ABSTRACT
Human cytomegalovirus (HCMV), a betaherpesvirus, can cause life-threatening disease in immunocompromised individuals. Viral envelope glycoproteins that mediate binding to and penetration into target cells have been identified previously. In contrast, cellular proteins supporting HCMV during entry are largely unknown. In order to systematically identify host genes affecting initial steps of HCMV infection, a targeted RNA interference screen of 96 cellular genes was performed in endothelial cells by use of a virus strain expressing the full set of known glycoprotein H and L (gH/gL) complexes. The approach yielded five proviral host factors from different protein families and eight antiviral host factors, mostly growth factor receptors. The tetraspanin CD151 was uncovered as a novel proviral host factor and was analyzed further. Like endothelial cells, fibroblasts were also less susceptible to HCMV infection after CD151 depletion. Virus strains with different sets of gH/gL complexes conferring either broad or narrow cell tropism were equally impaired. Infection of CD151-depleted cells by a fluorescent virus with differentially labeled capsid and envelope proteins revealed a role of CD151 in viral penetration but not in adsorption to the cell. In conclusion, the tetraspanin CD151 has emerged as a novel host factor in HCMV entry and as a putative antiviral target.
IMPORTANCE At present, the events at the virus-cell interface and the cellular proteins involved during the HCMV entry steps are scarcely understood. In this study, several host factors with putative roles in this process were identified. The tetraspanin CD151 was discovered as a previously unrecognized proviral host factor for HCMV and was found to support viral penetration into the target cells. The findings of this study shed light on the cellular contribution during the initial steps of HCMV infection and open a new direction in HCMV research.
INTRODUCTION
Human cytomegalovirus (HCMV) is a member of the Betaherpesvirinae subfamily of herpesviruses and can cause severe disease in immunocompromised individuals. The systemic disease pattern is reflected by the ability of the virus to infect a remarkably wide variety of cell types, including endothelial cells and fibroblasts (1–6). In infected individuals, vascular endothelial cells are considered to have a pivotal function in hematogenous dissemination of HCMV and viral spread (7–13).
HCMV strains with deleterious mutations in the UL128, UL130, or UL131A gene, including several prototypic laboratory-adapted strains, grow well in fibroblast cultures but are unable to replicate efficiently in a variety of other physiologically relevant cell types, such as endothelial or epithelial cells (14–19). The replication defect observed with cell culture-adapted strains in vitro in many cell types that are infected by clinical strains in vivo has been shown to be due to a failure during the entry process (14, 17–19). The UL128, UL130, and UL131A genes encode accessory proteins of one variant of the glycoprotein H and L (gH/gL) complexes, located in the virion envelope (14, 20, 21). gH/gL complexes are involved in virus-host interactions and in the modulation of the membrane fusion activity of glycoprotein B of several human herpesviruses (reviewed in references 22, 23, and 24). Recent work suggests that the gH/gL/pUL128/pUL130/pUL131A complex of HCMV determines broad cell tropism, while another variant of the complex, composed of gH, gL, and gO, regulates glycoprotein B fusion activity (25). This contrasts with previous assumptions that both gH/gL complexes may serve similar functions during entry into different target cells by binding to cell type-specific host factors, for example, entry receptors (reviewed in reference 7).
Host factors contributing to an entry step after attachment of HCMV have been reported, including epidermal growth factor receptor (EGFR) and beta1- or beta3-integrins (26–31), but the actual contributions of these factors have not yet been resolved fully (7).
RNA interference (RNAi) screens, in which individual proteins are depleted in a systematic manner, have become a valuable tool for identifying putative cellular factors in viral infections. A plethora of studies using many different viruses have been published in recent years, demonstrating the intrinsic power of the approach (32–36). RNAi screens can be carried out on a genome-wide scale or on a focused, small or medium scale, termed “targeted” screening (32, 33). Targeted screens concentrate on a biological process of interest or on preselected protein families and allow more complex assays and more robust data collection (32, 33).
In this work, we aimed to identify host factors contributing to initial steps of HCMV infection by a systematic targeted RNAi screening approach. Cellular genes encoding plasma membrane proteins from carefully selected protein families were addressed individually with a focused library of small interfering RNAs (siRNAs). Here we show that the tetraspanin CD151, identified in our RNAi screen, promotes HCMV infection in both endothelial cells and fibroblasts and acts during the penetration step of viral entry. This is, to our knowledge, the first report of a functional contribution by a tetraspanin to initial events in HCMV infection.
MATERIALS AND METHODS
Virus strains and stocks.
HCMV strains TB40/E and TB40/F were originally isolated by propagating a sample from a bone marrow transplant patient in human umbilical vein endothelial cells (HUVEC) and human foreskin fibroblasts (HFFs), respectively (37). The application of the dually labeled virus TB40-BACKL7-UL32eGFP-UL100mCherry has been described previously (38).
Virus stocks were generated by collecting supernatants from infected HFF cultures at 5 to 7 days postinfection (p.i.). The supernatants were cleared by centrifugation at 3,345 × g for 10 min to remove cell debris and were stored at −80°C until use. Stock titration was performed by serial dilution assays.
Cell culture.
EA.hy926 cells (ATCC CRL-2922) were purchased from LGC Standards, Wesel, Germany (39), and cultured in Dulbecco's modified Eagle's medium supplemented with GlutaMAX (Life Technologies GmbH, Darmstadt, Germany), 10% fetal bovine serum (PAA, Pasching, Austria), and 100 μg/ml gentamicin (Life Technologies). HEC-LTTs are derived from HUVEC and carry doxycycline-inducible expression cassettes for the simian virus 40 (SV40) large T antigen and the human telomerase catalytic subunit (40). HEC-LTTs were cultured in endothelial growth medium (EGM Bullet kit; Lonza Sales Ltd., Basel, Switzerland) supplemented with 2 μg/ml doxycycline (Sigma-Aldrich, St. Louis, MO) in culture vessels coated with 0.1% gelatin (Sigma-Aldrich). In order to halt cell proliferation, HEC-LTTs were deprived of doxycycline 1 day before infection (41).
Human foreskin fibroblasts were cultured in minimum essential medium (MEM) supplemented with GlutaMAX (Life Technologies), 5% fetal bovine serum, 0.5 ng/ml basic fibroblast growth factor (bFGF) (Life Technologies), and 100 μg/ml gentamicin. During experiments, bFGF was omitted to avoid interference with virus binding to heparan sulfate proteoglycans (HSPGs).
siRNAs, plasmids, and transfection.
The siRNA custom library used in the RNAi screen was purchased from Dharmacon, Inc., and was complemented with Erbb2, CD9, and CD151 siRNAs from Qiagen (Hilden, Germany) or Sigma-Aldrich (see Table S1 in the supplemental material). Mixtures (“pools”) of four individual siRNAs per target gene were used during the screen. Individual CD151 siRNAs were purchased from Sigma-Aldrich and had the following sequences: CD151-1(CDS), 5′-CAUGUGGCACCGUUUGCCU(dT)(dT)-3′; and CD151-2(3′UTR), 5′-CACAUACAGGUGCUCAAUAAA(dT)(dT)-3′ (42). Sequences of individual CD151 siRNAs were analyzed in silico by using standard nucleotide Basic Local Alignment Search Tool (BLAST) searches (https://blast.ncbi.nlm.nih.gov), with human herpesvirus 5 as a reference sequence in order to exclude likely off-targets with high homology in the HCMV genome (arbitrarily defined as continuous stretches of >13 complementary nucleotides). A positive-control siRNA directed against the shared exon 2 coding sequence of HCMV immediate early antigens 1 and 2 (UL122/123) [IE siRNA; 5′-CCGAGGAUUGCAACGAGAA(U)(U)-3′] was designed using the Web-based siDesign Center by Dharmacon and the open reading frame region from positions 1 to 256 of the UL122/UL123 sequence of TB40/E-BAC4. Results were filtered for a low seed frequency in the human genome, a GC content of 40 to 60%, and at least 3 mismatches in the human and HCMV genomes. As negative controls, untransfected cells, a “nontargeting” pool of 4 siRNAs (siGENOME nontargeting pool 2; Dharmacon), and a siRNA (siGENOME RISC-free control siRNA; Dharmacon) which is not incorporated into the RNA-induced silencing complex (RISC) were used. siRNAs were reverse transfected by use of Lipofectamine RNAiMAX reagent (Life Technologies) according to the manufacturer's instructions. The efficiency of siRNA delivery was tested using a fluorescently labeled siRNA designed to target the human lamin A/C genes (Dharmacon). The siRNA did not obviously affect the cellular phenotype. Therefore, this siRNA was suitable for use during optimization of the transfection protocols for monitoring siRNA uptake by fluorescence microscopy.
For infection assays, transfections were carried out in duplicate in 96-well microtiter plates. Controls were included on every plate. Cells were seeded at a density of 10,000 cells per well, and the final siRNA concentration was 50 nM. Infection was performed at a multiplicity of infection (MOI) of 0.5 to 1 48 h after siRNA transfection. Infection efficiency was analyzed at 1 day p.i. by detection of viral immediate early antigens as described below.
For immunoblot analysis of CD151 protein levels or immunostaining of CD151 on the cell surface, siRNA transfections were carried out in 24-well plates or 8-well chamber μ-slides (Ibidi, Martinsried, Germany), respectively, with 50 nM siRNA and a cell density comparable to that for the assays in the 96-well format.
The plasmid pRR47 (a kind gift from Bodo Plachter, Mainz, Germany) (43) was used to transiently express viral immediate early antigens in the absence of infection. Cotransfection of the plasmid and siRNA was performed with DharmaFECT Duo transfection reagent (Dharmacon, Inc.) according to the manufacturer's instructions for forward transfections. For the complementation assay, the plasmids pCR3:eGFP and pEGFP-C1/CD151wt were used to express enhanced green fluorescent protein (EGFP) and an EGFP-CD151 fusion protein, respectively. For simplicity, the terms GFP and CD151-GFP are used throughout the paper and in the figures.
