ABSTRACT
The wide phenotypic variability of prion diseases is thought to depend on the interaction of a host genotype with prion strains that have self-perpetuating biological properties enciphered in distinct conformations of the misfolded prion protein PrPSc. This concept is largely based on indirect approaches studying the effect of proteases or denaturing agents on the physicochemical properties of PrPSc aggregates. Furthermore, most data come from studies on rodent-adapted prion strains, making current understanding of the molecular basis of strains and phenotypic variability in naturally occurring diseases, especially in humans, more limited. To fill this gap, we studied the effects of guanidine hydrochloride (GdnHCl) and heating on PrPSc aggregates extracted from 60 sporadic Creutzfeldt-Jakob disease (CJD) and 6 variant CJD brains. While denaturation curves obtained after exposure of PrPSc to increasing GdnHCl concentrations showed similar profiles among the 7 CJD types analyzed, PrPSc exposure to increasing temperature revealed significantly different and type-specific responses. In particular, MM1 and VV2, the most prevalent and fast-replicating CJD types, showed stable and highly resistant PrPSc aggregates, whereas VV1, a rare and slowly propagating type, revealed unstable aggregates that easily dissolved at low temperature. Taken together, our results indicate that the molecular interactions mediating the aggregation state of PrPSc, possibly enciphering strain diversity, are differently targeted by GdnHCl, temperature, and proteases. Furthermore, the detected positive correlation between the thermostability of PrPSc aggregates and disease transmission efficiency makes inconsistent the proposed hypothesis that a decrease in conformational stability of prions results in an increase in their replication efficiency.
IMPORTANCE Prion strains are defined as infectious isolates propagating distinctive phenotypic traits after transmission to syngeneic hosts. Although the molecular basis of prion strains is not fully understood, it is largely accepted that variations in prion protein conformation drive the molecular changes leading to the different phenotypes. In this study, we exposed abnormal prion protein aggregates encompassing the spectrum of human prion strains to both guanidine hydrochloride and thermal unfolding. Remarkably, while exposure to increasing temperature revealed significant strain-specific differences in the denaturation profile of the protein, treatment with guanidine hydrochloride did not. The findings suggest that thermal and chemical denaturation perturb the structure of prion protein aggregates differently. Moreover, since the most thermostable prion protein types were those associated with the most prevalent phenotypes and most rapidly and efficiently transmitting strains, the results suggest a direct correlation between strain replication efficiency and the thermostability of prion protein aggregates.
INTRODUCTION
Prion diseases are invariably fatal neurodegenerative disorders of humans and other mammals, characterized by tissue deposition of aggregates of a misfolded, beta-sheet-rich, and partially protease-resistant isoform (PrPSc) of the cellular prion protein (PrPC). In prion diseases, misfolded PrPSc, originating exogenously or spontaneously, is thought to template the structural conversion of the host-encoded PrPC in an autocatalytic process (1, 2). Intriguingly, a wealth of recent evidence indicates that proteinaceous seeds serving as self-propagating prion-like agents may represent a common pathogenetic mechanism in most, if not all, neurodegenerative diseases (3).
Despite their relative rarity, prion diseases show a wide spectrum of clinical and pathological phenotypes with significant heterogeneity in disease duration, symptomatology, and distribution or type of brain lesions. The current classification of sporadic Creutzfeldt-Jakob disease (sCJD), the most common human prion disease, includes six major disease phenotypes that strongly correlate at the molecular level with the genotype at the polymorphic codon 129 (methionine [M] or valine [V]) of the PRNP gene (which encodes PrPC) and two PrPSc profiles or types (type 1 and type 2) comprising distinctive physicochemical properties such as size after protease treatment (respectively, 21 and 19 kDa) and glycoform ratio (4, 5). Recent studies in animal models have shown that phenotypic variations among sCJD phenotypes are largely maintained after transmission into genetically defined hosts, suggesting that different prion strains are the main cause of this diversity (6–11). Although it is well established that PrPC conversion into PrPSc involves consistent changes in the secondary structure with part of the α-helical structure turning into a β-sheet (12, 13), a complete structural characterization of PrPSc has been hampered by the propensity of the misfolded protein to form highly aggregated and detergent-insoluble polymers. Consequently, due to the limited data available from direct structural studies (14, 15), the putative central role of PrPSc tertiary or quaternary structure in determining the molecular basis of prion strains is not yet clearly demonstrated. Several experimental data, however, indirectly support this hypothesis, both in yeast (16) and in mammals. For example, it is largely believed that the heterogeneity in the fragment profile of proteinase K (PK)-digested PrPSc, which distinguishes at least some of the known prion strains, is the direct consequence of PrPSc aggregates having distinct conformations (17–21). Similarly, sedimentation profiles and protease sensitivity have been used as indirect markers of PrPSc structure and have shown a correlation with strain-specific transmission properties (22–27). More recently, studies with rodent-adapted, cloned prion strains demonstrated that also the conformational stability of PrPSc, measured indirectly either by inducing a progressive denaturation of the protein with the chaotropic salt guanidine hydrochloride (GdnHCl) or by exposing the protein to increasing temperatures in the presence of sodium dodecyl sulfate (SDS), may vary among different strains (28–30). Attempts have also been made to correlate PrPSc conformational stability to strain-specific properties such as replication rates, although with conflicting, opposite results in murine and hamster models (28–30).
Overall, despite the wealth of experimental data collected in rodents and some evidence obtained in humans suggesting that phenotypic diversity is somehow related to distinct PrPSc isoforms with distinct structures, the conformational spectrum of these isoforms and their relationship to the issue of prions strains are still poorly understood. In particular, a systematic analysis of the conformational stability of PrPSc aggregates across the spectrum of human prions is still lacking. Previous studies focused on the comparison between sCJD subtypes MM1 and MM2 (31–33) or between variant CJD (vCJD) and the most common sCJD type MM1 (34). Furthermore, only the unfolding induced by GdnHCl was addressed, whereas the thermostability of PrPSc has never been explored in CJD.
To add insights to the intriguing relationship among PrPSc molecular types, the conformational stability of abnormal PrPSc aggregates, and the phenotypic expression of disease, we have evaluated both guanidine-induced unfolding and thermostability of PrPSc across the whole spectrum of currently characterized human CJD strains.
MATERIALS AND METHODS
Patients and tissues.
We studied brain tissues from 60 cases of sCJD and 6 cases of vCJD. sCJD tissues included the whole spectrum of pure phenotypic variants recognized by current classification (5): 12 MM1, 9 VV1, 10 MV 2K, 12 VV2, 7 MM2-cortical (MM 2C), and 4 MM2-thalamic (MM 2T). In addition, 6 brains of sCJD MM 1 + 2C, the most common sCJD subtype with mixed histopathologic features and the cooccurrence of PrPSc types 1 and 2, were analyzed. Each sCJD brain was classified as a “pure” or “mixed” type based on the results of histopathological examination, PrP immunohistochemistry, and PrPSc typing in multiple brain regions, according to Parchi et al. (5). Unfixed brain tissues were obtained at autopsy and kept frozen at −80°C until use. All samples used in this study were taken from the cerebral cortex of the frontal lobe.
Antibodies.
The following monoclonal mouse antibodies, immunoreactive with human PrP, were used: 3F4 at 1:30,000, which recognizes residues 106 to 110 (35); 12B2, at 1:8,000, which binds residues 89 to 93 (36); and SAF60 at 1:2,000, which reacts with residues 157 to 161 (37). In addition, the PrPSc type 2-specific polyclonal antibody T2 (1:5,000), which binds residues 97 to 103 (7), and the rabbit antiserum 2301 (1:3,000) to human PrP residues 220 to 231 were used.
Preparation of THs.
