-expressing cells and suggest that SUMO conferred resistance to VSV in an IFN-independent manner. To further confirm these results, HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells were treated with an anti-IFNAR1 MAb targeting the extracellular domain of the IFNAR1 chain of the human IFN-α/β receptor before VSV infection. The anti-IFNAR1 MAb inhibits the binding and biological activity of type I IFN (23). We show that the antibody reduced both IFN-β-induced STAT1 expression and anti-VSV activity (Fig. 3B). In contrast, this antibody did not alter the anti-VSV activity conferred by SUMO1 or SUMO3 expression (Fig. 3C). As STAT1 is the central transcription factor required for the biological responses of all types of IFN, we also determined the effect of its downregulation on SUMO1- or SUMO3-induced VSV resistance (Fig. 3D). The capacity of SUMO1 or SUMO3 to inhibit VSV protein synthesis was still maintained in cells depleted of STAT1 (Fig. 3D), further demonstrating that the anti-VSV effect of SUMO1 or SUMO3 is independent of IFN.
FIG 3.
SUMO confers resistance to VSV in an IFN-independent way. (A) HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells were infected with VSV at an MOI of 1 for 4 h or 8 h. Total RNA was extracted, and mRNA encoding IFN-α, IFN-β, IFN-λ1, IFN-λ2/λ3, IFN-γ, and RPL13A was quantified by qRT-PCR. Mean values and standard deviations from three independent experiments are shown. (B) HeLa-wt cells were left untreated or treated for 24 h with 100 units/ml of IFN-β in the absence or the presence of 20 μg/ml of purified anti-IFNAR1 antibody (Ab) before infection with VSV at an MOI of 1 for 8 h. (C) HeLa-wt, HeLa-SUMO1 (HeLa-S1), and HeLa-SUMO3 (HeLa-S3) cells were infected with VSV at an MOI of 1 for 8 h in the absence or the presence of 20 μg/ml of anti-IFNAR1 antibody. (D) STAT1 depletion does not alter the anti-VSV effect of SUMO. HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells were untreated (control [Ctrl]) or transfected with scrambled siRNA (Sc) or STAT1-specific siRNA (STAT1). Two days later, the cells were infected with VSV at an MOI of 1 for 8 h. Cell extracts were analyzed by Western blotting and revealed by antibodies directed against STAT1, VSV proteins, and actin.
Effect of SUMO on MxA localization, expression, and oligomerization.
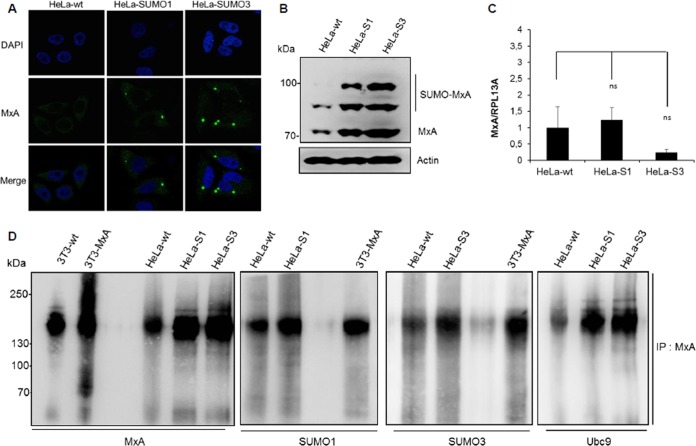
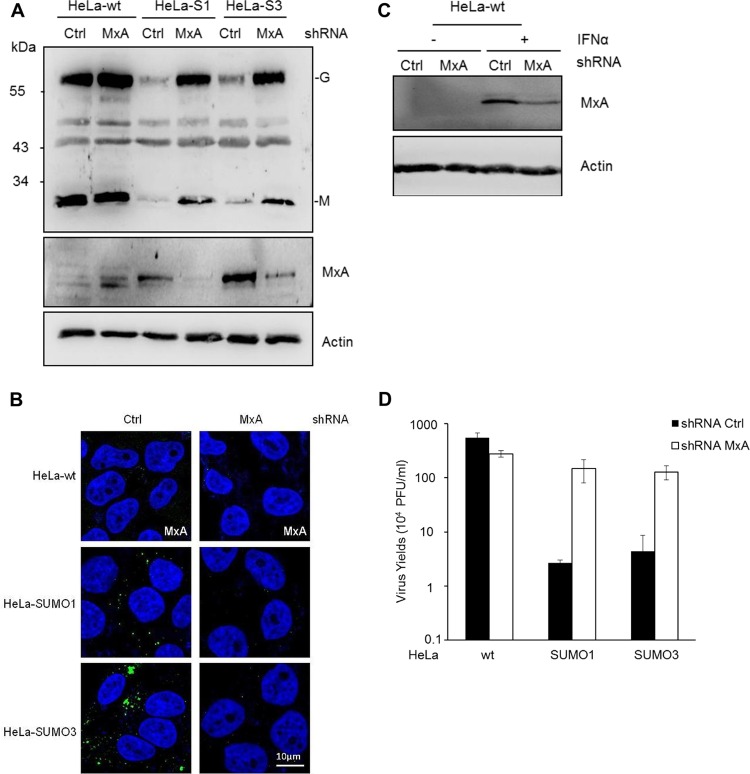
Among the IFN-stimulated gene (ISG) restriction factors exhibiting intrinsic anti-VSV activity (13), only MxA is capable of inhibiting VSV primary transcription (16). Our results demonstrate that VSV inhibition in SUMO-expressing cells was also at the level of primary transcription (Fig. 2B). Therefore, we focused on MxA and assessed its localization, expression, and oligomerization in SUMO-expressing cells. Immunofluorescence studies revealed that MxA was not detected in HeLa-wt cells. In contrast, in SUMO1- and SUMO3-expressing cells, the protein was found to be expressed and to localize in dots in the cytoplasm (Fig. 4A). The results of Western blot analysis of extracts isolated from these cells corroborated the immunofluorescence data and showed that MxA protein expression is much higher in SUMO1- and SUMO3-expressing cells than wt cells (Fig. 4B). It should be noted that the upper bands found to be migrating over 70 kDa in extracts from SUMO1- and SUMO3-expressing cells (Fig. 4B) may be attributed to SUMO-modified versions of the MxA protein, as we have previously shown that MxA is conjugated to SUMO at lysine 48 (17). The increase in the level of MxA protein expression in SUMO-expressing cells detected by Western blotting was confirmed by the use of two different rabbit anti-MxA antibodies (Fig. 4B and data not shown); however, only the antibodies from Proteintech were efficient in staining endogenous MxA by immunofluorescence. In contrast, the level of MxA mRNA was not increased by SUMO expression (Fig. 4C), thus suggesting that the effect of SUMO on MxA could be at the protein level, specifically, by affecting its stabilization.
