Abstract
The role of microRNAs (miRNAs) in infectious diseases is becoming more and more apparent, and the use of miRNAs as a diagnostic tool and their therapeutic application has become the major focus of investigation. The aim of this study was to identify miRNAs involved in the immune signaling of macrophages in response to Arcobacter (A.) butzleri infection, an emerging foodborne pathogen causing gastroenteritis. Therefore, primary human macrophages were isolated and infected, and miRNA expression was studied by means of RNAseq. Analysis of the data revealed the expression of several miRNAs, which were previously associated with bacterial infections such as miR-155, miR-125, and miR-212. They were shown to play a key role in Toll-like receptor signaling where they act as fine-tuners to establish a balanced immune response. In addition, miRNAs which have yet not been identified during bacterial infections such as miR-3613, miR-2116, miR-671, miR-30d, and miR-629 were differentially regulated in A. butzleri-infected cells. Targets of these miRNAs accumulated in pathways such as apoptosis and endocytosis – processes that might be involved in A. butzleri pathogenesis. Our study contributes new findings about the interaction of A. butzleri with human innate immune cells helping to understand underlying regulatory mechanisms in macrophages during infection.
Keywords: Arcobacter butzleri, macrophages, microRNAs, RNA interference, immune signaling
Introduction
Phagocytic and antigen-presenting cells of the innate immune system such as macrophages or dendritic cells immediately recognize invasive microorganisms, which are able to overcome anatomic host barriers such as epithelial surfaces and enter the organism. The undisturbed function of phagocytes in this first line of host defense is pivotal for a successful immune response towards infection.
The effective eradication of pathogens is underlain by several signaling events such as Toll-like-receptor (TLR) signaling; many of these are regulated in a miRNA-dependent manner [1]. MicroRNAs (miRNAs) are small noncoding RNAs, which negatively influence gene expression by degradation of mRNA or inhibition of protein translation. MiRNA dysregulation in many cases leads to disease such as cancer or metabolic disorder. As a consequence, the use of miRNAs as a diagnostic tool and also the therapeutic application has become the major focus of investigation [2]. The role of miRNAs in infectious diseases has become more and more apparent, and there are numerous reports about their involvement in viral and parasite infection. The contribution of miRNAs in bacterial diseases has been explored less. However, over the last years, several studies reported an increasing evidence of miRNA-mediated host response towards bacterial infection [3, 4]. Upon infection, miRNAs tightly control immune reactions, involving both miRNA-mediated unspecific cellular responses as well as pathogen-dependent regulation of a specific miRNA expression profile [5].
Pathogenic species within the family of Campylobacteraceae belong to the leading causes for severe gastroenteritis worldwide. Within this family, Arcobacter (A.) butzleri is an emerging zoonotic pathogen and has recently been associated with cases of diarrhea, peritonitis, and bacteremia, but its relevance associated with disease still remains to be evaluated [6]. Due to missing routine diagnostic and standardized isolation methods, the incidence of A. butzleri-associated diseases cannot be properly specified. Nevertheless, Arcobacter spp. are known to be the fourth most common pathogenic group isolated from stool specimens of patients with acute enteritis in several prevalence studies conducted in Europe [7–9].
Understanding underlying regulatory mechanisms that occur in host cells upon Arcobacter infection is indispensable to evaluate the pathogenicity and to develop strategies to diagnose and combat A. butzleri-induced diseases. Thus, the aim of this study was to identify miRNAs expressed in innate immune cells in response to an A. butzleri infection. For that purpose, primary human macrophages were isolated from three different donors and infected with A. butzleri. Subsequent to infection, expression of miRNAs was studied by means of RNAseq.
Materials and methods
Bacterial strain and culture condition
A. butzleri reference strain CCUG30485 (Culture Collection University of Göteborg, Sweden) was cultivated and prepared for infection experiments as described previously [10].
Isolation and cultivation of primary human macrophages
Buffy coats of healthy human donors were obtained at the German Red Cross in Berlin Wannsee. Blood mononuclear cells were isolated by Ficoll-Paque centrifugation and subsequent attachment to cell-culture flask surface. Cells were cultivated for 5 days in Gibco macrophage SFM medium including 10 μg/ml gentamycin (Biochrom) and 50 ng/ml M-CSF (PAN Biotech) (M-CSF stimulus was added for 2 days).
To determine the percentage of CD14+ cells in the isolated cell population, fluorescence-activated cell sorting (FACS) was performed using a FACSCalibur flow cytometer (Becton Dickinson GmbH). The CD14 antibody was purchased at Santa Cruz Biotechnology and used at a concentration of 1:100. The secondary antibody (IgG2a Goat Anti-Mouse, PE labeled, Southern Biotech) was used at 1:200.
If FACS analysis proved that the cell population contained more than 80% CD14+ cells, 6 × 105 macrophages in 1.5 ml medium per well were seeded in six-well plates and used for infection experiments. Cells were incubated for another 24 h at 37 °C and 5% CO2 before further experimental use. Monocytes were isolated from three different human donors, and infection experiments were reproduced in three independent experimental setups.
Infection experiments
Approximately 4–6 × 107 bacterial cells were inoculated on 4–6 × 105 primary human macrophages (multiplicity of infection [MOI] = 100) and incubated at 37 °C and 5% CO2. Noninfected cells served as a negative control. After 3 h of infection, cells were washed three times with phosphate-buffered solution (PBS) and incubated with fresh media containing 300 μg/ml gentamycin for another 2 h to remove remaining extracellular bacteria. Samples were taken 1 h, 5 h, and 24 h after infection. For the 24 h time point, cells were treated with 20 μg/ml gentamycin for the remaining incubation time. For RNA extraction, cells were washed three times with PBS, lysed with RNA lysis buffer (mirVANA, Life Technologies) and total RNA was isolated according to the manufacturer’s instruction.
Quality control of isolated RNA
Quantity and quality of RNA were first determined by measuring absorbance at 260 and 280 nm with a Nano Drop 1000 spectrophotometer according to the manufacturer’s instructions (Thermo Scientific). Samples were further analyzed for their RNA integrity with an Agilent 2100 BioAnalyzer and RNA 6000 Nano Kits (Agilent) according to the manufacturer’s protocol. RNA with integrity value (RIN) of ≥9 was used for further investigation.
