ABSTRACT
An Autographa californica nucleopolyhedrovirus-encoded microRNA (miRNA), AcMNPV-miR-1, downregulates the ac94 gene, reducing the production of infectious budded virions and accelerating the formation of occlusion-derived virions. In the current study, four viruses that constitutively overexpress AcMNPV-miR-1 were constructed to further explore the function of the miRNA. In addition to the ac94 gene, two new viral gene targets (ac18 and ac95) of AcMNPV-miR-1 were identified, and the possible interacting proteins were verified and tested. In the context of AcMNPV-miR-1 overexpression, ac18 was slightly upregulated, and ac95 was downregulated. Several interacting proteins were identified, and a functional pathway for AcMNPV-miR-1 was deduced. AcMNPV-miR-1 overexpression decreased budded virus infectivity, reduced viral DNA replication, accelerated polyhedron formation, and promoted viral infection efficiency in Trichoplusia ni larvae, suggesting that AcMNPV-miR-1 restrains virus infection of cells but facilitates virus infection of larvae.
IMPORTANCE Recently, microRNAs (miRNAs) have been widely reported as moderators or regulators of mammalian cellular processes, especially disease-related pathways in humans. However, the roles played by miRNAs encoded by baculoviruses, which infect numerous beneficial insects and agricultural pests, have rarely been described. To explore the actions of virus-encoded miRNAs, we investigated an miRNA encoded by Autographa californica nucleopolyhedrovirus (AcMNPV-miR-1). We previously identified this miRNA through the exogenous addition of AcMNPV-miR-1 mimics. In the current study, we constitutively overexpressed AcMNPV-miR-1 and analyzed the resultant effects to more comprehensively assess what is indeed the function of this miRNA during viral infection. In addition, we widely explored the target genes for the miRNA in the viral and host genomes and proposed a possible functional network for AcMNPV-miR-1, which provides a better general understanding of virus-encoded miRNAs. In brief, our study implied that AcMNPV-miR-1 constrains viral replication and cellular infection but enhances larval infection.
INTRODUCTION
MicroRNAs (miRNAs) are short RNAs ∼22 nucleotides (nt) in length that are derived from transcripts that fold back on themselves to form distinctive hairpin structures (1). miRNAs modulate the stability and/or translational potential of their mRNA targets (2, 3) to achieve regulatory control over virtually every biological process. In many cases, a single miRNA can regulate abundant targets (4, 5), and one mRNA can interact with several miRNAs (6). Therefore, the network formed between miRNAs and their target mRNAs is comprehensive and complex.
Over the last decade, there have been many developments in the understanding of virus-encoded miRNAs. In 2005, miRNAs encoded by simian vacuolating virus 40 (SV40) were reported to reduce the expression of viral T antigens (7). Since then, several virus-encoded miRNAs have been reported. For example, miR-BART, which is encoded by Epstein-Barr virus (EBV), was demonstrated to modify transcription of the BALF5 gene (8). Additionally, the insect Heliothis virescens ascovirus-encoded miRNA HvAV-miR-1, derived from the major capsid protein gene, was shown to downregulate the expression of viral DNA polymerase I (9). In a recent review, viral miRNAs were further summarized as moderators or mediators of virus-host interactions and were suggested to play a role in the immune response (10). These findings indicate that virus-encoded miRNAs may be vital for the attack and counterattack mechanisms that exist between hosts and viruses.
Autographa californica nucleopolyhedrovirus (AcMNPV) is a model species of the Baculoviridae family, which contains viruses that infect numerous beneficial insects and agricultural pests. Baculovirus-encoded miRNAs have been shown to participate in several activities during viral infection (11, 12). In our previous study, we identified the AcMNPV-encoded miRNA known as AcMNPV-miR-1. This miRNA is 20 nt in length and is encoded in the reverse strand of the ac94 gene coding sequence (CDS). The precursor of this miRNA is 58 nt in length and is transcribed at an early stage of virus infection. The mature AcMNPV-miR-1 accumulates at approximately 12 h postinfection (h p.i.) and downregulates the mRNA level of the viral gene ac94 (ODV-E25), an early gene required for infectious budded virion (BV) production and occlusion-derived virion (ODV) formation (13). In the presence of an AcMNPV-miR-1 mimic, we demonstrated a reduction in the infectivity of BVs and an increase in the formation of ODVs (14). However, the mechanism by which endogenous AcMNPV-miR-1 acts during a natural viral infection is unknown. In the current study, we further explored the function of AcMNPV-miR-1 and detected an extensive array of activities in which this miRNA is involved.
MATERIALS AND METHODS
Viruses and cell lines.
The bacmid bMON14272 (Invitrogen), which contains an AcMNPV genome, was maintained in DH10B cells as previously described (15). Spodoptera frugiperda IPLB-Sf21-AE clonal isolate 9 (Sf9) insect cells were cultured at 27°C in Grace's medium (Gibco) containing 10% fetal bovine serum (Gibco). The viral inoculum was allowed to adsorb to cells for 1 h of infection, or the bacmid was applied to cells for 4 h of transfection at 27°C. BV titers were determined by a 50% tissue culture infective dose (TCID50) endpoint dilution assay in Sf9 cells as previously described (16). Time zero was defined as the time when the viral inoculum was replaced.
Construction of viruses overexpressing AcMNPV-miR-1.
To generate viruses that constitutively overexpressed AcMNPV-miR-1, four donor plasmids (pFB1-1×-GP, pFB1-2×-GP, pFB1-4×-GP, and pFB1-8×-GP) were constructed as follows. A 199-bp fragment containing one copy of the AcMNPV-miR-1 precursor was synthesized (Beijing Genomics Institute-ShenZhen Co. [BGI-ShenZhen]) and ligated downstream of the IE-1 promoter and upstream of the SV40 poly(A) sequence in the pBlueScript plasmid. Then, the three-component fragment was digested by SacI and XhoI and inserted into a pFastBac1-PH-GFP (where PH is polyhedrin, and GFP is green fluorescent protein [pFB1-GP]) plasmid to generate the donor plasmid pFB1-1×-GP. To generate the plasmid pFB1-2×-GP, one copy of the AcMNPV-miR-1 precursor fragment was digested with BamHI and PstI and ligated into the pFB1-1×-GP plasmid that had been digested with PstI and BglII. The pFB1-4×-GP and pFB1-8×-GP donor plasmids were constructed in a similar manner. A nonrelated artificial stem-loop structure in pcDNA6.2 was used as a negative control (NC), and the donor plasmid pFB1-NC-GP was constructed by inserting the control sequence into the pFB1-PH-GFP plasmid. The donor plasmids were transformed into DH10B cells containing the bacmid bMON14272 and the helper plasmid pMON7124. As a result, five viruses (vAcNC-PH-GFP [NC], vAc1×-PH-GFP [1×], vAc2×-PH-GFP [2×], vAc4×-PH-GFP [4×], and vAc8×-PH-GFP [8×]) were generated. A wild-type (WT) control virus, vAcWT-PH-GFP (WT), was generated by the insertion of polh and gfp into bMON14272.
RT-qPCR analysis of AcMNPV-miR-1 expression.
RNA samples were extracted using an miRNA extraction kit (Ambion) and reverse transcribed using a stem-loop primer and a PrimeScript RT reagent kit (TaKaRa) in a 20-μl reaction mixture. The reverse transcription (RT) product was used to perform quantitative real-time PCR (qPCR) with a real-time PCR detection system (LightCycler 480; Roche).
The 10-μl qPCR mixture contained 1 μl of cDNA, 5 μl of 2× SYBR Premix Ex Taq II (TaKaRa), 0.75 μM primers, and nuclease-free water to make up the total volume. The PCR was conducted on a LightCycler 480 system (Roche) with an 8-min denaturation at 95°C, followed by 40 cycles of 10 s at 95°C and 30 s at 60°C. Then, a program of 95°C for 1 s, 65°C for 15 s, and 95°C continuously was used to obtain melting curves. The RT-qPCR data for each sample were calculated using the 2−ΔΔCT (where CT is threshold cycle) method (17). A 5S rRNA reverse transcript that was generated from random primers (TaKaRa) was used as a reference. Each reaction was performed in at least three independent experiments in triplicate.
Quantitative real-time PCR analysis of viral DNA copies.
To determine the number of genomic DNA copies, qPCR was performed using SYBR Premix Ex Taq II (TaKaRa). Primers (data not shown) for gp41, a gene unique to AcMNPV, were used for amplification on a LightCycler 480 real-time PCR system (Roche). The qPCR mixture and cycles were the same as those described above. The concentration of viral DNA genome copies within each sample was calculated using a standard curve generated from a dilution series of AcMNPV bacmid DNA. Each of the reactions was performed in three independent experiments in triplicate.