After removing any residual white matter from the cortical tissue sample, 50 to 100 mg of gray matter was homogenized at 20% (wt/vol) in TN-NP40 (100 mM Tris, 130 mM NaCl, 0.5% Nonidet P-40) at pH 7.4 (38) for the guanidine assay or at 10% (wt/vol) in LB100 (100 mM Tris, 100 mM NaCl, 10 mM EDTA, 0.5% Nonidet P-40, 0.5% sodium deoxycholate) at pH 6.9 (39) for the thermosolubilization assay (TSA). In a subset of experiments having the specific purpose of reproducing a previously published protocol (32), a clearing spin of total brain homogenates (THs) at 3,000 rpm for 10 min was performed. Total protein concentration was measured using a standard colorimetric method based on bicinchoninic acid (Pierce Biotechnology, Rockford, IL, USA).
Guanidine-induced unfolding/refolding assays.
THs were adjusted to a protein concentration of 5.5 mg/ml before denaturation. Equal volumes of TH and GdnHCl solutions ranging from 0 to 4 M (final concentration, Δ[GdnHCl] = 0.25 M) were mixed and incubated for 1 h at 37°C at 300 rpm (Thermomixer Confort; Eppendorf). After the addition of PK at a final concentration of 8 U/ml, samples were reincubated for another 1 h at 37°C at 300 rpm. Protease treatment was terminated by the addition of phenylmethylsulfonyl fluoride (PMSF) at a final concentration of 3.6 mM. Samples were then precipitated in prechilled methanol for at least 3 h at −20°C, resuspended in sample buffer (final concentrations, 3% SDS, 4% β-mercaptoethanol, 10% glycerol, 2 mM EDTA, 62.5 mM Tris, pH 6.8), and boiled for 6 min. Appropriate GdnHCl working concentrations were obtained from serial dilution of an 8 M stock solution (Thermo Scientific Pierce, Protein Biology Products).
To monitor PrPSc refolding, after incubation with GdnHCl, samples were immediately diluted with 19 volumes of TN-NP40 and subsequently PK digested under the same working conditions as those specified above, precipitated in methanol, and resuspended in sample buffer. To monitor reproducibility, each treatment was repeated twice.
In a subset of samples from 6 MM1 and 6 MM 2C brains, we also performed the conformational stability assay (CSA), after incubation with GdnHCl, according to a previously published protocol (32). Briefly, after a clearing spin at 3,000 rpm for 10 min, aliquots of TH were mixed with aliquots of GdnHCl stock solution to have a final concentration of GdnHCl ranging from 0 to 4.0 M. After 1.5 h of incubation at room temperature, samples were precipitated with 5-fold prechilled methanol, centrifuged at 16,000 × g for 30 min at 4°C, and resuspended by sonication in 20 μl of LB100 (pH 8.0). Each aliquot was digested with 5 U/ml PK for 1 h at 37°C. The reaction was stopped with 2 mM PMSF.
Thermosolubilization assays.
TSA was performed according to the method of Bett and colleagues with minor modifications (30). Briefly, THs were digested with 8 U/ml PK for 1 h at 37°C with mild shaking (300 rpm). PK digestion was inactivated with PMSF (final concentration, 3.6 mM). Aliquots were mixed with an equal volume of loading buffer (final concentrations, 1.5% SDS, 2% β-mercaptoethanol, 5% glycerol, 1 mM EDTA, 31.25 mM Tris, pH 6.8) and heated to temperatures ranging from 25°C to 95°C (ΔT = 10°C) for 6 min with shaking in a thermomixer at 1,000 rpm before loading. To monitor reproducibility, each treatment was repeated twice.
Western blot and quantitative analyses of protein signal.
After boiling (or heating treatment), proteins were resolved in 13% polyacrylamide gels using a medium-sized gel electrophoresis apparatus (Criterion; Bio-Rad) and transferred to Immobilon-P membranes (Millipore Corp., Billerica, MA). After blocking in 10% nonfat milk in Tween-Tris-buffered saline, membranes were incubated with the primary antibody. After four washings in Tween-Tris-buffered saline, membranes were incubated for 1 h with an anti-mouse or an anti-rabbit secondary antibody conjugated to horseradish peroxidase (HRP; working dilution, 1:4,000 or 1:3,000; GE Healthcare) and washed again four times in Tween-Tris-buffered saline. The immunoreactive signal was detected by enhanced chemiluminescence (Immobilon Western Chemiluminescent HRP substrate; Merck Millipore) on an LAS 3000 camera (Fujifilm Corp., Tokyo, Japan). Western blot signals were measured by densitometry using the software AIDA (Image Data Analyzer v.4.15; Raytest Isotopenmessgeraete GmbH, Straubenhardt, Germany).
For guanidine unfolding assays, curves were obtained by plotting the percentage of protein remaining after denaturation and PK digestion treatments (with respect to the PK-digested but GdnHCl-untreated sample, i.e., 0 M, referred to as %PrPfold) against the corresponding guanidine concentration.
ED50 ([GdnHCl]50, e.g., the guanidine concentration needed to unfold 50% of PrPSc) for each sample was calculated from the equation describing the sigmoidal curve that best fitted the densitometric data (R2 ≥ 0.95). To compare groups, we also considered the percentage of protein remaining after denaturation plus PK digestion at [GdnHCl] of 2 M (referred to as %PrPfold2M) and the percentage of protein detected after refolding plus PK digestion at [GdnHCl] of 2 M (referred to as %PrPrefold2M). Refolded PrPSc was calculated by means of the formula (R − F)/N, where R is the PrPSc signal detected after GdnHCl incubation, dilution, and PK digestion, F is the PrPSc signal detected after GdnHCl incubation without dilution and PK digestion, and N is the PrPSc signal detected after PK digestion without GdnHCl addition.
For the TSA, we plotted the percentage of protein solubilized after heating treatment (with respect to the sample treated at 95°C, referred to as %PrPScmon) against the corresponding heating temperature. The ED50 (T50, e.g., the temperature needed to solubilize 50% of PrPSc) for each sample was calculated from the equation describing the sigmoidal curve that best fitted the data (R2 ≥ 0.95). To compare groups, we also considered the percentage of protein remaining after treatment at T of 35°C (referred to as %PrPScmon35°C) and at T of 75°C (referred to as %PrPScmon75°C).
Statistical analyses.
Statistical analyses were performed with Sigma Plot 12.5 (Systat Software Inc., Chicago, IL). One-way analysis of variance (ANOVA) followed by all pairwise multiple comparison procedures was used to look for significant differences in the chosen parameters among CJD variants.
RESULTS
Analysis of molecular mechanisms associated with GdnHCl-induced unfolding of human PrPSc.
It is well established that the exposure of PrPSc to increasing concentrations of GdnHCl progressively makes the protein more sensitive to protease digestion. Moreover, while a >3 M concentration of the chaotropic denaturant irreversibly modifies PrPSc structure and protease resistance, milder conditions allow the structural change to be reversible (38). Studies on PrPSc extracted from Syrian hamster brains also showed that GdnHCl-induced unfolding involves mainly the N-terminal portion of PrPSc, as suggested by the persistence of a PK-resistant, glycosylphosphatidylinositol (GPI)-linked, proteolytic fragment of about 16 kDa over a wide range of GdnHCl concentrations (40).
Before proceeding with the systematic analysis of the effects of GdnHCl-induced unfolding on human prions, we carried out preliminary studies to select the most appropriate experimental conditions. We first compared the effects of PK digestion when performed before and after GdnHCl dilution and unexpectedly found that the concentration of the denaturing agent that is required to fully digest PrP27-30 was significantly lower when PK was added to the undiluted sample. Indeed, after GdnHCl serial dilution we noticed a rapid partial recovery of PrPSc protease resistance. Only treatments with GdnHCl concentrations of 3.5 to 4 M irreversibly denatured the protein after solubilization and made it completely PK sensitive (Fig. 1). Given the observed rapid recovery of the original conformation of PrPSc and the notion that PK is not significantly inhibited by GdnHCl even at a relatively high concentration (41, 42), we chose to perform PK digestions in the undiluted denaturing agent (i.e., PK hydrolysis in buffer containing GdnHCl). To select the most appropriate antibody for monitoring the effect of GdnHCl on PrPSc PK sensitivity, we compared the 3F4 antibody, which recognizes a central PrP epitope (35), with antibodies against distinct C-terminal epitopes, which are known to recognize PrPSc truncated fragments, in addition to PrPSc 27-30 (20). In contrast to 3F4, which revealed only PrPSc 27-30, C-terminal antibodies also showed variable amounts of an 18-kDa fragment, consistent with PrPC-C1 (43), and of a 7- to 8-kDa fragment, likely generated by endogenous proteolysis of PrPC during the 1-h incubation at 37°C preceding PK digestion. Given the presence of these fragments in untreated control samples and the lack of any correlation between the relative amount of such fragments and the GdnHCl concentration, we concluded that (i) the generation of these PrPSc fragments is not denaturation dependent and (ii) central antibodies such as 3F4 are more appropriate than C-terminal antibodies to study the effect of GdnHCl on PrPSc structure.