RNAseq data analysis
Sequencing of small RNA samples was performed at the Institute of Clinical Molecular Biology at Christian-Albrechts-University Kiel, Germany using a HiSeq2500 device (Illumina) as described earlier [11].
The raw sequencing data were processed as described in detail in Leidinger et al.[12]. In brief, we first trimmed the 3′ adapter sequence with the FastX toolkit. For quantification (mapping against miRBase release 20) and prediction of novel miRNAs (mapping against human genome hg19), the miRDeep2 pipeline was applied [13]. We summarized the final read counts for known and novel miRNAs in an expression matrix and applied quantile normalization. For detecting deregulated miRNAs between different time points, we applied analysis of variance (ANOVA). For pair wise group comparisons, we applied two-tailed t-tests. All statistical calculations have been carried out using R version 3.0.2.
Pathway analysis
To unravel the role of differentially expressed miRNAs which have not yet been reported to play a role in bacterial infection, in silico prediction was used to identify mRNA targets (miRmap, miRmap score <80) [14]. The list of potential target genes was applied to the DAVID functional annotation tool [15, 16] to identify pathways in which these potential mRNA targets accumulate.
Validation of potential novel miRNAs
Sequences of three potential novel miRNA candidates were selected from the sequencing results, and expression in macrophages upon infection was validated by means of miR-Q, a miRNA-specific reverse transcription quantitative polymerase chain reaction (RT-qPCR) [17]. Oligonucleotides (Table 1) were designed and synthesized (Sigma-Aldrich), and miR-Q assays were performed as described earlier [17] with an annealing temperature of 62 °C. Parallel to amplification of the potential novel miRNAs, reference small RNAs SNORD44 and SNORD47 were amplified and used for normalization. To determine specificity of amplified PCR products, melt curve analysis and electrophoresis were performed subsequent to amplification.
Table 1.
Oligonucleotides used for validation of novel miRNAs
| Novel-miR | Sequence (annealing temperature: 62 °C) |
|---|---|
| RT6-novel-miR-259 5′-3′ | tgtcaggcaaccgtattcaccgtgagtggtGCTAGA |
| Short-novel-miR-259-rev 5′-3′ | cgtcagatgtccgagtagagggggaacggcgCTCTGACCTCTGACCCTCT |
| RT6-novel-miR-55 5′-3′ | tgtcaggcaaccgtattcaccgtgagtggtTCTGCG |
| Short-novel-miR-55-rev 5′-3′ | cgtcagatgtccgagtagagggggaacggcgAGGGCCTGCTCCCACCCCGC |
| RT-6-novel-miR-134 5′-3′ | tgtcaggcaaccgtattcaccgtgagtggtAACTCT |
| Short-novel-miR-134-rev 5′-3′ | cgtcagatgtccgagtagagggggaacggcgAAAAGCTGTCCACTGTAGA |
| cDNA | |
| Novel-miR-259 5′-3′ | GCTAGAGGGTCAGAGGTCAGAG |
| Novel-miR-55 5′-3′ | TCTGCGGGGTGGGAGCAGGCCCT |
| Novel-miR-134 5′-3′ | AACTCTACAGTGGACAGCTTTT |
MiR-Q analysis
The ΔΔCT method [18] was used to calculate the relative fold difference of miRNA expression levels compared to the negative control. To determine CT values, thresholds were set at 0.2.
Ethics
The local Ethics Commission at the Charité Berlin approved the study (EA1/092/14).
Results
FACS analysis proves high efficiency of macrophage isolation and enrichment from human buffy coats
Since monocytes only represent a small fraction of leukocytes (2–6%), an enrichment step was necessary to obtain a high percentage of monocytes in the leukocyte cell composition. For this purpose, monocytes were isolated from buffy coats by density centrifugation and subsequent adherence to cell culture flasks in a differentiation medium. Subsequent to cultivation, flow cytometry was performed to analyze CD14 expression, a cell surface marker for monocytes and macrophages, and to reveal the purity of the isolated monocyte population. For that purpose, cells were stained with a CD14 antibody as well as a secondary fluorochrome-labeled antibody and analyzed with a cytometer. The fluorochrome-stained cells were gated and analyzed for CD14 antibody staining. Macrophage fraction in cell culture isolated and cultivated from Donor 1 constituted 97.02% (Fig. S1, A); from Donor 2, 92.70% (Fig. S1, B); and Donor 3, 81.0% (Fig. S1, C).
Sequencing and bioinformatic analysis reveal several miRNAs to be regulated upon A. butzleri infection
Since miRNAs were reported to exhibit rapid dynamics in their temporal expression in response to bacterial pathogens [5, 19], RNA samples of infected macrophages were investigated at three different time points to assess changes in miRNA patterns during the course of infection. An ANOVA analysis of the RNAseq data revealed 14 annotated human miRNAs, which were significantly regulated upon A. butzleri infection. An additional t-test displayed significant differences in expression levels compared to the noninfected control (Table 2).
Table 2.
MiRNAs found to be regulated upon A. butzleri infection
| miR | Sequence | p value (different time points of infection compared to noninfected control) |
|---|---|---|
| 125a-3p | acaggugagguucuugggagcc | 0.038 (24 h vs. n.c.) |
| 155-3p | cuccuacauauuagcauuaaca | 0.035 (5 h vs. n.c.) |
| 155-5p | uuaaugcuaaucgugauaggggu | 0.02 (5 h vs. n.c.) |
| 212-3p | uaacagucuccagucacggcc | 0.034 (24 h vs. n.c.) |
| 181c-5p | aacauucaaccugucggugagu | 0.034 (24 h vs. n.c.) |
| 21-3p | caacaccagucgaugggcugu | 0.012 (24 h vs. n.c.) |
| 27a-5p | agggcuuagcugcuugugagca | 0.049 (1 h vs. n.c.) |
| let-7a-3p | cuauacaaucuacugucuuuc | 0.05 (24 h vs. n.c.) |
| 26b-5p | uucaaguaauucaggauaggu | 0.044 (1 h vs. n.c.), 0.048 (5 h vs. n.c.), 0.004 (24 h vs. n.c.) |
| 148b-3p | ucagugcacuacagaacuuugu | 0.004 (24 h vs. n.c.) |
| 3613-5p | uguuguacuuuuuuuuuuguuc | 0.018 (5 h vs. n.c.), 0.019 (24 h vs. n.c.) |
| 2116-3p | ccucccaugccaagaacuccc | 0.025 (5 h vs. n.c.), 0.032 (24 h vs. n.c.) |
| 671-3p | uccgguucucagggcuccacc | 0.003 (5 h vs. n.c.), 0.009 (24 h vs. n.c.) |
| 30d-3p | cuuucagucagauguuugcugc | 0.038 (5 h vs. n.c.) |
| 30d-5p | uguaaacauccccgacuggaag | 0.035 (24 h vs. n.c.) |
| 629-5p | uggguuuacguugggagaacu | 0.03 (5 h vs. n.c.) |
According to previous reports regarding regulatory functions of miRNAs, they could be grouped into miRNAs commonly affected by various bacterial agents and identified as key players in host innate immune response (Fig. 1) and those which have not been associated with bacterial diseases so far (Fig. 2). Following this, differentially expressed miRNA in these two groups will be presented.