Analysis of BV titers and production.
Sf9 cells were infected (1 PFU/cell) with the NC, WT, 1×, 2×, 4×, and 8× viruses. Supernatants containing BVs were harvested, and BV titers were determined in triplicate using a TCID50 endpoint dilution assay (16). To determine the total copies of BVs, genomic DNA in the BVs was extracted from the supernatants (200 μl) of infected cells at the times indicated in the legend to Fig. 2 using a MiniBEST Viral RNA/DNA Extraction Kit (TaKaRa) according to the manufacturer's instructions. The DNA was quantified using a NanoDrop 2000c spectrophotometer (Thermo Scientific). The qPCR was conducted as described above.
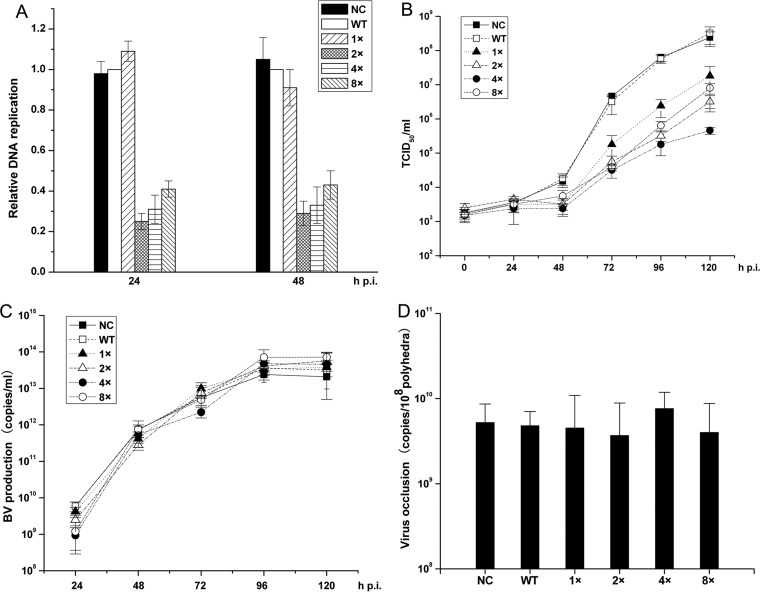
FIG 2.
Effects of AcMNPV-miR-1 overexpression on viral infection. (A) Relative amounts of DNA replication quantified by qPCR. The columns represent the amount of DNA normalized against the WT level. The DNA samples were derived from cells infected with NC, WT, 1×, 2×, 4×, and 8× viruses at 24 and 48 h p.i. (B) BV titer curves quantified by a TCID50 endpoint dilution assay. BVs were harvested from the supernatants of the infected cells at 0, 24, 48, 72, 96, and 120 h p.i. (C) BV production in cells infected by AcMNPV-miR-1 overexpression and WT viruses. BV production was determined by qPCR. (D) The quantities of viruses embedded in ODVs were determined by qPCR. A total of 10 million purified polyhedra were used for each sample. Each reaction was performed in triplicate for three independent experiments. The error bars represent the standard deviations.
Quantitative analysis of viral DNA replication.
To quantify viral DNA replication, qPCR was performed. Sf9 cells were infected in triplicate with the NC, WT, and the four overexpression viruses. The total DNA in each sample was purified using a Universal Genomic DNA Extraction Kit (TaKaRa) and resuspended in 100 μl of double-distilled water. Prior to qPCR, the DNA samples were quantified using a NanoDrop 2000c spectrophotometer (Thermo Scientific); 100 ng of DNA was used in each reaction mixture.
Polyhedron purification and viral DNA extraction.
Polyhedra were isolated from infected cells and purified as described by Braunagel and Summers (18). The concentration of the purified polyhedra was measured using a counting chamber (13). Briefly, 10 million polyhedra were incubated in polyhedron lysis buffer at 37°C for 15 min and centrifuged at 10,000 rpm for 5 min. The supernatant was supplemented with protease K (200 μg/ml) and incubated in a 55°C water bath for 2.5 h. Then, the mixture was incubated at 50°C for 30 min with 1% sodium dodecyl sulfate (SDS) and adequately mixed with an equal volume of Tris-phenol. The samples were centrifuged at 12,000 rpm (Sigma) for 10 min, and the pellets were washed with a mixture of phenol, chloroform, and isoamyl alcohol (25:24:1) and precipitated with 95% ethanol. After the samples were centrifuged at 12,000 rpm (Sigma) for 15 min and washed twice with 75% ethanol, the viral DNA was resuspended in 40 μl of double-distilled water and quantified using a NanoDrop 2000c spectrophotometer (Thermo Scientific).
TEM.
Transmission electron microscopy (TEM) samples were prepared as previously described (19) with some modifications. Infected Sf9 cells were dislodged with a rubber policeman and centrifuged at 3,000 × g for 5 min. Then, the cell pellet was fixed in 2.5% glutaraldehyde overnight at 4°C, washed three times with phosphate-buffered saline (PBS), and postfixed in 1% osmium tetroxide in PBS for 1 h at 4°C. The cells were washed three times with PBS, dehydrated in graded ethanol, and soaked in acetone. Spurr's medium was prepared using Spurr resin (Sigma-Aldrich) in gelatin capsules. Thin sections were stained with aqueous uranyl acetate and lead citrate and observed under a JEOL JEM-1400 transmission electron microscope at an accelerating voltage of 120 kV.
Insect bioassays.
To determine the 50% lethal dose (LD50), newly molted third-instar Trichoplusia ni (T. ni) larvae were reared individually at 27°C in 12-well plates. Then, the larvae were infected by allowing them to feed on a diet contaminated with concentrations of 0 (control), 2 × 102, 1 × 103, 2 × 103, and 1 × 104 polyhedral inclusion bodies (PIBs) of each virus as previously described (20, 21). A control experiment was conducted with an uncontaminated diet. Each experiment was repeated three times. Twenty-four larvae per dosage were used in the experiments. Mortality was recorded at 12-h intervals until the larvae died or pupated. The data were analyzed by probit analysis. The 50% lethal time (LT50) values were determined in T. ni larvae using droplet feeding assays as previously described (19). Orally infected larvae were maintained individually in the dark at 27°C, and mortality was recorded every 12 h until the larvae died or pupated. Twenty-four larvae were used per treatment group, and the experiments were repeated three times. The data were analyzed using a Kaplan-Meier estimator (22).
Dual-luciferase reporter assay.
To determine whether AcMNPV-miR-1 regulated the candidate target genes, the predicted binding site of each gene was amplified. Restriction sites for NotI and XhoI were added to the oligonucleotides at their 5′ and 3′ ends. The PCR products were digested with NotI and XhoI (TaKaRa) and inserted into a psicheck-2 vector (supplied by Promega) in a multiple cloning site (MCS) downstream of the Renilla luciferase gene according to the manufacturer's instructions. Selected plasmids with binding-site fragments were sequenced (BGI) and then quantified using a NanoDrop 2000c spectrophotometer (Thermo Scientific). The mutational sequence to each binding site was synthesized by BGI and cloned into the psicheck-2 vector using the method described above. The mutations were confirmed by sequencing.
Using the above strategies, for each possible target gene, two types of plasmids were constructed. For example, to verify that the ac18 gene is a target gene of AcMNPV-miR-1, the plasmid psicheck-ac18 was constructed by inserting the predicted AcMNPV-miR-1 binding-site segment from the ac18 transcript into the psicheck-2 vector (Promega) to generate a reporter gene with the AcMNPV-miR-1 target site. As a control, a psicheck-ac18-mut plasmid with a mutated binding site in the ac18 transcript was constructed. Additionally, an unrelated target sequence (Bombyx mori actin 3 [A3]) was inserted into the same vector as a universal control for all genes. Cel-miR-239b-5p, an miRNA from Caenorhabditis elegans, was used as the miRNA control for the AcMNPV-miR-1 mimic.
Because interactions between viral miRNAs and their hosts are complex (23), nonrelated cell lines were used to verify the possible targets of the miRNAs to avoid unknown virus-host interactions. In this study, the human embryo kidney 293 cell line was chosen. The 293 cells were a lab stock recovered from cryopreservation in liquid nitrogen and incubated in Dulbecco's modified Eagle's medium (DMEM; Gibco) containing 10% fetal bovine serum (Gibco) at 37°C in 5% CO2. Approximately 50% of the cells were seeded in a 96-well plate 1 day before transfection. The cells were cotransfected with 0.05 μg of each reconstructed plasmid and 50 nM miRNA mimic using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. A dual-luciferase assay was performed at 48 h posttransfection (h p.t.) using a dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions using a multimode reader (Tecan Infinite M200). Briefly, the cells were harvested using passive lysis buffer, and 20 μl of each cell lysate was transferred to an enzymatic reaction with a previously assessed background value (I0, where I is light intensity). For each reaction mixture, 50 μl of the LARII reagent from Promega's dual-luciferase assay kit was added automatically to the cell lysate, and the firefly luciferase activity (I1) was immediately measured. Then, 50 μl of Stop and Glo reagent was automatically added to the mixture, and the Renilla luciferase activity (I2) was assessed. The Renilla luciferase activity was normalized against the firefly luciferase activity, which was mediated by an independent promoter in the psicheck-2 vector. The ac94 gene was used as a positive control. The relative luciferase activity for each reconstructed reporter plasmid was calculated as follows: relative luciferase activity = RRAcMNPV-miR-1/RRNC, where RR is the raw ratio, calculated as (I2 − I0)/(I1 − I0).