Assessment of cell viability.
In the RNAi screen, the impact of siRNA transfection on cell viability was analyzed in replica samples in parallel with viral infection. The CellTiter-Blue cell viability assay (Promega) was applied, which makes use of the ability of viable cells to reduce the redox dye resazurin into the fluorescent product resorufin, indicating their metabolic capacity. Substrate conversion was detected after a 90-min incubation period by recording the fluorescence emitted by resorufin (excitation filter, 544/20 nm; and emission filter, 630/36 nm). Background fluorescence in blank wells was determined with CellTiter-Blue reagent in the absence of cells. Wells showing a reduction of fluorescence of >30% were excluded from further analysis.
Quantification of viral immediate early antigen expression by in situ enzyme-linked immunosorbent assay (ELISA).
Infected cell cultures in 96-well microtiter plates were fixed at 1 day p.i. with 80% acetone and incubated with primary mouse antibody E13 (Argene, Verniolle, France), diluted 1:1,000 in phosphate-buffered saline (PBS) with 0.5% milk powder, to detect immediate early antigens 1 and 2 (pUL122/123). A horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (sc-2055; Santa Cruz Biotechnology Inc., Dallas, TX) diluted 1:400 in PBS with 0.5% milk powder was used as the secondary antibody. The peroxidase substrate o-phenylenediamine (OPD) (Thermo Scientific) was added for 30 min, and the reaction was stopped by addition of 1 M sulfuric acid (1:1 ratio). Absorption was measured at 492 nm by use of a Chameleon microplate reader (Hidex, Turku, Finland). Mock-infected cells served as a background control.
Indirect immunofluorescence assay for detection of viral antigen and CD151.
Immediate early (IE) antigens 1 and 2 (pUL122/123) were detected by indirect immunofluorescence to quantify antigen-expressing cells either after infection with HCMV virions or after transient transfection of an IE antigen-encoding plasmid. For this purpose, cells cultured in 96-well microtiter plates were fixed with 80% acetone (Sigma-Aldrich) at 24 h p.i. Immunofluorescence detection was performed with the primary mouse antibody E13 (Argene, Verniolle, France) against viral IE antigens 1 and 2. A Cy3-conjugated goat polyclonal anti-mouse Ig F(ab′)2 antibody (Jackson ImmunoResearch) or an Alexa Fluor 488-conjugated goat anti-mouse Ig F(ab′)2 antibody (Life Technologies) served as the secondary antibody. Cell nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole; Sigma-Aldrich). A Zeiss Axio Observer D1 microscope (Zeiss, Jena, Germany) was used for visualization of fluorescence. IE-positive nuclei and DAPI-stained nuclei were counted. The ratio of IE antigen-expressing cells to the total number of cells was calculated for each image and normalized to the respective control. Three independent experiments were performed, with two replica wells each, and three images were taken per well. Mean values integrating all three experiments and the standard errors of the means were calculated.
To visualize siRNA efficiency, CD151 located on the cell surface was detected by indirect immunofluorescence. Living cells seeded in 8-well chamber μ-slides and the antibody were preincubated separately on ice for 30 min. A monoclonal CD151 antibody (clone 11G5a; AbD Serotec, Kidlington, United Kingdom) was added to the living cells and incubated for 90 min on ice. Cells were then fixed with 80% acetone, and the primary antibody was detected by use of a Cy3-conjugated goat polyclonal anti-mouse Ig F(ab′)2 antibody (Jackson ImmunoResearch). Cell nuclei were counterstained with DAPI, and a Zeiss Axio Observer D1 microscope (Zeiss, Jena, Germany) was used for visualization.
Complementation assay.
EA.hy926 cells were seeded in a 24-well format at a density of 40,000 cells per well. Cotransfection of plasmid and siRNA was performed with DharmaFECT Duo transfection reagent (Dharmacon, Inc.) according to the manufacturer's instructions for forward transfections. Infection was performed 48 h after transfection. The infection efficiency of transfected cells was analyzed at 1 day p.i. by flow cytometry as described below. The fraction of viral immediate early antigen-positive cells was calculated after gating for GFP-positive cells. Three independent experiments were performed, and the mean values were calculated to integrate the data from all three experiments.
Detection of viral immediate early antigen and CD151 by flow cytometry.
Cells were detached with 0.05% trypsin-EDTA, and for CD151 detection, a monoclonal CD151 antibody (clone 11G5a; AbD Serotec, Kidlington, United Kingdom) was added to living cells on ice. Staining of the nuclear immediate early antigen required permeabilization. Thus, cells were fixed with 4% paraformaldehyde and permeabilized with 80% ethanol followed by 0.4% saponin. The primary mouse antibody E13 (Argene, Verniolle, France) was used for detection of viral immediate early antigens 1 and 2. The primary antibodies were detected using an AF647-conjugated goat polyclonal anti-mouse IgG(H+L) antibody (Thermo Scientific). Cells were analyzed using a FACSCalibur (BD Biosciences) fluorescence-activated cell sorter.
Analysis of virus adsorption and penetration.
To analyze the efficiency of adsorption or penetration, a freshly produced infectious dual-fluorescence virus (TB40-BACKL7-UL32eGFP-UL100mCherry) was used. Cells were fixed at 30 min p.i. with 4% paraformaldehyde diluted in PBS and permeabilized by treating the fixed cells with 0.5% Nonidet P-40 diluted in PBS with 10% sucrose and 1% fetal bovine serum. Cell nuclei were counterstained with DAPI. Under a Zeiss Axio Observer D1 microscope, EGFP (pUL32)-positive and mCherry (gM/pUL100)-positive particles were counted. EGFP-positive particles lacking mCherry were considered to be nonenveloped, representing penetrated capsids, whereas dually fluorescent particles were considered to be enveloped. The penetration efficiency was calculated as the ratio of penetrated particles to the total number of particles. The data shown were generated by determining the means for three experiments. Five images were analyzed per condition in each experiment, with approximately 300 particles being counted per image.
Immunoblot analysis of CD151 protein levels.
The CD151 protein was detected by use of an affinity-purified polyclonal rabbit antibody raised against the recombinant large extracellular loop of CD151 (42). Cells were lysed by addition of RIPA buffer at 48 h posttransfection. SDS-PAGE was performed under nonreducing conditions because CD151 is not recognized by the antibody in the presence of reductants. A possible reason for this might be that antibody binding requires an intact tertiary structure of the large extracellular loop of CD151. The tertiary structure of tetraspanins is stabilized by disulfide bonds between conserved cysteines—as described for CD81 (44)—which are destroyed under reducing conditions. As a loading control for protein input, an unspecific band produced by the CD151 antibody was used.
RESULTS
RNAi screen for host factors in HCMV infection.
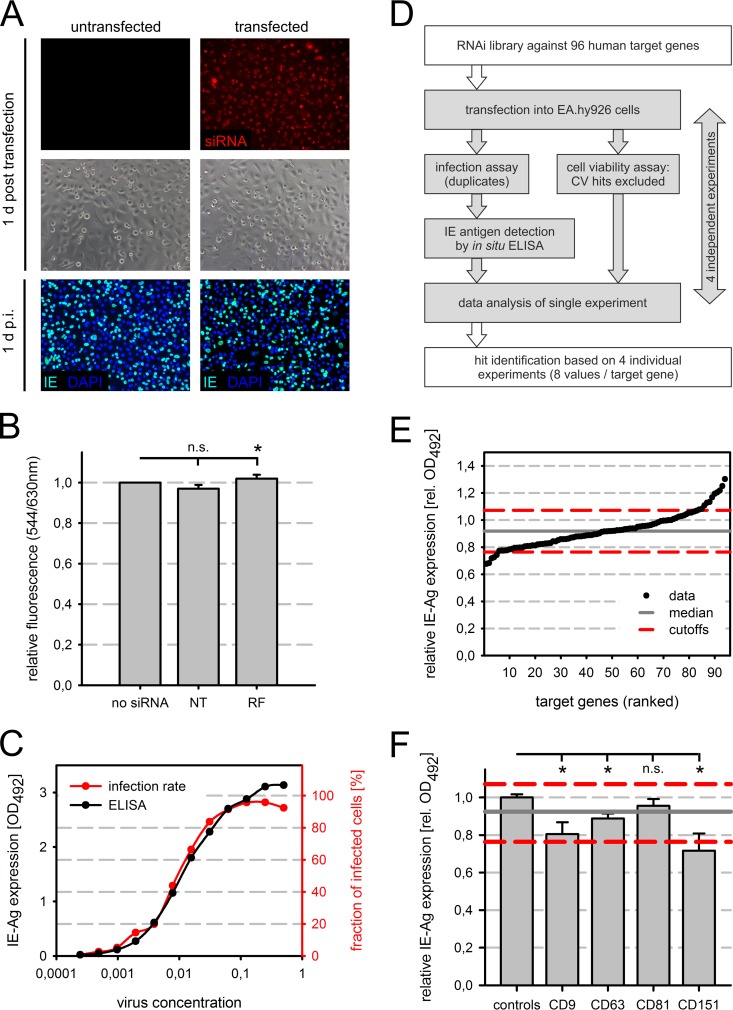
In an attempt to systematically identify host genes with a role during the initial phase of HCMV infection, an RNAi screen was carried out in the endothelial cell line EA.hy926. We previously showed that EA.hy926 cells support entry and expression of immediate early antigens of HCMV strains with a broad cell tropism, such as TB40/E, but not those of strains with narrow cell tropism, thus resembling primary endothelial cells with regard to the initial phase of infection (41). Therefore, EA.hy926 cells qualify for analysis of the viral entry mechanism in endothelial cells. Conditions for small interfering RNA (siRNA) transfection were first optimized to approximately 100% transfection efficiency as visualized with a fluorescently labeled siRNA (Fig. 1A, upper panels), while avoiding major cytotoxic effects. The latter was assessed coarsely by microscopic observation of the cell culture and by subsequent infection with HCMV strain TB40/E (Fig. 1A, middle and lower panels). Since the fluorescently labeled siRNA has human target sequences, the lamin A/C genes, its use as a negative control in the screen was excluded as a precaution. In order to test whether the established transfection procedure as such affected the HCMV infection, it was challenged with the negative-control siRNAs that we planned to use later in the screen. The siRNAs chosen for this purpose were a pool of four “nontargeting” (NT) siRNAs that lack a specific target in human cells and a “RISC-free” (RF) siRNA that is not incorporated by the RNA-induced silencing complex. The transfection procedure per se did not affect the infection efficiency of TB40/E (data not shown). Cell viability was also not reduced 48 h after transfection of these siRNAs (Fig. 1B). The infection efficiency of TB40/E was determined by measuring the expression level of viral major immediate early antigens (pUL122/123) with an in situ ELISA. The absorbance readout of the ELISA correlated accurately with the respective percentage of cells expressing the immediate early antigens in replica cultures (Fig. 1C). The correspondence relaxed at high virus concentrations, where the absorbance continued to increase while the fraction of infected cells approached its maximum. The ELISA readout yielded a good resolution of the virus dose around an MOI of 0.5 to 1 (approximately 40 to 63% infected cells), which was also what we aimed for in the RNAi screen (Fig. 1C).