Fig. 1.
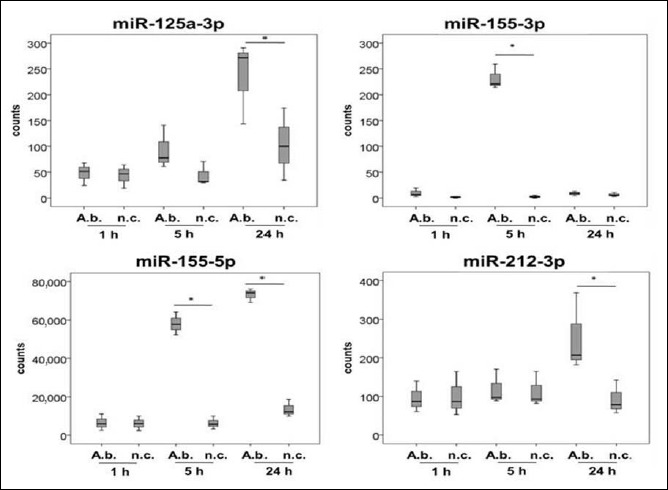
MiRNAs associated with bacterial diseases, expressed in primary human macrophages as a response to A. butzleri infection. Cells were infected at a MOI of 100. Samples were taken 1 h, 5 h, and 24 h after infection (A.b. = A. butzleri reference strain CCUG30485, n.c. = noninfected cells). Asterisks in figures summarize p values (*p < 0.05; **p < 0.01)
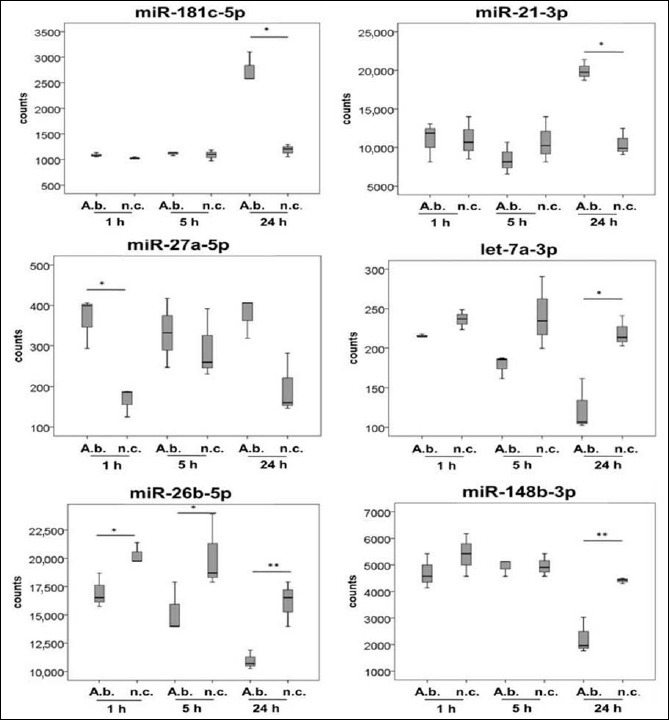
Fig. 2.
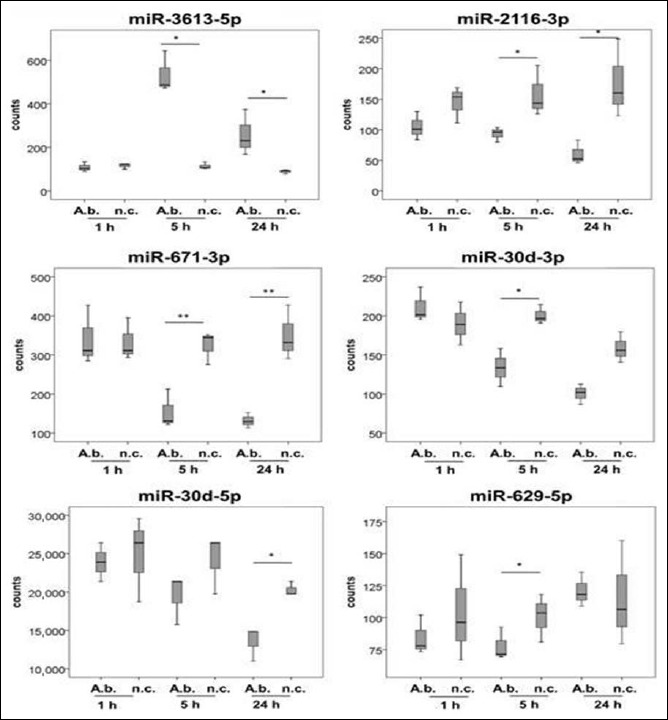
MiRNAs not yet associated with bacterial diseases, expressed in primary human macrophages as a response to A. butzleri infection. Cells were infected at a MOI of 100. Samples were taken 1 h, 5 h, and 24 h after infection. Asterisks in figures summarize p values (*p < 0.05; **p < 0.01)
MiRNAs associated with bacterial infections
MiR-125a, miR-155, miR-212, miR-181c, miR-21, miR-27a, let-7a, miR-26b, and miR-148b were differentially expressed in A. butzleri-infected human macrophages compared to the noninfected control and have previously been reported to play a role in host immune response towards bacterial infection (Fig. 1). Expression levels of miR-125a-3p were increased in 5 h and 24 h A. butzleri-infected cells compared to respective controls. A continuous increase could be observed over the time of infection although slightly elevated expression levels were also found in 24 h noninfected cells. Significant differences were calculated for expression levels in 24 h infected samples versus the respective negative control (p < 0.05). MiR-155-3p was significantly upregulated in infected macrophages 5 h postinfection (p < 0.05). Twenty-four hours after infection, the expression level was similar to those of noninfected cells, whereas for miR-155-5p, expression level at 24 h remained significantly elevated (p < 0.05). The expression level of neither miR-155-3p nor 5p was affected 1 h after infection. Overall, miR-155-5p was accumulated to a higher degree compared to miR-155-3p. MiR-212-3p was increased 24 h postinfection compared to the noninfected control as well as compared to the other time points of infection (p < 0.05). Similar to miR-212-3p, miR-181c-5p as well as miR-21-3p was found to be upregulated 24 h after A. butzleri infection compared to other time points of infection. MiR-27a-5p showed an increased expression 1 h after infection compared to the noninfected control (p < 0.05). MiRNA let-7a-3p (p < 0.05) and miR-26b-5p decreased over the time of infection (p < 0.05) whereas miR-148b-3p exhibited reduced expression levels 24 h after infection (p < 0.05), and 1 h and 5 h after infection, expression was not affected. Highest miRNA copy numbers were found for miR-155-5p.