FIG 4.
Effect of SUMO on MxA localization, expression, and oligomerization. (A) The immunofluorescence of MxA in HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells using rabbit anti-MxA antibodies is shown. (B) One hundred micrograms of protein extracts from HeLa-wt, HeLa-SUMO1 (HeLa-S1), and HeLa-SUMO3 (HeLa-S3) cells was analyzed by Western blotting using a 4 to 12% gradient gel, and the proteins were revealed with rabbit anti-MxA antibodies. (C) Total RNA was extracted from HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells, and mRNAs encoding MxA and RPL13A were quantified by RT-qPCR. Means and standard deviations from three independent experiments are shown. The Student t test was performed to determine the P value. ns, not significant. (D) SUMO enhances MxA oligomerization. Extracts from NIH 3T3-wt, NIH 3T3-MxA, HeLa-wt, HeLa-SUMO1, and HeLa SUMO3 cells were lysed, and proteins were cross-linked with DSS and immunoprecipitated (IP) using MxA antibodies. The eluates were subsequently analyzed by Western blotting using a 4 to 12% gradient gel, and proteins were revealed with anti-MxA, anti-SUMO1, anti-SUMO2/3, or anti-Ubc9 antibodies. The rabbit anti-MxA antibodies used recognize endogenous mouse Mx1 as well as human MxA.
It has been shown that oligomerization has a stabilizing effect on MxA protein (24) and that the MxA oligomerization capacity is important for its interaction with SUMO and Ubc9 (17). Therefore, we tested whether SUMO affects MxA oligomerization. To do this, extracts from NIH 3T3-wt, NIH 3T3-MxA, HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells were lysed, cross-linked, immunoprecipitated with MxA antibodies, and analyzed by Western blotting. Endogenous MxA in extracts from HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells was detected as oligomers, which became more abundant in HeLa cells expressing SUMO1 or SUMO3 or in NIH 3T3-MxA cells than in wild-type cells (Fig. 4D). Probing of the Western blot with anti-SUMO1, anti-SUMO2/3, or anti-Ubc9 antibodies indicated that SUMO1, SUMO3, and Ubc9 interacted with MxA oligomers (Fig. 4D). These results suggest that SUMO enhances MxA's capacity to oligomerize, thus protecting the MxA protein from degradation and providing a stable intracellular pool of MxA able to protect cells from viral infection.
MxA mediates the anti-VSV effect of SUMO.
In order to demonstrate the implication of MxA in the anti-VSV effect of SUMO, we silenced MxA by transducing HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells with lentiviral vectors expressing shRNA targeting MxA (shMxA). As a control, we also expressed a nontargeting shRNA. The shMxA was efficient in downregulating MxA protein expression in both HeLa-SUMO1 and HeLa-SUMO3 cells (Fig. 5A) and in decreasing cytoplasmic MxA dot staining (Fig. 5B). In addition, treatment with IFN-α2 increased the MxA levels in HeLa-wt cells, but the increase was much less pronounced in shMxA-transduced cells, indicating an effective knockdown of MxA (Fig. 5C). Remarkably, shRNA-mediated MxA knockdown rendered the SUMO1- and SUMO3-expressing cells more sensitive to VSV infection, as confirmed by the higher expression levels of VSV proteins (Fig. 5A) and the increased viral titer (Fig. 5D). The depletion of endogenous MxA in HeLa-wt cells infected at an MOI of 1 did not alter the expression of viral proteins (Fig. 5A); however, larger amounts of VSV proteins were expressed in cells depleted of MxA than in control cells upon infection at a lower MOI of 0.2 (data not shown).