PCR.
Each 20-μl PCR mixture included 1.0 μl of cDNA, 2 μl of 10× PCR buffer, 0.2 μl of 10 mM deoxynucleoside triphosphates (dNTPs; TaKaRa), 1.5 μl each of 10 μM forward and reverse primers, and 0.25 μl of Taq polymerase (TaKaRa). PCR was performed using a PTC-101TM Peltier thermal cycler (Bio-Rad) and 200-μl microtubes. The PCR program was as follows: 5 min at 95°C, followed by 30 cycles of 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C, with a final extension at 72°C for 8 min. In total, 3 μl of PCR product was resolved by agarose gel electrophoresis containing ethidium bromide and imaged under UV light using an AlphaImager EP system (Alpha InnoTech).
Construction of HA-tagged viruses.
Viruses with hemagglutinin (HA) tags on target genes were generated using ET recombination as previously described (19), with some modifications. To illustrate the above process, the ac18 gene will be used as an example. First, we ligated two SV40 poly(A) sequences (an HA tag was introduced prior to the stop codon in one of the sequences using the primer PstI-HA-SV40-F) and a Zeocin gene into a pBlueScript plasmid to construct the pBlue-HA-SV40-Zeocin-SV40 plasmid (see Fig. 4D). The two SV40 poly(A) sequences were inserted in opposite directions, and the Zeocin gene was inserted between them. Then, PCR products from the upstream (US) homology arm and the downstream (DS) homology arm were ligated into pBlue-HA-SV40-Zeocin-SV40 to generate pBlue-US-HA-SV40-Zeocin-SV40-DS. PCR amplification of the US and the DS homology arms was performed using the primers BamHI-ac18-F/PstI-ac18-R and EcoRI-ac17-F/HindIII-ac17-R (data not shown), respectively. Finally, the vector was digested with BamHI and HindIII, and the resulting linear US-HA-SV40-Zeocin-SV40-DS fragment was gel purified and suspended in distilled water.
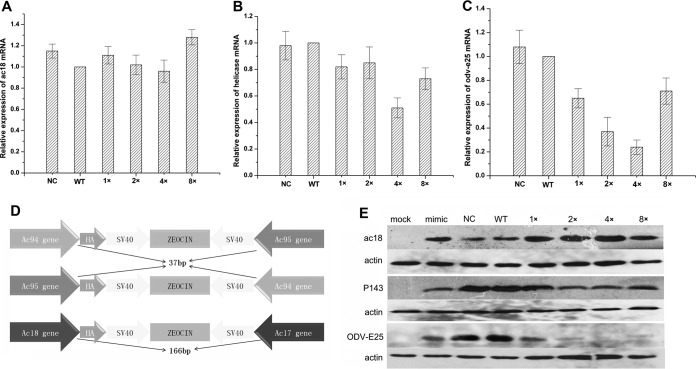
FIG 4.
The effect of AcMNPV-miR-1 on the expression of the target genes. Relative fold change in the expression levels of ac18 mRNA (A), ac95 mRNA (B), and ac94 mRNA (C) was investigated by qRT-PCR. The columns represent the fold changes in the PCR products. Total RNA samples were isolated from NC, WT, 1×, 2×, 4×, and 8× virus-infected Sf9 cells at 24 h p.i. (10 PFU/cell). The RNA level at each time point was normalized to that of the WT. 5S rRNA was used as a reference gene, and the 2−ΔΔCT method was used to calculate the RNA level of each sample. The quantification cycle value of the no-template controls (NC) was >40. The error bars represent the standard deviations. (D) Schematic diagram of the reconstructed viruses with HA tags on the ac94 gene, the ac95 gene, and the ac18 gene, as indicated. The HA tag was inserted into the open reading frame of each target gene upstream of the stop codon in the NC, WT, 1×, 2×, 4×, and 8× bacmids. (E) Regulation of Ac18, P143, and ODV-E25 following AcMNPV-miR-1 overexpression. Proteins were detected by Western blotting with an anti-HA antibody.
To facilitate homologous recombination, six types of DH10B cells (containing the NC, WT, 1×, 2×, 4×, or 8× bacmid) were cotransformed with pBAD-gbaA. The resulting clones were induced by the addition of l-arabinose to achieve competence and then electro-transformed with 1 μg of the purified US-HA-SV40-Zeocin-SV40-DS fragment. As a result, six viruses that included the ac18 gene with an HA tag were generated and named vAcac18-HA-NC-PH-GFP(NC-18:HA), vAcac18-HA-WT-PH-GFP(WT-18:HA), vAcac18-HA-1×-PH-GFP(1×-18:HA), vAcac18-HA-2×-PH-GFP(2×-18:HA), vAcac18-HA-4×-PH-GFP(4×-18:HA), and vAcac18-HA-8×-PH-GFP(8×-18:HA). The HA-tagged ac94 and ac95 viruses were constructed using the same protocol as described above.
Western blot assay.
Western blotting was performed to analyze the expression of the target genes in the Sf9 cells at 60 h p.i. with each reconstructed virus. The cells were collected, lysed in radioimmunoprecipitation assay (RIPA) buffer (Beyotime) for 30 min on ice, and centrifuged for 10 min at 10,000 × g at 4°C. The supernatants were collected and boiled at 100°C for 10 min with 5× protein loading buffer (TaKaRa). Equal volumes of each sample were resolved by 8% SDS-PAGE (for ac95) or 12% SDS-PAGE (for ac94 and ac18), electrophoretically transferred to nitrocellulose transfer membranes, and probed with a monoclonal HA antibody (1:1,000; Abcam) according to the manufacturer's instructions. A goat anti-mouse horseradish peroxidase (HRP)-conjugated antibody (1:8,000; Thermo) was used as a secondary antibody. Proteins were visualized using an enhanced chemiluminescence (ECL) system (Beyotime) according to the manufacturer's instructions.
Co-IP and mass spectrometric analysis.
Coimmunoprecipitations (co-IPs) were conducted using an agarose-conjugated anti-HA-tagged mouse monoclonal antibody (MAb; Abmart) according to the manufacturer's instructions as follows. The cells were collected, lysed in RIPA buffer (Beyotime) for 30 min on ice, and centrifuged for 10 min at 10,000 × g at 4°C. The supernatants were collected, and 20 to 50 μl of a 50% slurry of anti-HA agarose was added using a wide-bore pipette tip. The compound was incubated with gentle mixing overnight at 4°C. The beads were washed three times with 1 ml of lysis buffer and centrifuged for 3 min at 2,000 × g, and the supernatant was discarded. Finally, the HA-tagged proteins were eluted with elution buffer and resolved by 12% SDS-PAGE. The gel was then silver stained using a protocol (recommended by EMBL [www.ebi.ac.uk]) optimized for subsequent mass spectrometry (MS). Proteins of interest were excised from the gel and subjected to in-gel trypsin digestion. Briefly, the excised gel cubes were washed twice in 100 μl of 40% acetonitrile in 0.1 M NH4HCO3 and once in 100 μl of acetonitrile and hydrolyzed with 4 μl of trypsin (15 μg/ml in 0.05 M NH4HCO3 [Promega]) at 37°C overnight. After the samples were mixed with 10 μl of 0.5% acetonitrile and 0.2% acetic acid, they were sent to the Instrumental Analysis and Research Center (http://iarc.sysu.edu.cn/) of Sun Yat-sen University for mass spectrometric analysis. The mass spectra of the tryptic peptides of the derived proteins were obtained using an LTQ Orbitrap liquid chromatograph (LC)-MS (Germany). The results were analyzed using SEQUEST software and data from the NCBI and UniProt databases.
RESULTS
Endogenous expression of AcMNPV-miR-1 in viruses containing multiple copies of the miRNA.
