Abstract
The pathogen Leptospira interrogans is a highly motile spirochete that causes acute and fulminant infections in humans and other accidental hosts. Hematogenous dissemination is important for infection by the pathogen but remains poorly understood because few animal model studies have used sensitive tools to quantify the bacteria. We evaluated the kinetics of leptospiral infection in Golden Syrian hamsters by a sensitive quantitative real-time PCR (TaqMan) with lipl32 as the target gene. The dissemination and bacterial burden were measured after intraperitoneal infection with a high dose (108) or low dose (2.5 × 102) of leptospires. We also examined the conjunctival challenge route to mimic the natural history of infection. Quantification of leptospires in perfused animals revealed that pathogens were detected in all organs of intraperitoneally infected hamsters, including the eye and brain, within 1 h after inoculation of 108 virulent L. interrogans bacteria. Peaks of 105 to 108 leptospires per gram or per milliliter were achieved in blood and all tissues between day 4 and day 8 after intraperitoneal inoculation of high- and low-dose challenges, respectively, coinciding with macroscopic and histological changes. The conjunctival route resulted in a delay in the time to peak organ burden in comparison to intraperitoneal infection, indicating that although infection could be established, penetration efficiency was low across this epithelial barrier. Surprisingly, infection with a large inoculum of high-passage-number attenuated L. interrogans strains resulted in dissemination to all organs in the first 4 days postinfection, albeit with a lower burden, followed by clearance from the blood and organs 7 days postinfection and survival of all animals. These results demonstrate that leptospiral dissemination and tissue invasion occur. In contrast, development of a critical level of tissue burden and pathology are dependent on the virulence of the infecting strain.
INTRODUCTION
Leptospirosis is a neglected life-threatening disease occurring in a wide range of epidemiological settings with a higher incidence in low-income, tropical countries (1–3). The causative agents are a unique group of spirochetes divided into 10 pathogenic Leptospira species and >200 serovars (1, 2, 4, 5). Leptospires are able to establish acute disease in susceptible hosts and chronic carriage in the proximal kidney tubules of reservoir hosts and persist for weeks to months in the environment after excretion (1, 2, 6, 7). Leptospirosis is among the most important bacterial zoonoses worldwide. It causes substantial morbidity and mortality in diverse human populations exposed to the wide range of wild and domestic reservoir hosts living in close proximity to anthropogenically modified environments (3, 8).
Human infection most often results from contact with an environment contaminated with the urine of animals with chronic or acute infection. A broad spectrum of clinical manifestations may result, ranging from a self-limited febrile illness to potentially fatal infection characterized by liver dysfunction, bleeding, kidney failure, shock, and pulmonary hemorrhage (8, 9). In areas of endemicity, acute leptospirosis can account for more than 10% of hospitalizations for acute febrile illness (2), with leptospirosis outbreaks predictably occurring after periods of heavy rainfall and flooding (10). The major public health burden of leptospirosis involves its ability to cause severe clinical outcomes, with mortality rates varying from 10 to 70% among recognized cases (2, 3, 8).
Leptospires are highly motile spirochetes that penetrate abraded skin and mucous membranes and cross tissue barriers to disseminate hematogenously, resulting in a systemic infection (2, 5, 8, 11, 12). After dissemination, leptospiremia persists until the onset of the host immune response, which occurs within 2 weeks after exposure (13). Leptospires can be detected in the bloodstream within minutes after intraperitoneal (i.p.) inoculation (14) and are present in visceral organs 2 days later (13, 15–19), reaching a range of 106 to 107 organisms per milliliter of blood or per gram of tissue of patients and animals (16, 19, 20). In 1957 and 1964, Faine (17) and Green and Arean (18) studied the kinetics of the leptospiral infection using dark-field microscopy and culture, respectively. The advent of molecular biology techniques, such as real-time PCR, allows a new level of sensitivity and precision for these studies.
The development of leptospirosis and disease progression are influenced by the susceptibility of the host, the virulence of the infecting strain, and the initial inoculum dose for infection (8). In animal model studies, higher inoculum doses have resulted in shorter incubation periods and decreased survival in a dose-dependent manner (13, 21). Nonetheless, many questions remain regarding the correlation of the bacterial burden in tissues with the natural history of the disease (16, 20, 22).
Animal model studies are essential to understanding the biology, transmission, colonization, and pathogenesis of Leptospira spp. The Golden Syrian hamster is commonly employed as a model for acute leptospirosis due to its high susceptibility to leptospiral infection and because the clinical features mimic those of severe human infection (23). Although experimental inoculation is typically performed by intraperitoneal inoculation (24, 25), this route does not reflect natural transmission of the pathogen. Relatively few studies have examined challenge routes that mimic natural entry of leptospires via the skin or mucous membranes (15, 19, 26, 27). Although quantitative-PCR assays have been extensively evaluated as a diagnostic tool, few studies have applied the technique to pathogenesis studies in animal models of leptospirosis. In this study, we compared the kinetics of leptospiral infection in the hamster after high and low inoculum doses of distinct L. interrogans strains by intraperitoneal and ocular infection using a sensitive TaqMan quantitative real-time PCR (qPCR) assay.
MATERIALS AND METHODS
Bacterial strains and clones.
Leptospires were cultivated in liquid Ellinghausen-McCullough-Johnson-Harris (EMJH) medium (28, 29) supplemented with 1% rabbit serum. The cultures were kept up to 7 days at 30°C, reaching a log phase between 4 and 5 days of culture. Bacteria were counted in a Petroff-Hausser counting chamber (Fisher Scientific). Leptospira interrogans serovar Copenhageni strain Fiocruz L1-130, a virulent clinical isolate from Brazil (10, 30) with 4 and 8 passages in vivo and in vitro, respectively, was used for the spiking and kinetics experiments. Pathogenesis studies involved an attenuated Fiocruz L1-130 strain obtained after 42 in vitro passages (high passage) and L. interrogans serovar Canicola strain Kito attenuated by disruption of the clpB gene, as previously described (31).
Standard-curve and spiking experiments.
DNA from L. interrogans serovar Copenhageni strain Fiocruz L1-130 was extracted using the QIAamp DNA minikit (Qiagen, Valencia, CA). The DNA was quantified using the NanoDrop instrument (Thermo Fisher Scientific Inc., Waltham, MA). A genome size of 4.6 Mb was used to determine the genomic equivalent (GEq) concentration per microliter of the purified DNA (30). To generate a standard curve, serial dilutions of the DNA were made starting at 1 × 107 GEq to 1 × 100 GEq/5 μl. Standard-curve assays were performed in duplicate. The analytic sensitivity of the assay was calculated, and the lower limit of detection (LLOD) was determined to be the concentration at which 95% or more of the replicated reactions yielded a positive result.
Leptospires were added to 1 ml of water or EDTA-anticoagulated whole blood from uninfected hamsters to achieve a final concentration of 1 × 107 leptospires/ml. After spiking, serial 10-fold dilutions of 1 × 106 to 1 × 100 leptospires/ml were then performed using blood or water as the diluent. For tissues, kidneys and livers were acquired from uninfected hamsters. Fifty microliters of serial 10-fold dilutions of leptospires was spiked into 25 mg of kidney and liver before DNA extraction to achieve concentrations of 1 × 107 to 1 × 100 leptospires per gram of tissue. After spiking, the tissues were processed for DNA extraction as described below. Unspiked water and tissues were used as negative controls.
Hamster infection and necropsy.
All experiments were performed using 5- to 8-week-old male Syrian Golden hamsters. For the kinetics experiments with the Copenhageni strain, two groups of 15 animals each were inoculated intraperitoneally (i.p.) with either a high dose (108 leptospires) or a low dose (2.5 × 102 leptospires) in 1 ml of EMJH medium. For animals infected with a high-dose inoculum, three animals were euthanized at 1 h, 1 day, 3 days, and 4 days after infection. For the animals infected with a low-dose inoculum, three animals were euthanized at 3 days, 5 days, 8 days, and 11 days after infection. For each dose, one group of 3 animals remained as a positive control. As negative controls, two groups of 3 animals were injected intraperitoneally with 1 ml of EMJH medium and euthanized at either 4 or 11 days after injection for the high- and low-dose-inoculum experiments, respectively.