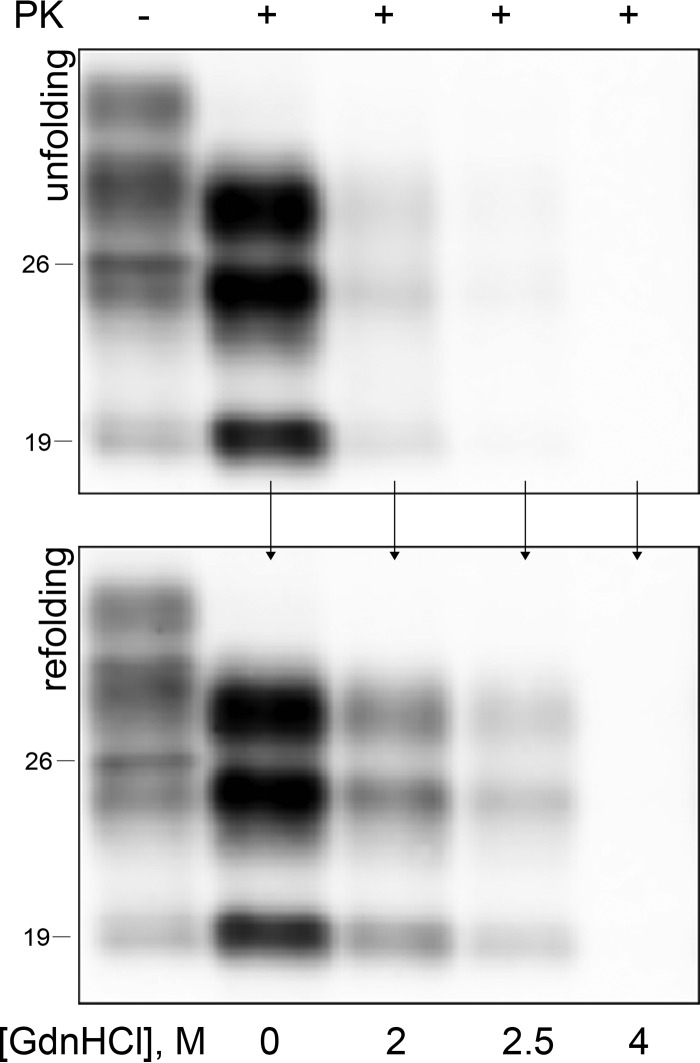
FIG 1.
Monitoring PrPSc refolding after GdnHCl-induced unfolding. Samples were exposed to increasing concentrations of GdnHCl and then PK digested immediately (upper panel) or after GdnHCl dilution (lower panel). PrPSc refolding is detected after GdnHCl dilution in samples exposed to up to 2.5 M GdnHCl.
PrPSc isolates associated with distinct human prion strains show similar denaturation curves after exposure to GdnHCl.
Guanidine denaturation curves showed similar sigmoid profiles in all CJD samples analyzed (Fig. 2A and B). The curve that best fitted the data (R2 ≥ 0.95) was a four-parameter sigmoid equation. Calculated values for [GdnHCl]50 ranged from 0.86 M (sCJDVV2) to 1.03 M (sCJDMM 2T) with no statistically significant differences among CJD types (Fig. 2C). A similar value trend was obtained by calculating the percentage of detectable PrPSc signal at a GdnHCl concentration of 2 M (%PrPfold2M) (Fig. 2D).
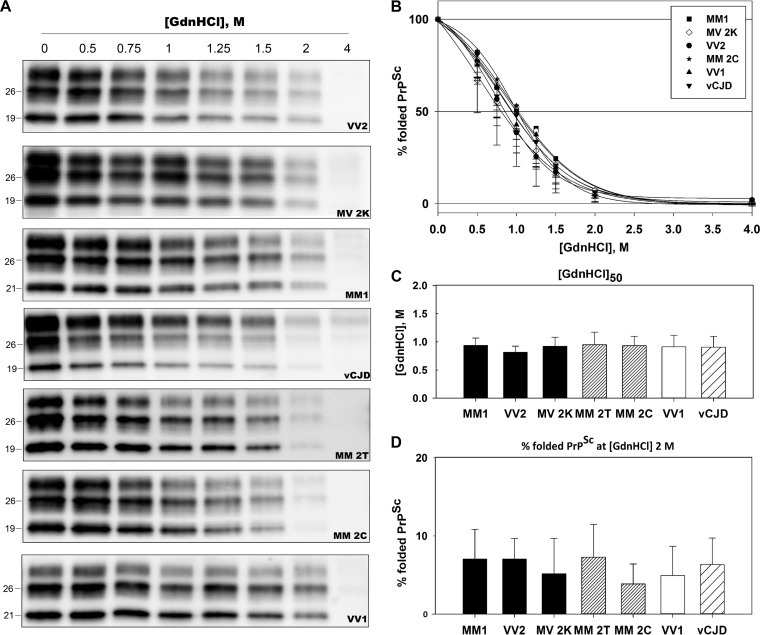
FIG 2.
GdnHCl-induced unfolding demonstrates a similar conformational stability of PrPSc aggregates across the spectrum of CJD prions. Brain homogenates were treated with increasing concentrations of GdnHCl and digested with PK, followed by SDS-PAGE and immunoblotting. Membranes were incubated with the primary antibody 3F4. (A) Representative immunoblots showing similar denaturation profiles among the seven CJD types analyzed. (B) Plots of GdnHCl-induced PrPSc unfolding for each sCJD type and vCJD. The y axis reports the percentage of folded PrPSc (e.g., percentage of PK-resistant PrPSc) relative to the untreated sample. Symbols represent data expressed as means ± standard deviations, and lines represent the sigmoid curves that best fit the data. (C) [GdnHCl]50 values for each CJD type, expressed as means ± standard deviations, indicating the GdnHCl molar concentration necessary to unfold 50% of untreated PrPSc. (D) Percentage of folded PrPSc at 2 M GdnHCl, expressed as means ± standard deviations.
To further look for strain-specific effects of GdnHCl, we also calculated the percentage of refolded PrPSc (%PrPrefold2M; see Materials and Methods) at a given, intermediate GdnHCl concentration (2 M). In agreement with our preliminary screening, PrPSc renaturation occurred in all groups analyzed. Calculated values ranged from 14% to 29% but, again, with no statistically significant differences among CJD types (Table 1).
TABLE 1.
Mean values of %PrPrefold2M for each CJD typea
| CJD type | %PrPrefold2M |
|---|---|
| MM1 | 24.06 ± 11.69 |
| VV2 | 21.32 ± 6.02 |
| MV 2K | 19.72 ± 15.53 |
| MM 2C | 24.53 ± 9.69 |
| MM 2T | 21.07 ± 10.73 |
| VV1 | 11.19 ± 5.18 |
| vCJD | 17.60 ± 10.25 |
All parameters are expressed as means ± standard deviations; no statistically significant differences were observed among CJD types.