MiRNAs not yet associated with bacterial infections
MiR-3613, miR-2116, miR-671, miR-30d, and miR-629 exhibited significant differences in expression levels in infected macrophages. So far, none of these miRNAs have been described being commonly expressed in response to bacterial infection in human host cells.
As indicated in Fig. 2, miR-3613-5p exhibited elevated expression levels 5 h and 24 h postinfection compared to respective controls (p < 0.05). Expression remained unaffected 1 h after infection. MiR-2116-3p and miR-671-3p were downregulated during infection 5 h and 24 h compared to respective controls (p < 0.05). Expression of miR-30d-3p decreased over the time of infection; 5 h after infection, expression was significantly reduced compared to the noninfected control (p < 0.05). Expression of miR-30d-5p exhibited a similar pattern as expression of miR-30d-3p did. Nevertheless, miR-30d-5p was accumulated to a much higher degree compared to miR-30d-3p. Expression of miR-629-5p was reduced 5 h after infection compared to noninfected cells (p < 0.05).
To unravel the role of these miRNAs, in silico prediction was used to identify mRNA targets and pathways in which they accumulate. Beneath metabolic pathways, target accumulation was found for apoptosis/p53 signaling (targets of miR-3613 and miR-2116), MAPK signaling (targets of miR-671 and miR-30d-5p), endocytosis (targets of miR-30d-3p and 30d-5p), regulation of actin cytoskeleton (targets of miR-30d-3p), and formation of the immunoproteasome (targets of miR-629). The results are summarized in Table 3. Full data of the pathway analysis is attached in the supplementary data.
Table 3.
Pathways with enriched mRNA targets potentially influenced by A. butzleri-induced miRNAs
| Pathway | miRNAs | mRNA targets | p value |
|---|---|---|---|
| Endocytosis | miR-30d-3p | SH3GL3, DNM3, FLT1, ERBB4, STAM2, VTA1, PRKCI, ARF6, HLA-B, KIT, ZFYVE20, RAB31, AP2B1, RAB11FIP2, CHMP1B, RAB22A, RAB11B, VPS36, RNF41 | 0.063 |
| miR-30d-5p | NEDD4, ACAP2, RAB11A, EEA1, NEDD4L, KIT, ARAP2, CHMP2B | 0.085 | |
| Apoptosis/p53 signaling pathway | miR-3613 | CDK6, RRM2B, ATR, SESN3 | 0.037 |
| miR-2116 | IRAK2, IRAK1, IRAK3, PRKAR2A, DFFA, PIK3CB, CASP8, CHP2, EXOG, PPP3R2, PIK3R1 | 0.076 | |
| EI24, CD82, RRM2, SERPINE1, CASP8, RCHY1, RRM2B, MDM4, PERP, CDK2, SESN3 | 0.017 | ||
| Regulation of actin cytoskeleton | miR-30d-3p | GNA13, FGF7, PIK3CB, ROCK2, DIAPH2, SSH2, GNA12, ITGA1, ACTN2, PPP1CB, NCKAP1, DOCK1, CHRM2, TIAM1, SOS1, SOS2, WASL, CRK, FGF2, MYLK, APC | 0.079 |
| MAPK signaling pathway | miR-671 | RASGRF1, FGF11, CACNB3, FGF1, CRK | 0.063 |
| miR-30d-5p | MAP3K7, CASP3, MAP3K5, TAOK1, MAP3K2, PLA2G12A, NF1, PLA2G2C, PPP3CA, FGF20 | 0.099 | |
| Formation of immunoproteasome | miR-629 | PSMA2, PSMB10, PSMD13, PSMB2, PSME4 | 0.023 |
Potential novel human miRNAs
Raw data analysis indicated potential novel miRNAs, which have not yet been described. To follow up the expression of respective transcripts, primers were designed and expression in the same macrophage samples used for RNAseq was investigated by means of miRNA specific RT-qPCR (miR-Q). Fold induction of potential novel-miR-55 compared to the negative control increased over the time of infection; this was the case for both sequencing data (Fig. 3) and RT-qPCR analysis (Fig. 4). However, sequencing data indicated a downregulation of novel-miR-55 in infected cells compared to respective negative controls, whereas in RT-qPCR data, novel-miR-55 was upregulated in infected cells. Transcript counts of potential novel-miRNAs in the analyzed samples are listed in Table 4. Potential novel-miR-134 was slightly upregulated 5 h and 24 h in RNAseq data (Fig. 3). RT-qPCR indicated an upregulation in 24 h infected samples (Fig. 4). Expression dynamics of potential novel-miR-259 correlated in comparing RNAseq and RT-qPCR analysis. Again, as described for novel-miR-55, RNAseq revealed a downregulation in infected cells (Fig. 3), whereas RT-qPCR indicated an upregulation (Fig. 4).
Fig. 3.