For the pathogenesis experiments, one group of six animals for each strain was inoculated intraperitoneally with 108 leptospires in 1 ml of EMJH medium. At 1 h and 4 days postinfection, subgroups of two animals were euthanized. The conjunctival (CJ) challenge route experiments were performed by centrifugation of 30-ml cultures of leptospires for 10 min at 3,000 × g to obtain an inoculum of 108 leptospires in 10 μl of EMJH medium, which was applied to the conjunctival membrane of the left eye. Animals in groups of four were infected, with two animals euthanized after 7 days of infection for each strain tested. For each experiment, the two remaining animals were included as positive controls for infection and were not euthanized until an endpoint was achieved.
All the animals were monitored twice daily for endpoints, including signs of disease and death, up to 30 days postinfection. Surviving animals after 30 days postinfection or moribund animals presenting with difficulty moving or breathing or signs of bleeding or seizure were immediately sacrificed by inhalation of CO2. Blood was collected by intracardiac puncture. Each hamster was perfused with 100 ml of saline solution to eliminate blood in the organs that might contribute to the quantitative-PCR signal. After perfusion, the heart, right pulmonary lobe, right dorsocaudal hepatic lobe, spleen, right kidney, and muscle of the right thigh were carefully removed. The brain was exposed by craniotomy, and the right eye was enucleated. All the tissues were collected into cryotubes and immediately placed into liquid nitrogen before being stored at −80°C until extraction. Sera were obtained by centrifugation of clotted blood at 1,000 × g for 15 min at room temperature and frozen at −20°C until analysis for the presence of antibodies against leptospires using the microscopic agglutination test (MAT), performed as previously described (10, 32). For the pathogenesis and conjunctival-route experiments, only blood, kidney, liver, lung, spleen, and eye were analyzed. All animal protocols were approved by the Committee for the Use of Experimental Animals of the Gonçalo Moniz Research Center, Fundação Oswaldo Cruz (Fiocruz), and Yale University.
DNA extraction.
Using scissors and scalpels, 200 μl of water or blood; 25 mg of heart, lung, liver, kidney cortex, muscle, brain, and eye; and 10 mg of spleen were aseptically collected. DNA was extracted using a DNeasy blood and tissue kit (Qiagen), following the manufacturer's instructions but using only a 100-μl elution volume. Previous results showed that there was inhibition of amplification of the DNA from the blood of the hamster obtained with the Qiagen kit (data not shown). For that reason, DNA was extracted from 200 μl blood using the Maxwell 16 Tissue DNA purification kit (Promega Corporation, Madison, WI), following the manufacturer's instructions and using a 200-μl elution volume.
Quantitative real-time PCR analysis.
The concentration of leptospires was quantified by a TaqMan-based quantitative-PCR assay using an ABI 7500 system (Thermo Fisher Scientific, Inc.) and Platinum Quantitative PCR SuperMix-UDG (Thermo Fisher Scientific, Inc.) (33). The lipL32 gene was amplified using a set of primers described previously (33, 34), LipL32-45F (5′-AAG CAT TAC CGC TTG TGG TG-3′) and LipL32-286R (5′-GAA CTC CCA TTT CAG CGA TT-3′), which amplify a fragment of 242 bp that was detected by the probe, LipL32-189P (6-carboxyfluorescein [FAM]-5′-AA AGC CAG GAC AAG CGC CG-3′-black hole quencher 1 [BHQ1]). The bacterial quantification was calculated based on the standard curve described above and was expressed as the number of leptospires per milliliter or gram of tissue. For that calculation, the amount of sample (200 μl of water or blood, 10 mg of spleen, and 25 mg of other tissues), the elution volume (100 μl), and the volume of template DNA used in the PCR assay (5 μl) were taken into consideration. To calculate the theoretical limit of the assay, we used the following formula: LLOD of the assay × elution factor × volume factor, where the elution factor is equal to 20 (100 μl/5 μl) and the volume factors for water/blood and liver/kidney are 5 (1,000 ml/200 μl) and 40 (1,000 g/25 mg), respectively.
As a control for PCR inhibitors and to monitor nucleic acid extraction efficiency, spiked and experimental specimens were tested for the presence of a hamster glyceraldehyde-3-phosphate dehydrogenase (GAPDH) housekeeping gene. The primer pair and probe were designed using Primer Express version 1.3 (Thermo Fisher Scientific, Inc.). The forward primer, GAPDH_F (5′-GGT GGA GCC AAG AGG GTC AT-3′), and the reverse primer, GAPDH_R (5′-GGT TCA CAC CCA TCA CAA ACA T-3′), were selected to amplify a fragment that was detected by the probe, GAPDH_P (FAM-5′-ATC TCC GCA CCT TCT GCT GAT GCC-3′-BHQ1). A sample with a threshold cycle (Ct) value of 19 ± 2 was considered positive and further analyzed by real-time PCR targeting lipL32. In the case of a negative sample, a new DNA extraction was performed.
The final PCR mixture for both sets of primers and probes was 10 μM each primer, 5 μM the specific probe, and 5 μl of DNA in a total volume of 25 μl. There was no addition of MgCl2 or passive reference dye. The amplification protocol consisted of 2 min at 50°C and 10 min at 95°C, followed by 45 cycles of amplification (95°C for 15s and 60°C for 1 min). A negative result was assigned when no amplification occurred or if the Ct was greater than 40 (34). A no-template control (NTC) that contained all the reagents described above but no DNA was also included to detect the presence of contaminating DNA. For each organ, the DNA was extracted from one sample and the real-time PCR was performed in duplicate. Considering the amount of tissue that was used for DNA extraction, an equation was applied to express the results as the number of leptospires per gram of tissue or per milliliter of blood or water.
Histopathology and immunohistochemistry studies.
Kidney, liver, and lung tissue sections were embedded in paraffin. Three- to 5-μm sections were stained with hematoxylin and eosin (H&E) and examined by light microscopy (Zeiss Axioskop), using previous standardized protocols for each organ. A semiquantitative analysis was made, ranking the presence of each parameter as negative (0), discrete (1), mild (2), or intense (3). The pathologist viewed the histopathological preparations without knowing the infection status of the animals. The following parameters were analyzed: hemorrhage, calcification, edema, atrophy, dilatation, necrosis, inflammation, hyaline degeneration, hydropic degeneration, and the presence of cylinders for kidney; hydropic degeneration, septation, apoptosis, fibrosis, hyaline degeneration, infiltrating mononuclear or polymorphonuclear cells, necrosis, steatosis, and loss of parenchymal architecture for liver; and alveolar and bronchial hemorrhage, bronchial hemorrhage, necrosis, edema, fibrosis, pneumonia, alveolar congestion, and septal thickening for lung.
Immunohistochemistry was performed to confirm the presence of Leptospira in tissue sections. Paraffin was removed from the tissue sections with xylene and ethanol. Slides were blocked by incubation with phosphate-buffered saline (PBS)-10% milk at room temperature for 20 min, followed by the addition of a 1:1,000 dilution of rabbit polyclonal antiserum to LipL32 or L. interrogans serovar Icterohaemorrhagiae strain RGA or normal rabbit serum (as a negative control) for 1 h at room temperature. The slides were treated with 0.3% hydrogen peroxide for 15 min at room temperature and then incubated at room temperature for 30 min with biotin-conjugated anti-rabbit secondary antibodies, followed by streptavidin peroxidase (Histostain-Bulk kit; Invitrogen). Enzyme reactions were developed using 3,3′-diaminobenzidine (Sigma-Aldrich, St. Louis, MO). The slides were examined by optical microscopy (Zeiss Axioskop).
Statistical analysis.
GraphPad Prism 5.0c (GraphPad Software, San Diego, CA) was used to perform all the statistical analysis. Fisher's exact test and analysis of variance (ANOVA) were applied to assess statistical differences between pairs of groups and multiple groups, respectively. A P value of <0.05 was considered significant.