FIG 5.
MxA mediates SUMO-induced VSV resistance. (A) Nontargeting control shRNA (Ctrl) and shRNA targeting MxA stably expressed in HeLa-wt, HeLa-SUMO1 (HeLa-S1) and HeLa-SUMO3 (HeLa-S3) cells were infected with VSV at an MOI of 1 for 8 h. Twenty micrograms of protein extracts from infected cells was analyzed by Western blotting for VSV proteins, MxA, and actin. (B) Immunofluorescence using rabbit anti-MxA antibodies was carried out with HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells stably expressing control shRNA and shMxA. (C) The knockdown of IFN-induced MxA by shRNA in HeLa-wt cells is shown using mouse monoclonal anti-MxA antibody (which does not recognize endogenous MxA). (D) The supernatants from infected cells were used to determine virus yields. Mean values and standard deviations from three independent experiments are shown.
Taken together, our results show that the MxA protein was highly stabilized through its oligomerization in cells expressing SUMO1 or SUMO3 and that its knockdown in SUMO-expressing cells abrogated the anti-VSV effect of SUMO, thus demonstrating that MxA is the key mediator of the SUMO-induced resistance to VSV.
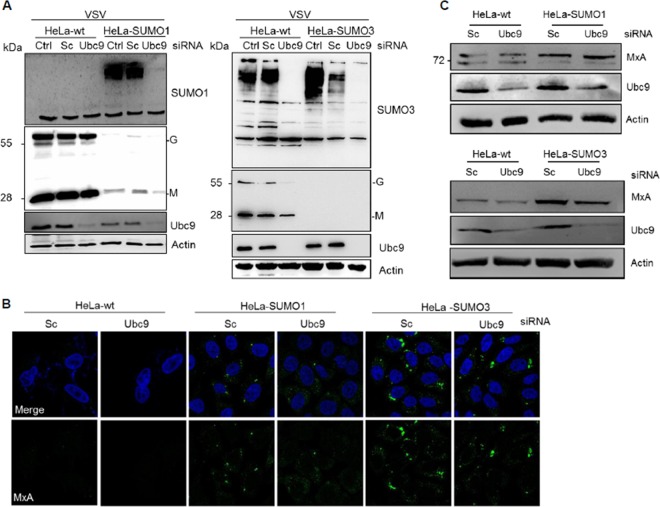
SUMO confers resistance to VSV in cells depleted of Ubc9.
In a reverse experiment, we examined the impact of the inhibition of SUMOylation on SUMO-induced VSV resistance. For this purpose, HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells were depleted of Ubc9 (Fig. 6A), the unique E2-conjugating enzyme, and subsequently infected with VSV at an MOI of 2 for 8 h. Analysis of the cell extracts was carried out by Western blotting, and the data revealed that the knockdown of Ubc9 was accompanied, as expected, by a decrease in the amounts of SUMO-conjugated proteins in all cell extracts (Fig. 6A). Ubc9 depletion in wt cells did not alter VSV protein synthesis and, unexpectedly, did not result in a reduced SUMO1-induced (Fig. 6A, left) or SUMO3-induced (Fig. 6A, right) anti-VSV effect. Consequently, we analyzed the effect of Ubc9 depletion on MxA localization (Fig. 6B) and protein expression (Fig. 6C) in SUMO-expressing cells. Immunofluorescence analysis revealed that in SUMO1- and SUMO3-expressing cells, the MxA staining was similar in cells depleted of Ubc9 and cells transfected with scrambled siRNA (Fig. 6B). Western blot analysis showed that the MxA protein was still expressed in SUMO-expressing cells depleted of Ubc9 (Fig. 6C) and able to confer VSV resistance (Fig. 6A). Therefore, depletion of Ubc9 decreased new SUMO conjugation to substrates and had no effect on the already existing pool of MxA stabilized in SUMO-expressing cells.
FIG 6.
SUMO confers resistance to VSV in cells depleted of Ubc9. (A) HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells, nontransfected cells (control [Ctrl]), and cells transfected with scrambled siRNA (Sc) or siRNA targeting Ubc9 were infected with VSV at an MOI of 2 for 8 h. Cell extracts were analyzed by Western blotting for Ubc9, VSV proteins, SUMO1, SUMO2/3, and actin. (B) Immunofluorescence staining for MxA in HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells depleted of Ubc9. (C) MxA protein expression in HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells depleted of Ubc9. Cell extracts from the assay whose results are presented in panel A were analyzed by Western blotting for MxA, Ubc9, and actin.