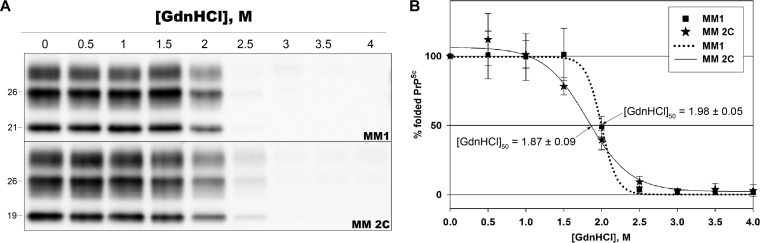
In order to exclude that the divergent results previously obtained by Cali et al. (32) on MM1 and MM 2C prions would reflect only a difference in the methodology used, we also repeated the experiments in a subgroup of samples by using their protocol (32). The results obtained confirmed the lack of remarkable differences between MM1 and MM 2C (Fig. 3). In particular, the [GdnHCl]50 values obtained in both groups (1.98 ± 0.05 M and 1.87 ± 0.09 M, respectively) were intermediate between those previously observed in MM 2C (1.42 M) and MM1 (2.76 M) (32).
FIG 3.
The conformational stability assay shows only subtle differences between sCJDMM1 and MM 2C prions. (A) Representative immunoblots of CSA, performed according to the protocol described by Cali et al. (32) on MM1 and MM 2C prions. Membranes were incubated with the primary antibody 3F4. (B) Plots of GdnHCl-induced PrPSc unfolding data sets for MM1 and MM 2C prions. The y axis reports the percentage of folded PrPSc (e.g., percentage of PK-resistant PrPSc) relative to the untreated sample. Symbols represent data expressed as means ± standard deviations, and lines represent the sigmoid curves that best fit the data. [GdnHCl]50 is expressed as means ± standard deviations.
PrPSc aggregates associated with distinct human prion strains show a divergent response to thermal solubilization.
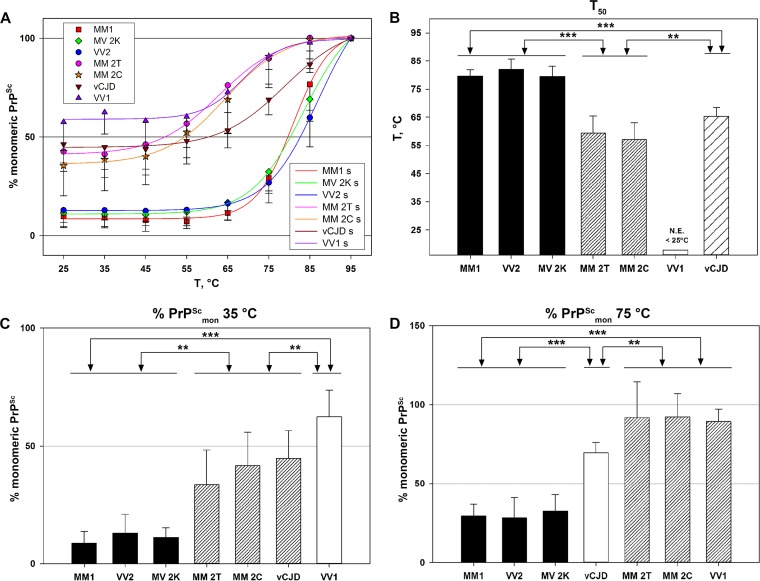
It has been shown that the exposure of PK digested PrPSc to a thermal gradient in the presence of SDS induces a progressive “solubilization” of protein aggregates that can be measured by a semiquantitative immunoblot analysis of monomeric PrPSc (30). By applying this experimental approach to the full spectrum of human prions, we found that, at variance with the GdnHCl assay, the specific profiles of the calculated solubilization curve and of values for T50, %PrPScmon35°C, and %PrPScmon75°C varied significantly according to the CJD type (Fig. 4 and 5). Overall, while PrPSc aggregates in sCJD VV1 and to a lesser extent in MM 2T or MM 2C showed a relatively high sensitivity to thermal solubilization, those associated with MM1, VV2, and MV2K were significantly more resistant. Hence, on the basis of the analyzed parameters, CJD types could be grossly classified in three groups: resistant (MM1, VV2, MV2K), sensitive (vCJD, MM2C, and MM2T), and highly sensitive (VV1) to thermal solubilization. A further heterogeneity was observed within the sensitive group with vCJD prions showing a more “resistant” profile at the highest temperatures than MM 2T and MM 2C (Fig. 5D).
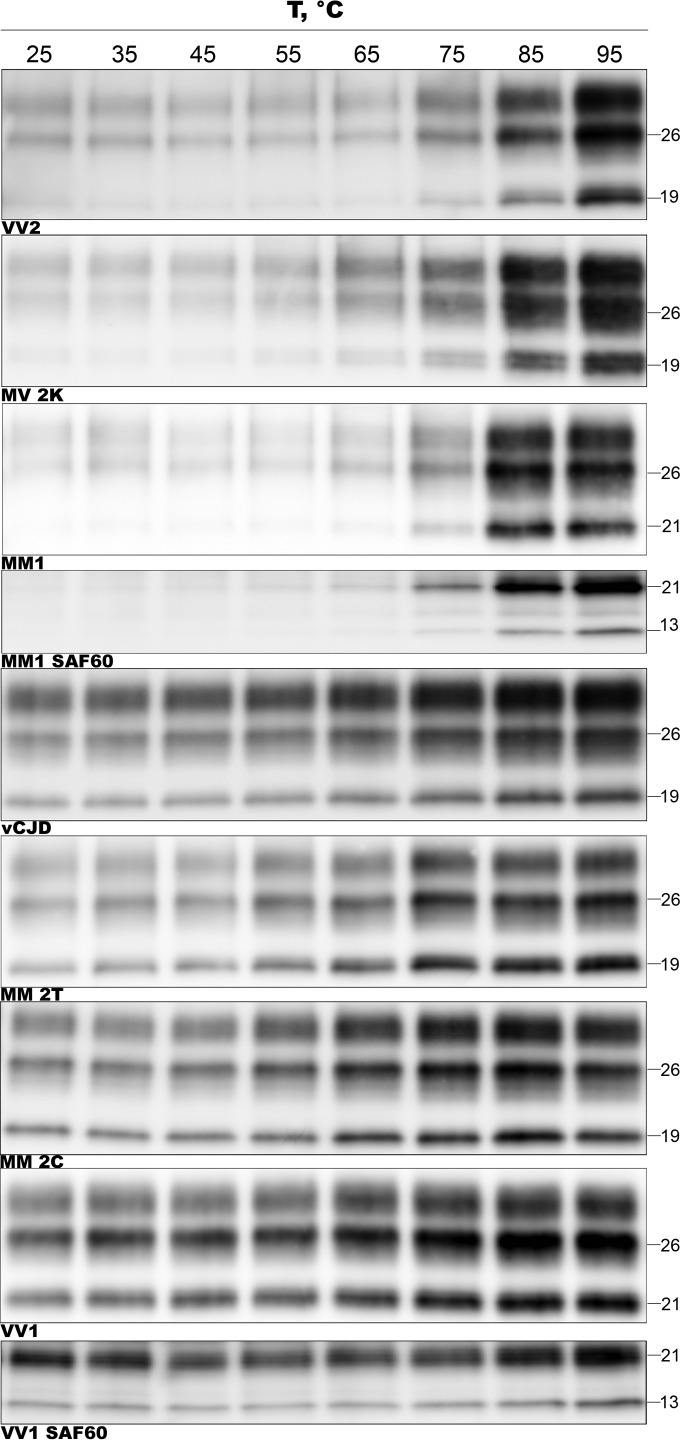
FIG 4.
CJD types show remarkable differences in the thermal stability of PK-digested PrPSc aggregates. PK-digested brain samples were subjected to increasing temperatures, followed by SDS-PAGE and immunoblotting. Representative immunoblots for each CJD type, probed with the primary antibodies 3F4 (all types) and SAF60 (only for sCJD MM1 and VV1, as labeled), are shown.
FIG 5.