FIG 1.
Identification of host factors in initial steps of HCMV infection by RNAi screen. (A) A fluorescently labeled siRNA (red) was transfected into EA.hy926 cells, and uptake was monitored at 1 day (d) posttransfection. The cells were subsequently infected with HCMV strain TB40/E at 48 h posttransfection, and infected cells were visualized at 1 day p.i. by immunostaining of viral immediate early antigens (IE) pUL122 and pUL123 (green). Nuclei were stained with DAPI (blue). (B) Metabolic conversion of resazurin to the fluorescent molecule resorufin served as an indicator of cell viability. EA.hy926 cells were transfected with a nontargeting pool (NT) or RISC-free (RF) siRNA and compared to untransfected cells. Raw data for duplicates from four experiments (n = 8) were standardized based on the untransfected cells. (C) In situ ELISA was employed to measure immediate early antigen (IE-Ag) levels in infected cells. Serial virus dilutions were prepared and used to infect EA.hy926 cells. In order to test whether the ELISA results could serve as an indirect readout for the fraction of infected EA.hy926 cells determined by IE-Ag detection via immunofluorescence, both readouts were performed in parallel. The ELISA data (black) were plotted with the percentages of infected cells (red) in the same graph to show the degree of correlation. (D) Schematic overview of the RNAi screening strategy. Four individual experiments (gray) were carried out with an RNAi library encompassing 96 human target genes. The data from all experiments (two replicates per experiment) were analyzed collectively to determine “hits.” (E) Ranking of the targeted genes according to their corresponding ELISA results for hit identification. In both panels E and F, the median for all samples is shown as a solid gray line, and the cutoffs (median ± 2 standard deviations [for the negative controls, i.e., untransfected cells, NT siRNA-transfected cells, and RF siRNA-transfected cells]) are shown as dashed red lines. (F) The screen results for the four tetraspanin genes covered by the library (CD9, CD63, CD81, and CD151) are shown and compared to the mean for the negative controls (untransfected cells, NT siRNA-transfected cells, and RF siRNA-transfected cells). Error bars show standard errors of the means. n.s., not significant; *, significant (P < 0.05; Student's t test); rel. OD492, relative optical density at 492 nm.
A search of the existing literature revealed that most host factors reported to be involved in entry of other human viruses preferentially belong to only a few distinct protein families, such as growth factor receptors, integrins, chemokine receptors, or the immunoglobulin superfamily (reviewed in reference 45). In addition, recent studies showed that tetraspanins, which form a family of membrane scaffold proteins or four-span membrane proteins, have a role in the entry processes of a number of viruses, such as influenza A virus, hepatitis C virus, and human papillomavirus (HPV) (46, 47). We thus reasoned that cellular proteins contributing to the HCMV entry process have a high probability of being members of these protein families. Based on this assumption, the RNAi library used in this study was designed to systematically target members of these preselected protein families (see Table S1 in the supplemental material).
In the RNAi screening experiment, cells were transfected with an arrayed library of siRNAs corresponding to 96 human target genes for 48 h and then infected with the HCMV strain TB40/E (Fig. 1D). Equimolar mixtures (“pools”) of four individual siRNAs directed against different target sites within the same gene were employed to limit undesired off-target effects by individual siRNAs. By using mixtures of four siRNAs, the concentration of each individual siRNA is reduced to 25%, which is supposed to decrease the efficacy of each single siRNA while maintaining a high overall efficacy of the pool. Therefore, a common specific target of all four siRNAs of the pool should be regulated more effectively than any inadvertent target of a single siRNA.
On each cell culture plate, untransfected cells and cells transfected with control siRNA (NT or RF) were included as negative controls and for data processing in order to account for plate-to-plate variation. As a positive control for inhibition of infection, an siRNA (IE) directly targeting the shared coding sequence of viral major immediate early genes 1 and 2 (UL122 and UL123) was included. The screen was repeated four times in duplicate to generate a robust data set of eight replicates in total (Fig. 1D). Additionally, each screen cycle was accompanied by a cell viability assay performed on replica cultures at the time of infection to exclude false-positive hits caused by cytotoxicity of the respective siRNA or by cell loss. Data obtained with siRNAs that caused a pronounced decrease in viability (below 70% of the negative-control levels) were excluded from the final analysis as invalid data sets. The cutoff was chosen arbitrarily, with the intention that siRNAs not be excluded prematurely from the valid data merely because they affected cell proliferation. Among the 96 siRNA pools tested, only two pools were found to decrease cell viability below the cutoff: those directed against the integrins ITGAV and ITGB5, with residual cell viabilities of 51% and 68%, respectively. It should be noted that integrins function as adhesion molecules, and therefore the decreased signal in the cell viability assay might be caused by detachment and loss of cells rather than actual cell death, since the assay as such does not resolve the cell number and the level of metabolic activity. In either case, the conclusion regarding the validity of the data remains the same. The ELISA data for the remaining 94 valid data sets were processed by subtracting the background measured in uninfected cells and normalizing the data to the mean for the negative controls (NT or RF siRNA-transfected and untransfected cells) on a per-plate basis. Finally, the arithmetic mean for eight replicates was determined (Fig. 1E; see Table S2 in the supplemental material).
Promoting and inhibitory host genes of high confidence, termed hits, were defined by setting upper and lower cutoffs. To qualify as a hit, the sample mean was demanded to differ from the median for all samples by twice the standard deviation of the negative controls (Fig. 1E). Statistical significance was assessed using two-sided Student's t test (see Table S2 in the supplemental material). This procedure resulted in five candidate genes promoting (“proviral”) and eight candidate genes counteracting (“antiviral”) HCMV infection (P < 0.05) (Table 1). The proviral class encompassed members from different protein families (tetraspanins, integrins, and chemokine receptors), while antiviral hits were predominantly growth factor receptors (Table 1). The cell viability of the hits ranged from 90.9% to 111.7%. Cell viability values for all siRNAs are indicated in Table S2.
TABLE 1.
Pro- and antiviral genes identified in the RNAi screen
| Gene category and name | Relative IE expression (rel. OD492)a | SD | P value | Gene description/full gene name | Other name(s) | Classification |
|---|---|---|---|---|---|---|
| Proviral genes | ||||||
| CXCR5 | 0.68 | 0.12 | 0.00010 | Chemokine (C-X-C motif) receptor 5 | BLR1, CD185, MDR15 | Chemokine receptor |
| ITGB4 | 0.68 | 0.09 | 0.00002 | Integrin beta 4 | CD104 | Integrin |
| CD151 | 0.72 | 0.17 | 0.00240 | CD151 molecule (Raph blood group) | GP27, MER2, RAPH, SFA1, PETA-3, TSPAN24 | Tetraspanin |
| CCRL2 | 0.73 | 0.07 | 0.00001 | Chemokine (C-C motif) receptor-like 2 | HCR, CKRX, CRAM, ACKR5, CCRL2, CRAM-A, CRAM-B | Chemokine receptor |
| ITGA11 | 0.74 | 0.09 | 0.00007 | Integrin alpha 11 | HsT18964 | Integrin |
| Antiviral genes | ||||||
| CD34 | 1.11 | 0.08 | 0.00837 | CD34 molecule | Adhesion molecule | |
| GPR126 | 1.13 | 0.09 | 0.00446 | Adhesion G protein-coupled receptor G6 | ADGRG6, APG1, DREG, VIGR, PS1TP2 | Adhesion molecule |
| FLT1 | 1.17 | 0.12 | 0.00522 | Fms-related tyrosine kinase 1 | FLT, FLT-1, VEGFR1, VEGFR-1 | Growth factor receptor |
| EGFR | 1.19 | 0.15 | 0.00798 | Epidermal growth factor receptor | ERBB, HER1, mENA, ERBB1, PIG61, NISBD2 | Growth factor receptor |
| CCR4 | 1.20 | 0.17 | 0.01131 | Chemokine (C-C motif) receptor 4 | CKR4, K5-5, CD194, CMKBR4, ChemR13, CC-CKR-4, HGCN:14099 | Chemokine receptor |
| ENG | 1.22 | 0.23 | 0.03337 | Endoglin | END, HHT1, ORW1, CD105 | Glycoprotein, growth factor coreceptor |
| KDR | 1.25 | 0.21 | 0.01120 | Kinase insert domain receptor | FLK1, CD309, VEGFR, VEGFR2 | Growth factor receptor |
| FGFR1 | 1.30 | 0.20 | 0.00381 | Fibroblast growth factor receptor 1 | CEK, FLG, HH2, OGD, FLT2, KAL2, BFGFR, CD331, FGFBR, FLT-2, HBGFR, N-SAM, FGFR-1, HRTFDS, bFGF-R-1 | Growth factor receptor |
rel. OD492, relative optical density at 492 nm.