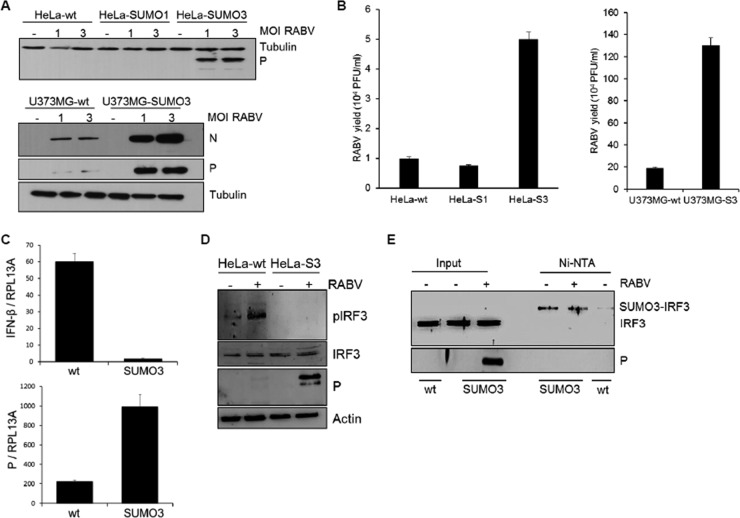
Expression of SUMO3 renders cells more sensitive to rabies virus infection.
To further elucidate the effects of SUMO on viral infection, we carried out another set of experiments using RABV, another member of the Rhabdoviridae family. HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells (Fig. 7A, top, and B, left) and U373MG-wt and U373MG-SUMO3 cells (Fig. 7A, bottom, and B, right) were infected with RABV at different MOIs for 24 h, the cell extracts were analyzed by Western blotting for RABV P expression (Fig. 7A), and the supernatants were used to determine the RABV titers (Fig. 7B). The results show that SUMO3 expression but not SUMO1 expression rendered U373-MG cells more sensitive to RABV than wt cells, as significantly higher levels of RABV P (Fig. 7A, bottom) were synthesized, in line with the enhanced viral growth (Fig. 7B, right). Furthermore, it should be noted that the HeLa cells, which are not permissive to RABV, became sensitive to infection in the presence of SUMO3 (Fig. 7A, top, and B, left). Analysis by qRT-PCR of the IFN-β mRNA produced upon RABV infection revealed that IFN-β mRNA was synthesized in wt cells but not in cells expressing SUMO3 (Fig. 7C). This inhibition of IFN synthesis was due to a lower level of phosphorylated IRF3 in RABV-infected SUMO3 cells than in infected wt cells (Fig. 7D). Next, we investigated whether IRF3 was SUMOylated in SUMO3-expressing cells, which may result in its lower level of activation upon RABV infection. We performed an Ni-NTA purification of extracts from HeLa-wt and HeLa-SUMO3 cells either not infected or infected with RABV (Fig. 7E). As expected, RABV P was expressed in the input of the extracts isolated from the infected SUMO3 cells. Ni-NTA purification revealed that IRF3 was highly conjugated to SUMO3 at a level similar to that found in noninfected or RABV-infected SUMO3 cells (Fig. 7E). The results also demonstrate that SUMO1 expression did not affect RABV infection (Fig. 7A). We were not able to detect IRF3 modification by SUMO1 in Ni-NTA-purified extracts from HeLa-SUMO1 cells (data not shown). This observation is in line with the notion that SUMO1 conjugation is significantly more difficult to detect than SUMO2/3 conjugation (25).
FIG 7.
Expression of SUMO3 renders cells more sensitive to RABV infection. (A) HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells (top) and U373MG-wt and U373MG-SUMO3 cells (bottom) were infected with RABV at different MOIs for 24 h, and cell extracts were analyzed by Western blotting for RABV P and tubulin. (B) The supernatants from cells infected with RABV at an MOI of 3 were used to determine RABV titers. Mean values and standard deviations from three independent experiments are shown. (C) HeLa-wt and HeLa-SUMO3 cells were infected with RABV at an MOI of 3 for 24 h, total RNA was extracted, and mRNAs encoding IFN-β, RABV P, and RPL13A were quantified by qRT-PCR. Mean values and standard deviations from three independent experiments are shown. (D) Extracts from HeLa-wt and HeLa-SUMO3 (HeLa-S3) cells infected with RABV at an MOI of 3 for 24 h were analyzed by Western blotting for IRF3, phosphorylated IRF3 (pIRF3), RABV P, and actin. (E) Cell extracts from uninfected HeLa-wt cells and from HeLa-SUMO3 cells uninfected or infected with RABV at an MOI of 3 for 24 h were purified on Ni-NTA-agarose beads. Western blotting analyses were performed with the inputs using anti-IRF3 and anti-RABV P antibodies and the purified extracts using anti-IRF3 antibodies.