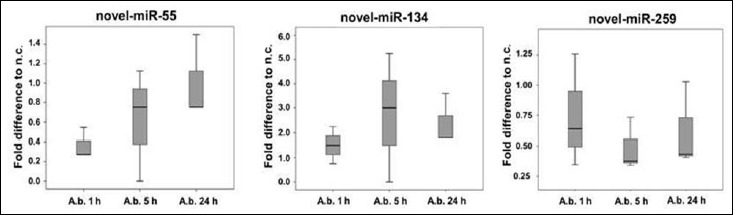
RNA sequences were analyzed for potential novel miRNA sequences using mirDeep2 algorithms [13]
Fig. 4.
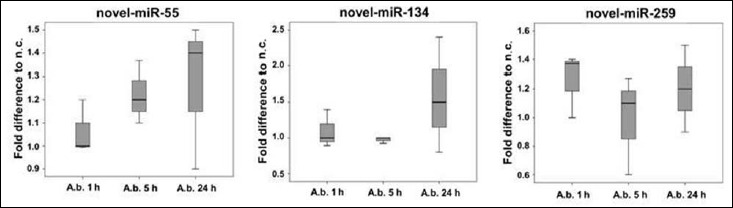
Expression of respective novel sequences in macrophage samples were followed up by means of RT-qPCR (miR-Q)
Table 4.
Mean counts of potential novel-miRNA transcripts in infected cells (A.b.) and controls (n.c.) determined by RNAseq
| miRNA | A.b. 1 h | n.c. 1 h | A.b. 5 h | n.c. 5 h | A.b. 24 h | n.c. 24 h |
|---|---|---|---|---|---|---|
| Novel-miR-55 | 1.3 | 3.7 | 1.7 | 2.7 | 0.7 | 1.3 |
| Novel-miR-134 | 2 | 1.3 | 3.7 | 1.3 | 4 | 1.7 |
| Novel-miR-259 | 43.3 | 58 | 27 | 56 | 23 | 37 |
Discussion
Several studies have shown that miRNAs are key regulators of innate immunity [19–21] and play a prominent role in the specific host response towards bacterial pathogens [22, 23]. We have previously demonstrated that A. butzleri induce a pro-inflammatory response in human macrophages and have limited ability for intracellular survival [10]. Since nothing is yet known about underlying regulatory mechanisms such as miRNA expression in response to infection, we challenged primary human macrophages with A. butzleri and analyzed miRNA expression by means of RNAseq.
Bioinformatic investigation of generated sequencing data revealed several miRNAs, which were differentially expressed in infected cells compared to the noninfected control. Some of these miRNAs have not yet been reported to play a role in the host response towards bacterial infection. However, most of the identified miRNAs belonged to a set of candidates, which were also expressed in immune cells in response to bacterial infection or LPS stimulus in previous studies (miR-155, miR-21, miR-125, miR-212, let-7a, miR-181c, miR-26b, miR-148a, and miR-27a) [24–28].
Among these, miR-155, miR-21, miR-125, and let-7a play a central role in the immune response towards bacterial pathogens [3, 25]. They are mainly regulated in a Toll-like receptor dependent manner and, in turn, target different components of the TLR signaling cascade including TLRs themselves as well as downstream signaling proteins such as MyD88 and transcription factors such as NFκB as well as cytokines. MiRNAs can thereby control strength, location, and timing of TLR response [19], which makes them crucial for a balanced immune homeostasis and protects the host from uncontrolled inflammatory conditions. Notably, expression of miR-146 did not display significant differences in A. butzleri-infected cells compared to the noninfected control, even though it is reported to act as a key regulator of TLR signaling in immune cells in response to various bacterial pathogens such as Campylobacter concisus, Helicobacter pylori, or Salmonella enterica [4, 29]. This might be due to expression dynamics in the course of infection. Nevertheless, in THP-1-derived macrophages infected with C. concisus (another member of the family of Campylobacteraceae), miR-146a was upregulated 6 h after infection [29].
Interestingly, some differentially expressed miRNAs identified in macrophages in response to A. butzleri have not been reported yet as regulators of host response to infection (miR-3613, miR-2116, miR-671, miR-30d-3p, miR-30d-5p, and miR-629-5p). In silico analysis employing miRmap software and DAVID functional annotation tool revealed that potential targets of these miRNAs (besides putative targets in metabolism and cancer) were enriched in pathways such as apoptosis/DNA damage, MAPK signaling, endocytosis, regulation of actin cytoskeleton, and formation of the immunoproteasome. Except for miR-3613, all miRNAs that were not yet associated with bacterial infections were downregulated during Arcobacter challenge.
In case of apoptosis, possibly involved miR-2116 was downregulated 24 h after infection. MiRmap revealed the initiator caspase 8 as a potential target. Therefore, an increased caspase activity due to the loss of miRNA-inhibition leading to onset of apoptosis in the course of infection could be hypothesized. The induction of apoptosis in macrophages to enhance virulence has been shown for several bacterial pathogens such as Salmonella spp. or Shigella flexneri [30] and could favor the establishment of A. butzleri infection by impairing phagocytosis and eradication by immune cells. The regulatory role of miRNAs in apoptosis is well known [31], and the modulation of host miRNA expression to attenuate macrophage function has been demonstrated for pathogens such as Mycobacterium avium [4, 23]. Although we were able to demonstrate the initial activation of initiator as well as effector caspases in THP-1-derived macrophages in response to A. butzleri infection in a previous study [10], DNA damage and end-point apoptosis could not be detected. However, this could be different in primary human macrophages and needs to be the matter of deeper investigations. In case of endocytosis, possibly involved miR-30d-3p and miR-30d-5p were downregulated in A. butzleri-challenged cells, suggesting a miRNA-mediated boost of endocytosis in response to Arcobacter infection.
In addition, three potential novel miRNAs (novel-miR-55, novel-miR-134, and novel-miR-259) were identified in A. butzleri-infected macrophages and expression was followed up by RT-qPCR. Nevertheless, further experiments are necessary to validate the expression of the novel miRNAs and their functional role.
Overall, the miRNA response of macrophages towards A. butzleri infection differed from recent findings regarding infection of THP-1 cells with C. concisus [29]. There were no overlaps in miRNA expression in both studies underlining the observation that differences in expression levels and profiles depend on the stimulus and cell type [19]. This study suggests that macrophage function is controlled by a specific set of miRNAs during A. butzleri infection. These miRNAs may not only constitute a potential therapeutic target but also function as promising diagnostic marker for the infection.