As one of the most potent HCMV-promoting host factors, the tetraspanin CD151 (also called TSPAN24 or PETA-3) was identified with a mean relative IE antigen expression of 72% and a mean cell viability of 93% (Fig. 1F; Table 1). siRNAs directed against two other tetraspanins, CD9 and CD63, also significantly reduced IE antigen expression, but this reduction was too weak to fulfill the requirements for being a hit (Fig. 1F; see Table S2 in the supplemental material). In contrast, tetraspanin CD81 siRNA did not affect IE antigen expression (Fig. 1F; see Table S2). Due to its novelty as a putative HCMV host factor, CD151 was selected for further analyses in this study.
CD151 siRNA efficiently depletes CD151 protein and does not directly target immediate early transcripts.
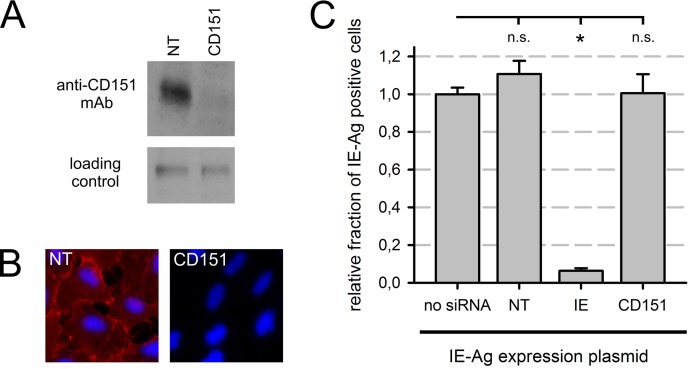
To investigate whether the CD151 protein is expressed at detectable levels in EA.hy926 cells and whether CD151 protein expression is efficiently regulated by CD151 siRNA, immunoblot analysis was performed. Cells were treated with CD151 or NT control siRNA and harvested at 48 h posttransfection. The CD151 protein was readily detectable in cells treated with NT control siRNA (Fig. 2A). CD151 siRNA virtually abolished CD151 protein expression within 48 h, demonstrating the efficiency of the siRNA (Fig. 2A).
FIG 2.
Regulation by CD151 siRNA was efficient and specific. (A) CD151 protein levels were analyzed by Western blot analysis of EA.hy926 cells 48 h after transfection with CD151 or nontargeting (NT) siRNA. A nonspecific band produced by the CD151 antibody served as a loading control. (B) Live EA.hy926 cells were immunostained for CD151 using a Cy3-conjugated secondary antibody (red) after siRNA treatment (NT or CD151 siRNA). After fixation, nuclei were counterstained with DAPI (blue). (C) The indicated siRNAs were transiently cotransfected into EA.hy926 cells with a plasmid conferring immediate early antigen (IE-Ag) expression in the absence of infection. Error bars show standard errors of the means. n.s., not significant; *, significant (P < 0.05; Student's t test).
Immunostaining of living cells confirmed CD151 localization on the surfaces of cells treated with NT control siRNA (Fig. 2B). The individual cells showed comparable staining intensities, suggesting that the CD151 protein was expressed at homogenous levels in the culture. Consistent with the results of immunoblot analysis, CD151 siRNA treatment reduced CD151 surface expression to almost undetectable amounts (Fig. 2B). Together, these data reveal CD151 expression on EA.hy926 cells and efficient depletion of CD151 levels by siRNA.
CD151 siRNA sequences had been analyzed in silico prior to synthesis to avoid potential off-target sites. In order to cross-check that CD151 siRNA does not directly interfere with HCMV immediate early antigen expression, EA.hy926 cells were cotransfected with siRNA and with a plasmid containing the HCMV major immediate early locus to express the immediate early antigens in the absence of infection. Unlike the IE siRNA, intended to target the encoded immediate early transcripts, neither the nontargeting siRNA nor the CD151 siRNA reduced the number of cells expressing immediate early antigens (Fig. 2C). This result demonstrates that CD151 siRNA does not directly affect HCMV immediate early transcripts. Instead, depletion of the CD151 protein seems to inhibit initial events of the HCMV infection upstream of IE gene expression.
CD151 is a valid proviral hit and promotes initial events of infection.
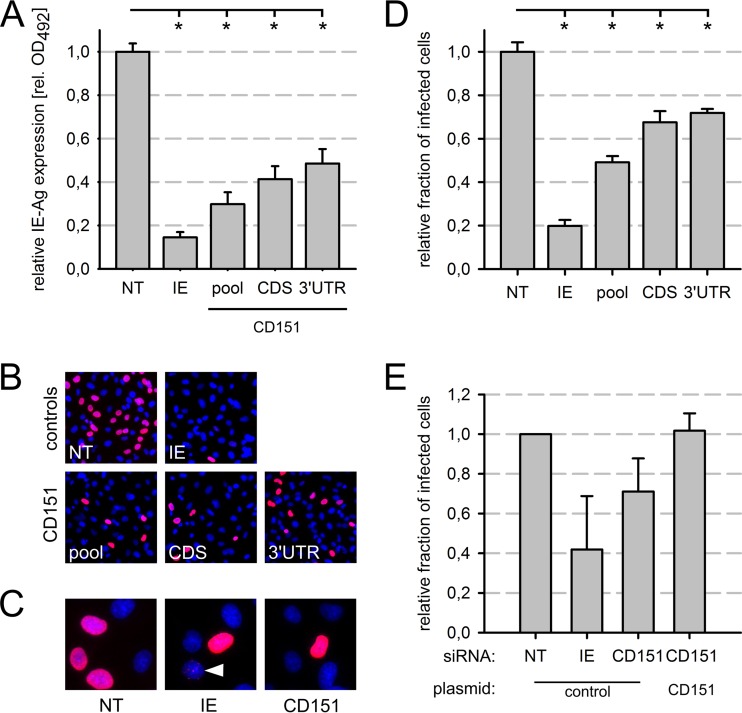
In order to exclude the possibility that inhibition of HCMV infection was due to residual off-target activity of one single siRNA of the siRNA pool, two individual CD151 siRNAs, targeting either the coding sequence (CDS) or the 3′ untranslated region (3′ UTR) of the CD151 transcript, were tested. The single siRNAs were compared to an equimolar mixture of the two siRNAs and the NT control siRNA. Transfected cells were infected with HCMV TB40/E at 48 h posttransfection, and after another 24 h, immediate early antigen expression was quantified by in situ ELISA. Both CD151 siRNAs as well as the corresponding siRNA pool, an equimolar mixture of the two CD151 siRNAs, significantly inhibited viral immediate early antigen expression more than 50% compared to that with the NT control siRNA (Fig. 3A). The inhibition was even more pronounced than in the RNAi screen, where IE antigen expression was reduced to 72% of the control level (Table 1). This was likely due to a higher transfection efficiency in this series of experiments, since the IE siRNA was also more efficient in the validation experiment than in the screen (15% versus 32% residual IE antigen expression, respectively) (Fig. 3A; see Table S2 in the supplemental material). Note that the inhibition was significantly weaker with all CD151 siRNAs than with siRNAs directed against immediate early antigens (15% residual expression) (Fig. 3A). Taken together, these results supported the specificity of CD151 siRNAs and a promoting effect of CD151 on HCMV immediate early gene expression.
FIG 3.
CD151 siRNAs specifically reduced the infection of EA.hy926 cells. (A to D) Individual CD151 siRNAs from the siRNA pool used in the screen or an equimolar mixture thereof were transfected 48 h prior to infection. Immediate early antigen (IE-Ag) expression was measured by ELISA (A), and the fraction of infected cells was determined by indirect immunofluorescence staining of IE-Ag (B and D) with a primary antibody and a Cy3-conjugated secondary antibody (red). Nuclei were counterstained with DAPI (blue). (C) Magnified images showing pan-nuclear (NT, IE, and CD151 siRNAs) or punctate (IE siRNA) (white arrowhead) expression pattern of IE-Ag. siRNAs: NT, nontargeting pool; IE, immediate early antigen; CDS, CD151 coding sequence; 3′ UTR, CD151 3′ untranslated region; pool, equimolar mix of both CD151 siRNAs. (E) A CD151 siRNA that targets the 3′ UTR of the CD151 gene was cotransfected with a plasmid encoding either GFP (as a control) or a CD151-GFP transgene resistant to the siRNA. NT siRNA was used as a reference, and IE siRNA was used as a positive control. The data were standardized to the negative control (NT siRNA plus GFP plasmid). The graph shows mean values for three independent experiments. Error bars show standard errors of the means. n.s., not significant; *, significant (P < 0.05; Student's t test).
The in situ ELISA used in the screen does not distinguish between a reduction in the fraction of infected cells and a reduction in the amount of immediate early antigen produced per cell. In order to investigate whether CD151 siRNAs affect the fraction of HCMV-infected cells, immediate early antigen expression was analyzed by immunofluorescence staining of fixed cells (Fig. 3B to D). Cells showing the typical pan-nuclear staining were counted. In cells treated with single CD151 siRNAs, the infection efficiency was significantly reduced, by 23 to 28%, and the CD151 siRNA pool led to a reduction of 51% compared to the level with the NT control (Fig. 3B and D). siRNA directed against viral immediate early transcripts resulted in an overall decrease of antigen-positive cells, as well as the occurrence of a punctate staining pattern in some nuclei, which was observed only rarely with any other siRNA at 24 h p.i. (Fig. 3C). In conclusion, a reduction in the number of infected cells accounts—at least in part—for the decrease in immediate early antigen levels in CD151-depleted cultures.