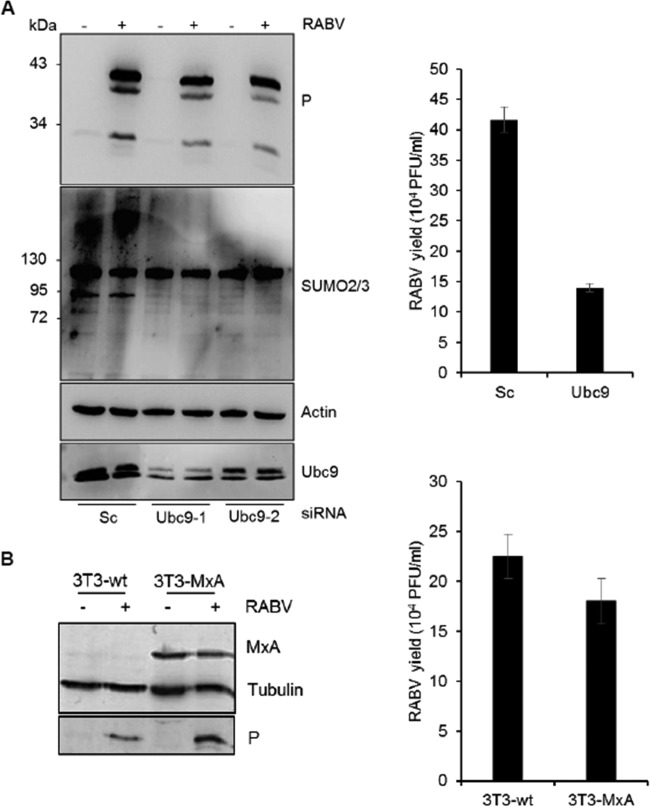
From these results, we conclude that the conjugation of IRF3 to SUMO3 reduced both its activation and IFN production upon RABV infection, thereby rendering the cells more susceptible to RABV infection. A reverse experiment was carried out to reduce the levels of cellular SUMOylation through the knockdown of endogenous Ubc9 in wt cells using siRNA targeting Ubc9 (Fig. 8A). This resulted in a small and reproductive reduction of RABV P synthesis (Fig. 8A, left) and RABV replication (Fig. 8A, right). In addition, it can be observed that the SUMOylation in wt cells remained unchanged between RABV-infected and noninfected cells and, as expected, decreased when Ubc9 was depleted (Fig. 8A, left).
FIG 8.
Effect of Ubc9 depletion in wt cells and MxA overexpression on RABV infection. (A) U373MG cells transfected with scrambled siRNA (Sc) or siRNA targeting Ubc9-1 or Ubc9-2 were infected with RABV at an MOI of 1 for 24 h. Cell extracts were analyzed by Western blotting for Ubc9, RABV P, SUMO2/3, and actin (left), and the supernatants from cells transfected with scrambled siRNA or siRNA targeting Ubc9-1 were used for the determination of titers (right). Mean values and standard deviations from three independent experiments are shown. (B) NIH 3T3 cells transfected with the empty vector or overexpressing MxA were infected with RABV at an MOI of 1 for 24 h; cell extracts were analyzed by Western blotting for RABV P, MxA, and tubulin (left), and the supernatants were used for the determination of the titers (right). Mean values and standard deviations from three independent experiments are shown.
To further understand the differential effect of SUMO on VSV and RABV, the effect of human MxA expression on RABV replication was tested. NIH 3T3 cells transfected with the empty vector or overexpressing MxA were infected with RABV (Fig. 8). Protein extracts from uninfected or infected cells were analyzed by Western blotting for RABV protein expression at 1 day postinfection. The level of RABV P expression (Fig. 8B, left) and the viral titers (Fig. 8B, right) were similar in infected control NIH 3T3 and NIH 3T3-MxA cells, indicating that MxA did not confer resistance to RABV, as previously shown (26).
DISCUSSION
SUMOylation is associated with the replication of a variety of viruses with regard to the modification of viral proteins and the modulation of SUMOylated cellular proteins implicated in antiviral defense (2, 27–30). The covalent attachment of the small SUMO moiety to substrate proteins has recently emerged as a main regulatory system, leading to dramatic changes in the SUMO proteome (31–34) with consequences on IFN responses and antiviral defense mechanisms (9, 10, 27).
SUMOylation is known to regulate the signaling pathway governing IFN production, as virus-mediated IRF3 and IRF7 SUMOylation attenuates IFN-β synthesis (12). We also show in this study that SUMO expression results in the SUMOylation of IRF3, subsequently decreasing the virus-induced activation of IRF3 and leading to inhibition of RABV-induced IFN-β production. Furthermore, SUMO expression inhibited the increase in the levels of IFN-α, IFN-β, IFN-λ1, IFN-λ2/λ3, and IFN-γ mRNAs in VSV-infected cells. Accordingly, it has recently been reported that SUMOylation deficiency in Ubc9−/− cells causes an enhancement of IFN-β production after the engagement of Toll-like receptors compared to that in wild-type cells (9). Taken together, these results show that SUMO significantly reduces the level of IFN production.
As expected, the decrease in the level of RABV-induced IFN production by SUMO renders cells more sensitive to viral infection. Intriguingly, although SUMO reduced the amount of VSV-induced IFN, the cells were resistant to viral infection. We show in this study that the stable expression of SUMO1, SUMO2, or SUMO3 in human cell lines did not affect VSV entry but did inhibit primary viral mRNA transcription, thereby blocking viral protein synthesis and viral production, as well as protecting the cells from virus-induced cell lysis. SUMO-induced VSV resistance was independent of IFN, as it was maintained in cells depleted of STAT1 or in cells treated with anti-IFNAR1 antibody. These results highlight the differences between RABV and VSV, two members of the Rhabdoviridae family, and further define the role of SUMO in viral infection.