Comparative analysis of the divergent thermal stability of CJD prions. (A) Plots of TSA data sets for each sCJD type and vCJD. The y axis reports the percentage of PrPSc in the monomeric state at each tested temperature relative to the sample treated at 95°C. Symbols represent the data, expressed as means ± standard deviations, and lines represent the sigmoid curves (s) that best fit the data. (B) T50 values for each tested CJD group, expressed as means ± standard deviations, indicating the temperature necessary to unfold 50% of PrPSc relative to the sample treated at 95°C. N.E., not estimable. (C) Percentage of monomeric PrPSc at 35°C expressed as means ± standard deviations. (D) Percentage of monomeric PrPSc at 75°C expressed as means ± standard deviations. In panels B to D, the triple asterisk (***) indicates a P value of <0.001 for all pairwise multiple comparisons between the groups; the double asterisk (**) indicates a P value of <0.001 for all pairwise multiple comparisons with the following exceptions: vCJD versus MM 2C (P < 0.005) and vCJD versus MM 2T (P = 0.184, not significant) (B); VV2 versus MM 2T (P < 0.002), and VV1 versus vCJD (P < 0.005) (C); MM 2C versus vCJD (P < 0.005), VV1 versus vCJD (P = 0.016), and MM 2T versus vCJD (P = 0.029) (D).
To exclude the possibility that the observed heterogeneity in the thermostability of PrPSc aggregates derives from conformational changes that are limited to the 3F4 binding region, we reanalyzed a subgroup of MM1 and VV1 samples using the monoclonal antibody (MAb) SAF60. The thermosolubilization curves calculated from the immunoblots labeled with SAF60 fully matched those obtained using 3F4 (Fig. 4). In addition, the 13-kDa C-terminal fragment that is visualized by this antibody (20) in addition to PrP27-30 showed a solubilization kinetics that paralleled that of PrP27-30 in each CJD type (e.g., more thermostable in MM1 than in VV1) (Fig. 4). The latter observation strongly suggests that PrPSc aggregates in CJD MM1 and VV1 include both fragments.
Finally, we plotted the solubilization curves for each of the three PrPSc glycoforms and found a similar thermosolubilization kinetics for each of them with only a minor trend toward a preferential solubilization of the di- and monoglycosylated forms (data not shown).
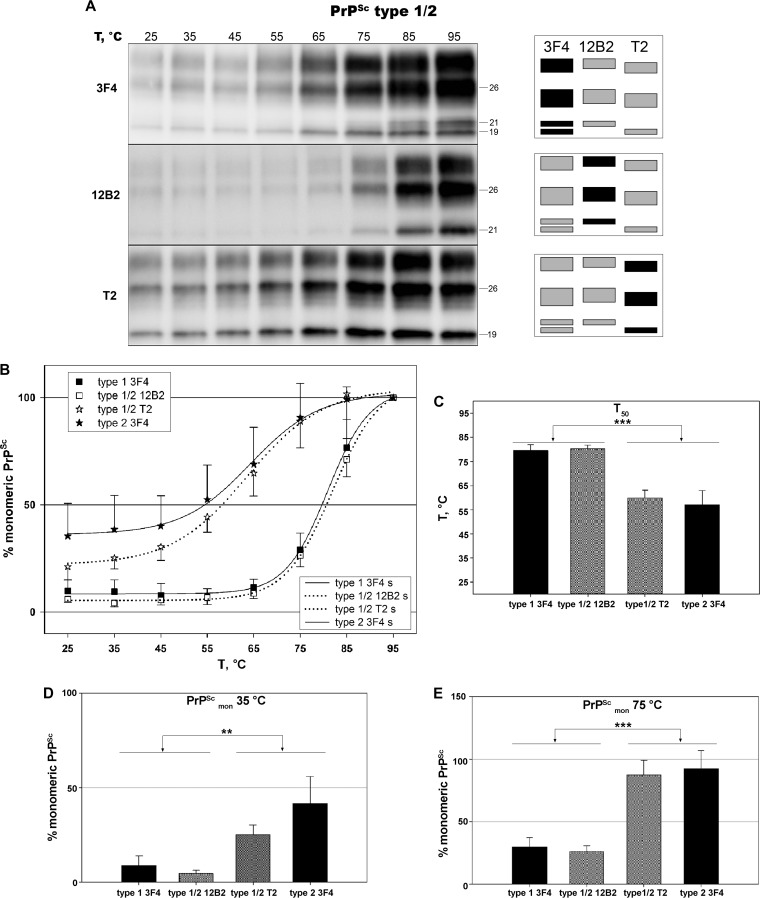
The cooccurrence of PrPSc types in mixed sCJD phenotypes does not alter/affect the thermal solubilization properties of coexisting isoforms.
It is well known that PrPSc types 1 and 2 coexist within the same brain in about 35% of sCJD cases (5). Accordingly, mixed phenotypes have been considered distinct subtypes in current sCJD classification (44). However, the critical question of whether the cooccurrence of PrPSc types in the brain simply reflects the neutral coexistence of two prion strains forming independent protein aggregates or in contrast represents interacting strains forming mixed aggregates with distinct physicochemical properties remains unanswered. To investigate this issue, we selected 6 cortical samples from sCJD MM1 + 2C containing a significant amount of both types (e.g., with the less-represented type 2 being between 30 and 50% of the total PrPSc signal). We calculated the solubilization curves for these samples using three different antibodies. More specifically, we monitored the solubilization trend of the two PrPSc types simultaneously using 3F4 and analyzed PrPSc types 1 and 2 separately, despite their cooccurrence, with the type-specific antibodies 12B2 and T2 (Fig. 6A). Remarkably, the curves generated with the 12B2 and T2 antibodies fully matched or paralleled very closely those of the corresponding pure phenotype (Fig. 6B). Accordingly, no significant statistical differences were observed in the calculated T50, %PrPScmon35°C, and %PrPScmon75°C values between mixed and pure samples for both PrPSc types 1 and 2. Moreover, statistical analysis confirmed the differences between PrPSc types 1 and 2 when analyzed in mixed samples using the type-specific antibodies (Fig. 6C, D, and E).
FIG 6.
In vivo cooccurring MM1 and MM 2C prions maintain the distinctive thermostability of the corresponding pure CJD type. PK-digested brain samples were subjected to increasing temperatures, followed by SDS-PAGE and immunoblotting. (A) Representative immunoblots of TSA performed on an MM1 + 2C sample, probed with primary antibodies 3F4, 12B2, and T2. (B) Plots of temperature solubility assay data sets for PrPSc types 1 and 2, when cooccurring prions (MM1 + 2C) are compared to those of the corresponding pure type (MM1 and MM 2C, respectively). Type-specific antibodies 12B2 and T2 have been used to obtain accurate separate measurements of type 1 and type 2 signals in mixed samples. Symbols represent data, expressed as means ± standard deviations, and lines represent the sigmoid curves (s) that best fit the data. (C) Comparison of T50 values between pure and mixed variants. The triple asterisk (***) indicates a statistically significant difference between groups at P values of <0.001. (D) Comparison of the percentage of monomeric PrPSc at 35°C between pure and mixed variants. The double asterisk (**) indicates a P value of <0.05 between groups with the exception of type 1 3F4 versus type 1/2 T2 (not significant). (E) Comparison of the percentages of monomeric PrPSc at 75°C between pure and mixed variants. The triple asterisk (***) indicates a P value of <0.001 between groups.
DISCUSSION
The analysis of GdnHCl-induced unfolding by either the conformation-dependent immunoassay (CDI), which measures the extent of epitope exposure, or the conformational stability assay (CSA), which instead monitors the progressive loss of PK resistance after exposure to increasing concentrations of GdnHCl, has been extensively applied to the study of PrPSc molecules. CDI analysis of PrPSc unfolding in different murine prion strains yielded unique binding profiles, suggesting that each strain is related to a specific PrPSc conformation (45). Similarly, the stability of PrPSc aggregates measured by CSA or by a thermal stability assay (TSA) was found to be strain dependent and inversely correlated with the capacity to induce a rapidly lethal disease in murine models (30). At variance with experimentally cloned rodent prion strains, however, less numerous and less conclusive data have been obtained with PrPSc preparations extracted from naturally occurring prion diseases, especially in humans. In the few studies performed to date, sCJD MM1 PrPSc was shown to be more stable than MM 2C PrPSc by both CDI and CSA, and MM1 PrPSc was shown to have a moderately higher stability than vCJD PrPSc by CDI (32–34). In the present study, we have carried out the first systematic analysis of PrPSc conformational stability in a large series of brain samples across the whole spectrum of human sCJD and vCJD prions. Our results failed to reveal significant strain-specific differences in the GdnHCl denaturation curve of PrPSc aggregates. Thus, despite the limitations related to the methodology being rather crude, the data suggest that the intermolecular interactions modulating the divergent PrPSc aggregation propensity among CJD types (27) are not strongly targeted by GdnHCl.