RESULTS
Standard-curve and spiking experiments.
Three independent qPCR assays were performed on duplicate serial dilutions of genomic DNA isolated from L. interrogans serovar Copenhageni strain Fiocruz L1-130. In our experiments, the LLOD, at which 95% of the tested samples were positive, was 10 GEq, with a linear correlation coefficient (R2) of 0.998 (data not shown). Serial dilutions of the same batch of genomic DNA were used for subsequent qPCR experiments.
We were able to determine the theoretical limit of detection of our assay. With an LLOD of 10 GEq, leptospires (in 5 μl), water, and blood would have a theoretical limit of 1 × 103 leptospires/ml, and kidney and liver would have a theoretical limit of 8 × 103 leptospires/g. Water and blood spiked with leptospires showed an LLOD of 1 × 102 leptospires/ml, whereas kidney and liver had an LLOD of 1 × 104 leptospires/g (see Fig. S1 in the supplemental material). These data indicate that the assay has better performance with liquids than with solid tissues (see Fig. S1 in the supplemental material). Water and tissues from control animals that were used as donors for the spiking experiments were negative by qPCR (data not shown). The gapdh real-time PCR results showed amplification in all samples tested, and all the NTC samples were negative.
Kinetics of dissemination of hamsters infected with different dose inocula.
Consistent with previous results (24), the mean 50% lethal dose (LD50) for strain Fiocruz L1-130 was 45.9 leptospires in hamster when using i.p. injection (data not shown). Doses as high as 108 leptospires are normally used for experimental infection, and the low dose used here was chosen to be approximately 5 times the LD50. Pilot experiments were performed to determine the optimal time points for sample collection during infection. Time point selection also took into account the time to death after infection and the first time point at which a detectable number of leptospires per gram of tissue was achieved.
To control for the possibility of discrepant quantification results when collecting tissue samples from different parts of the same specimen, tissues were sampled from three different sites in the same organ (kidney, liver, and lung) from animals infected at either high or low doses. No site-specific differences were observed in any tissue tested (see Table S1 in the supplemental material). These data show that leptospires are distributed equally throughout these organs, indicating that a single DNA extraction yields quantification results applicable to the entire organ. Furthermore, there were no differences in the results between the right and left kidneys (data not shown).
Hamsters infected i.p. with 108 leptospires (high dose) were euthanized 1 h and 1, 3, and 4 days after infection. The positive-control animals died 5 to 6 days postinfection. Hamsters infected i.p. with 2.5 × 102 leptospires (low dose) were euthanized at time points 3, 5, 8, and 11 days after infection, while the positive-control animals died 9 to 11 days postinfection. Although there was a delay in the detection of leptospires in tissues after a low infection dose compared to a high infection dose, the number of organisms increased exponentially in tissues after 8 days, reaching high levels of tissue burden equivalent to those observed in animals infected at a high dose (Fig. 1).
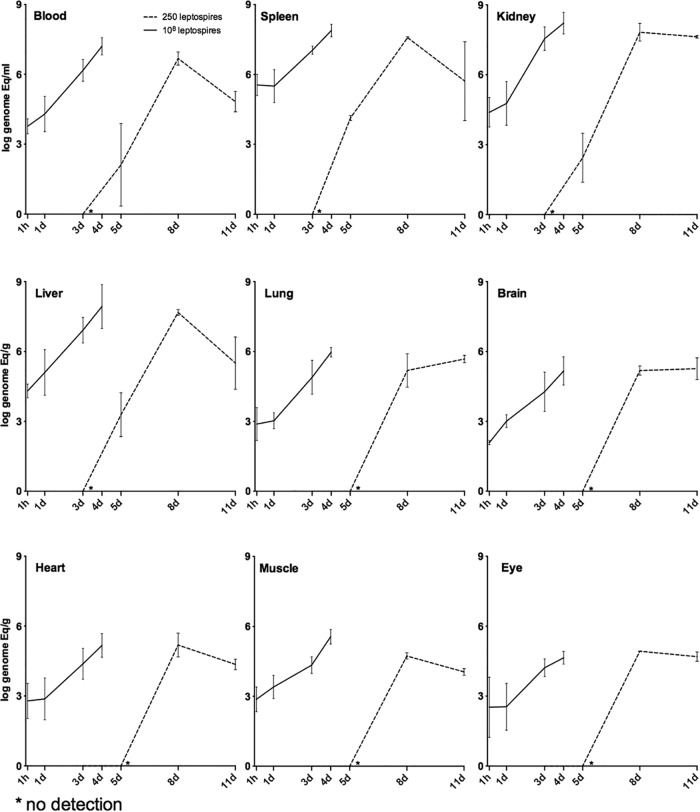
FIG 1.
Kinetics of leptospirosis dissemination in tissues taken from hamsters infected i.p. with 108 and 2.5 × 102 leptospires of the strain Fiocruz L1-130. The bacterial load for each tissue was calculated based on the mean result for three perfused hamsters for each time point. Each line represents the mean result for the bacterial load (logarithmic scale) of three independent experiments. The error bars represent the standard deviations. Animals infected with 108 leptospires (solid lines) were analyzed at 1 h and 1, 3, and 4 days postinfection. Animals infected with 2.5 × 102 leptospires (dashed lines) were analyzed at 3, 5, 8, and 11 days postinfection.
After high-dose infection, all the tissues analyzed were positive after 1 h for the presence of Leptospira, with a range of 1.2 × 102 to 7.6 × 102 leptospires/g for brain, heart, eye, muscle, and lung (Fig. 1) and 5.8 × 103 to 2.4 × 105 leptospires/g for blood, liver, and kidney (Fig. 1). The spleen was the organ that showed the highest number of leptospires (3.5 × 105) per gram after 1 h of infection (Fig. 1). We observed a statistically significant difference in the bacterial burden in all tissues between the first and the third day postinfection (P < 0.005). The bacterial burden continued to increase in vivo, as demonstrated by the observation that after 4 days, kidney, liver, and spleen reached levels of 1.65 × 108, 8.5 × 107, and 7.72 × 107 leptospires/g, respectively, with a difference of 4 log units between the first and last time points (Fig. 1). The eye had the lowest tissue load, 2 × 104 leptospires/g, and also the smallest differences among time points. Blood was found to contain 1.6 × 107 leptospires/ml, and the burden for the other tissues on the last day sampled was between 1.4 × 105 and 9.3 × 105 leptospires/g, with no decrease in burden.
After a low infection dose, no bacteria were detected in any tissues 3 days postinfection, indicating that after low-dose inoculation, a few days are required before the bacteria can reach a detectable number of organisms in any tissue. At 5 days after infection, 1.3 × 102 leptospires/ml were detected in blood, with positive detection also in the kidney, liver, and spleen (2.74 × 102, 1.94 × 103, and 1.36 × 104 leptospires/g, respectively) (Fig. 1). After 8 days of infection, all the tissues were positive, with a burden ranging from 5.4 × 104 leptospires/g for muscle to 6.7 × 107 leptospires/g in the kidney, with a statistical difference between the day 5 and day 8 time points in all tissues (P < 0.0001) (Fig. 1). On day 11 all the analyzed tissues except lung and brain exhibited a decrease in the number of detected leptospires compared to day 8, which was not a statistically significant difference (P = 0.084) (Fig. 1). In tissues such as blood, liver, and spleen, the marked decrease in burden could be associated with the appearance of agglutinating antibodies, as 50% of the animals had serum MAT titers ranging from 1:25 to 1:100. No animals infected with a high dose of leptospires developed antibodies detectable by MAT.
Necropsy and histopathology analyses.
At necropsy, animals inoculated with high and low doses of leptospires showed clinical signs and macroscopic lesions 3 and 8 days postinfection, respectively, culminating in death 2 to 3 days later. The most common clinical signs observed were anorexia, depression, and hemorrhage just before death. In terms of gross pathology observed at necropsy, animals showed a small amount of jaundice in the liver and abdominal muscles, enlargement of the spleen, and hemorrhage of the kidneys. Localized hemorrhage of the lungs was the most prominent feature observed at necropsy (Fig. 2) and was most likely related to infection rather than CO2 inhalation, considering that negative-control animals did not have these lesions (data not shown). Kidney, liver, and lung histopathology revealed no differences between animals at 1 and 3 days postinfection with high-dose inoculum and 5 and 8 days postinfection with low-dose inoculum.