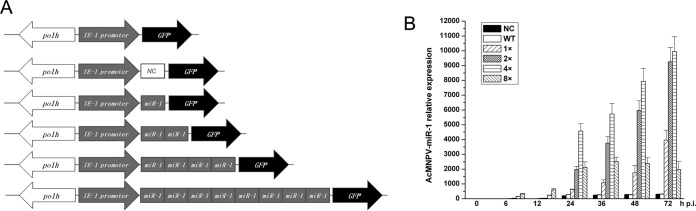
Four viruses, named 1×, 2×, 4×, and 8×, containing, respectively, 1, 2, 4, and 8 additional copies of precursor AcMNPV-miR-1 downstream of an IE-1 promoter, were constructed using a Bac-to-Bac system to explore the function of AcMNPV-miR-1. A negative control (NC) virus was modified by inserting a nonrelated stem-loop structure, and a wild-type (WT) virus was used as a blank control. SYBR green-based qRT-PCR with stem-loop RT primers (24, 25) was used to analyze the expression levels of AcMNPV-miR-1 in Sf9 cells infected with the six reconstructed viruses (NC, WT, 1×, 2×, 4×, and 8×) (26).
Because AcMNPV-miR-1 had been previously validated to accumulate at 12 h p.i. and to reach a maximum level at approximately 24 h p.i (14), we collected RNA samples from cells infected with the NC, WT, 1×, 2×, 4×, and 8× viruses (10 PFU/cell) at 0, 6, 12, 24, 48 and 72 h p.i.
AcMNPV-miR-1 expression levels were similar between the NC and WT groups at all time points tested. For the 1× virus, AcMNPV-miR-1 expression was first detected at 12 h p.i., while for each of the other three overexpression viruses, AcMNPV-miR-1 expression was first detected at 6 h p.i. The RNA levels rose consistently over time and increased by 100- to 1,000-fold by 72 h p.i. for all four overexpression viruses (Fig. 1). The increase in AcMNPV-miR-1 levels was correlated with the numbers of precursor miRNA copies (Fig. 1A) in the reconstructed viruses (i.e., 1×, 2×, 4×, and 8× viruses) at 6 and 12 h p.i. At 24 h p.i., AcMNPV-miR-1 expression continued to increase gradually at the same rate as the precursor copies in the 1×, 2×, and 4× viruses. For the 8× viruses, the AcMNPV-miR-1 levels no longer increased after 24 h p.i. Overall, we demonstrated that AcMNPV-miR-1 was successfully expressed from our reconstructed overexpression viruses, enabling us to assess the biological changes that occur in the presence of high levels of endogenous AcMNPV-miR-1 and providing insight into the roles played by AcMNPV-miR-1 during viral infection.
FIG 1.
Construction of recombinant viruses and analysis of AcMNPV-miR-1 expression. (A) Schematic diagram of the reconstructed overexpression viruses (1×, 2×, 4×, and 8×) and the control WT and NC viruses. The gray rectangles represent the copies of the AcMNPV-miR-1 precursor, and the arrows represent the indicated genes. Recombination was conducted using the Bac-to-Bac system. (B) Relative expression of AcMNPV-miR-1 was determined by qPCR. The columns represent the fold change in AcMNPV-miR-1 expression normalized to that of 5S rRNA expression. RNA samples were isolated from infected Sf9 cells at 0, 6, 12, 24, 36, 48, and 72 h p.i., and the 2−ΔΔCT method was used to calculate the RNA level in each sample. The quantification cycle value of the no-template control (NTC) was >40. Each reaction was performed in triplicate in at least three independent experiments. The error bars represent the standard deviations.
AcMNPV-miR-1 overexpression suppressed viral DNA replication.
To explore how AcMNPV-miR-1 overexpression affects viral DNA replication, qPCR was performed. To accomplish this, total cellular DNA was isolated from cells infected (10 PFU/cell) with the NC, WT, 1×, 2×, 4×, and 8× viruses at 24 and 48 h p.i.
The amount of viral DNA in the 1× virus-infected cells was comparable to that in the NC- and WT-infected cells, while the replication levels in the 2×, 4×, and 8× virus-infected cells were substantially lower (Fig. 2A). The lowest level of replication was found in the 2× virus-infected cells, which was about a quarter of the level in WT-infected cells. Replication was slightly increased from the level with the 2× virus in 4× and 8× virus-infected cells (P > 0.05 compared with 2×, except P = 0.037 for 8× virus at 24 h p.i.). When the 2×, 4×, and 8× viruses were considered together as an influence of high AcMNPV-miR-1 expression and compared to the results produced by the 1×, WT, and NC viruses, which did not increase the expression of this miRNA to the same extent, a relationship emerged in which decreased DNA replication was reflected by increased levels of AcMNPV-miR-1.
AcMNPV-miR-1 overexpression reduced BV infectivity.
To determine whether AcMNPV-miR-1 overexpression affected infectious BV production, Sf9 cells were infected with the NC, WT, 1×, 2×, 4×, and 8× viruses (1 PFU/cell). A TCID50 endpoint dilution assay and qPCR were performed to determine BV titers and quantities at 0, 24, 48, 72, 96, and 120 h p.i. As shown in Fig. 2B, the NC and WT groups showed almost equal BV titers at each time point. In contrast, the four overexpression groups started at levels similar to the level of the NC or WT group at 24 h p.i. and then exhibited decreased infectious BV levels from 48 h p.i. (Fig. 2B) through 120 h p.i. The 4× group showed the lowest infectious BV levels, which were nearly 1,000-fold less than those of the NC or WT group. However, the BV production curves that were determined by qPCR did not show any differences among the six groups (Fig. 2C). This result indicated that AcMNPV-miR-1 overexpression downregulated BV infection by reducing BV infectivity but not by reducing BV production.
AcMNPV-miR-1 overexpression expedited polyhedron formation.
To determine whether AcMNPV-miR-1 overexpression affected viral morphogenesis, infected Sf9 cells were analyzed by TEM (19). The Sf9 cells were randomly divided into six groups and infected (10 PFU/cell) with the NC, WT, 1×, 2×, 4×, and 8× viruses. The cells were harvested at 36 and 72 h p.i., and typical sections were analyzed.
No significant differences in viral morphogenesis were observed among the six groups. However, at 36 h p.i., an increased number of polyhedra was observed in the cells infected with the overexpression viruses compared to levels in those infected with the NC or WT virus (Table 1) based on random evaluations of 40 cells per group by TEM. This difference was especially evident in the 2× and 4× groups. In the 2× group, 23.9% of the evaluated cells contained polyhedra, with an average of 5.1 polyhedra per cell. In the 4× group, 28.4% of the evaluated cells contained polyhedra, with an average of 8.2 polyhedra per cell. In the control NC group and the WT group, 2.1% and 1.7% of the evaluated cells contained polyhedra, respectively, with an average of 1.3 polyhedra per cell and 1.2 polyhedra per cell, respectively. The 1× (7.1% cells containing polyhedra with 3.3 polyhedra per cell) and 8× groups (9.9% cells containing polyhedra with 3.4 polyhedra per cell) exhibited an intermediary state between the 2×/4×-infected and the NC/WT-infected cells. All six groups exhibited similar phenotypes at 72 h p.i. (Table 1). This result indicated that AcMNPV-miR-1 overexpression enhanced the polyhedron formation speed but did not finally increase total polyhedron production.
TABLE 1.
Polyhedra and ODVs observed under TEM
| Virus | Infection profile at 36 h p.i.a |
Infection profile at 72 h p.i. |
||||
|---|---|---|---|---|---|---|
| % of cells containing polyhedrab | No. of polyhedra per cellc | No. of viruses per polyhedrond | % of cells containing polyhedra | No. of polyhedra per cell | No. of viruses per polyhedron | |
| NC | 2.1 | 1.3 | 76.4 | 93.5 | 14.4 | 91.2 |
| WT | 1.7 | 1.2 | 88.2 | 89.7 | 15.2 | 90.6 |
| 1× | 7.1 | 3.3 | 71.3 | 94.1 | 13.8 | 87.6 |
| 2× | 23.9 | 5.1 | 79.2 | 92.7 | 12.5 | 83.3 |
| 4× | 28.4 | 8.2 | 81.6 | 88.6 | 13.9 | 87.3 |
| 8× | 9.9 | 3.4 | 72.2 | 90.4 | 12.9 | 89.1 |
Underlining indicates the increased polyhedron numbers in the cells of the 2× and 4× groups at 36 h p.i.
We randomly chose 40 cells for each sample and calculated the percentage of cells containing polyhedra.
Forty cells containing polyhedra were counted, and counts were averaged in each sample.
Thirty polyhedra were observed, and the total amount of viruses was divided by 30.
To investigate the number of viruses embedded in each polyhedron, Sf9 cells were infected (10 PFU/cell) with the NC, WT, 1×, 2×, 4×, and 8× viruses and harvested at 96 h p.i. We conducted qPCR assays using DNA extracted from 10 million purified polyhedra per group. Similar viral DNA quantities were detected for all six viruses (Fig. 2D), consistent with our results from TEM examination (Table 1). These results indicated that AcMNPV-miR-1 overexpression accelerated the formation of polyhedra but did not influence the total amount of polyhedra or affect the quantity of viruses embedded within them.