Fig. S1.
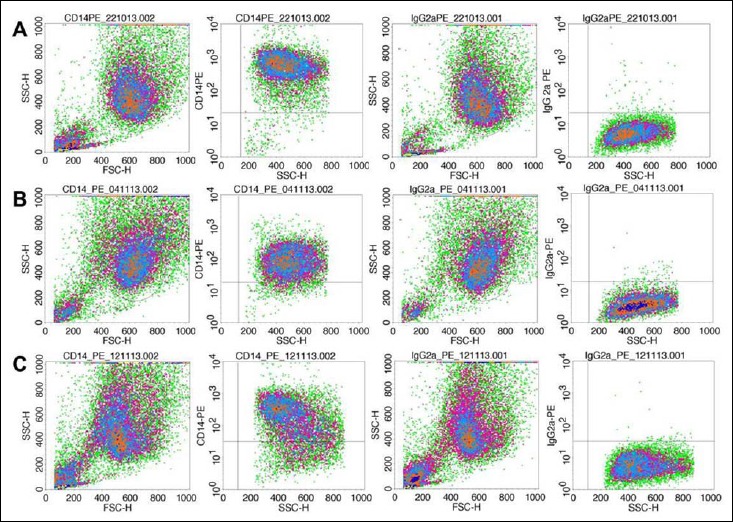
FACS plots CD14, primary human macrophages. (A) FACS plot indicates 97.82% CD14+ of gated cells with 0.8% unspecific binding of the secondary antibody (IgG2a) isolated from Donor 1. (B) Isolated primary cells of Donor 2 yield 93.51% CD14+ cells minus 0.81% due to unspecific binding of the secondary antibody. (C) 81.45% CD14+ cells isolated from Donor 3 with 0.45% unspecific binding of the secondary antibody
Table S1.
DAVID functional annotation list of A. butzleri-induced miRNAs and potentially influenced pathways
| miRNA | Pathway | Count | p value | Genes | List total |
|---|---|---|---|---|---|
| 3613-5p | Glycerolipid metabolism | 4 | 0.012 | ALDH7A1, AKR1B1, GPAM, ALDH9A1 | 56 |
| Ascorbate and aldarate metabolism | 3 | 0.014 | ALDH7A1, UGT2B15, ALDH9A1 | 56 | |
| p53 signaling pathway | 4 | 0.037 | CDK6, RRM2B, ATR, SESN3 | 56 | |
| Tryptophan metabolism | 3 | 0.069 | ALDH7A1, CYP1B1, ALDH9A1 | 56 | |
| Pyruvate metabolism | 3 | 0.069 | ALDH7A1, AKR1B1, ALDH9A1 | 56 | |
| Steroid hormone biosynthesis | 3 | 0.088 | CYP1B1, CYP7A1, UGT2B15 | 56 | |
| 2116-3p | Wnt signaling pathway | 21 | 0.003 | FZD8, DVL3, WNT10B, ROCK1, ROCK2, NLK, CAMK2G, CSNK1A1L, CHP2, PPP3R2, FZD7, PRKCB, WNT2, MAP3K7, SENP2, EP300, PRICKLE2, NFAT5, FRAT2, WNT7A, NFATC3 | 352 |
| B cell receptor signaling pathway | 12 | 0.013 | MAPK1, CR2, PIK3CB, SOS2, NFAT5, CHP2, PPP3R2, VAV2, NFATC3, PIK3R1, PRKCB, BTK | 352 | |
| p53 signaling pathway | 11 | 0.017 | EI24, CD82, RRM2, SERPINE1, CASP8, RCHY1, RRM2B, MDM4, PERP, CDK2, SESN3 | 352 | |
| ErbB signaling pathway | 12 | 0.036 | MAPK1, CDKN1B, EREG, PIK3CB, CAMK2G, BTC, CBL, GAB1, SOS2, ELK1, PIK3R1, PRKCB | 352 | |
| Melanogenesis | 13 | 0.039 | DVL3, FZD8, ADCY1, WNT10B, ADCY2, CAMK2G, CREB1, FZD7, PRKCB, WNT2, MAPK1, EP300, WNT7A | 352 | |
| VEGF signaling pathway | 10 | 0.072 | MAPK1, PIK3CB, NFAT5, CHP2, MAPKAPK3, PPP3R2, PLA2G2D, NFATC3, PIK3R1, PRKCB | 352 | |
| Apoptosis | 11 | 0.076 | IRAK2, IRAK1, IRAK3, PRKAR2A, DFFA, PIK3CB, CASP8, CHP2, EXOG, PPP3R2, PIK3R1 | 352 | |
| Neurotrophin signaling pathway | 14 | 0.084 | IRAK2, IRAK1, PIK3CB, CAMK2G, IRAK3, MAPK1, YWHAG, PRDM4, NTRK2, SOS2, GAB1, SH2B3, MAPK7, PIK3R1 | 352 | |
| Prostate cancer | 11 | 0.085 | FGFR1, MAPK1, CDKN1B, EP300, PIK3CB, CREB1, SOS2, NKX3-1, CREB5, CDK2, PIK3R1 | 352 | |
| Fc epsilon RI signaling pathway | 10 | 0.087 | MAPK1, GAB2, PIK3CB, SOS2, VAV2, PRKCE, PLA2G2D, PIK3R1, PRKCB, BTK | 352 | |
| Long-term potentiation | 9 | 0.094 | MAPK1, ADCY1, EP300, GRIN2B, CAMK2G, PPP1R1A, CHP2, PPP3R2, PRKCB | 352 | |
| Pathways in cancer | 30 | 0.097 | FGFR1, FGF16, EGLN1, WNT2, PAX8, CASP8, SOS2, NKX3-1, RALA, PIK3R1, FZD8, DVL3, WNT10B, EPAS1, PIK3CB, CBL, SKP2, FGF23, MECOM, DAPK2, CDK2, FZD7, CTNNA2, PRKCB, MAPK1, CDKN1B, EP300, ITGA6, PIAS2, WNT7A | 352 | |
| 671-3p | MAPK signaling pathway | 5 | 0.063 | RASGRF1, FGF11, CACNB3, FGF1, CRK | 30 |
| 30d-3p | Wnt signaling pathway | 19 | 0.011 | FZD8, TBL1XR1, DVL3, VANGL1, ROCK2, CAMK2G, SMAD2, FZD4, FZD6, MAP3K7, CSNK2A1, CSNK1E, CACYBP, FRAT1, MAPK8, SIAH1, PRKACB, PLCB1, APC | 340 |
| Prion diseases | 7 | 0.026 | C8A, EGR1, NCAM2, IL1B, HSPA5, PRKACB, PRNP | 340 | |