As far as CSA is concerned, we introduced some variations in the protocol to minimize PrPSc refolding, namely, by performing the PK digestion step without removing or diluting GdnHCl. Given that PrPSc refolding is paralleled by an increase in PK resistance of the protein, it is expected that GdnHCl dilution before PK treatment would increase the calculated [GdnHCl]50, and, indeed, [GdnHCl]50 values in previous studies were greater than 1.5 M, whereas our mean values ranged from 0.86 M to 1.03 M. In order to exclude the possibility that the discrepant results with a previous study (32) concerning MM1 and MM 2C prions could be due to a difference in the methodology used, we repeated the study of GdnHCl-induced PrPSc unfolding using the original protocol (32). While, as expected, the changed procedure led to an increase in [GdnHCl]50 values, the results confirmed the lack of significant differences in the GdnHCl denaturation curve between MM1 and MM 2C sCJD prions.
Another approach that has been used to characterize prion strains in mice (30, 46), although never applied to the study of CJD prions, focuses on the denaturing effect of heating in the presence of SDS. As with GdnHCl, this assay likely measures mainly the propensity of PK-digested PrPSc aggregates to depolymerize. At variance with GdnHCl, however, the exposure to increasing temperatures revealed significantly different responses among sCJD types with MM1, VV2, and MV 2K PrPSc forming mainly stable, highly resistant aggregates, the VV1 type comprising highly unstable aggregates that easily dissolve at a relatively low temperature, and MM 2C, MM 2T, and vCJD prions exhibiting an intermediate behavior.
The relatively high level of PrPSc that is solubilized at relatively low temperatures in some prion types is intriguing and may suggest that these agents comprise different forms of PrPSc. Consistently, an increasing number of studies support the idea that a variable proportion of abnormal PrP is made by a soluble, poorly aggregated, and fairly PK-sensitive form, although its specific role and relevance for disease pathogenesis remain controversial (27, 47). Our approach, focusing on PK-treated PrPSc, did not allow us to address in depth the issue of the influence of this putative “fully PK-sensitive PrPSc” on thermostability profile; nevertheless, the lack of an obvious correlation between sedimentation profile (as determined in reference 27) and thermostability (present work) of PK-treated PrPSc suggests that factors other than size also contribute to the thermostability of PrPSc aggregates in CJD.
The present results, when combined with the growing knowledge on human prion strains, provide novel insights into the relationships between the physicochemical properties of PrPSc aggregates and disease characteristics such as incidence, phenotypic expression, and transmission properties (5, 7, 8, 10, 11, 48–50). In particular, PrPSc thermostability appears to largely, although not entirely, correlate with prion replication efficiency expressed either by the attack rate and incubation time after experimental transmission in the most compatible host genotype or by the relative incidence and duration of clinical disease (Table 2). In this regard, the comparison between VV2 and VV1 human prions appears to be particularly illustrative. Indeed, compared to VV1, VV2 prions are more common, cause a more rapid disease (5), are more readily transmissible (10), and produce PrPSc aggregates with higher thermostability, PK resistance, and aggregation propensity (Table 2) (27). Thus, at variance with earlier observations in murine and yeast prions (PSI+) suggesting that a low PrPSc aggregate stability, by favoring the fragmentation of PrPSc aggregates, would result in a higher prion replication rate, in humans the thermostability of PrPSc seems to positively correlate with disease severity and “virulence” in the most compatible host genotype. It is noteworthy that our observation is also consistent with data obtained with hamster-adapted prion strains, showing that short-incubation-period strains are characterized by a higher conformational stability of PrPSc and a more efficient replication (28, 29).
TABLE 2.
Comparison of epidemiological and clinical data, results of experimental transmission, and biochemical PrPSc properties of CJD strainse
| CJD type | Strain | Incidence (% of total sCJD cases) | Mean disease duration (mo) | Attack rate (%)a,b | Days of incubationa,b | T50 (°C) (this study) | PK resistance ED50 (U/ml)c (reference 27) |
|---|---|---|---|---|---|---|---|
| VV2 | V2 | 15 (5) | 6.3d (5) | 100 (7, 8, 10) | 274 ± 4 (10), 302 ± 9 (7, 8) | 82.05 ± 3.70 | 4.137 ± 3.562 |
| MV 2K | V2 | 8 (5) | 6.3d (5) | 100 (8, 10) | 288 ± 3 (10), 329 ± 3 (8) | 79.48 ± 3.63 | 1.407 ± 0.892 |
| MM1 | M1 | 40 (5) | 4.0 (5) | 100 (7, 8, 10) | 446 ± 3 (10) 467 ± 24 (7, 8) | 79.66 ± 2.32 | 0.093 ± 0.068 |
| vCJD | BSE | Rare (48) | 14 (48) | 83 (49), 100 (49) | 540 ± 41 (49), 668 ± 22 (49) | 65.26 ± 3.19 | 5.192 ± 2.378 |
| MM 2T | M 2T | 1 (5) | 15.5 (5) | 93 (11) | 535 ± 32 (11) | 59.41 ± 6.04 | 0.134 ± 0.089 |
| VV1 | V1 | 1 (5) | 15.3 (5) | 50 (10) | 568 ± 0 (10) | <25 | 0.034 ± 0.028 |
| MM 2C | M 2C | 1 (5) | 20.0 (5) | 0 (8, 10, 11, 50) | NAf (8, 10, 11, 50) | 57.11 ± 5.96 | 0.276 ± 0.210 |
Data are from PrP-humanized knock-in mice expressing normal levels of human PrPC.
Data for each strain refer to those obtained in recipient animals carrying the most compatible PRNP 129 genotype.
ED50 is the PK concentration needed to digest 50% of PrPSc.
The reported mean disease duration refers only to VV2, the group with the most compatible 129 genotype to the V2 strain.
Relevant references are in parentheses. In the last 3 columns, values are means ± standard deviations.
NA, not applicable.
Our results also reveal a relatively high thermostability, comparable to that of VV2, for MM1 and MV 2K prions. For the latter, the results are consistent with data from transmission studies showing that sCJD VV2 and MV 2K are linked to the same prion strain (V2) (9), whereas the result obtained with MM1 further confirms the positive correlation between PrPSc thermostability and strain “virulence” (Table 2). However, the degree of PK resistance of PrPSc aggregates in MM1 is not as high as in VV2 and MV 2K (27), indicating that the two variables are not necessarily directly correlated (Table 2). Our findings that PrPSc aggregates in vCJD show a very high resistance to PK digestion (27) despite being less thermostable than those from sCJD MM1 prions also support this conclusion. It has been suggested that PK itself may act as a disaggregating agent by eliminating PrP monomers (51) and thus changing the equilibrium between monomers and polymers in favor of monomers. Likewise, GdnHCl or heating exposure also acts as a disaggregating agent. Thus, the divergent responses that we obtained using these three methods suggest that the propensity of PrPSc to disaggregate is significantly affected by the type of treatment, which is also in line with evidence indicating that temperature and GdnHCl destabilize the folded structure of proteins by means of distinct molecular mechanisms (52–54).
In addition to the analyses on pure phenotypes, we performed TSA on six cases with a mixed phenotype, carrying MM at PRNP codon 129 and showing the cooccurrence of PrPSc types 1 and 2 in the brain. In particular, we addressed the unsolved question of whether the cooccurrence of PrPSc types in the brain simply reflects the neutral coexistence of two prion strains forming independent protein aggregates or, in contrast, interacting strains forming mixed aggregates with novel specific properties. At variance with the findings of a previous study (32) arguing that the coexistence of types 1 and 2 in the same anatomical region may confer special conformational characteristics to the mixed PrPSc type aggregate, the results that we obtained using type-specific antibodies support the former hypothesis. Indeed, properties of each individual type, when coexisting, exactly fit those of the corresponding pure type, suggesting a lack of interaction between the two PrPSc types. The discrepant results between the two studies are difficult to explain given that they were obtained using different approaches (CSA versus TSA). Unfortunately, we could not address the issue further, since we failed to reveal a significant difference in PrPSc conformational stability between the pure MM1 and MM 2C types using CSA.