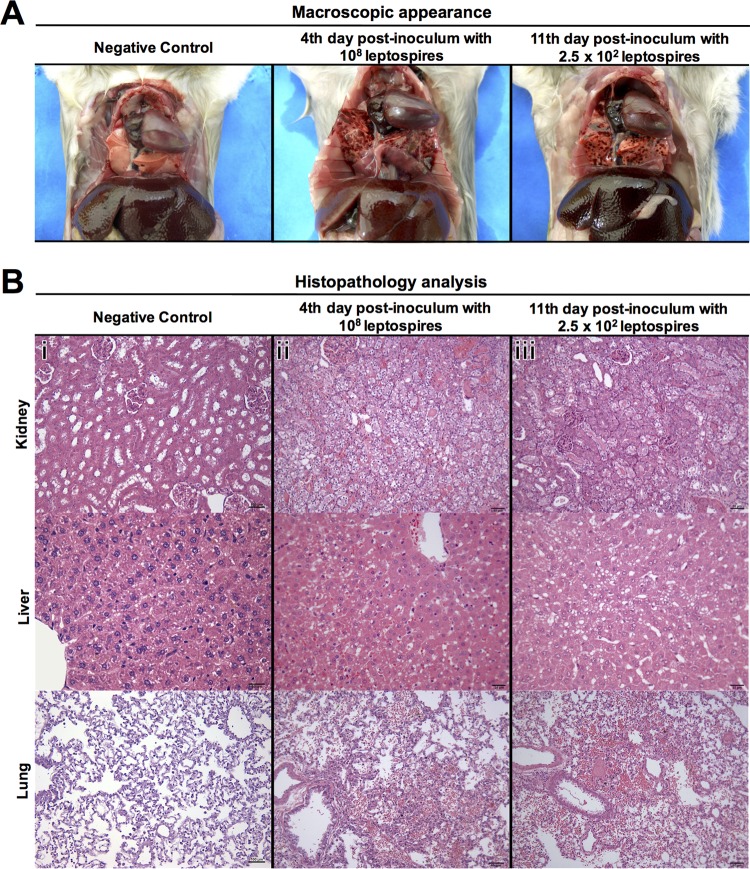
FIG 2.
Pathology of hamsters infected intraperitoneally with strain Fiocruz L1-130. (A) Representative photographs of gross examination of a negative-control hamster and hamsters infected with 108 leptospires (high dose) or 2.5 × 102 leptospires (low dose) at days 4 and 11 postchallenge, respectively. Infected hamsters had evident localized hemorrhage of the lungs. (B) Histopathology analysis showing representative photomicrographs of hematoxylin- and eosin-stained sections of kidney, liver, and lung of a negative-control animal (i) and animals infected with a high dose (ii) or a low dose (iii) of leptospires at days 4 and 11 postchallenge, respectively. The histopathology photomicrographs were taken at a magnification of ×400. Infected animals had similar histopathological features, with kidneys showing mild hyaline degeneration and hemorrhage, livers with mild loss of parenchymal architecture and steatosis, and lungs with hemorrhage. Bars, 100 μm for tissues of negative-control animal and 50 μm for tissues of animals infected with a high or a low dose, with exception of liver, which has a scale bar of 25 μm.
Analysis of hematoxylin- and eosin-stained sections of the kidney, liver, and lung revealed similar histopathological features of leptospirosis at the time of maximal tissue damage in animals infected with high or low doses of Leptospira (Fig. 2). In the kidney, discrete hemorrhage, inflammation, and mild hyaline degeneration were observed in animals at the last two time points for both doses. In the liver, animals infected with lower doses showed discrete hydropic degeneration and infiltrating mononuclear cells. In animals infected with either the high- or low-dose inoculum, we also observed discrete hepatic necrosis, mild steatosis, and intense loss of parenchymal architecture. In the lungs, mild to intense alveolar hemorrhage and discrete bronchial hemorrhage were observed in all the animals at later stages of infection at a high or low dose (Fig. 2 and Table 1). The localization of leptospires in tissues, as determined by immunohistochemistry, was found to be intercellular in the liver, in the kidney interstitium and tubules, and in the alveolar septa of the lungs (Fig. 3); 70% of the lung sections analyzed were negative for leptospires.
TABLE 1.
Analysis of major histopathological alterations in hamsters infected with 108 or 2.5 × 102 leptospires according to days postinfection for each dose of inoculum
| Tissue and histopathology | Resulta |
|||||||
|---|---|---|---|---|---|---|---|---|
| 108 leptospires i.p. |
2.5 × 102 leptospires i.p. |
|||||||
| Control | Day postinfection |
Control | Day postinfection |
|||||
| 1 | 3 | 4 | 5 | 8 | 12 | |||
| Kidney | ||||||||
| Hemorrhage | 0, 0, 0 | 0, 0, 0 | 1, 1, 0 | 0, 0, 0 | 1, 1, 1 | 1, 0, 2 | 1, 0, 1 | 0, 0, 0 |
| Inflammation | 0, 0, 0 | 0, 0, 0 | 0, 0, 0 | 1, 0, 0 | 0, 0, 0 | 0, 0, 0 | 0, 0, 1 | 0, 0, 1 |
| Hyaline degeneration | 0, 2, 1 | 1, 1, 0 | 0, 1, 1 | 1, 1, 1 | 1, 1, 1 | 2, 2, 2 | 2, 3, 3 | 2, 2, 1 |
| Hydropic degeneration | 0, 0, 0 | 0, 0, 1 | 0, 0, 0 | 1, 1, 1 | 0, 0, 0 | 0, 0, 0 | 0, 1, 1 | 3, 1, 3 |
| Cylinders | 1, 1, 1 | 0, 0, 0 | 0, 1, 1 | 1, 1, 0 | 1, 1, 0 | 1, 1, 0 | 2, 1, 2 | 1, 0, 1 |
| Liver | ||||||||
| Hydropic degeneration | 0, 0, 0 | 0, 0, 0 | 0, 0, 0 | 0, 0, 0 | 0, 0, 0 | 0, 0, 0 | 2, 2, 0 | 0, 0, 0 |
| Infiltrating mononuclear cells | 0, 0, 0 | 0, 0, 0 | 0, 0, 0 | 1, 0, 0 | 1, 0, 0 | 0, 0, 0 | 1, 0, 0 | 1, 0, 0 |
| Necrosis | 0, 0, 0 | 0, 0, 0 | 0, 0, 1 | 0, 0, 1 | 0, 0, 0 | 0, 0, 0 | 0, 0, 1 | 0, 0, 0 |
| Steatosis | 0, 0, 0 | 0, 0, 0 | 2, 1, 0 | 2, 1, 1 | 0, 0, 0 | 0, 0, 0 | 0, 1, 0 | 2, 1, 1 |
| Loss of parenchymal architecture | 0, 0, 0 | 2, 2, 1 | 0, 0, 0 | 0, 0, 0 | 0, 0, 1 | 2, 2, 2 | 2, 3, 3 | 1, 1, 1 |
| Lung | ||||||||
| Alveolar hemorrhage | 0, 0, 0 | 0, 0, 0 | 1, 2, 2 | 1, 2, 1 | 0, 0, 0 | 0, 0, 0 | 0, 2, 1 | 3, 2, 2 |
| Bronchial hemorrhage | 0, 0, 0 | 0, 0, 0 | 1, 1, 0 | 1, 1, 1 | 0, 0, 0 | 1, 0, 0 | 1, 1, 1 | 1, 1, 1 |
| Alveolar congestion | 0, 0, 0 | 0, 0, 0 | 1, 1, 0 | 2, 1, 1 | 0, 0, 0 | 0, 0, 1 | 1, 1, 1 | 2, 2, 1 |
| Septal thickening | 0, 0, 0 | 1, 1, 1 | 1, 1, 0 | 1, 1, 1 | 0, 0, 0 | 1, 1, 1 | 1, 1, 1 | 1, 1, 0 |
The results are shown as scores for 3 randomly chosen animals analyzed individually on each day. For comparison and analysis, the semiquantitative ranking was transformed into numbers: negative, 0; discrete, 1; mild, 2; and intense, 3.