Unlike the IFN-mediated innate immune response, intrinsic antiviral resistance (known as intrinsic immunity) is mediated by endogenously expressed antiviral proteins (defined as restriction factors), which confer viral resistance in a direct manner (in the absence of IFN synthesis) by a variety of inhibitory mechanisms. These constitutive antiviral proteins can be further induced by viral infection and/or IFN treatment. Individually, these antiviral proteins can interfere with a particular stage of the VSV life cycle (13). For example, IFN-induced transmembrane (IFITM) proteins block viral entry (35), cholesterol 25-hydroxylase (CH25h) impairs the virus cell fusion step by inducing cellular membrane changes (36), MxA inhibits primary transcription (15, 16), ISG20 (a 3′-5′ exonuclease) degrades single-stranded viral RNA (37), the PML protein inhibits secondary transcription (38, 39), IFN-induced proteins with tetratricopeptide repeats (IFIT) (40) inhibit viral translation, and tetherin prevents the release of virions from the cell (35).
Among the numerous anti-VSV restriction factors, only MxA was shown to inhibit VSV primary transcription (13, 16), and we found that the inhibition of VSV in SUMO-expressing cells was also at the level of primary transcription. As such, we analyzed the role of MxA within this system. We have previously shown that MxA is conjugated to SUMO at lysine 48 (17). We demonstrated in the study described in this report that the MxA protein is highly stabilized in cells expressing SUMO1 or SUMO3 and that its depletion in SUMO-expressing cells blocks the anti-VSV effect of SUMO, thus signifying that MxA is a key mediator of SUMO-induced resistance to VSV. To further confirm the role of SUMO in VSV resistance, Ubc9 was silenced in HeLa-wt, HeLa-SUMO1, and HeLa-SUMO3 cells. Ubc9 depletion decreased the level of SUMO-conjugated proteins but did not alter VSV protein synthesis in wt cells and did not reduce SUMO-induced VSV resistance because MxA protein levels were stably maintained in SUMO-expressing cells depleted of Ubc9.
GTPase activity and oligomerization are required for MxA antiviral activity (41–43), but the precise mechanism of antiviral action differs between viruses. The MxA protein forms an oligomeric complex via the stalk domain. We previously reported that MxA interacts with SUMO via its CID-GED (stalk) domain and that the MxA oligomerization capacity is important for its interaction with SUMO and Ubc9 (17). Accordingly, monomeric mutant MxA-L612K has a reduced interaction with SUMO and Ubc9 (17). Importantly, the MxA-L612K mutant is rapidly degraded in cells compared to wild-type MxA, demonstrating that the self-assembly of the MxA protein is critical for protein stability (24). Our finding suggests that SUMO enhances MxA oligomers, thus providing a stable intracellular pool of MxA that is available to fight invading viruses.
A large number of studies have shown that viruses interact with host SUMO systems in various ways, with no apparent unifying theme or mechanism being found to exist. VSV and RABV (both of which are RNA viruses that replicate in the cytoplasm) did not alter global cellular protein SUMOylation (data not shown). Furthermore, the knockdown of Ubc9 did not alter VSV protein synthesis but slightly inhibited RABV replication. Infection with herpes simplex virus 1 (a DNA virus that replicates in the nucleus), on the other hand, causes a loss of SUMO conjugates; however, the depletion of Ubc9 has no subsequent effect on virus replication (44). In contrast, infection with the influenza A virus (an RNA virus that replicates in the nucleus) causes a global increase in SUMO conjugates (45), and the depletion of Ubc9 decreases overall viral replication efficiency (46). Overall, depending on the infecting virus, the SUMO system may affect viral replication either negatively or positively, or it may have no effect.
In conclusion, although IFN production was highly reduced in VSV-infected SUMO-expressing cells, the intrinsic anti-VSV activity of SUMO mediated by MxA was not affected. However, in the case of RABV, the lower level of IFN synthesis rendered SUMO3-expressing cells more sensitive to viral infection than wt cells, even though MxA was stabilized in SUMO-expressing cells, since its expression did not confer resistance to RABV (26; this study). Further experiments are required to determine whether the replication process of other viral families can be inhibited by SUMO expression via the stabilization of MxA or other restriction factors. Our research within this field will advance our knowledge and understanding of the relationship between the SUMO pathway and virus life cycles and infection and will also positively contribute to the development of effective antiviral treatments.
ACKNOWLEDGMENTS
We thank Francois-Loïc Cosset, Pierre Eid, and Jovan Pavlovic for sharing materials. We are very grateful to Gianfranco Pancino for the critical reading of the manuscript. We also thank the FACS and Cell Sorting Platform (S2C) of the Saints Pères Center, Paris Descartes University, for fluorescence-activated cell sorting (FACS) analyses.