In order to test whether the depletion of CD151 prior to infection causes the observed inhibition of infection, a complementation assay was performed. EA.hy926 cells were cotransfected with an siRNA targeting the endogenous CD151 3′ UTR together with a CD151 expression plasmid. The plasmid expresses the CD151 coding sequence with a heterologous 3′ UTR and therefore lacks the siRNA binding site. Thus, CD151 expression from the plasmid is resistant to regulation by the siRNA. In order to visualize transgene expression and to facilitate detection of the transfected cells, a CD151-green fluorescent protein (GFP) fusion construct was used. A GFP-encoding plasmid served as a control. Because the transfection and infection efficiencies were low in the cotransfection experiments with these plasmids, fluorescence-activated cell sorting was applied after IE immunostaining to determine the fractions of GFP- and IE antigen-expressing cells. IE and CD151 siRNAs cotransfected with the control plasmid both inhibited infection to degrees similar to those observed in the previous experiments (Fig. 3E). Full recovery of infection was observed in cells cotransfected with CD151 siRNA and the CD151-GFP plasmid, in contrast to the GFP control cells (Fig. 3E). However, the recovery effect did not pass the statistical test for significance, which might be due to the variable siRNA efficiencies obtained. In conclusion, these data further support a role for CD151 in HCMV infection rather than an off-target effect of the CD151 siRNAs.
The proviral role of CD151 in infection is observed with different cell types and virus strains.
The cell line HEC-LTT is derived from human umbilical vein endothelial cells (HUVEC) (40). Proliferation of HEC-LTT cells can be controlled by switching between silent and active states of transgenes encoding the SV40 T antigen and human telomerase. Recently, we adopted the cell line as a cell culture model for endothelial cell infection by HCMV (41). Like primary HUVEC, HEC-LTTs are susceptible to highly endotheliotropic virus strains and release comparable amounts of infectious virus, but they scarcely allow infection by poorly endotheliotropic strains (41).
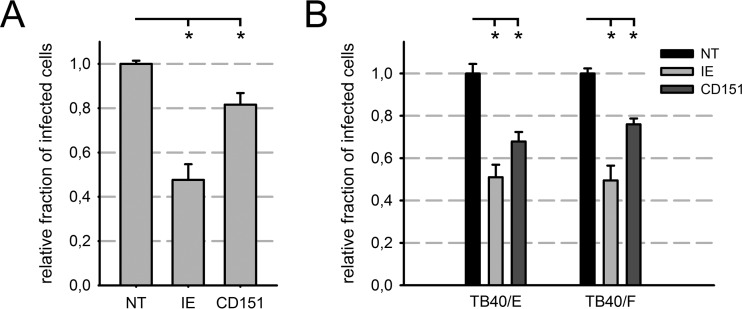
In order to exclude the possibility that the inhibition of HCMV infection by CD151 siRNA is restricted to EA.hy926 cells, HEC-LTTs were transfected with CD151 siRNA and NT control siRNA and infected with HCMV strain TB40/E. Infection efficiency was determined by immunofluorescence detection of immediate early antigens (pUL122/123). CD151 siRNA caused a significant reduction (18%) in HEC-LTTs compared to cells treated with NT control siRNA. IE siRNA again led to a more pronounced reduction than that with CD151 siRNA (52%) (Fig. 4A). The effect of the CD151 siRNA was repeatedly milder than that in EA.hy926 cells (Fig. 2D and 4A). This might have been due to a lower transfection efficiency in HEC-LTTs, since the IE siRNA was also less efficient in HEC-LTTs than in EA.hy926 cells.
FIG 4.
The contribution of CD151 to HCMV infection was not cell type restricted. (A and B) Relative fractions of HCMV-infected HEC-LTTs (A) or HFFs (B) as measured by immunofluorescence staining of immediate early antigens. Cells were transfected with a pool of CD151 siRNAs or control siRNAs, as indicated, 48 h prior to infection. NT siRNA-treated cells served as a reference, and IE siRNA was used as a positive control. (A) HEC-LTTs were infected with HCMV strain TB40/E. (B) HFFs were infected with a highly endotheliotropic (TB40/E) or poorly endotheliotropic (TB40/F) strain. Error bars show standard errors of the means. *, significant (P < 0.05; Student's t test).
To investigate whether CD151 function is specific to endothelial cells or also relevant in other cell types, siRNA-transfected primary HFFs were infected with TB40/E. IE siRNA significantly inhibited infection, by 49% (Fig. 4B). CD151 siRNA caused a significant reduction in the fraction of cells infected with TB40/E, by 32%, compared to the NT siRNA control. Thus, CD151 function during the initial steps of HCMV infection is not confined to endothelial cells.
Highly and poorly endotheliotropic HCMV strains differ in their repertoire of gH/gL glycoprotein complexes on the virion surface, which presumably leads to different virus-host interactions during viral entry. In order to analyze if the function of CD151 is restricted to highly endotheliotropic HCMV, infection of siRNA-treated fibroblasts with the poorly endotheliotropic strain TB40/F was examined in comparison to infection with the highly endotheliotropic strain TB40/E. The percentage of cells infected by TB40/F was markedly decreased in HFFs treated with CD151 siRNA (76%), as well as those treated with IE siRNA (50%), compared to the NT siRNA control (Fig. 4B). This observation suggests that CD151 functions comprehensively and regardless of the composition of the viral gH/gL complexes.
CD151 promotes viral penetration into the cell.
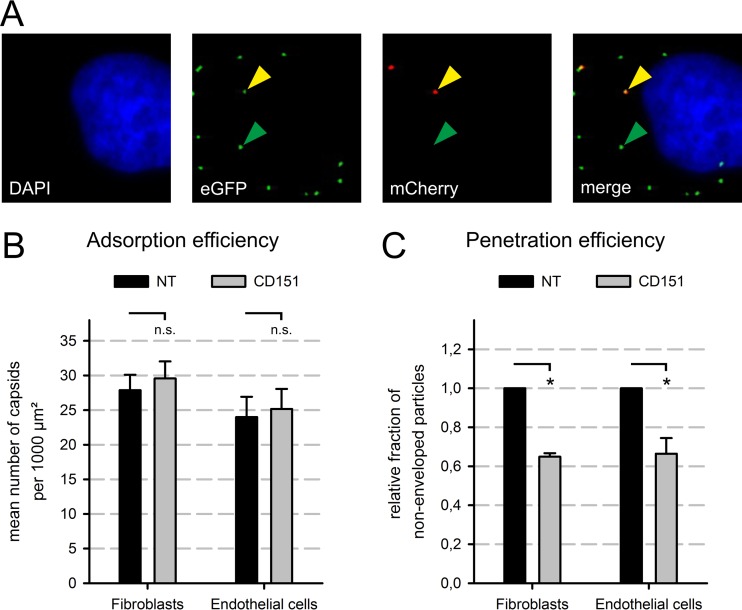
CD151 depletion lowered the fraction of cells that initiated expression of IE antigens 1 and 2 (Fig. 2B and C), which are among the first viral genes expressed in infected cells (48–50), but IE expression from a plasmid was unaltered by CD151 siRNA (Fig. 3C). These findings indicated that CD151 contributes to initial events of HCMV infection, such as adsorption to the cell, fusion with a cellular membrane, or nuclear translocation. To analyze whether viral adsorption or penetration was affected by CD151, a dual-fluorescence virus was used (38). In this virus, the capsid-associated tegument protein pp150 (pUL32) is labeled with enhanced green fluorescent protein (EGFP), and the membrane-associated glycoprotein M (pUL100) is tagged with the red fluorescent protein mCherry (38). Thus, virions possess a red fluorescent envelope and a green fluorescent capsid. This allows discrimination between dually fluorescent enveloped capsids and singly fluorescent nonenveloped viral particles, which have undergone membrane fusion and, as a consequence, have lost the red fluorescence (Fig. 5A). To analyze adsorption, images were taken by means of fluorescence microscopy, and the total number of green fluorescent viral particles was counted, representing both enveloped and nonenveloped capsids. We found that siRNA-mediated CD151 depletion did not impair viral adsorption to the cell in either fibroblasts or endothelial cells (Fig. 5B). This was not unexpected, as adsorption is thought to be mediated by unspecific binding to abundant heparan sulfate proteoglycans (HSPGs) on the cell surface (51, 52). In order to examine the penetration efficiency of the virus, the fraction of green fluorescent particles lacking a red fluorescence signal were determined. CD151 depletion by siRNA significantly decreased the fraction of nonenveloped capsids in the population of viral particles, and hence, the penetration efficiency compared to that with the NT siRNA control was 65% for HFFs and 68% for HEC-LTTs (Fig. 5C). In conclusion, CD151 promotes viral penetration into both fibroblasts and endothelial cells.
FIG 5.
CD151 depletion affected viral penetration but not adsorption. (A) Exemplary images showing fluorescence signals in a cell infected with the dual-fluorescence virus strain TB40-BACKL7-UL32eGFP-UL100mCherry. Green signals were interpreted as EGFP-labeled capsids (green arrowheads) and red signals as mCherry-labeled viral envelopes. In the merged image, particles showing both green and red signals appear yellow (yellow arrowhead). The cell nucleus was counterstained with DAPI (blue). (B and C) Fibroblasts (HFFs) and endothelial cells (HEC-LTTs) were transfected for 48 h with CD151 or NT siRNA and then inoculated with the dual-fluorescence HCMV strain TB40-BACKL7-UL32eGFP-UL100mCherry at an MOI of 20 to 80. Cells were fixed after 30 min. Fluorescence images were taken and analyzed by determining the total number of green signals (all capsids, regardless of the presence or absence of envelope) (B) and the fraction of green-only signals, without red (nonenveloped particles) (C), to determine the total number of adsorbed virus particles (B) and the fraction of successfully penetrated particles (C). In each experiment, >800 particles were counted for each condition. The penetration efficiency was standardized to the NT control level. Error bars show standard errors of the means. n.s., not significant; *, significant (P < 0.05; Student's t test).
DISCUSSION
Identification of cellular proteins that contribute to entry of HCMV into target cells is a relevant issue, as each of these factors provides a potential target for antiviral strategies. Here we report that the tetraspanin CD151 is a previously unrecognized proviral host factor which contributes to HCMV penetration.