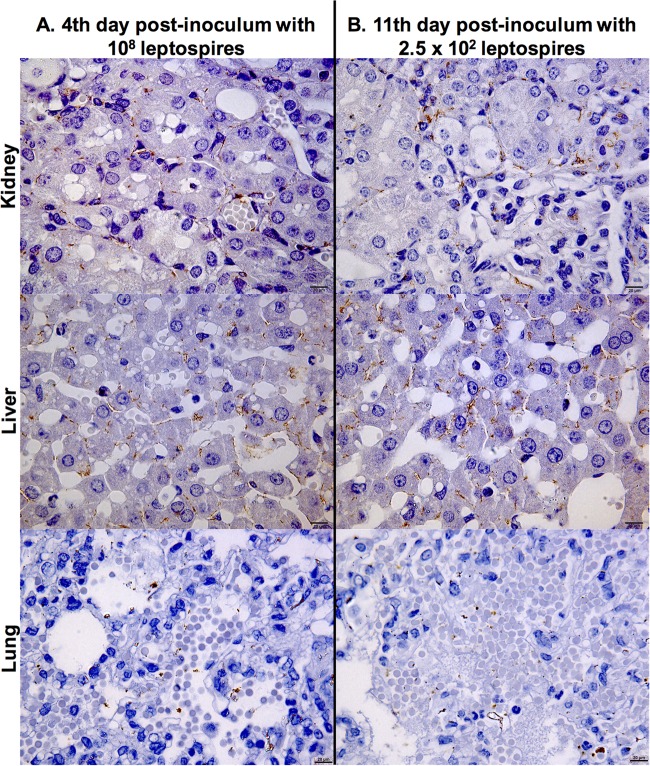
FIG 3.
Representative photomicrographs of immunohistochemically stained sections of kidney, liver, and lung from hamsters infected intraperitoneally with strain Fiocruz L1-130. Shown are stained tissues of an animal infected with 108 leptospires (high dose) at day 4 postchallenge (A) and an animal infected with 2.5 × 102 leptospires at day 11 postchallenge (B). Detection was performed with monoclonal antiserum specific for LipL32. The photomicrographs were taken at a magnification of ×1,000 and show whole leptospires and degraded cells in kidney tubules, interstitium of the liver, and alveolar septa of the lung. Bar, 20 μm.
Mimicking the natural route of infection by conjunctival instillation.
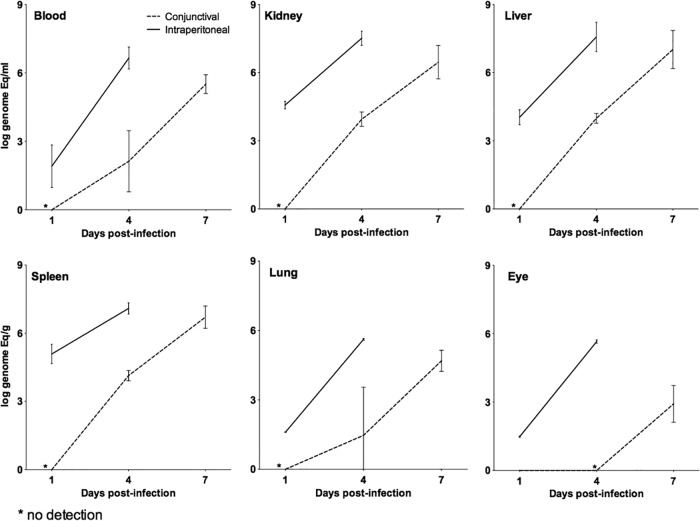
Infection kinetics in animals challenged with 108 leptospires by the CJ and i.p. routes were compared (Fig. 4). Blood, kidney, liver, spleen, lung, and eye tissues were analyzed. Both infection routes caused death of the animals, but the time to death in animals challenged by the CJ route was 3 to 4 days longer. All tissues of animals challenged by the i.p. route were positive at the 1-day time point, with burdens ranging from 3 × 101 to 3.5 × 105 leptospires/g. These numbers increased exponentially for all tissues by 4 days after infection, reaching 4.2 × 105 to 3.3 × 107 leptospires/g (Fig. 4). All these results are similar to those of the previous experiments. In contrast, leptospires were undetectable in all tissues from animals at the 1-day time point after infection by the CJ route. However, the tissue burdens at the 4- and 7-day time points were as high as 1.4 × 104 leptospires/g and 1 × 107 leptospires/g, respectively. These results indicate that, although there was a delay in the time to death of the animals infected by the i.p. route compared to those infected by the CJ route, the bacterial loads in the tissues analyzed were similar on the days prior to death for both routes. Furthermore, the leptospiral burdens in the eyes of animals infected by the CJ route were lower than those in the i.p.-challenged animals, and detection was possible only after 7 days of infection, reaching 8.3 × 102 leptospires/g (Fig. 4).
FIG 4.
Kinetics of dissemination of leptospires in tissues from a hamster infected with 108 leptospires of strain Fiocruz L1-130 comparing the i.p. and CJ routes of infection. The bacterial load for each tissue was calculated based on the mean result for two perfused hamsters for each time point. Each line represents the mean (logarithmic scale) of two independent experiments. The error bars represent the standard deviations. Animals infected by the i.p. route (straight lines) were analyzed after 1 and 4 days postinfection. Animals infected by the CJ route of infection (dashed lines) were analyzed 1, 4, and 7 days postinfection.
Bacterial burden as a marker for lethality.
The mean bacterial burden observed in each tissue for the different routes of infection (infected at a low or high dose) was used to calculate the threshold of the leptospiral burden that could be related to lethality (Table 2). Kidney, liver, and spleen showed a threshold of 2 × 107 to 3 × 107 per gram, whereas tissues like heart, lung, muscle, and brain showed a threshold of 1 × 105 to 2 × 105 leptospires/g. The threshold for blood was 3 × 106 leptospires/ml (Table 2).
TABLE 2.
Lethality and bacterial burden in hamsters
| Tissue | Avg bacterial burdena (GEq) |
|||
|---|---|---|---|---|
| Intraperitoneal |
Conjunctival (108) (day 7 p.i.) | Total avgb (log ± SD) | ||
| 108 (day 4 p.i.) | 2.5 × 102 (day 8 p.i.) | |||
| Blood | 1.6 × 107 | 5.0 × 106 | 3.16 × 105 | 3.2 × 106 (6.5 ± 0.87) |
| Spleen | 7.9 × 107 | 4.0 × 107 | 5.01 × 106 | 2.5 × 107 (7.4 ± 0.62) |
| Kidney | 1.6 × 108 | 6.3 × 107 | 3.16 × 106 | 3.2 × 107 (7.5 ± 0.88) |
| Liver | 7.9 × 107 | 5.0 × 107 | 1.00 × 107 | 3.2 × 107 (7.5 ± 0.47) |
| Lung | 1.0 × 106 | 1.6 × 105 | 5.01 × 104 | 2.0 × 105 (5.3 ± 0.65) |
| Brain | 1.6 × 105 | 1.6 × 105 | ND | 1.6 × 105 (5.2 ± 0.00) |
| Heart | 1.6 × 105 | 1.6 × 105 | ND | 1.6 × 105 (5.2 ± 0.00) |
| Muscle | 4.0 × 105 | 5.0 × 104 | ND | 1.4 × 105 (5.1 ± 0.63) |
| Eye | 5.0 × 104 | 7.9 × 104 | 7.94 × 102 | 1.6 × 104 (4.2 ± 1.10) |
The average leptospire burdens in different tissues were measured before death to calculate the bacterial-load threshold at which lethality occurs. The mean average was calculated based on the mean of the bacterial-load result for three (i.p.) and two (CJ) independent experiments. p.i., postinfection; ND, not determined.