REFERENCES
- 1.Kerscher O, Felberbaum R, Hochstrasser M. 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.Boggio R, Chiocca S. 2006. Viruses and sumoylation: recent highlights. Curr Opin Microbiol 9:430–436. doi: 10.1016/j.mib.2006.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson ES. 2004. Protein modification by SUMO. Annu Rev Biochem 73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 4.Geiss-Friedlander R, Melchior F. 2007. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 5.Hay RT. 2007. SUMO-specific proteases: a twist in the tail. Trends Cell Biol 17:370–376. doi: 10.1016/j.tcb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Ulrich HD. 2009. The SUMO system: an overview. Methods Mol Biol 497:3–16. doi: 10.1007/978-1-59745-566-4_1. [DOI] [PubMed] [Google Scholar]
- 7.Geoffroy MC, Hay RT. 2009. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Mol Cell Biol 10:564–568. doi: 10.1038/nrm2707. [DOI] [PubMed] [Google Scholar]
- 8.Dasso M. 2008. Emerging roles of the SUMO pathway in mitosis. Cell Div 3:5. doi: 10.1186/1747-1028-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decque A, Joffre O, Magalhaes JG, Cossec JC, Blecher-Gonen R, Lapaquette P, Silvin A, Manel N, Joubert PE, Seeler JS, Albert ML, Amit I, Amigorena S, Dejean A. 2016. Sumoylation coordinates the repression of inflammatory and anti-viral gene-expression programs during innate sensing. Nat Immunol 17:140–149. doi: 10.1038/ni.3342. [DOI] [PubMed] [Google Scholar]
- 10.Maarifi G, Maroui MA, Dutrieux J, Dianoux L, Nisole S, Chelbi-Alix MK. 2015. Small ubiquitin-like modifier alters IFN response. J Immunol 195:2312–2324. doi: 10.4049/jimmunol.1500035. [DOI] [PubMed] [Google Scholar]
- 11.Chang TH, Kubota T, Matsuoka M, Jones S, Bradfute SB, Bray M, Ozato K. 2009. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog 5:e1000493. doi: 10.1371/journal.ppat.1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubota T, Matsuoka M, Chang TH, Tailor P, Sasaki T, Tashiro M, Kato A, Ozato K. 2008. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J Biol Chem 283:25660–25670. doi: 10.1074/jbc.M804479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blondel D, Maarifi G, Nisole S, Chelbi-Alix MK. 2015. Resistance to Rhabdoviridae infection and subversion of antiviral responses. Viruses 7:3675–3702. doi: 10.3390/v7072794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haller O, Stertz S, Kochs G. 2007. The Mx GTPase family of interferon-induced antiviral proteins. Microbes Infect 9:1636–1643. doi: 10.1016/j.micinf.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Pavlovic J, Zurcher T, Haller O, Staeheli P. 1990. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J Virol 64:3370–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staeheli P, Pavlovic J. 1991. Inhibition of vesicular stomatitis virus mRNA synthesis by human MxA protein. J Virol 65:4498–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brantis-de-Carvalho CE, Maarifi G, Goncalves Boldrin PE, Zanelli CF, Nisole S, Chelbi-Alix MK, Valentini SR. 2015. MxA interacts with and is modified by the SUMOylation machinery. Exp Cell Res 330:151–163. doi: 10.1016/j.yexcr.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 18.Raux H, Iseni F, Lafay F, Blondel D. 1997. Mapping of monoclonal antibody epitopes of the rabies virus P protein. J Gen Virol 78(Pt 1):119–124. doi: 10.1099/0022-1317-78-1-119. [DOI] [PubMed] [Google Scholar]
- 19.Raux H, Obiang L, Richard N, Harper F, Blondel D, Gaudin Y. 2010. The matrix protein of vesicular stomatitis virus binds dynamin for efficient viral assembly. J Virol 84:12609–12618. doi: 10.1128/JVI.01400-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Negre D, Mangeot PE, Duisit G, Blanchard S, Vidalain PO, Leissner P, Winter AJ, Rabourdin-Combe C, Mehtali M, Moullier P, Darlix JL, Cosset FL. 2000. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Ther 7:1613–1623. doi: 10.1038/sj.gt.3301292. [DOI] [PubMed] [Google Scholar]
- 21.Patton JT, Davis NL, Wertz GW. 1984. N protein alone satisfies the requirement for protein synthesis during RNA replication of vesicular stomatitis virus. J Virol 49:303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang XT, Samuel CE. 1987. Mechanism of interferon action. Interferon alpha inhibits vesicular stomatitis virus primary transcript accumulation in P1/eIF-2 alpha protein kinase-deficient human fibroblast cells. J Biol Regul Homeost Agents 1:157–165. [PubMed] [Google Scholar]
- 23.Giron-Michel J, Weill D, Bailly G, Legras S, Nardeux PC, Azzarone B, Tovey MG, Eid P. 2002. Direct signal transduction via functional interferon-alphabeta receptors in CD34+ hematopoietic stem cells. Leukemia 16:1135–1142. doi: 10.1038/sj.leu.2402492. [DOI] [PubMed] [Google Scholar]