Tetraspanins are a family of glycoproteins characterized by four transmembrane domains and other structural features (53–55), and they are highly conserved in multicellular eukaryotes (56). Because they are integral membrane proteins, they are enriched in distinct plasma membrane microdomains, also termed the tetraspanin web (57, 58), where they associate with a multitude of partner proteins and with each other in a lateral manner (reviewed in references 59, 60, and 61). Tetraspanin-enriched microdomains have been reported to be involved in a wide range of cellular functions, including adhesion, membrane fusion, and signaling processes (reviewed in references 62 and 63). Notably, several tetraspanins contribute to viral infections in the human host, but only a few tetraspanins, including CD151, CD81, CD63, and CD9, have been implicated in viral entry processes (64–72; reviewed in references 46 and 47). A role of CD151 in the viral entry process of HPV has been demonstrated for three oncogenic types of HPV and in different cell types (42, 65, 66, 73). Recombinant large extracellular domains of CD151 and other tetraspanins were able to inhibit infection of macrophages by human immunodeficiency virus type 1 (HIV-1) (74), though the mechanism is not yet fully understood. To our knowledge, a contribution of CD151 to infection by HCMV or other herpesviruses has not been reported to date.
In this work, siRNA-induced depletion of CD151 from endothelial cells prior to infection with the HCMV strain TB40/E reduced IE antigen expression, and immunofluorescence data demonstrated that this was due at least in part to a decreased fraction of infected cells. IE antigen expression from a plasmid in the absence of infection was not altered by CD151 siRNA. Thus, an unintended direct depletion of IE proteins by off-target siRNA regulation of IE transcripts could be excluded. Together, the results point to a function of CD151 prior to initiation of viral gene expression.
By analyzing fluorescent viral particles during entry into target cells, we showed that HCMV adsorbs at normal levels to cells depleted of CD151. Therefore, the initial attachment of HCMV to heparan sulfate proteoglycans (tethering) (51) appears to occur independently of CD151. The general view of herpesvirus entry postulates specific binding of the viral particle to a cellular entry receptor, and possibly to additional host proteins following viral attachment, thereby promoting fusion of the virion envelope with a cellular membrane and release of the capsid into the cytoplasm (penetration). As a measure of penetration efficiency, we visualized the fusion-induced loss of the viral envelope by using a virus strain with a differentially labeled capsid and envelope. Penetration efficiency was significantly reduced in CD151-depleted cells. This suggests that CD151 promotes penetration by contributing to entry receptor binding, membrane fusion, or release of the capsid. A postattachment function of CD151 in HCMV entry is in line with the reported role of CD151 in HPV infection, where depletion of CD151 resulted in reduced endocytosis and infection without affecting viral adsorption to the cell (42, 65).
The promoting function of CD151 in HCMV entry was observed in both endothelial cells and fibroblasts. This opposes a role of CD151 in regulation of viral cell tropism. Instead, this result speaks in favor of a common entry-related function across different cell types, and the contribution of CD151 is likely independent of the repertoire of gH/gL complexes of the incoming virus. Indeed, two virus strains, TB40/E and TB40/F, the latter of which lacks the gH/gL/pUL128/pUL130/pUL131A complex (18, 75), were equally affected by CD151 depletion. A broadly relevant role of CD151 in HCMV infection requires a ubiquitous CD151 expression pattern as a prerequisite, since clinical HCMV isolates are able to enter almost all tested cell types in vivo and in vitro (reviewed in references 2 and 7). In fact, previous analyses by others revealed ubiquitous expression of human CD151 in almost any cell type and tissue (76, 77).
Meanwhile, the actual molecular function of CD151 in HCMV entry remains a matter of speculation. It is nevertheless remarkable that tetraspanins, especially CD151, interact with proteins related to most of the putative HCMV entry factors that have been described in the literature or identified in our RNAi screen: growth factor receptors, integrins, chemokine receptors, and members of the immunoglobulin superfamily (60, 73). In accordance with the reported functions of tetraspanins in recruiting cellular proteins to membrane microdomains, a role of CD151 as an organizer of viral entry platforms appears likely. By structural analyses, the tetraspanins CD81 and uroplakin have been shown to protrude from the membrane by only 3.5 and 5 nm, respectively (44, 78). Large partner proteins, which are assembled by tetraspanins in membrane microdomains, are considered to protrude beyond the tetraspanins into the extracellular space, where they are accessible to incoming viral particles. In support of a role for CD151 in the formation of viral entry platforms is the observation that HPV16 endocytosis depends on the recruitment of CD151 to tetraspanin-enriched microdomains and their association with distinct cellular proteins (42, 73). Beyond that, CD151 appears to have other functions, as it was shown to modulate adhesion-dependent Ras signaling, which affects the actin cytoskeleton (79, 80), and antibody inhibition assays hint at a possible role of CD151 in regulation of egg-sperm fusion (81). Therefore, CD151 might serve in infection-activated signal transduction or in regulation of endocytic or fusion processes during HCMV entry. Further studies are needed to clarify the mechanism of CD151 function in HCMV infection.
Tetraspanins have been described as “gateways for infection” for many different intracellular pathogens: viruses, bacteria, fungi, and protozoa (reviewed in reference 46). Therefore, strategies for targeting tetraspanins are being discussed. These include disruption of tetraspanin-enriched microdomains and depletion of individual tetraspanins by either antibodies, siRNAs, or small molecules, for example, the soluble large extracellular domain or aptamers (82, 83). It remains to be seen whether an effective inhibition of CD151 functions related to HCMV infection can be separated sufficiently from unwanted side effects caused by interference with other biological functions of CD151.
Although the CD151 protein was hardly detectable in CD151 siRNA-treated cells by immunofluorescence and immunoblotting techniques, infection with HCMV was not completely abolished. In all data sets, the inhibitory effect of the CD151 siRNA was weaker than the effect of the IE siRNA, and the extent of the inhibition caused by the CD151 siRNA correlated with the efficiency of the IE siRNA in the same experiment. Therefore, the partial IE antigen expression in CD151 siRNA-treated cells might have been due to a minor amount of residual CD151 protein that was below the detection limit of our experiments. Another possible explanation is that HCMV, possibly via gH/gL complexes, might interact with a variety of cellular partners, and some of these interactions may take place independently of CD151. This would be analogous to human herpes simplex virus type 1 glycoprotein D, which has been shown to bind several unrelated host proteins or receptors (84–90). Alternatively, an as yet unidentified tetraspanin may act redundantly to CD151 and partially compensate for CD151 depletion. In the human genome, 33 tetraspanin genes have been identified to date (56). Of these, 23 were reported to be coexpressed in human umbilical vein endothelial cells, including CD151 (91). It will be interesting to systematically test the remaining members of the tetraspanin family for redundancy with CD151 or an additional function in HCMV infection.
In conclusion, we have identified several pro- or antiviral host factors acting during the initial steps of HCMV infection, including the novel host factor CD151. Our data provide a resource for future studies striving for a better understanding of virus-host interactions during viral entry and for potential new targets of innovative antiviral strategies. CD151 promoted the penetration of HCMV into different physiologically relevant target cell types, and it will be interesting to further investigate the underlying molecular mechanism.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bodo Plachter and Fedor Berditchevski for kindly providing reagents and Fatima Boukhallouk for excellent technical support.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00145-16.