Total average for each tissue was calculated based on the mean average of bacterial burden after intraperitoneal (high and low dose) and conjunctival infection.
Use of real-time PCR for pathogenesis studies.
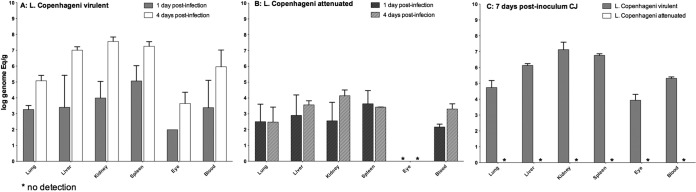
The infection kinetics of two different attenuated strains were analyzed. One of these attenuated strains was a clone of a Copenhageni strain passaged 42 times in vitro, and the other was a clpB mutant obtained by transposon mutagenesis from a virulent strain of serovar Canicola (31). In both cases, the inoculum was 108 leptospires, and samples were collected 1 and 4 days after challenge. The bacterial burden 1 day after intraperitoneal infection for the culture-attenuated Copenhageni strain was similar to that of the virulent strain (Fig. 5A and B). However, 4 days after challenge, the burdens of leptospires were statistically different between hamsters infected with the two strains (P = 0.014), with the attenuated strain showing a lower burden in all analyzed tissues (Fig. 5A and B). The results were similar for the clpB mutant and wild-type strains; after 1 day of infection, the burdens in the tissues were similar (see Fig. S2A and B in the supplemental material), while the leptospire burdens in all tissues after 4 days were significantly lower for the mutant than for the wild-type strain (see Fig. S2A and B in the supplemental material). All the control animals infected with the virulent Copenhageni and Canicola strains died after 5 to 6 days of infection, while animals infected with the attenuated strains survived. Analysis of tissues from animals that survived after 30 days revealed that only animals infected with the clpB mutant were chronically infected in their kidneys, with a bacterial burden of 108 leptospires/g with the ability to be reisolated from the kidney and retain the attenuated status upon reinfection (data not shown). Of note, for the nonvirulent clones, leptospires were not detected in the eyes of infected animals at any time point.
FIG 5.
Kinetics of the dissemination of leptospires in tissues from hamsters infected intraperitoneally and conjunctivally with 108 leptospires. Animals were infected with the virulent wild-type strain Copenhageni (A) and compared with animals infected with the attenuated clone of the strain Copenhageni obtained after 42 in vitro passages (B) using the i.p. route (A and B) and the CJ route (C). Analysis of the tissues was performed 1 and 4 days after i.p. infection (A and B) and 7-days after CJ infection (C), using the mean result for two perfused hamsters for all the strains. Each bar represents the mean (logarithmic scale) of the results of two independent experiments. The error bars represent the standard deviations.
When the attenuated strains Copenhageni (Fig. 5C) and Canicola (see Fig. S2C in the supplemental material) were inoculated via the conjunctival route, no leptospires were detected in any tissues at 7 days after challenge, whereas the parental strains were detected in every tissue. The leptospiral burden in tissues was similar to that found in previous experiments involving the conjunctival challenge route for the virulent Copenhageni strain (Fig. 4) with the control animals 8 and 9 days postinfection. All the control animals infected with the attenuated strains Canicola and Copenhageni survived, and tissues were negative 30 days after challenge. These data indicate that attenuation resulting from either passage in vitro or clpB gene disruption affected the overall ability of the cells to cause disease, either by increasing their susceptibility to the innate immune system or, most likely, by disrupting their ability to actively penetrate mucous membranes.
DISCUSSION
Pathogenic leptospires are motile, life-threatening spirochetes that readily disseminate to all tissues (1, 2). Little is known about the rates of spread to various tissues and their consequences for the pathogenesis of the disease. Recent studies have shown that different routes of infection result in changes in dissemination kinetics (15, 19). However, given the use of different animal models and methods of detection of the agent, major questions remain regarding the effect of the dose on dissemination and the relevance of this approach to understanding pathogenesis. Here, we demonstrated that the maximum tissue burden was independent of the challenge route and dose. On the other hand, the time to maximum tissue burden was dependent on both the challenge route and dose. These findings are of great importance to our understanding of the natural history of leptospirosis and its pathogenesis, as well as to leptospirosis vaccine development and testing.
No specific tissue tropism was observed during the dissemination and multiplication phases of leptospiral infection. Our results indicate that pathogenic leptospires rapidly invade the bloodstream and are subsequently distributed to all body tissues even before multiplication (Fig. 1), a finding in accordance with previous studies (35, 36). As noted in previous studies, the burden is higher in highly vascularized tissues, such as the spleen, liver, and kidney, which receive greater blood flow than other organs (18, 37). After i.p. challenge, leptospires were detected in the brains and eyes of animals infected with high and low doses by 1 h and 8 days, respectively (Fig. 1). The ability of leptospires to cross the blood brain barrier, indicated by the presence of leptospires in the brain, and other endothelial barriers formed by tight junctions is consistent with the results of previous animal and human studies (18, 38, 39).
The intraperitoneal and conjunctival challenge routes have fundamental differences. Intraperitoneal infection bypasses tissue barriers and host immune defenses, particularly after high challenge doses (40), by providing direct access of the peritoneal cavity to the bloodstream via lymphatic and thoracic ducts. In contrast, the conjunctival challenge route requires bacteria to adhere to and penetrate mucous membranes, a more natural route of leptospiral infection (41). The conjunctival route is of interest in identifying mechanisms of tissue penetration that might be targeted by leptospiral vaccines. Although the dose required to cause disease in naturally occurring infections is unknown, it is likely to be lower than the 108 dose used in this study. This difference may be explained by the expression of virulence factors occurring only in the reservoir host or in the environment that do not occur in vitro. Nevertheless, our results are similar to those of previous studies with animal models (19, 26), in which higher numbers of leptospires were needed to cause disease by conjunctival inoculation. While both the intraperitoneal (higher and lower doses) and conjunctival inoculation routes ultimately lead to overwhelming infection, the dissemination, multiplication, and spread of leptospires are delayed under conjunctival inoculation (Fig. 4). Our findings are consistent with those of a previous study (19), which showed that leptospires inoculated by the conjunctival route caused systemic infection detectable within 4 days after challenge, with a bacterial burden similar to but occurring later than that in animals infected by the intraperitoneal route. Additional investigations are needed to examine different mucous membrane routes of infection, the ability of leptospires to penetrate intact and abraded skin, and environmental factors contributing to mucous membrane penetration.
We demonstrated here that the inoculation dose is an important factor contributing to the kinetics of infection and disease severity. Intraperitoneal injection using a high dose (108) of leptospires resulted in dissemination to all tissues, including the eye and brain, at levels ranging from 102 to 105 leptospires/g within 1 h after infection. In contrast, intraperitoneal injection with a lower inoculum dose (2.5 × 102) was associated with longer times to leptospiral detection in tissues (Fig. 1) and clinical-disease development. Animal studies with strain Fiocruz L1-130 found an inverse correlation between the dose of infection and the time to death (24). Similarly, the natural history of the disease in humans varies from an asymptomatic form to acute and severe disease (1, 8, 10). In addition, factors such as the dose, serovar, and genetics of the strain or host may contribute to the outcome (8). Our findings suggest that the inoculum dose is an important factor that contributes to the outcome and severity of disease, particularly in areas where a single serovar is the cause of the majority of the cases identified (10).