ACKNOWLEDGMENT
We thank Barbara Polischi for her excellent technical assistance.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Prusiner SB. 1998. Prions. Proc Natl Acad Sci U S A 95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieschke J, Weber P, Sarafoff N, Beekes M, Giese A, Kretzschmar H. 2004. Autocatalytic self-propagation of misfolded prion protein. Proc Natl Acad Sci U S A 101:12207–12211. doi: 10.1073/pnas.0404650101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker LC, Jucker M. 2015. Neurodegenerative diseases: expanding the prion concept. Annu Rev Neurosci 38:87–103. doi: 10.1146/annurev-neuro-071714-033828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parchi P, Giese A, Capellari S, Brown P, Schulz-Schaeffer W, Windl O, Zerr I, Budka H, Kopp N, Piccardo P, Poser S, Rojiani A, Streichemberger N, Julien J, Vital C, Ghetti B, Gambetti P, Kretzschmar H. 1999. Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann Neurol 46:224–233. [PubMed] [Google Scholar]
- 5.Parchi P, Strammiello R, Notari S, Giese A, Langeveld JPM, Ladogana A, Zerr I, Roncaroli F, Cras P, Ghetti B, Pocchiari M, Kretzschmar H, Capellari S. 2009. Incidence and spectrum of sporadic Creutzfeldt-Jakob disease variants with mixed phenotype and co-occurrence of PrPSc types: an updated classification. Acta Neuropathol 118:659–671. doi: 10.1007/s00401-009-0585-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nonno R, Di Bari MA, Cardone F, Vaccari G, Fazzi P, Dell'Omo G, Cartoni C, Ingrosso L, Boyle A, Galeno R, Sbriccoli M, Lipp H-P, Bruce M, Pocchiari M, Agrimi U. 2006. Efficient transmission and characterization of Creutzfeldt-Jakob disease strains in bank voles. PLoS Pathog 2:e12. doi: 10.1371/journal.ppat.0020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi A, Sakuma N, Matsuura Y, Mohri S, Aguzzi A, Kitamoto T. 2010. Experimental verification of a traceback phenomenon in prion infection. J Virol 84:3230–3238. doi: 10.1128/JVI.02387-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi A, Teruya K, Matsuura Y, Shirai T, Nakamura Y, Yamada M, Mizusawa H, Mohri S, Kitamoto T. 2015. The influence of PRNP polymorphisms on human prion disease susceptibility: an update. Acta Neuropathol 130:159–170. doi: 10.1007/s00401-015-1447-7. [DOI] [PubMed] [Google Scholar]
- 9.Parchi P, Cescatti M, Notari S, Schulz-Schaeffer WJ, Capellari S, Giese A, Zou W-Q, Kretzschmar H, Ghetti B, Brown P. 2010. Agent strain variation in human prion disease: insights from a molecular and pathological review of the National Institutes of Health series of experimentally transmitted disease. Brain 133:3030–3042. doi: 10.1093/brain/awq234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop MT, Will RG, Manson JC. 2010. Defining sporadic Creutzfeldt-Jakob disease strains and their transmission properties. Proc Natl Acad Sci U S A 107:12005–12010. doi: 10.1073/pnas.1004688107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moda F, Suardi S, Di Fede G, Indaco A, Limido L, Vimercati C, Ruggerone M, Campagnani I, Langeveld J, Terruzzi A, Brambilla A, Zerbi P, Fociani P, Bishop MT, Will RG, Manson JC, Giaccone G, Tagliavini F. 2012. MM2-thalamic Creutzfeldt-Jakob disease: neuropathological, biochemical and transmission studies identify a distinctive prion strain. Brain Pathol 22:662–669. doi: 10.1111/j.1750-3639.2012.00572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caughey BW, Dong A, Bhat KS, Ernst D, Hayes SF, Caughey WS. 1991. Secondary structure analysis of the scrapie-associated protein PrP 27-30 in water by infrared spectroscopy. Biochemistry 30:7672–7680. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- 13.Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE. 1993. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci U S A 90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caughey B, Raymond GJ, Bessen RA. 1998. Strain-dependent differences in beta-sheet conformations of abnormal prion protein. J Biol Chem 273:32230–32235. doi: 10.1074/jbc.273.48.32230. [DOI] [PubMed] [Google Scholar]
- 15.Safar JG, Xiao X, Kabir ME, Chen S, Kim C, Haldiman T, Cohen Y, Chen W, Cohen ML, Surewicz WK. 2015. Structural determinants of phenotypic diversity and replication rate of human prions. PLoS Pathog 11:e1004832. doi: 10.1371/journal.ppat.1004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. 2004. Conformational variations in an infectious protein determine prion strain differences. Nature 428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 17.Bessen RA, Marsh RF. 1994. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol 68:7859–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Telling GC, Parchi P, DeArmond SJ, Cortelli P, Montagna P, Gabizon R, Mastrianni J, Lugaresi E, Gambetti P, Prusiner SB. 1996. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science 274:2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 19.Parchi P, Zou W, Wang W, Brown P, Capellari S, Ghetti B, Kopp N, Schulz-Schaeffer WJ, Kretzschmar HA, Head MW, Ironside JW, Gambetti P, Chen SG. 2000. Genetic influence on the structural variations of the abnormal prion protein. Proc Natl Acad Sci U S A 97:10168–10172. doi: 10.1073/pnas.97.18.10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Notari S, Strammiello R, Capellari S, Giese A, Cescatti M, Grassi J, Ghetti B, Langeveld JPM, Zou W-Q, Gambetti P, Kretzschmar HA, Parchi P. 2008. Characterization of truncated forms of abnormal prion protein in Creutzfeldt-Jakob disease. J Biol Chem 283:30557–30565. doi: 10.1074/jbc.M801877200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguzzi A, Heikenwalder M, Polymenidou M. 2007. Insights into prion strains and neurotoxicity. Nat Rev Mol Cell Biol 8:552–561. doi: 10.1038/nrm2204. [DOI] [PubMed] [Google Scholar]
- 22.Bessen RA, Marsh RF. 1992. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J Virol 66:2096–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzaban S, Friedlander G, Schonberger O, Horonchik L, Yedidia Y, Shaked G, Gabizon R, Taraboulos A. 2002. Protease-sensitive scrapie prion protein in aggregates of heterogeneous sizes. Biochemistry 41:12868–12875. doi: 10.1021/bi025958g. [DOI] [PubMed] [Google Scholar]
- 24.Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B. 2005. The most infectious prion protein particles. Nature 437:257–261. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tixador P, Herzog L, Reine F, Jaumain E, Chapuis J, Le Dur A, Laude H, Béringue V. 2010. The physical relationship between infectivity and prion protein aggregates is strain-dependent. PLoS Pathog 6:e1000859. doi: 10.1371/journal.ppat.1000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laferrière F, Tixador P, Moudjou M, Chapuis J, Sibille P, Herzog L, Reine F, Jaumain E, Laude H, Rezaei H, Béringue V. 2013. Quaternary structure of pathological prion protein as a determining factor of strain-specific prion replication dynamics. PLoS Pathog 9:e1003702. doi: 10.1371/journal.ppat.1003702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saverioni D, Notari S, Capellari S, Poggiolini I, Giese A, Kretzschmar HA, Parchi P. 2013. Analyses of protease resistance and aggregation state of abnormal prion protein across the spectrum of human prions. J Biol Chem 288:27972–27985. doi: 10.1074/jbc.M113.477547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ayers JI, Schutt CR, Shikiya RA, Aguzzi A, Kincaid AE, Bartz JC. 2011. The strain-encoded relationship between PrP replication, stability and processing in