AcMNPV-miR-1 overexpression improves viral infection efficiency in T. ni larvae.
To explore how AcMNPV-miR-1 overexpression affects viral infection of larvae, the infectivities of the NC, WT, 1×, 2×, 4×, and 8× viruses were determined for newly molted third-instar T. ni larvae in LD50 and LT50 bioassays. No obvious differences in the LD50 values were observed among the six viruses (Table 2). However, the LT50 values of the six viruses did differ, in the following order: WT (101.8 h) ≈ NC (103.7 h) > 1× (91.7 h) > 2× (77.3 h) ≈ 4× (82.2 h). The difference observed between the LT50 values corresponding to the WT and NC viruses was not significant (P > 0.05), and no difference was noted between the 2× and 4× groups (P > 0.05). The 2× virus killed half of the larvae within a time frame that was approximately 25 h shorter than that of the WT virus, suggesting that AcMNPV-miR-1 overexpression promoted viral infection efficiency in T. ni larvae.
TABLE 2.
Dose-mortality and time-mortality tests in third-instar T. ni larvaea
| Virus | LD50 (no. of PIBs/larva) |
LT50 (h) |
||||
|---|---|---|---|---|---|---|
| Value for the group | 95% fiducial limit |
Value for the group | 95% fiducial limit |
|||
| Lower | Upper | Upper | Lower | |||
| NC | 2.59 × 103 | 1.98 × 103 | 3.20 × 103 | 103.7 | 110.9 | 96.5 |
| WT | 2.31 × 103 | 1.76 × 103 | 2.86 × 103 | 101.8 | 109.4 | 94.3 |
| 1× | 2.42 × 103 | 1.95 × 103 | 2.89 × 103 | 91.7 | 97.5 | 85.9 |
| 2× | 2.23 × 103 | 1.67 × 103 | 2.79 × 103 | 77.3 | 86.2 | 68.4 |
| 4× | 2.88 × 103 | 2.47 × 103 | 3.29 × 103 | 82.2 | 90.1 | 74.3 |
| 8× | 2.63 × 103 | 2.09 × 103 | 3.17 × 103 | 96.9 | 104.7 | 89.1 |
Twenty-four larvae were used per treatment group, and the experiments were repeated three times. The data were analyzed using the Kaplan-Meier estimator. The extremely shortened LT50 values for the 2× and 4× viruses are underlined.
ac18 and ac95 are targets of AcMNPV-miR-1.
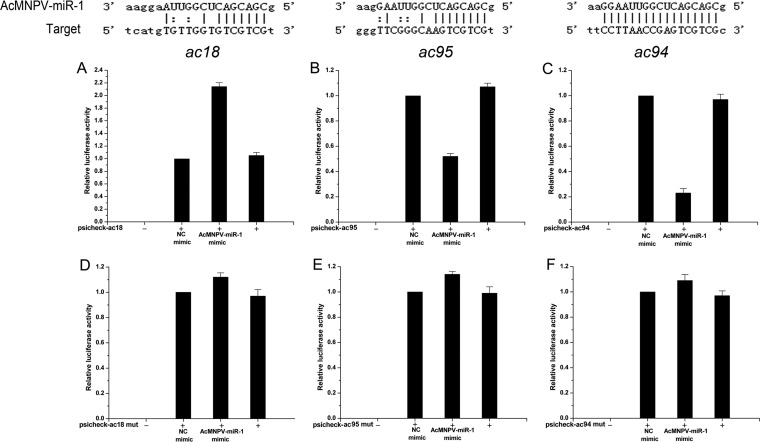
Identifying the target genes of AcMNPV-miR-1 is essential (27) for understanding how this miRNA functions during viral infection. We previously validated ac94 as a target gene of AcMNPV-miR-1 (14). To identify additional target genes, mRNA sequences from AcMNPV and Bombyx mori, as well as the AcMNPV genome, were examined using the MiRanda (28) program under loose conditions (minimum free energy [MFE] above −20.00 kcal/mol and MiRanda score above 120.00), which generated six new viral target gene candidates (ac95, ac18, ac135, ac30, ac66, and ac11) after selection with the miRNA seed sequence (6). Combined with the seven previously predicted candidate target genes (ac15, ac131, ac34, ac82, ac101, ac55, and ac64) (14), which were not detected in the previous study, a total of 13 genes (Table 3) were experimentally examined using a dual-luciferase reporter assay with the Renilla luciferase gene as a reporter and the firefly luciferase gene as a reference. Only two (ac18 and ac95) of the examined genes were regulated by the AcMNPV-miR-1 mimic.
TABLE 3.
The predicted viral targets of AcMNPV-miR-1
| GenBank accession no. | Scorea | MFE (kcal/mol)a | Binding site (nt)b | Target gene ORF (size [nt])c |
|---|---|---|---|---|
| NP_054124.1 | 190.00 | −41.38 | 75–94 | ac94 (687) |
| NP_054044.1 | 161.00 | −21.10 | 397–416 | ac15 (1,521) |
| NP_054161.1 | 155.00 | −23.83 | 283–300 | ac131 (759) |
| NP_054112.1 | 151.00 | −22.26 | 348–368 | ac82 (543) |
| NP_054063.1 | 149.00 | −21.10 | 490–511 | ac34 (648) |
| NP_054131.1 | 144.00 | −21.29 | 225–245 | ac101 (1,086) |
| NP_054125.1 | 138.00 | −18.80 | 2757–2777 | ac95 (3,666) |
| NP_054085.1 | 134.00 | −19.69 | 57–77 | ac55 (222) |
| NP_054047.1 | 133.00 | −17.02 | 657–677 | ac18 (1,062) |
| NP_054165.1 | 132.00 | −17.21 | 645–668 | ac135 (900) |
| NP_054059.1 | 130.00 | −19.21 | 249–269 | ac30 (1,392) |
| NP_054096.1 | 129.00 | −15.44 | 127–147 | ac66 (2,427) |
| NP_054040.1 | 128.00 | −14.90 | 729–794 | ac11 (1,023) |
| NP_054094.1 | 126.00 | −23.55 | 517–537 | ac64 (909) |
The score and MFE (mean free energy) were calculated by MiRanda, version 3.3a. The score represents the possibility of the target gene using bioinformatics method.
Binding site was counted from the 5′ end of the target gene sequence searched using accession numbers in the NCBI database.
ORF, open reading frame.
A 2-fold increase in reporter activity was detected for the psicheck-ac18 plasmid when the AcMNPV-miR-1 mimic was used versus the level with the NC mimic (Fig. 3A), while the AcMNPV-miR-1 mimic did not affect the reporter expression of the control psicheck-ac18-mut plasmid (Fig. 3D). The reporter gene for ac95 was suppressed by approximately 26% by the AcMNPV-miR-1 mimic (Fig. 3B), while the reporter expression of the psicheck-ac95-mut control plasmid was almost the same for the AcMNPV-miR-1 mimic and the NC mimic (Fig. 3E). A miRNA Cel-miR-239b-5p mimic from Caenorhabditis elegans did not affect reporter activity and was used as the NC mimic. Expression from the psicheck-A3 (a nonrelated gene) plasmid was not affected by the AcMNPV-miR-1 mimic. Based on these results, we also performed dual-luciferase reporter assays for ac18 and ac95 in Sf9 cells, a representative insect cell line (data not shown). The AcMNPV-miR-1 mimic upregulated ac18 reporter activity and downregulated ac95 reporter activity, consistent with the results obtained from 293 cells. Therefore, AcMNPV-miR-1 targeted the ac18 and ac95 genes, as well as the previously reported ac94 gene.
FIG 3.
Target sites of AcMNPV-miR-1 and dual-luciferase reporter assay showing the regulation of the target genes. In total, a 50 nM concentration of the AcMNPV-miR-1 mimic was used in each system; the same concentration of the Cel-miR-239b-5p mimic was used as a negative control. Luciferase reporter plasmid constructs containing the predicted binding sites of the ac18 gene (A), ac95 gene (B), and the positive-control ac94 gene (C) were cotransfected with miRNA mimics into 293 cells. A multimode reader (Tecan Infinite M200) was used to assay the luciferase activities at 48 h p.t. As a control, the activities of reporter plasmids with mutations in the ac18 gene (D), ac95 gene (E), and ac94 gene (F) were detected in the same manner. All six graphs represent the mean values of three independent experiments performed in triplicate.
mRNA levels of target genes.