| Insulin signaling pathway | 16 | 0.034 | IRS2, PIK3CB, PHKB, PRKAB2, PRKCI, MKNK1, PPP1CB, SORBS1, SOS1, PRKAR1A, SOS2, MAPK8, PRKAA2, PRKACB, CRK, AKT3 | 340 | |
| Renal cell carcinoma | 10 | 0.041 | EPAS1, PIK3CB, SOS1, GAB1, SOS2, TGFA, EGLN1, TCEB1, CRK, AKT3 | 340 | |
| Colorectal cancer | 11 | 0.051 | FZD8, DVL3, PIK3CB, SOS1, SOS2, MAPK8, SMAD2, FZD4, AKT3, FZD6, APC | 340 | |
| Alanine, aspartate, and glutamate metabolism | 6 | 0.052 | ADSS, GOT1, GFPT1, GLS, GAD1, PPAT | 340 | |
| ErbB signaling pathway | 11 | 0.062 | CDKN1B, ERBB4, PIK3CB, CAMK2G, SOS1, GAB1, SOS2, TGFA, MAPK8, CRK, AKT3 | 340 | |
| Endocytosis | 19 | 0.063 | SH3GL3, DNM3, FLT1, ERBB4, STAM2, VTA1, PRKCI, ARF6, HLA-B, KIT, ZFYVE20, RAB31, AP2B1, RAB-11FIP2, CHMP1B, RAB22A, RAB11B, VPS36, RNF41 | 340 | |
| Melanogenesis | 12 | 0.064 | FZD8, DVL3, GNAI3, GNAQ, CAMK2G, CREB1, CREB3L3, KIT, PRKACB, PLCB1, FZD4, FZD6 | 340 | |
| Regulation of actin cytoskeleton | 21 | 0.079 | GNA13, FGF7, PIK3CB, ROCK2, DIAPH2, SSH2, GNA12, ITGA1, ACTN2, PPP1CB, NCKAP1, DOCK1, CHRM2, TIAM1, SOS1, SOS2, WASL, CRK, FGF2, MYLK, APC | 340 | |
| Neuroactive ligand–receptor interaction | 24 | 0.085 | F2RL2, GABRG1, PTGER2, TACR3, RXFP1, GABRB3, CCKBR, OPRK1, GLRA3, LEPR, NPY2R, GRIN2A, P2RY13, PRLR, CHRM2, NMUR1, CNR1, HTR7, NPFFR2, ADRA2C, GLP2R, HTR2C, GABRQ, GABRP | 340 | |
| One carbon pool by folate | 4 | 0.086 | MTHFD2, MTHFR, DHFR, MTHFD1L | 340 | |
| 30d-5p | Limonene and pinene degradation | 3 | 0.033 | ALDH2, LCLAT1, YOD1 | 106 |
| Ether lipid metabolism | 4 | 0.035 | PLA2G12A, LCLAT1, PLA2G2C, PAFAH1B2 | 106 | |
| Natural killer cell mediated cytotoxicity | 7 | 0.056 | CASP3, TNFRSF10B, NFAT5, PPP3CA, SH2D1B, SH3BP2, LCP2 | 106 | |
| ABC transporters | 4 | 0.062 | ABCC9, ABCG5, ABCD2, ABCC4 | 106 | |
| Endocytosis | 8 | 0.085 | NEDD4, ACAP2, RAB11A, EEA1, NEDD4L, KIT, ARAP2, CHMP2B | 106 | |
| Amyotrophic lateral sclerosis (ALS) | 4 | 0.096 | CASP3, MAP3K5, DERL1, PPP3CA | 106 | |
| MAPK signaling pathway | 10 | 0.099 | MAP3K7, CASP3, MAP3K5, TAOK1, MAP3K2, PLA2G12A, NF1, PLA2G2C, PPP3CA, FGF20 | 106 | |
| 629-5p | Wnt signaling pathway | 11 | 0.003 | PLCB3, TCF7, SFRP2, VANGL2, NFAT5, LRP6, FZD3, SMAD2, MAPK10, NFATC2, MYC | 120 |
| ErbB signaling pathway | 8 | 0.004 | NRG4, GRB2, GAB1, MAPK10, MAP2K7, ABL2, MYC, AKT3 | 120 | |
| Colorectal cancer | 7 | 0.013 | TCF7, GRB2, FZD3, SMAD2, MAPK10, MYC, AKT3 | 120 | |
| Proteasome | 5 | 0.023 | PSMA2, PSMB10, PSMD13, PSMB2, PSME4 | 120 | |
| Neurotrophin signaling pathway | 8 | 0.025 | YWHAZ, GRB2, GAB1, MAPK10, FOXO3, MAP2K7, AKT3, CALM1 | 120 | |
| Endometrial cancer | 5 | 0.033 | TCF7, GRB2, FOXO3, MYC, AKT3 | 120 | |
| Insulin signaling pathway | 8 | 0.038 | PRKAR2A, PTPRF, TSC1, GRB2, MAPK10, INSR, AKT3, CALM1 | 120 | |
| Acute myeloid leukemia | 5 | 0.046 | TCF7, GRB2, MYC, STAT3, AKT3 | 120 | |
| Sphingolipid metabolism | 4 | 0.062 | SGMS2, KDSR, CERK, GAL3ST1 | 120 | |
| Adipocytokine signaling pathway | 5 | 0.071 | MAPK10, ADIPOQ, STAT3, AKT3, ACSL6 | 120 | |
| Tight junction | 7 | 0.093 | CLDN8, RAB3B, MAGI2, MYH11, CLDN2, TJP2, AKT3 | 120 |
Acknowledgements
We are grateful to Barbara Kutz-Lohroff for excellent technical assistance. This work was funded by the German Research Foundation (DFG) through SFB852.