REFERENCES
- 1.Sinzger C, Grefte A, Plachter B, Gouw AS, The TH, Jahn G. 1995. Fibroblasts, epithelial cells, endothelial cells and smooth muscle cells are major targets of human cytomegalovirus infection in lung and gastrointestinal tissues. J Gen Virol 76:741–750. doi: 10.1099/0022-1317-76-4-741. [DOI] [PubMed] [Google Scholar]
- 2.Sinzger C, Digel M, Jahn G. 2008. Cytomegalovirus cell tropism, p 63–83. In Shenk T, Stinski M (ed), Human cytomegalovirus. Springer, Berlin, Germany. [DOI] [PubMed] [Google Scholar]
- 3.Roberts WH, Sneddon JM, Waldman J, Stephens RE. 1989. Cytomegalovirus infection of gastrointestinal endothelium demonstrated by simultaneous nucleic acid hybridization and immunohistochemistry. Arch Pathol Lab Med 113:461–464. [PubMed] [Google Scholar]
- 4.Muhlemann K, Miller RK, Metlay L, Menegus MA. 1992. Cytomegalovirus infection of the human placenta: an immunocytochemical study. Hum Pathol 23:1234–1237. doi: 10.1016/0046-8177(92)90290-J. [DOI] [PubMed] [Google Scholar]
- 5.Sinzger C, Muntefering H, Loning T, Stoss H, Plachter B, Jahn G. 1993. Cell types infected in human cytomegalovirus placentitis identified by immunohistochemical double staining. Virchows Arch A Pathol Anat Histopathol 423:249–256. doi: 10.1007/BF01606887. [DOI] [PubMed] [Google Scholar]
- 6.Bissinger AL, Sinzger C, Kaiserling E, Jahn G. 2002. Human cytomegalovirus as a direct pathogen: correlation of multiorgan involvement and cell distribution with clinical and pathological findings in a case of congenital inclusion disease. J Med Virol 67:200–206. doi: 10.1002/jmv.2208. [DOI] [PubMed] [Google Scholar]
- 7.Adler B, Sinzger C. 2013. Cytomegalovirus inter-strain variance in cell-type tropism, p 297–321. In Reddehase MJ. (ed), Cytomegaloviruses: from molecular pathogenesis to intervention, vol I, chapter 17 Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 8.Grundy JE, Lawson KM, MacCormac LP, Fletcher JM, Yong KL. 1998. Cytomegalovirus-infected endothelial cells recruit neutrophils by the secretion of C-X-C chemokines and transmit virus by direct neutrophil-endothelial cell contact and during neutrophil transendothelial migration. J Infect Dis 177:1465–1474. doi: 10.1086/515300. [DOI] [PubMed] [Google Scholar]
- 9.Percivalle E, Revello MG, Vago L, Morini F, Gerna G. 1993. Circulating endothelial giant cells permissive for human cytomegalovirus (HCMV) are detected in disseminated HCMV infections with organ involvement. J Clin Invest 92:663–670. doi: 10.1172/JCI116635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahl M, Siegel-Axel D, Stenglein S, Jahn G, Sinzger C. 2000. Efficient lytic infection of human arterial endothelial cells by human cytomegalovirus strains. J Virol 74:7628–7635. doi: 10.1128/JVI.74.16.7628-7635.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adler B, Sinzger C. 2009. Endothelial cells in human cytomegalovirus infection: one host cell out of many or a crucial target for virus spread? Thromb Haemost 102:1057–1063. doi: 10.1160/TH09-04-0213. [DOI] [PubMed] [Google Scholar]
- 12.Grefte A, Blom N, van der Giessen M, van Son W, The TH. 1993. Ultrastructural analysis of circulating cytomegalic cells in patients with active cytomegalovirus infection: evidence for virus production and endothelial origin. J Infect Dis 168:1110–1118. doi: 10.1093/infdis/168.5.1110. [DOI] [PubMed] [Google Scholar]
- 13.Grefte A, van der Giessen M, van Son W, The TH. 1993. Circulating cytomegalovirus (CMV)-infected endothelial cells in patients with an active CMV infection. J Infect Dis 167:270–277. doi: 10.1093/infdis/167.2.270. [DOI] [PubMed] [Google Scholar]
- 14.Adler B, Scrivano L, Ruzcics Z, Rupp B, Sinzger C, Koszinowski U. 2006. Role of human cytomegalovirus UL131A in cell type-specific virus entry and release. J Gen Virol 87:2451–2460. doi: 10.1099/vir.0.81921-0. [DOI] [PubMed] [Google Scholar]
- 15.Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, Wagner M, Gallina A, Milanesi G, Koszinowski U, Baldanti F, Gerna G. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J Virol 78:10023–10033. doi: 10.1128/JVI.78.18.10023-10033.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patrone M, Secchi M, Fiorina L, Ierardi M, Milanesi G, Gallina A. 2005. Human cytomegalovirus UL130 protein promotes endothelial cell infection through a producer cell modification of the virion. J Virol 79:8361–8373. doi: 10.1128/JVI.79.13.8361-8373.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryckman BJ, Jarvis MA, Drummond DD, Nelson JA, Johnson DC. 2006. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J Virol 80:710–722. doi: 10.1128/JVI.80.2.710-722.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinzger C, Hahn G, Digel M, Katona R, Sampaio KL, Messerle M, Hengel H, Koszinowski U, Brune W, Adler B. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J Gen Virol 89:359–368. doi: 10.1099/vir.0.83286-0. [DOI] [PubMed] [Google Scholar]
- 19.Wang D, Shenk T. 2005. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J Virol 79:10330–10338. doi: 10.1128/JVI.79.16.10330-10338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryckman BJ, Rainish BL, Chase MC, Borton JA, Nelson JA, Jarvis MA, Johnson DC. 2008. Characterization of the human cytomegalovirus gH/gL/UL128-131 complex that mediates entry into epithelial and endothelial cells. J Virol 82:60–70. doi: 10.1128/JVI.01910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang D, Shenk T. 2005. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci U S A 102:18153–18158. doi: 10.1073/pnas.0509201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. 2011. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat Rev Microbiol 9:369–381. doi: 10.1038/nrmicro2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisenberg RJ, Atanasiu D, Cairns TM, Gallagher JR, Krummenacher C, Cohen GH. 2012. Herpes virus fusion and entry: a story with many characters. Viruses 4:800–832. doi: 10.3390/v4050800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stampfer SD, Heldwein EE. 2013. Stuck in the middle: structural insights into the role of the gH/gL heterodimer in herpesvirus entry. Curr Opin Virol 3:13–19. doi: 10.1016/j.coviro.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou M, Lanchy JM, Ryckman BJ. 2015. Human cytomegalovirus gH/gL/gO promotes the fusion step of entry into all cell types, whereas gH/gL/UL128-131 broadens virus tropism through a distinct mechanism. J Virol 89:8999–9009. doi: 10.1128/JVI.01325-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan G, Nogalski MT, Yurochko AD. 2009. Activation of EGFR on monocytes is required for human cytomegalovirus entry and mediates cellular motility. Proc Natl Acad Sci U S A 106:22369–22374. doi: 10.1073/pnas.0908787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feire AL, Koss H, Compton T. 2004. Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proc Natl Acad Sci U S A 101:15470–15475. doi: 10.1073/pnas.0406821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Huang DY, Huong SM, Huang ES. 2005. Integrin alphavbeta3 is a coreceptor for human cytomegalovirus. Nat Med 11:515–521. doi: 10.1038/nm1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Huong SM, Chiu ML, Raab-Traub N, Huang ES. 2003. Epidermal growth factor receptor is a cellular receptor for human cytomegalovirus. Nature 424:456–461. doi: 10.1038/nature01818. [DOI] [PubMed] [Google Scholar]
- 30.Cobbs CS, Soroceanu L, Denham S, Zhang W, Britt WJ, Pieper R, Kraus MH. 2007. Human cytomegalovirus induces cellular tyrosine kinase signaling and promotes glioma cell invasiveness. J Neurooncol 85:271–280. doi: 10.1007/s11060-007-9423-2. [DOI] [PubMed] [Google Scholar]
- 31.Isaacson MK, Feire AL, Compton T. 2007. Epidermal growth factor receptor is not required for human cytomegalovirus entry or signaling. J Virol 81:6241–6247. doi: 10.1128/JVI.00169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kilcher S, Mercer J. 2014. Next generation approaches to study virus entry and infection. Curr Opin Virol 4:8–14. doi: 10.1016/j.coviro.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Panda D, Cherry S. 2012. Cell-based genomic screening: elucidating virus-host interactions. Curr Opin Virol 2:784–792. doi: 10.1016/j.coviro.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Houzet L, Jeang KT. 2011. Genome-wide screening using RNA interference to study host factors in viral replication and pathogenesis. Exp Biol Med (Maywood) 236:962–967. doi: 10.1258/ebm.2010.010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirsch AJ. 2010. The use of RNAi-based screens to identify host proteins involved in viral replication. Future Microbiol 5:303–311. doi: 10.2217/fmb.09.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cherry S. 2009. What have RNAi screens taught us about viral-host interactions? Curr Opin Microbiol 12:446–452. doi: 10.1016/j.mib.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinzger C, Schmidt K, Knapp J, Kahl M, Beck R, Waldman J, Hebart H, Einsele H, Jahn G. 1999. Modification of human cytomegalovirus tropism through propagation in vitro is associated with changes in the viral genome. J Gen Virol 80:2867–2877. doi: 10.1099/0022-1317-80-11-2867. [DOI] [PubMed] [Google Scholar]
- 38.Sampaio KL, Jahn G, Sinzger C. 2013. Applications for a dual fluorescent human cytomegalovirus in the analysis of viral entry. Methods Mol Biol 1064:201–209. doi: 10.1007/978-1-62703-601-6_14. [DOI] [PubMed] [Google Scholar]
- 39.Edgell CJ, McDonald CC, Graham JB. 1983. Permanent cell line expressing human factor VIII-related antigen established by hybridization. Proc Natl Acad Sci U S A 80:3734–3737. doi: 10.1073/pnas.80.12.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.May T, Butueva M, Bantner S, Markusic D, Seppen J, MacLeod RA, Weich H, Hauser H, Wirth D. 2010. Synthetic gene regulation circuits for control of cell expansion. Tissue Eng Part A 16:441–452. doi: 10.1089/ten.tea.2009.0184. [DOI] [PubMed] [Google Scholar]
- 41.Lieber D, Hochdorfer D, Stoehr D, Schubert A, Lotfi R, May T, Wirth D, Sinzger C. 2015. A permanently growing human endothelial cell line supports productive infection with human cytomegalovirus under conditional cell growth arrest. Biotechniques 59:127–136. doi: 10.2144/000114326. [DOI] [PubMed] [Google Scholar]
- 42.Scheffer KD, Gawlitza A, Spoden GA, Zhang XA, Lambert C, Berditchevski F, Florin L. 2013. Tetraspanin CD151 mediates papillomavirus type 16 endocytosis. J Virol 87:3435–3446. doi: 10.1128/JVI.02906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stamminger T, Puchtler E, Fleckenstein B. 1991. Discordant expression of the immediate-early 1 and 2 gene regions of human cytomegalovirus at early times after infection involves posttranscriptional processing events. J Virol 65:2273–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kitadokoro K, Bordo D, Galli G, Petracca R, Falugi F, Abrignani S, Grandi G, Bolognesi M. 2001. CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J 20:12–18. doi: 10.1093/emboj/20.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flint SJ, Racaniello VR, Rall GF, Skalka AM. 2003. Principles of virology: molecular biology, pathogenesis, and control of animal viruses, 2nd ed American Society for Microbiology, Washington, DC. [Google Scholar]