The temporal transcriptional patterns of ac18, ac94, and ac95 were determined by qRT-PCR using total RNA extracted from the NC, WT, 1×, 2×, 4×, and 8× virus-infected (10 PFU/cell) Sf9 cells at 24 h p.i. As shown in Fig. 4A, AcMNPV-miR-1 overexpression did not affect the ac18 mRNA level. For ac95 mRNA, approximately 50% suppression was detected in the 4× virus-infected cells although the mRNA levels in the 1×, 2×, and 8×virus-infected cells were close to the level measured in the WT virus-infected cells (Fig. 4B). The ac94 mRNA level was downregulated to 65%, 37%, 24%, and 71% of the control level in the 1×, 2×, 4×, and 8× virus-infected cells, respectively (Fig. 4C). These results indicated that AcMNPV-miR-1 overexpression did not affect the ac18 mRNA level but significantly downregulated the ac94 mRNA level and that only extremely high expression of this miRNA (as in the 4× virus-infected cells) will decrease the ac95 mRNA level.
Protein levels of the target genes.
To investigate AcMNPV-miR-1 regulation at the translational level, an HA tag was inserted into the ac18, ac94, and ac95 genes prior to the stop codon in the NC, WT, 1×, 2×, 4×, and 8× bacmids via the λ Red recombination system (Fig. 4D). This led to the generation of six viruses with HA tags on the ac18 gene (NC-18:HA, WT-18:HA, 1×-18:HA, 2×-18:HA, 4×-18:HA, and 8×-18:HA), six viruses with HA tags on the ac94 gene (NC-94:HA, WT-94:HA, 1×-94:HA, 2×-94:HA, 4×-94:HA, and 8×-94:HA), and six viruses with HA tags on the ac95 gene (NC-95:HA, WT-95:HA, 1×-95:HA, 2×-95:HA, 4×-95:HA, and 8×-95:HA). Protein expression levels for ac18, ac94, and ac95 were determined by Western blotting using proteins extracted from Sf9 cells infected (10 PFU/cell) with the 18 HA-tagged viruses. Mock-infected cells were used as negative controls. Positive-control samples included cells transfected with the AcMNPV-miR-1 mimic and infected with the WT-18:HA, WT-94:HA, and WT-95:HA viruses at 8 h p.t. All protein samples were harvested at 48 h p.i.
As shown in Fig. 4E, ac18 protein expression was slightly upregulated by the overexpression of AcMNPV-miR-1 or its mimic. In contrast, P143 (the protein encoded by ac95) expression was reduced in cells infected with 2×-95:HA, 4×-95:HA, and 8×-95:HA although cells infected with 1×-95:HA showed a level of P143 expression comparable to the levels observed in cells infected with NC-95:HA and WT-95:HA. As expected, ODV-E25 (the protein encoded by ac94) expression was also downregulated by AcMNPV-miR-1 overexpression. This effect was more extreme than the downregulation of P143, with almost no ODV-E25 signal being detected in 4×-94:HA or 8×-94:HA virus-infected cells. The 1×-94:HA and 2×-94:HA virus-infected cells exhibited a gradually decreasing trend of ODV-E25 expression (Fig. 4E). These results were in agreement with those produced by the dual-luciferase reporter assay.
AcMNPV-miR-1 target genes might be involved in pathways concerning virus infection.
The function of AcMNPV-miR-1 was further explored based on the tk;4interactions that form between its target proteins and other proteins. We used reconstructed viruses with HA-tagged ac18, ac94, or ac95 for co-IP followed by LC-MS analysis. The results (Table 4) showed that Ac18 likely interacted with ODV-E25, a protein encoded by another target gene (ac94), in addition to FP25 (encoded by ac61) and various host heat shock proteins (HSPs), including HSP 21.4, HSP 70, and HSP 90. As expected, ODV-E25 also interacted with Ac18, as well as FP25, in addition to Ac126 (chitinase), Ac135 (P35), and Ac142 (P49). Therefore, changes in the expression of Ac18 and/or ODV-E25 might affect pathways related to the interacting proteins, thereby affecting viral infection. To our disappointment, the co-IPs of P143 did not show any significant signals.
TABLE 4.
Protein interactions tested by co-IP
| Proteina | AcMNPV ORFb | Protein designation | Predicted molecular mass (kDa) | Scorec | Coverage (%)d |
|---|---|---|---|---|---|
| Ac18 | 61 | FP25 | 25.2 | 46.65 | 42.06 |
| 94 | ODV-E25 | 25.5 | 33.85 | 43.86 | |
| ODV-E25 | 18 | 40.8 | 46.19 | 30.88 | |
| 61 | FP25 | 25.2 | 3.36 | 9.35 | |
| 126 | Chitinase | 61.3 | 30.41 | 14.88 | |
| 135 | P35 | 34.8 | 2.63 | 3.34 | |
| 142 | P49 | 55.4 | 5.43 | 5.45 |
HA-tagged proteins were identified by LC-MS.
ORF, open reading frame.
The score was given by SEQUEST software. Matched proteins with a confidence level of more than 95% were considered significant.
Percentage of identified peptides (with confidence of >95%) covered on the protein sequence.
DISCUSSION
In this study, we explored the function of AcMNPV-miR-1 and demonstrated that AcMNPV-miR-1 overexpression decreased viral DNA replication and BV infectivity but promoted polyhedron formation and virus infection efficiency in T. ni larvae. In addition to ac94, two new target genes, ac18 and ac95, were also identified in this study. The former was slightly upregulated, and the latter was downregulated. Based on these results, a possible functional pathway for this miRNA was implied.
We first constructed viruses that constitutively overexpressed AcMNPV-miR-1 and found that AcMNPV-miR-1 expression (Fig. 1) consistently increased with the number of miRNA precursor copies at 6 and 12 h p.i. After that, the trend remained in cells that were infected with the 1×, 2×, and 4× viruses. However, in the 8× virus-infected cells, AcMNPV-miR-1 accumulation substantially slowed after 24 h p.i., plateauing instead of increasing as with the other overexpression viruses. The limited quantity of AcMNPV-miR-1 present in the 8× virus-infected cells at late time points might have resulted because the transcripts of the precursor became so numerous that dysfunctional biogenesis occurred. Such dysfunction possibly affects the miRNA targeting process subsequently, resulting in the abnormal actions of the 8× virus in terms of target gene regulation and the unique viral phenotype compared to the phenotypes of the other reconstructed viruses. Additionally, these results could reflect the existence of an upper limit for AcMNPV-miR-1 expression, wherein once the expression reaches a ceiling (such as in the 8× virus-infected cells after 24 h p.i.), other mechanisms might be activated and adjust the level of this miRNA.
Virus-encoded miRNAs that regulate virus replication are a universal phenomenon. miR-BART2, encoded by EBV, was shown to downregulate the expression of the DNA polymerase BALF5 to inhibit viral replication (8). In the Heliothis virescens ascovirus, HvAV-miR-1 regulates viral replication by targeting DNA polymerase I (9). In our study, we also found that AcMNPV-miR-1 overexpression could affect viral DNA replication.
We found that P143 expression was substantially downregulated in 2×, 4×, and 8× virus-infected cells (Fig. 4E). ac95 has been reported to encode a helicase protein (29, 30) that interacts with lef-3 (31–33), and both genes are associated with viral DNA replication (34, 35). Therefore, a decrease in P143 expression would lead to the suppression of DNA replication, which was consistent with our qRT-PCR results showing the reduced DNA replication in the 2×, 4×, and 8× viruses (Fig. 2A). Overall, our results indicated that AcMNPV-miR-1 overexpression suppressed viral DNA replication by downregulating P143 expression.
Although P143 expression and viral DNA replication were primarily downregulated by AcMNPV-miR-1 overexpression, both of these characteristics exhibited a slightly increasing trend as AcMNPV-miR-1 copies increased (P > 0.05). This finding suggested that AcMNPV-miR-1 levels in 2× virus-infected cells were the most propitious for P143 downregulation. It is likely that an opposing mechanism exists that upregulates P143 expression when P143 is reduced below a threshold level. Alternatively, an miRNA threshold limit (36) might exist for P143 expression. Once the threshold has been exceeded, a feedback signal could activate other control regulatory pathways and restore P143 expression to a relatively normal level.
The ac95 mRNA level was not reduced to the same extent as the ac94 mRNA level in the cells infected with the overexpression viruses although P143 expression was substantially reduced, which might be due to the different interaction mechanisms that exist between the miRNA and targets. AcMNPV-miR-1 is not as perfectly matched to the ac95 mRNA binding site (with a lower predicted matching score of only 126.00) (Table 3) as it is to the ac94 mRNA binding site (score of 190.00); therefore, AcMNPV-miR-1 might not induce small interfering RNA (siRNA)-like cleavage at the ac95 binding site and would mainly hinder the translation of P143.