Manifestations of disease are related to the tissue bacterial burden. The relationship between the leptospiral tissue burden, lesion formation, and death, in which the first appearance of macroscopic hemorrhage in the animals coincided with a defined leptospiral burden, has been described previously (8). Another study found that the time of death coincided with a higher leptospiral tissue burden (42). However, regardless of the inoculum size and the length of the course of infection, we found that the threshold burdens of leptospires in all tissues, above which macroscopic lesions appeared and animals were near death, were comparable. In both cases, this disease threshold of leptospiral burden was over 107 leptospires/g in tissues such as kidney, liver, spleen, and blood (Fig. 1). Similar to what was found in other studies (15, 19), our final bacterial burdens before death (Fig. 1 and 4 and Table 2) were similar among the different doses and routes of infection. Furthermore, there were no major differences between the doses and routes of infection when comparing the macroscopic and histopathological changes in different tissues (Fig. 2). Although our calculated threshold for lethality was as high as 106 leptospires/ml in blood and 107 leptospires/g in kidney, liver, and spleen, our findings corroborate those of previous studies in human patients with clinical leptospirosis (16, 20, 43), which showed that a threshold burden of 104 to 106 leptospires/ml in the blood was associated with poor outcomes.
Attenuated mutants of pathogenic L. interrogans can disseminate in tissues but cannot multiply or cause the death of infected animals after a high inoculum dose. An important finding of our study was that after 1 day of infection with a high intraperitoneal dose of two different attenuated strains there were no statistical differences between the abilities of virulent and attenuated organisms to penetrate and spread from the peritoneal cavity to the bloodstream. In contrast, attenuated organisms were found at lower levels in multiple organs by day 4 after injection and had been almost completely cleared from blood and organs by day 30. A previous study (19) in guinea pigs showed no dissemination of the same in vitro-attenuated strain Fiocruz L1-130 in guinea pigs, indicating that the assay used in our study had greater sensitivity. Dissemination and rapid clearance of attenuated organisms had been noted by Faine in studies published in 1957 (13). Our findings corroborate these results, suggesting that a critical threshold number of leptospires in tissues is required to cause lesions and death in experimental animals. Moreover, that threshold has to be reached before the host immune system can prevent the establishment of severe disease. Interestingly, while both mutants were unable to actively penetrate mucous membranes or cause death, only the clpB mutant was able to establish renal colonization, suggesting a fundamental difference between the mutants. Our data validate the use of this assay for the identification and characterization of mutants that have lost the ability to penetrate, disseminate, and/or multiply in tissues. Furthermore, our data suggest that it is possible to avoid using high numbers of animals to study the kinetics of dissemination, because blood kinetics alone, which can be determined with relatively few animals per group, can serve as a surrogate for the entire dissemination process.
In summary, our results demonstrate that both pathogenic and attenuated leptospires rapidly disseminate within 1 hour after challenge, and this early dissemination is independent of virulence. Regardless of the inoculum dose and the challenge route, the burden in target tissues is more important for pathogenesis than the ability of the pathogen to disseminate. However, the burden of infection is correlated with disease outcomes, as has been shown in humans (16, 20, 43). The quantitative real-time PCR assay, which we have standardized and described here, may be useful in tracking mutant strains during the infection process to identify genes involved in penetration, dissemination, and multiplication in the host. Furthermore, this method can be used to verify the effectiveness of vaccines in preventing dissemination and colonization. Regardless of the application, our results and findings indicate that this approach is an important tool for the study of leptospirosis, an important yet neglected disease, the biology and pathogenesis of which remain to be fully unveiled and understood.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (grants R01 AI052473, R01 AI034431, U01 AI0088752, R25 TW009338, R01 TW009504, and D43 TW00919) and the National Council of Scientific and Technological Development (CNPq), Brazilian Ministry of Science and Technology.
We are thankful to Amie Shei for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00094-16.
REFERENCES
- 1.Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, Levett PN, Gilman RH, Willig MR, Gotuzzo E, Vinetz JM, Peru-United States Leptospirosis Consortium . 2003. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis 3:757–771. doi: 10.1016/S1473-3099(03)00830-2. [DOI] [PubMed] [Google Scholar]
- 2.McBride AJ, Athanazio DA, Reis MG, Ko AI. 2005. Leptospirosis. Curr Opin Infect Dis 18:376–386. doi: 10.1097/01.qco.0000178824.05715.2c. [DOI] [PubMed] [Google Scholar]
- 3.Costa F, Hagan JE, Calcagno J, Kane M, Torgerson P, Martinez-Silveira MS, Stein C, Abela-Ridder B, Ko AI. 2015. Global morbidity and mortality of leptospirosis: a systematic review. PLoS Negl Trop Dis 9:e0003898. doi: 10.1371/journal.pntd.0003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourhy P, Collet L, Brisse S, Picardeau M. 2014. Leptospira mayottensis sp. nov., a pathogenic species of the genus Leptospira isolated from humans. Int J Syst Evol Microbiol 64:4061–4067. doi: 10.1099/ijs.0.066597-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adler B. 2015. History of leptospirosis and leptospira. Curr Top Microbiol Immunol 387:1–9. doi: 10.1007/978-3-662-45059-8_1. [DOI] [PubMed] [Google Scholar]
- 6.Mwachui MA, Crump L, Hartskeerl R, Zinsstag J, Hattendorf J. 2015. Environmental and behavioural determinants of leptospirosis transmission: a systematic review. PLoS Negl Trop Dis 9:e0003843. doi: 10.1371/journal.pntd.0003843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pappas G, Papadimitriou P, Siozopoulou V, Christou L, Akritidis N. 2008. The globalization of leptospirosis: worldwide incidence trends. Int J Infect Dis 12:351–357. doi: 10.1016/j.ijid.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Ko AI, Goarant C, Picardeau M. 2009. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol 7:736–747. doi: 10.1038/nrmicro2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trevejo RT, Rigau-Perez JG, Ashford DA, McClure EM, Jarquin-Gonzalez C, Amador JJ, de los Reyes JO, Gonzalez A, Zaki SR, Shieh WJ, McLean RG, Nasci RS, Weyant RS, Bolin CA, Bragg SL, Perkins BA, Spiegel RA. 1998. Epidemic leptospirosis associated with pulmonary hemorrhage—Nicaragua, 1995. J Infect Dis 178:1457–1463. doi: 10.1086/314424. [DOI] [PubMed] [Google Scholar]
- 10.Ko AI, Galvao Reis M, Ribeiro Dourado CM, Johnson WD Jr, Riley LW. 1999. Urban epidemic of severe leptospirosis in Brazil. Salvador Leptospirosis Study Group. Lancet 354:820–825. [DOI] [PubMed] [Google Scholar]
- 11.Inada R, Ido Y, Hoki R, Kaneko R, Ito H. 1916. The etiology, mode of infection, and specific therapy of Weil's disease (spirochaetosis icterohaemorrhagica). J Exp Med 23:377–402. doi: 10.1084/jem.23.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barocchi MA, Ko AI, Reis MG, McDonald KL, Riley LW. 2002. Rapid translocation of polarized MDCK cell monolayers by Leptospira interrogans, an invasive but nonintracellular pathogen. Infect Immun 70:6926–6932. doi: 10.1128/IAI.70.12.6926-6932.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faine S. 1957. Virulence in Leptospira. I. Reactions of guinea-pigs to experimental infection with Leptospira icterohaemorrhagiae. Br J Exp Pathol 38:1–7. [PMC free article] [PubMed] [Google Scholar]