- 24.Janzen C, Kochs G, Haller O. 2000. A monomeric GTPase-negative MxA mutant with antiviral activity. J Virol 74:8202–8206. doi: 10.1128/JVI.74.17.8202-8206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitoh H, Hinchey J. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem 275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 26.Leroy M, Pire G, Baise E, Desmecht D. 2006. Expression of the interferon-alpha/beta-inducible bovine Mx1 dynamin interferes with replication of rabies virus. Neurobiol Dis 21:515–521. doi: 10.1016/j.nbd.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 27.Sloan E, Tatham MH, Groslambert M, Glass M, Orr A, Hay RT, Everett RD. 2015. Analysis of the SUMO2 proteome during HSV-1 infection. PLoS Pathog 11:e1005059. doi: 10.1371/journal.ppat.1005059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pampin M, Simonin Y, Blondel B, Percherancier Y, Chelbi-Alix MK. 2006. Cross talk between PML and p53 during poliovirus infection: implications for antiviral defense. J Virol 80:8582–8592. doi: 10.1128/JVI.00031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hilgarth RS, Murphy LA, Skaggs HS, Wilkerson DC, Xing H, Sarge KD. 2004. Regulation and function of SUMO modification. J Biol Chem 279:53899–53902. doi: 10.1074/jbc.R400021200. [DOI] [PubMed] [Google Scholar]
- 30.Wimmer P, Schreiner S, Dobner T. 2012. Human pathogens and the host cell SUMOylation system. J Virol 86:642–654. doi: 10.1128/JVI.06227-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamoliatte F, Caron D, Durette C, Mahrouche L, Maroui MA, Caron-Lizotte O, Bonneil E, Chelbi-Alix MK, Thibault P. 2014. Large-scale analysis of lysine SUMOylation by SUMO remnant immunoaffinity profiling. Nat Commun 5:5409. doi: 10.1038/ncomms6409. [DOI] [PubMed] [Google Scholar]
- 32.Tammsalu T, Matic I, Jaffray EG, Ibrahim AF, Tatham MH, Hay RT. 2014. Proteome-wide identification of SUMO2 modification sites. Sci Signal 7:rs2. doi: 10.1126/scisignal.2005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hendriks IA, D'Souza RC, Yang B, Verlaan-de Vries M, Mann M, Vertegaal AC. 2014. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat Struct Mol Biol 21:927–936. doi: 10.1038/nsmb.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galisson F, Mahrouche L, Courcelles M, Bonneil E, Meloche S, Chelbi-Alix MK, Thibault P. 2011. A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Mol Cell Proteomics 10:M110.004796. doi: 10.1074/mcp.M110.004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weidner JM, Jiang D, Pan XB, Chang J, Block TM, Guo JT. 2010. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J Virol 84:12646–12657. doi: 10.1128/JVI.01328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu SY, Aliyari R, Chikere K, Li G, Marsden MD, Smith JK, Pernet O, Guo H, Nusbaum R, Zack JA, Freiberg AN, Su L, Lee B, Cheng G. 2013. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity 38:92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Espert L, Degols G, Gongora C, Blondel D, Williams BR, Silverman RH, Mechti N. 2003. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. J Biol Chem 278:16151–16158. doi: 10.1074/jbc.M209628200. [DOI] [PubMed] [Google Scholar]
- 38.Chelbi-Alix MK, Quignon F, Pelicano L, Koken MH, de The H. 1998. Resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J Virol 72:1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El Asmi F, Maroui MA, Dutrieux J, Blondel D, Nisole S, Chelbi-Alix MK. 2014. Implication of PMLIV in both intrinsic and innate immunity. PLoS Pathog 10:e1003975. doi: 10.1371/journal.ppat.1003975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fensterl V, Wetzel JL, Ramachandran S, Ogino T, Stohlman SA, Bergmann CC, Diamond MS, Virgin HW, Sen GC. 2012. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog 8:e1002712. doi: 10.1371/journal.ppat.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao S, von der Malsburg A, Paeschke S, Behlke J, Haller O, Kochs G, Daumke O. 2010. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 465:502–506. doi: 10.1038/nature08972. [DOI] [PubMed] [Google Scholar]
- 42.Ponten A, Sick C, Weeber M, Haller O, Kochs G. 1997. Dominant-negative mutants of human MxA protein: domains in the carboxy-terminal moiety are important for oligomerization and antiviral activity. J Virol 71:2591–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pitossi F, Blank A, Schroder A, Schwarz A, Hussi P, Schwemmle M, Pavlovic J, Staeheli P. 1993. A functional GTP-binding motif is necessary for antiviral activity of Mx proteins. J Virol 67:6726–6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boutell C, Cuchet-Lourenco D, Vanni E, Orr A, Glass M, McFarlane S, Everett RD. 2011. A viral ubiquitin ligase has substrate preferential SUMO targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. PLoS Pathog 7:e1002245. doi: 10.1371/journal.ppat.1002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pal S, Santos A, Rosas JM, Ortiz-Guzman J, Rosas-Acosta G. 2011. Influenza A virus interacts extensively with the cellular SUMOylation system during infection. Virus Res 158:12–27. doi: 10.1016/j.virusres.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 46.Wu CY, Jeng KS, Lai MM. 2011. The SUMOylation of matrix protein M1 modulates the assembly and morphogenesis of influenza A virus. J Virol 85:6618–6628. doi: 10.1128/JVI.02401-10. [DOI] [PMC free article] [PubMed] [Google Scholar]