AcMNPV-miR-1 produced clear downregulation of ac94 (ODV-E25). ODV-E25, located in the envelopes of BVs and ODVs (13), is required for infectious BV production and ODV formation. Knockout of ac94 could destroy BV infectivity and affect ODV formation (37). Our results showed that decreased BV infectivity accompanied downregulated ac94 expression. Therefore, by regulating ac94, AcMNPV-miR-1 potentially reduced BV infectivity.
However, the accelerated polyhedron formation induced by the overexpression of AcMNPV-miR-1, which downregulated ODV-E25, appeared contradictory to a previous report that ac94 knockout suppressed ODV formation (37). Our results from the co-IP assay might offer some alternative explanations in addition to our previous hypothesis that “the cleavage of ac94 mRNA is responsible for ODV formation” (14). Because both Ac18 and ODV-E25 were found to interact with FP25, they are both likely associated with the function of FP25. It has been reported that knockout of FP25 resulted in a reduction in polyhedrin protein and p10 promoter transcript expression (38). Therefore, increased expression of Ac18 and decreased expression of ODV-E25 might affect the FP25 pathway, thereby promoting polyhedron formation.
Additionally, we did not observe an obvious acceleration in ODV formation when multiple copies of the AcMNPV-miR-1 precursor were present; this finding differs from our previous report using the miRNA mimic (14). This discrepancy might be due to differences in miRNA administration. Transcripts containing multiple copies of the miRNA precursor undergo the entire process of endogenous miRNA maturation, after which the mature miRNA molecules target their corresponding mRNAs. In the internalization course of miRNA formation, a number of factors could affect this process and impact the final phenotype. In contrast, transfection of an miRNA mimic has been shown to result in direct joining to the miRNA-RNP complex (miRNP) (6). In this case, the amount of the mimic increased by >1,000,000-fold over the amount of the original mature miRNA. The instantaneous appearance of such a tremendous quantity of mimic miRNA in cells surely results in various cytopathologic effects.
AcMNPV-miR-1 was also validated to accelerate ac18 translation using both a dual-luciferase assay and Western blot analysis. Although the upregulation of miRNA target genes is not common and although the mechanisms associated with such upregulation are unknown, some cases have been reported (3, 39). For instance, miR-122 was reported to upregulate hepatitis C virus (HCV) gene translation by increasing its viral association with the 40S subunit, thereby influencing polysome formation (40, 41). miR-10a was shown to enhance the translation of ribosomal proteins by alleviating their TOP-mediated translational repression (42). There might be a possible mode of Ac18 upregulation. Once Ac18 was combined with AcMNPV-miR-1, the three-dimensional (3D) structure of ac18 mRNA was disturbed, and the resulting complex could induce the upregulation of ac18 mRNA translation.
Ac18 has been reported to affect virus infection efficiency in T. ni larvae (43). Knockout of ac18 delayed the LT50, suggesting that the shortening of LT50 following AcMNPV-miR-1 overexpression was probably due to upregulation of Ac18 although the functional pathway of Ac18 remains unknown. Co-IP analysis demonstrated that Ac18 interacted with three HSPs, which may provide clues regarding the mechanism. These three HSPs were originally shown to defend against infection but have recently been demonstrated to support virus replication and assembly via interactions with viral proteins (44–47). Thus, by interacting with HSPs, Ac18 may act as a moderator of HSP-associated virus-host interplay (48–50). The HSPs themselves may play defensive roles; however, with the appearance of Ac18, their roles may become advantageous for the virus. Therefore, although an absence of ac18 may not be fatal to the virus, it may disturb and hinder the physiological process of viral infection.
miRNAs modulate an immense and complex regulatory network of gene expression that affects a broad spectrum of developmental and cellular processes (51, 52), including cell proliferation (53), cell division (54), cell differentiation (55), metabolism (56), viral infection (57), etc. Furthermore, virus-encoded miRNAs are believed to be primarily associated with the longevity of infected cells, the immune response and lytic cycle in the host, and other noncanonical functions (58). In our study, AcMNPV-miR-1 was found to participate in a large functional network of viral gene regulation and take part in the viral life cycle and in virus-host interactions.
From a macroscopic viewpoint, AcMNPV-miR-1 may be advantageous with respect to the viral infection efficiency of larvae. Once a larva is infected with AcMNPV, reductions in viral DNA load and infectious BV number would most likely prolong the time frame that viruses could exist in the larva. As a result, greater numbers of polyhedra and ODVs could be assembled. Combined with accelerated polyhedron formation and more efficient larval infectivity, the released polyhedra could likely infect other larvae more easily and effectively.
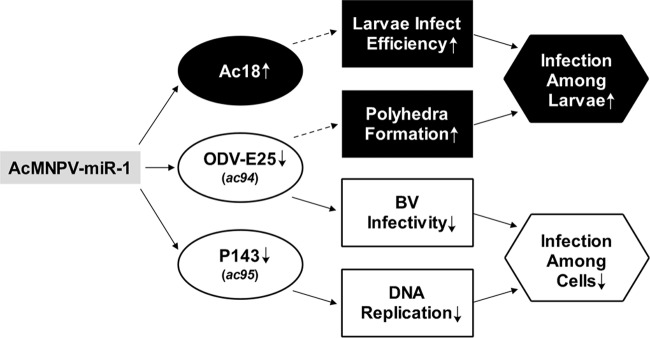
The viral infection cycle can be divided into two stages: (i) infectious BV production and infection of cells in different tissues and (ii) ODV formation and infection of additional larvae. Our results suggest that AcMNPV-miR-1 plays a role in facilitating the transition from the first stage to the second stage by regulating genes that alleviate cell infection in the first stage and that enhance larval infection in the second stage (Fig. 5). This phenomenon subsequently augments the spread of the virus in the natural environment.
FIG 5.
The functional network concerning AcMNPV-miR-1. Inferences based on previous reports are denoted by dashed arrows.
ACKNOWLEDGMENTS
This research was supported by grant 30970139 from the National Natural Science Foundation of China and grant 2015A030313100 from the Natural Science Foundation of Guangdong Province.
REFERENCES
- 1.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Bushati N, Cohen SM. 2007. MicroRNA functions. Annu Rev Cell Dev Biol 23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 3.Leung AK, Sharp PA. 2010. MicroRNA functions in stress responses. Mol Cell 40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hafner M, Lianoglou S, Tuschl T, Betel D. 2012. Genome-wide identification of miRNA targets by PAR-CLIP. Methods 58:94–105. doi: 10.1016/j.ymeth.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arvey A, Larsson E, Sander C, Leslie CS, Marks DS. 2010. Target mRNA abundance dilutes microRNA and siRNA activity. Mol Syst Biol 6:363. doi: 10.1038/msb.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. 2005. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 8.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. 2004. Identification of virus-encoded microRNAs. Science 304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 9.Hussain M, Taft RJ, Asgari S. 2008. An insect virus-encoded microRNA regulates viral replication. J Virol 82:9164–9170. doi: 10.1128/JVI.01109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hussain M, Asgari S. 2014. MicroRNAs as mediators of insect host-pathogen interactions and immunity. J Insect Physiol 70:151–158. doi: 10.1016/j.jinsphys.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Singh CP, Singh J, Nagaraju J. 2012. A baculovirus-encoded MicroRNA (miRNA) suppresses its host miRNA biogenesis by regulating the exportin-5 cofactor Ran. J Virol 86:7867–7879. doi: 10.1128/JVI.00064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh CP, Singh J, Nagaraju J. 2014. bmnpv-miR-3 facilitates BmNPV infection by modulating the expression of viral P6.9 and other late genes in Bombyx mori. Insect Biochem Mol Biol 49:59–69. doi: 10.1016/j.ibmb.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Yuan M, Huang Z, Wei D, Hu Z, Yang K, Pang Y. 2011. Identification of Autographa californica nucleopolyhedrovirus ac93 as a core gene and its requirement for intranuclear microvesicle formation and nuclear egress of nucleocapsids. J Virol 85:11664–11674. doi: 10.1128/JVI.05275-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu M, Wang J, Deng R, Xiong P, Liang H, Wang X. 2013. A microRNA encoded by Autographa californica nucleopolyhedrovirus regulates expression of viral gene ODV-E25. J Virol 87:13029–13034. doi: 10.1128/JVI.02112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luckow VA, Lee SC, Barry GF, Olins PO. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J Virol 67:4566–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emery VC. 1992. Baculovirus expression vectors: choice of expression vector. Methods Mol Biol 8:287–307. [DOI] [PubMed] [Google Scholar]