- 14.Levett PN. 2001. Leptospirosis. Clin Microbiol Rev 14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coutinho ML, Matsunaga J, Wang LC, de la Pena Moctezuma A, Lewis MS, Babbitt JT, Aleixo JA, Haake DA. 2014. Kinetics of Leptospira interrogans infection in hamsters after intradermal and subcutaneous challenge. PLoS Negl Trop Dis 8:e3307. doi: 10.1371/journal.pntd.0003307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segura ER, Ganoza CA, Campos K, Ricaldi JN, Torres S, Silva H, Cespedes MJ, Matthias MA, Swancutt MA, Lopez Linan R, Gotuzzo E, Guerra H, Gilman RH, Vinetz JM, Peru-United States Leptospirosis Consortium . 2005. Clinical spectrum of pulmonary involvement in leptospirosis in a region of endemicity, with quantification of leptospiral burden. Clin Infect Dis 40:343–351. doi: 10.1086/427110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faine S. 1957. Virulence in leptospira. II. The growth in vivo of virulent Leptospira icterohaemorrhagiae. Br J Exp Pathol 38:8–14. [PMC free article] [PubMed] [Google Scholar]
- 18.Green JH, Arean VM. 1964. Virulence and distribution of Leptospira icterohaemorrhagiae in experimental guinea pig infections. Am J Vet Res 25:264–267. [PubMed] [Google Scholar]
- 19.Lourdault K, Aviat F, Picardeau M. 2009. Use of quantitative real-time PCR for studying the dissemination of Leptospira interrogans in the guinea pig infection model of leptospirosis. J Med Microbiol 58:648–655. doi: 10.1099/jmm.0.008169-0. [DOI] [PubMed] [Google Scholar]
- 20.Truccolo J, Serais O, Merien F, Perolat P. 2001. Following the course of human leptospirosis: evidence of a critical threshold for the vital prognosis using a quantitative PCR assay. FEMS Microbiol Lett 204:317–321. doi: 10.1111/j.1574-6968.2001.tb10904.x. [DOI] [PubMed] [Google Scholar]
- 21.Adler B, Faine S. 1976. Susceptibility of mice treated with cyclophosphamide to lethal infection with Leptospira interrogans serovar pomona. Infect Immun 14:703–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agampodi SB, Matthias MA, Moreno AC, Vinetz JM. 2012. Utility of quantitative polymerase chain reaction in leptospirosis diagnosis: association of level of leptospiremia and clinical manifestations in Sri Lanka. Clin Infect Dis 54:1249–1255. doi: 10.1093/cid/cis035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haake DA. 2006. Hamster model of leptospirosis. Curr Protoc Microbiol Chapter 12:Unit 12E.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva EF, Santos CS, Athanazio DA, Seyffert N, Seixas FK, Cerqueira GM, Fagundes MQ, Brod CS, Reis MG, Dellagostin OA, Ko AI. 2008. Characterization of virulence of Leptospira isolates in a hamster model. Vaccine 26:3892–3896. doi: 10.1016/j.vaccine.2008.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marinho M, Oliveira IS Jr, Monteiro CM, Perri SH, Salomao R. 2009. Pulmonary disease in hamsters infected with Leptospira interrogans: histopathologic findings and cytokine mRNA expressions. Am J Trop Med Hyg 80:832–836. [PubMed] [Google Scholar]
- 26.Bolin CA, Alt DP. 2001. Use of a monovalent leptospiral vaccine to prevent renal colonization and urinary shedding in cattle exposed to Leptospira borgpetersenii serovar hardjo. Am J Vet Res 62:995–1000. doi: 10.2460/ajvr.2001.62.995. [DOI] [PubMed] [Google Scholar]
- 27.Truccolo J, Charavay F, Merien F, Perolat P. 2002. Quantitative PCR assay to evaluate ampicillin, ofloxacin, and doxycycline for treatment of experimental leptospirosis. Antimicrob Agents Chemother 46:848–853. doi: 10.1128/AAC.46.3.848-853.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellinghausen HC Jr, McCullough WG. 1965. Nutrition of Leptospira pomona and growth of 13 other serotypes: a serum-free medium employing oleic albumin complex. Am J Vet Res 26:39–44. [PubMed] [Google Scholar]
- 29.Johnson RC, Harris VG. 1967. Differentiation of pathogenic and saprophytic letospires. I. Growth at low temperatures. J Bacteriol 94:27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nascimento AL, Ko AI, Martins EA, Monteiro-Vitorello CB, Ho PL, Haake DA, Verjovski-Almeida S, Hartskeerl RA, Marques MV, Oliveira MC, Menck CF, Leite LC, Carrer H, Coutinho LL, Degrave WM, Dellagostin OA, El-Dorry H, Ferro ES, Ferro MI, Furlan LR, Gamberini M, Giglioti EA, Goes-Neto A, Goldman GH, Goldman MH, Harakava R, Jeronimo SM, Junqueira-de-Azevedo IL, Kimura ET, Kuramae EE, Lemos EG, Lemos MV, Marino CL, Nunes LR, de Oliveira RC, Pereira GG, Reis MS, Schriefer A, Siqueira WJ, Sommer P, Tsai SM, Simpson AJ, Ferro JA, Camargo LE, Kitajima JP, Setubal JC, Van Sluys MA. 2004. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J Bacteriol 186:2164–2172. doi: 10.1128/JB.186.7.2164-2172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lourdault K, Cerqueira GM, Wunder EA Jr, Picardeau M. 2011. Inactivation of clpB in the pathogen Leptospira interrogans reduces virulence and resistance to stress conditions. Infect Immun 79:3711–3717. doi: 10.1128/IAI.05168-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faine S, Adler B, Bolin C, Perolat P (ed). 1999. Leptospira and leptospirosis. MediSci, Melbourne, Australia. [Google Scholar]
- 33.Stoddard RA. 2013. Detection of pathogenic Leptospira spp. through real-time PCR (qPCR) targeting the LipL32 gene. Methods Mol Biol 943:257–266. doi: 10.1007/978-1-60327-353-4_17. [DOI] [PubMed] [Google Scholar]
- 34.Stoddard RA, Gee JE, Wilkins PP, McCaustland K, Hoffmaster AR. 2009. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn Microbiol Infect Dis 64:247–255. doi: 10.1016/j.diagmicrobio.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 35.Thompson JC, Manktelow BW. 1989. Pathogenesis of renal lesions in haemoglobinaemic and non-haemoglobinaemic leptospirosis. J Comp Pathol 101:201–214. doi: 10.1016/0021-9975(89)90066-2. [DOI] [PubMed] [Google Scholar]
- 36.Athanazio DA, Silva EF, Santos CS, Rocha GM, Vannier-Santos MA, McBride AJ, Ko AI, Reis MG. 2008. Rattus norvegicus as a model for persistent renal colonization by pathogenic Leptospira interrogans. Acta Trop 105:176–180. doi: 10.1016/j.actatropica.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 37.Gulati OP, Ponard G. 1980. Cardiac output and regional blood flow studies in golden hamsters. Experientia 36:984–985. doi: 10.1007/BF01953835. [DOI] [PubMed] [Google Scholar]
- 38.Brown PD, Carrington DG, Gravekamp C, van de Kemp H, Edwards CN, Jones SR, Prussia PR, Garriques S, Terpstra WJ, Levett PN. 2003. Direct detection of leptospiral material in human postmortem samples. Res Microbiol 154:581–586. doi: 10.1016/S0923-2508(03)00166-9. [DOI] [PubMed] [Google Scholar]
- 39.Romero EC, Billerbeck AE, Lando VS, Camargo ED, Souza CC, Yasuda PH. 1998. Detection of Leptospira DNA in patients with aseptic meningitis by PCR. J Clin Microbiol 36:1453–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ratet G, Veyrier FJ, Fanton d'Andon M, Kammerscheit X, Nicola MA, Picardeau M, Boneca IG, Werts C. 2014. Live imaging of bioluminescent Leptospira interrogans in mice reveals renal colonization as a stealth escape from the blood defenses and antibiotics. PLoS Negl Trop Dis 8:e3359. doi: 10.1371/journal.pntd.0003359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evangelista KV, Coburn J. 2010. Leptospira as an emerging pathogen: a review of its biology, pathogenesis and host immune responses. Future Microbiol 5:1413–1425. doi: 10.2217/fmb.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van den Ingh TS, Hartman EG. 1986. Pathology of acute Leptospira interrogans serotype icterohaemorrhagiae infection in the Syrian hamster. Vet Microbiol 12:367–376. doi: 10.1016/0378-1135(86)90086-6. [DOI] [PubMed] [Google Scholar]
- 43.Hochedez P, Theodose R, Olive C, Bourhy P, Hurtrel G, Vignier N, Mehdaoui H, Valentino R, Martinez R, Delord JM, Herrmann C, Lamaury I, Cesaire R, Picardeau M, Cabie A. 2015. Factors associated with severe leptospirosis, Martinique, 2010-2013. Emerg Infect Dis 21:2221–2224. doi: 10.3201/eid2112.141099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.