- 46.Monk PN, Partridge LJ. 2012. Tetraspanins: gateways for infection. Infect Disord Drug Targets 12:4–17. doi: 10.2174/187152612798994957. [DOI] [PubMed] [Google Scholar]
- 47.van Spriel AB, Figdor CG. 2010. The role of tetraspanins in the pathogenesis of infectious diseases. Microbes Infect 12:106–112. doi: 10.1016/j.micinf.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 48.Chambers J, Angulo A, Amaratunga D, Guo H, Jiang Y, Wan JS, Bittner A, Frueh K, Jackson MR, Peterson PA, Erlander MG, Ghazal P. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J Virol 73:5757–5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blanton RA, Tevethia MJ. 1981. Immunoprecipitation of virus-specific immediate-early and early polypeptides from cells lytically infected with human cytomegalovirus strain AD169. Virology 112:262–273. doi: 10.1016/0042-6822(81)90631-0. [DOI] [PubMed] [Google Scholar]
- 50.Stinski MF. 1978. Sequence of protein synthesis in cells infected by human cytomegalovirus: early and late virus-induced polypeptides. J Virol 26:686–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Compton T, Nowlin DM, Cooper NR. 1993. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology 193:834–841. doi: 10.1006/viro.1993.1192. [DOI] [PubMed] [Google Scholar]
- 52.Kari B, Gehrz R. 1992. A human cytomegalovirus glycoprotein complex designated gC-II is a major heparin-binding component of the envelope. J Virol 66:1761–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horejsi V, Vlcek C. 1991. Novel structurally distinct family of leucocyte surface glycoproteins including CD9, CD37, CD53 and CD63. FEBS Lett 288:1–4. doi: 10.1016/0014-5793(91)80988-F. [DOI] [PubMed] [Google Scholar]
- 54.Levy S, Nguyen VQ, Andria ML, Takahashi S. 1991. Structure and membrane topology of TAPA-1. J Biol Chem 266:14597–14602. [PubMed] [Google Scholar]
- 55.Oren R, Takahashi S, Doss C, Levy R, Levy S. 1990. TAPA-1, the target of an antiproliferative antibody, defines a new family of transmembrane proteins. Mol Cell Biol 10:4007–4015. doi: 10.1128/MCB.10.8.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang S, Yuan S, Dong M, Su J, Yu C, Shen Y, Xie X, Yu Y, Yu X, Chen S, Zhang S, Pontarotti P, Xu A. 2005. The phylogenetic analysis of tetraspanins projects the evolution of cell-cell interactions from unicellular to multicellular organisms. Genomics 86:674–684. doi: 10.1016/j.ygeno.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 57.Levy S, Shoham T. 2005. Protein-protein interactions in the tetraspanin web. Physiology (Bethesda) 20:218–224. doi: 10.1152/physiol.00015.2005. [DOI] [PubMed] [Google Scholar]
- 58.Rubinstein E, Le Naour F, Lagaudriere-Gesbert C, Billard M, Conjeaud H, Boucheix C. 1996. CD9, CD63, CD81, and CD82 are components of a surface tetraspan network connected to HLA-DR and VLA integrins. Eur J Immunol 26:2657–2665. doi: 10.1002/eji.1830261117. [DOI] [PubMed] [Google Scholar]
- 59.Boucheix C, Rubinstein E. 2001. Tetraspanins. Cell Mol Life Sci 58:1189–1205. doi: 10.1007/PL00000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rubinstein E, Charrin S, Tomlinson M. 2013. Organisation of the tetraspanin web, p 47–90. In Berditchevski F, Rubinstein E (ed), Tetraspanins, vol 9 Springer, Dordrecht, Netherlands. [Google Scholar]
- 61.Yanez-Mo M, Barreiro O, Gordon-Alonso M, Sala-Valdes M, Sanchez-Madrid F. 2009. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol 19:434–446. doi: 10.1016/j.tcb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 62.Fanaei M, Monk PN, Partridge LJ. 2011. The role of tetraspanins in fusion. Biochem Soc Trans 39:524–528. doi: 10.1042/BST0390524. [DOI] [PubMed] [Google Scholar]
- 63.Hemler ME. 2005. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol 6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 64.Homsi Y, Schloetel JG, Scheffer KD, Schmidt TH, Destainville N, Florin L, Lang T. 2014. The extracellular delta-domain is essential for the formation of CD81 tetraspanin webs. Biophys J 107:100–113. doi: 10.1016/j.bpj.2014.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spoden G, Freitag K, Husmann M, Boller K, Sapp M, Lambert C, Florin L. 2008. Clathrin- and caveolin-independent entry of human papillomavirus type 16—involvement of tetraspanin-enriched microdomains (TEMs). PLoS One 3:e3313. doi: 10.1371/journal.pone.0003313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spoden G, Kuhling L, Cordes N, Frenzel B, Sapp M, Boller K, Florin L, Schelhaas M. 2013. Human papillomavirus types 16, 18, and 31 share similar endocytic requirements for entry. J Virol 87:7765–7773. doi: 10.1128/JVI.00370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. 1998. Binding of hepatitis C virus to CD81. Science 282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 68.Farquhar MJ, Harris HJ, McKeating JA. 2011. Hepatitis C virus entry and the tetraspanin CD81. Biochem Soc Trans 39:532–536. doi: 10.1042/BST0390532. [DOI] [PubMed] [Google Scholar]
- 69.Li G, Endsley MA, Somasunderam A, Gbota SL, Mbaka MI, Murray JL, Ferguson MR. 2014. The dual role of tetraspanin CD63 in HIV-1 replication. Virol J 11:23. doi: 10.1186/1743-422X-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ooi YS, Stiles KM, Liu CY, Taylor GM, Kielian M. 2013. Genome-wide RNAi screen identifies novel host proteins required for alphavirus entry. PLoS Pathog 9:e1003835. doi: 10.1371/journal.ppat.1003835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kurzeder C, Koppold B, Sauer G, Pabst S, Kreienberg R, Deissler H. 2007. CD9 promotes adeno-associated virus type 2 infection of mammary carcinoma cells with low cell surface expression of heparan sulphate proteoglycans. Int J Mol Med 19:325–333. [PubMed] [Google Scholar]
- 72.Konig R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L, Ideker T, Garcia-Sastre A, Young JA, Palese P, Shaw ML, Chanda SK. 2010. Human host factors required for influenza virus replication. Nature 463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scheffer KD, Berditchevski F, Florin L. 2014. The tetraspanin CD151 in papillomavirus infection. Viruses 6:893–908. doi: 10.3390/v6020893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ho SH, Martin F, Higginbottom A, Partridge LJ, Parthasarathy V, Moseley GW, Lopez P, Cheng-Mayer C, Monk PN. 2006. Recombinant extracellular domains of tetraspanin proteins are potent inhibitors of the infection of macrophages by human immunodeficiency virus type 1. J Virol 80:6487–6496. doi: 10.1128/JVI.02539-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Straschewski S, Patrone M, Walther P, Gallina A, Mertens T, Frascaroli G. 2011. Protein pUL128 of human cytomegalovirus is necessary for monocyte infection and blocking of migration. J Virol 85:5150–5158. doi: 10.1128/JVI.02100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sincock PM, Mayrhofer G, Ashman LK. 1997. Localization of the transmembrane 4 superfamily (TM4SF) member PETA-3 (CD151) in normal human tissues: comparison with CD9, CD63, and alpha5beta1 integrin. J Histochem Cytochem 45:515–525. doi: 10.1177/002215549704500404. [DOI] [PubMed] [Google Scholar]
- 77.Uhlén M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F. 2015. Proteomics. Tissue-based map of the human proteome. Science 347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 78.Min G, Wang H, Sun TT, Kong XP. 2006. Structural basis for tetraspanin functions as revealed by the cryo-EM structure of uroplakin complexes at 6-A resolution. J Cell Biol 173:975–983. doi: 10.1083/jcb.200602086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sawada S, Yoshimoto M, Odintsova E, Hotchin NA, Berditchevski F. 2003. The tetraspanin CD151 functions as a negative regulator in the adhesion-dependent activation of Ras. J Biol Chem 278:26323–26326. doi: 10.1074/jbc.C300210200. [DOI] [PubMed] [Google Scholar]
- 80.Hong IK, Jeoung DI, Ha KS, Kim YM, Lee H. 2012. Tetraspanin CD151 stimulates adhesion-dependent activation of Ras, Rac, and Cdc42 by facilitating molecular association between beta1 integrins and small GTPases. J Biol Chem 287:32027–32039. doi: 10.1074/jbc.M111.314443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ziyyat A, Rubinstein E, Monier-Gavelle F, Barraud V, Kulski O, Prenant M, Boucheix C, Bomsel M, Wolf JP. 2006. CD9 controls the formation of clusters that contain tetraspanins and the integrin alpha 6 beta 1, which are involved in human and mouse gamete fusion. J Cell Sci 119:416–424. doi: 10.1242/jcs.02730. [DOI] [PubMed] [Google Scholar]
- 82.Hassuna N, Monk PN, Moseley GW, Partridge LJ. 2009. Strategies for targeting tetraspanin proteins: potential therapeutic applications in microbial infections. BioDrugs 23:341–359. doi: 10.2165/11315650-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hemler ME. 2008. Targeting of tetraspanin proteins—potential benefits and strategies. Nat Rev Drug Discov 7:747–758. doi: 10.1038/nrd2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Montgomery RI, Warner MS, Lum BJ, Spear PG. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427–436. doi: 10.1016/S0092-8674(00)81363-X. [DOI] [PubMed] [Google Scholar]
- 85.Whitbeck JC, Peng C, Lou H, Xu R, Willis SH, Ponce de Leon M, Peng T, Nicola AV, Montgomery RI, Warner MS, Soulika AM, Spruce LA, Moore WT, Lambris JD, Spear PG, Cohen GH, Eisenberg RJ. 1997. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J Virol 71:6083–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krummenacher C, Nicola AV, Whitbeck JC, Lou H, Hou W, Lambris JD, Geraghty RJ, Spear PG, Cohen GH, Eisenberg RJ. 1998. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J Virol 72:7064–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 88.Warner MS, Geraghty RJ, Martinez WM, Montgomery RI, Whitbeck JC, Xu R, Eisenberg RJ, Cohen GH, Spear PG. 1998. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology 246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 89.Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. 1998. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J Virol 72:9992–10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Spear PG. 2004. Herpes simplex virus: receptors and ligands for cell entry. Cell Microbiol 6:401–410. doi: 10.1111/j.1462-5822.2004.00389.x. [DOI] [PubMed] [Google Scholar]
- 91.Bailey RL, Herbert JM, Khan K, Heath VL, Bicknell R, Tomlinson MG. 2011. The emerging role of tetraspanin microdomains on endothelial cells. Biochem Soc Trans 39:1667–1673. doi: 10.1042/BST20110745. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.