- 17.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braunagel SC, Summers MD. 1994. Autographa californica nuclear polyhedrosis virus, PDV, and ECV viral envelopes and nucleocapsids: structural proteins, antigens, lipid and fatty acid profiles. Virology 202:315–328. doi: 10.1006/viro.1994.1348. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Wang J, Deng R, Zhang Q, Yang K, Wang X. 2005. vlf-1 deletion brought AcMNPV to defect in nucleocapsid formation. Virus Genes 31:275–284. doi: 10.1007/s11262-005-3242-3. [DOI] [PubMed] [Google Scholar]
- 20.Li G, Wang J, Deng R, Wang X. 2008. Characterization of AcMNPV with a deletion of ac68 gene. Virus Genes 37:119–127. doi: 10.1007/s11262-008-0238-9. [DOI] [PubMed] [Google Scholar]
- 21.Eldridge R, Li Y, Miller LK. 1992. Characterization of a baculovirus gene encoding a small conotoxinlike polypeptide. J Virol 66:6563–6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalbfleisch JD, Prentice RL. 2002. The statistical analysis of failure time data, 2nd ed John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- 23.Cullen BR. 2006. Viruses and microRNAs. Nat Genet 38(Suppl):S25–S30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 24.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. 2005. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ro S, Park C, Jin J, Sanders KM, Yan W. 2006. A PCR-based method for detection and quantification of small RNAs. Biochem Biophys Res Commun 351:756–763. doi: 10.1016/j.bbrc.2006.10.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu H, He JH, Xiao ZD, Zhang QQ, Chen YQ, Zhou H, Qu LH. 2010. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology 52:1431–1442. doi: 10.1002/hep.23818. [DOI] [PubMed] [Google Scholar]
- 27.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 28.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. 2003. MicroRNA targets in Drosophila. Genome Biol 5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heldens JG, Liu Y, Zuidema D, Goldbach RW, Vlak JM. 1997. Characterization of a putative Spodoptera exigua multicapsid nucleopolyhedrovirus helicase gene. J Gen Virol 78:3101–3114. doi: 10.1099/0022-1317-78-12-3101. [DOI] [PubMed] [Google Scholar]
- 30.McDougal VV, Guarino LA. 2000. The Autographa californica nuclear polyhedrosis virus p143 gene encodes a DNA helicase. J Virol 74:5273–5279. doi: 10.1128/JVI.74.11.5273-5279.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Y, Carstens EB. 1998. A baculovirus single-stranded DNA binding protein, LEF-3, mediates the nuclear localization of the putative helicase P143. Virology 247:32–40. doi: 10.1006/viro.1998.9235. [DOI] [PubMed] [Google Scholar]
- 32.Yu M, Carstens EB. 2012. Choristoneura fumiferana multiple nucleopolyhedrovirus LEF-3-P143 complex can complement DNA replication and budded virus in an AcMNPV LEF-3-P143 double knockout bacmid. J Gen Virol 93:383–388. doi: 10.1099/vir.0.036699-0. [DOI] [PubMed] [Google Scholar]
- 33.Evans JT, Rosenblatt GS, Leisy DJ, Rohrmann GF. 1999. Characterization of the interaction between the baculovirus ssDNA-binding protein (LEF-3) and putative helicase (P143). J Gen Virol 80:493–500. doi: 10.1099/0022-1317-80-2-493. [DOI] [PubMed] [Google Scholar]
- 34.McDougal VV, Guarino LA. 2001. DNA and ATP binding activities of the baculovirus DNA helicase P143. J Virol 75:7206–7209. doi: 10.1128/JVI.75.15.7206-7209.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu G, Carstens EB. 1999. Site-directed mutagenesis of the AcMNPV p143 gene: effects on baculovirus DNA replication. Virology 253:125–136. doi: 10.1006/viro.1998.9485. [DOI] [PubMed] [Google Scholar]
- 36.Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. 2011. MicroRNAs can generate thresholds in target gene expression. Nat Genet 43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L, Hu X, Xiang X, Yu S, Yang R, Wu X. 2012. Autographa californica multiple nucleopolyhedrovirus odv-e25 (Ac94) is required for budded virus infectivity and occlusion-derived virus formation. Arch Virol 157:617–625. doi: 10.1007/s00705-011-1211-9. [DOI] [PubMed] [Google Scholar]
- 38.Harrison RL, Jarvis DL, Summers MD. 1996. The role of the AcMNPV 25K gene, “FP25,” in baculovirus polh and p10 expression. Virology 226:34–46. doi: 10.1006/viro.1996.0625. [DOI] [PubMed] [Google Scholar]
- 39.Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. 2014. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics 2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niepmann M. 2009. Activation of hepatitis C virus translation by a liver-specific microRNA. Cell Cycle 8:1473–1477. doi: 10.4161/cc.8.10.8349. [DOI] [PubMed] [Google Scholar]
- 41.Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M. 2008. MicroRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J 27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orom UA, Nielsen FC, Lund AH. 2008. MicroRNA-10a binds the 5′ UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Wu W, Li Z, Yuan M, Feng G, Yu Q, Yang K, Pang Y. 2007. ac18 is not essential for the propagation of Autographa californica multiple nucleopolyhedrovirus. Virology 367:71–81. doi: 10.1016/j.virol.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 44.Burch AD, Weller SK. 2005. Herpes simplex virus type 1 DNA polymerase requires the mammalian chaperone hsp90 for proper localization to the nucleus. J Virol 79:10740–10749. doi: 10.1128/JVI.79.16.10740-10749.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dutta D, Bagchi P, Chatterjee A, Nayak MK, Mukherjee A, Chattopadhyay S, Nagashima S, Kobayashi N, Komoto S, Taniguchi K, Chawla-Sarkar M. 2009. The molecular chaperone heat shock protein-90 positively regulates rotavirus infection. Virology 391:325–333. doi: 10.1016/j.virol.2009.06.044. [DOI] [PubMed] [Google Scholar]
- 46.Schultz KL, Friesen PD. 2009. Baculovirus DNA replication-specific expression factors trigger apoptosis and shutoff of host protein synthesis during infection. J Virol 83:11123–11132. doi: 10.1128/JVI.01199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lyupina YV, Zatsepina OG, Timokhova AV, Orlova OV, Kostyuchenko MV, Beljelarskaya SN, Evgen'ev MB, Mikhailov VS. 2011. New insights into the induction of the heat shock proteins in baculovirus infected insect cells. Virology 421:34–41. doi: 10.1016/j.virol.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 48.Zhao Y, Kurian D, Xu H, Petherbridge L, Smith LP, Hunt L, Nair V. 2009. Interaction of Marek's disease virus oncoprotein Meq with heat-shock protein 70 in lymphoid tumour cells. J Gen Virol 90:2201–2208. doi: 10.1099/vir.0.012062-0. [DOI] [PubMed] [Google Scholar]
- 49.Lyupina YV, Dmitrieva SB, Timokhova AV, Beljelarskaya SN, Zatsepina OG, Evgen'ev MB, Mikhailov VS. 2010. An important role of the heat shock response in infected cells for replication of baculoviruses. Virology 406:336–341. doi: 10.1016/j.virol.2010.07.039. [DOI] [PubMed] [Google Scholar]
- 50.Couturier M, Buccellato M, Costanzo S, Bourhis JM, Shu Y, Nicaise M, Desmadril M, Flaudrops C, Longhi S, Oglesbee M. 2010. High affinity binding between Hsp70 and the C-terminal domain of the measles virus nucleoprotein requires an Hsp40 co-chaperone. J Mol Recognit 23:301–315. doi: 10.1002/jmr.982. [DOI] [PubMed] [Google Scholar]
- 51.Ambros V. 2004. The functions of animal microRNAs. Nature 431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 52.Kloosterman WP, Plasterk RH. 2006. The diverse functions of microRNAs in animal development and disease. Dev Cell 11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 53.Cheng AM, Byrom MW, Shelton J, Ford LP. 2005. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res 33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shcherbata HR, Hatfield S, Ward EJ, Reynolds S, Fischer KA, Ruohola-Baker H. 2006. The MicroRNA pathway plays a regulatory role in stem cell division. Cell Cycle 5:172–175. doi: 10.4161/cc.5.2.2343. [DOI] [PubMed] [Google Scholar]
- 55.Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S, Harel-Bellan A. 2006. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol 8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 56.Filipowicz W. 2005. RNAi: the nuts and bolts of the RISC machine. Cell 122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 57.Mollaie HR, Monavari SH, Arabzadeh SA, Shamsi-Shahrabadi M, Fazlalipour M, Afshar RM. 2013. RNAi and miRNA in viral infections and cancers. Asian Pac J Cancer Prev 14:7045–7056. doi: 10.7314/APJCP.2013.14.12.7045. [DOI] [PubMed] [Google Scholar]
- 58.Kincaid RP, Sullivan CS. 2012. Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog 8:e1003018. doi: 10.1371/journal.ppat.1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]