References
- 1.Alam M, O’Neill LA: MicroRNAs and the resolution phase of inflammation in macrophages. Eur J Immunol 41, 2482–2485 (2011) [DOI] [PubMed] [Google Scholar]
- 2.Broderick JA, Zamore PD: MicroRNA therapeutics. Gene Ther 18, 1104–1110 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eulalio A, Schulte L, Vogel J: The mammalian microRNA response to bacterial infections. RNA Biol 9, 742–750 (2012) [DOI] [PubMed] [Google Scholar]
- 4.Staedel C, Darfeuille F: MicroRNAs and bacterial infection. Cell Microbiol 15, 1496–1507 (2013) [DOI] [PubMed] [Google Scholar]
- 5.Siddle KJ, Tailleux L, Deschamps M, Loh YH, Deluen C, Gicquel B, Antoniewski C, Barreiro LB, Farinelli L, Quintana-Murci L: Bacterial infection drives the expression dynamics of microRNAs and their isomiRs. PLoS Genet 11, e1005064 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Figueras MJ, Levican A, Pujol I, Ballester F, Rabada Quilez MJ, Gomez-Bertomeu F: A severe case of persistent diarrhoea associated with Arcobacter cryaerophilus but attributed to Campylobacter sp. and a review of the clinical incidence of Arcobacter spp. New Microb New Infect 2, 31–37 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prouzet-Mauléon V, Labadi L, Bouges N, Ménard A, Mégraud F: Arcobacter butzleri: underestimated enteropathogen. Emerg Infect Dis 12, 307–309 (2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van den Abeele AM, Vogelaers D, Van Hende J, Houf K: Prevalence of Arcobacter species among humans, Belgium, 2008–2013. Emerg Infect Dis 20, 1731–1734 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vandenberg O, Dediste A, Houf K, Ibekwem S, Souayah H, Cadranel S, et al. : Arcobacter species in Humans. Emerg Infect Dis 10, 1863-1867 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.zur Bruegge J, Hanisch C, Einspanier R, Alter T, Gölz G, Sharbati S: Arcobacter butzleri induces a pro-inflammatory response in THP-1 derived macrophages and has limited ability for intracellular survival. Int J Med Microbiol 304, 1209–1211 (2014) [DOI] [PubMed] [Google Scholar]
- 11.Keller A, Leidinger P, Steinmeyer F, Stähler C, Franke A, Hemmrich-Stanisak G, Ruprecht K: Comprehensive analysis of microRNA profiles in multiple sclerosis including next-generation sequencing. Mult Scler 20, 295–303 (2013) [DOI] [PubMed] [Google Scholar]
- 12.Leidinger P, Backes C, Deutscher S, Schmitt K, Mueller SC, Frese K, Haas J, Ruprecht K, Paul F, Stähler C, Lang CJ, Meder B, Bartfai T, Meese E, Keller A: A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol 14, R78 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N: miR Deep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 40, 37 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vejnar CE, Zdobnov EM: MiRmap: comprehensive prediction of microRNA target repression strength. Nucleic Acids Res 40, 11673–11683 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang DW, Sherman BT, Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc 4, 44–57 (2009) [DOI] [PubMed] [Google Scholar]
- 16.Huang DW, Sherman BT, Lempicki RA: Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharbati-Tehrani S, Kutz-Lohroff B, Bergbauer R, Scholven J, Einspanier R: miR-Q: a novel quantitative RT-PCR approach for the expression profiling of small RNA molecules such as miRNAs in a complex sample. BMC Mol Biol 9, 34 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfaffl W: A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29, e45 (2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Neill LA, Sheedy FJ, McCoy CE: MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol 11, 163–175 (2011) [DOI] [PubMed] [Google Scholar]
- 20.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D: Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol 10, 111–122 (2010) [DOI] [PubMed] [Google Scholar]
- 21.Taganov KD, Boldin MP, Chang KJ, Baltimore D: NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA 103, 12481–12486 (2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoeke L, Sharbati J, Pawar K, Keller A, Einspanier R, Sharbati S: Intestinal Salmonella typhimurium infection leads to miR-29a induced caveolin 2 regulation. PLoS ONE 8, e67300 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharbati J, Lewin A, Kutz-Lohroff B, Kamal E, Einspanier R, Sharbati S: Integrated microRNA-mRNA-analysis of human monocyte derived macrophages upon Mycobacterium avium subsp. hominissuis infection. PLoS ONE 6, e20258 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Zhan Z, Xu L, Ma F, Li D, Guo Z, Li N, Cao X: MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIα. J Immunol 185, 7244–7251 (2010) [DOI] [PubMed] [Google Scholar]
- 25.Maudet C, Mano M, Eulalio A: MicroRNAs in the interaction between host and bacterial pathogens. FEBS Lett 588, 4140–4147 (2014) [DOI] [PubMed] [Google Scholar]
- 26.Nahid MA, Yao B, Dominguez-Gutierrez PR, Kesavalu L, Satoh M, Chan EK: Regulation of TLR2-mediated tolerance and cross-tolerance through IRAK4 modulation by miR-132 and miR-212. J Immunol 190, 1250–1263 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharbati S, Sharbati J, Hoeke L, Bohmer M, Einspanier R: Quantification and accurate normalisation of small RNAs through new custom RT-qPCR arrays demonstrates Salmonella-induced microRNAs in human monocytes. BMC Genomics 13, 23 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu SC, Yang JC, Rau CS, Chen YC, Lu TH, Lin MW, Tzeng SL, Wu YC, Wu CJ, Hsieh CH: Profiling circulating microRNA expression in experimental sepsis using cecal ligation and puncture. PLoS ONE 8, e77936 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaakoush NO, Deshpande NP, Man SM, Burgos-Portugal JA, Khattak FA, Raftery MJ, Wilkins MR, Mitchell HM: Transcriptomic and proteomic analyses reveal key innate immune signatures in the host response to the gastrointestinal pathogen Campylobacter concisus. Infect Immun 83, 832–845 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Navarre WW, Zychlinsky A: Pathogen-induced apoptosis of macrophages: a common end for different pathogenic strategies. Cell Microbiol. 2, 265–273 (2000) [DOI] [PubMed] [Google Scholar]
- 31.Jovanovic M, Hengartner MO: miRNAs and apoptosis: RNAs to die for. Oncogene 25, 6176–6187 (2006) [DOI] [PubMed] [Google Scholar]