neurons is predictive of the incubation period of disease. PLoS Pathog 7:e1001317. doi: 10.1371/journal.ppat.1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez-Montalban N, Makarava N, Savtchenko R, Baskakov IV. 2011. Relationship between conformational stability and amplification efficiency of prions. Biochemistry 50:7933–7940. doi: 10.1021/bi200950v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bett C, Joshi-Barr S, Lucero M, Trejo M, Liberski P, Kelly JW, Masliah E, Sigurdson CJ. 2012. Biochemical properties of highly neuroinvasive prion strains. PLoS Pathog 8:e1002522. doi: 10.1371/journal.ppat.1002522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pirisinu L, Di Bari M, Marcon S, Vaccari G, D'Agostino C, Fazzi P, Esposito E, Galeno R, Langeveld J, Agrimi U, Nonno R. 2010. A new method for the characterization of strain-specific conformational stability of protease-sensitive and protease-resistant PrP. PLoS One 5:e12723. doi: 10.1371/journal.pone.0012723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cali I, Castellani R, Alshekhlee A, Cohen Y, Blevins J, Yuan J, Langeveld JPM, Parchi P, Safar JG, Zou W-Q, Gambetti P. 2009. Co-existence of scrapie prion protein types 1 and 2 in sporadic Creutzfeldt-Jakob disease: its effect on the phenotype and prion-type characteristics. Brain 132:2643–2658. doi: 10.1093/brain/awp196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim C, Haldiman T, Cohen Y, Chen W, Blevins J, Sy M-S, Cohen M, Safar JG. 2011. Protease-sensitive conformers in broad spectrum of distinct PrPSc structures in sporadic Creutzfeldt-Jakob disease are indicator of progression rate. PLoS Pathog 7:e1002242. doi: 10.1371/journal.ppat.1002242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi YP, Peden AH, Gröner A, Ironside JW, Head MW. 2010. Distinct stability states of disease-associated human prion protein identified by conformation-dependent immunoassay. J Virol 84:12030–12038. doi: 10.1128/JVI.01057-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou W-Q, Langeveld J, Xiao X, Chen S, McGeer PL, Yuan J, Payne MC, Kang H-E, McGeehan J, Sy M-S, Greenspan NS, Kaplan D, Wang G-X, Parchi P, Hoover E, Kneale G, Telling G, Surewicz WK, Kong Q, Guo J-P. 2010. PrP conformational transitions alter species preference of a PrP-specific antibody. J Biol Chem 285:13874–13884. doi: 10.1074/jbc.M109.088831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langeveld JPM, Jacobs JG, Erkens JHF, Bossers A, van Zijderveld FG, van Keulen LJM. 2006. Rapid and discriminatory diagnosis of scrapie and BSE in retro-pharyngeal lymph nodes of sheep. BMC Vet Res 2:19. doi: 10.1186/1746-6148-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Féraudet C, Morel N, Simon S, Volland H, Frobert Y, Créminon C, Vilette D, Lehmann S, Grassi J. 2005. Screening of 145 anti-PrP monoclonal antibodies for their capacity to inhibit PrPSc replication in infected cells. J Biol Chem 280:11247–11258. doi: 10.1074/jbc.M407006200. [DOI] [PubMed] [Google Scholar]
- 38.Kocisko DA, Come JH, Priola SA, Chesebro B, Raymond GJ, Lansbury PT, Caughey B. 1994. Cell-free formation of protease-resistant prion protein. Nature 370:471–474. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- 39.Notari S, Capellari S, Giese A, Westner I, Baruzzi A, Ghetti B, Gambetti P, Kretzschmar HA, Parchi P. 2004. Effects of different experimental conditions on the PrPSc core generated by protease digestion: implications for strain typing and molecular classification of CJD. J Biol Chem 279:16797–16804. doi: 10.1074/jbc.M313220200. [DOI] [PubMed] [Google Scholar]
- 40.Kocisko DA, Lansbury PT, Caughey B. 1996. Partial unfolding and refolding of scrapie-associated prion protein: evidence for a critical 16-kDa C-terminal domain. Biochemistry 35:13434–13442. doi: 10.1021/bi9610562. [DOI] [PubMed] [Google Scholar]
- 41.Hilz H, Wiegers U, Adamietz P. 1975. Stimulation of proteinase K action by denaturing agents: application to the isolation of nucleic acids and the degradation of “masked” proteins. Eur J Biochem 56:103–108. doi: 10.1111/j.1432-1033.1975.tb02211.x. [DOI] [PubMed] [Google Scholar]
- 42.Poulsen JW, Madsen CT, Young C, Poulsen FM, Nielsen ML. 2013. Using guanidine-hydrochloride for fast and efficient protein digestion and single-step affinity-purification mass spectrometry. J Proteome Res 12:1020–1030. doi: 10.1021/pr300883y. [DOI] [PubMed] [Google Scholar]
- 43.Chen SG, Teplow DB, Parchi P, Teller JK, Gambetti P, Autilio-Gambetti L. 1995. Truncated forms of the human prion protein in normal brain and in prion diseases. J Biol Chem 270:19173–19180. doi: 10.1074/jbc.270.32.19173. [DOI] [PubMed] [Google Scholar]
- 44.Parchi P, Strammiello R, Giese A, Kretzschmar H. 2011. Phenotypic variability of sporadic human prion disease and its molecular basis: past, present, and future. Acta Neuropathol 121:91–112. doi: 10.1007/s00401-010-0779-6. [DOI] [PubMed] [Google Scholar]
- 45.Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. 1998. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med 4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 46.Bett C, Kurt TD, Lucero M, Trejo M, Rozemuller AJ, Kong Q, Nilsson KPR, Masliah E, Oldstone MB, Sigurdson CJ. 2013. Defining the conformational features of anchorless, poorly neuroinvasive prions. PLoS Pathog 9:e1003280. doi: 10.1371/journal.ppat.1003280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silva CJ, Vázquez-Fernández E, Onisko B, Requena JR. 2015. Proteinase K and the structure of PrPSc: the good, the bad and the ugly. Virus Res 207:120–126. doi: 10.1016/j.virusres.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 48.Diack AB, Head MW, McCutcheon S, Boyle A, Knight R, Ironside JW, Manson JC, Will RG. 2014. Variant CJD. 18 years of research and surveillance. Prion 8:286–295. doi: 10.4161/pri.29237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeuchi A, Kobayashi A, Ironside JW, Mohri S, Kitamoto T. 2013. Characterization of variant Creutzfeldt-Jakob disease prions in prion protein-humanized mice carrying distinct codon 129 genotypes. J Biol Chem 288:21659–21666. doi: 10.1074/jbc.M113.470328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kobayashi A, Matsuura Y, Iwaki T, Iwasaki Y, Yoshida M, Takahashi H, Murayama S, Takao M, Kato S, Yamada M, Mohri S, Kitamoto T. 2016. Sporadic Creutzfeldt-Jakob disease MM1+2C and MM1 are identical in transmission properties. Brain Pathol 26:95–101. doi: 10.1111/bpa.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Masel J, Jansen VA. 1999. The kinetics of proteinase K digestion of linear prion polymers. Proc Biol Sci 266:1927–1931. doi: 10.1098/rspb.1999.0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alonso DO, Dill KA. 1991. Solvent denaturation and stabilization of globular proteins. Biochemistry 30:5974–5985. doi: 10.1021/bi00238a023. [DOI] [PubMed] [Google Scholar]
- 53.Pica A, Russo Krauss I, Castellano I, Rossi M, La Cara F, Graziano G, Sica F, Merlino A. 2012. Exploring the unfolding mechanism of γ-glutamyltranspeptidases: the case of the thermophilic enzyme from Geobacillus thermodenitrificans. Biochim Biophys Acta 1824:571–577. doi: 10.1016/j.bbapap.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 54.Mahapa A, Mandal S, Biswas A, Jana B, Polley S, Sau S, Sau K. 2015. Chemical and thermal unfolding of a global staphylococcal virulence regulator with a flexible C-terminal end. PLoS One 10:e0122168. doi: 10.1371/journal.pone.0122168. [DOI] [PMC free article] [PubMed] [Google Scholar]