Abstract
Bioluminescent reporter assays have been widely used to study the effect of Toxoplasma gondii on host gene expression. In the present study, we extend these studies by engineering novel reporter cell lines containing a gamma-activated sequence (GAS) element driving firefly luciferase (FLUC). In RAW264.7 macrophages, T. gondii type I strain (GT1) infection blocked interferon gamma (IFN-γ)-induced FLUC activity to a significantly greater extent than infection by type II (ME49) and type III (CTG) strains. Quantitative trait locus (QTL) analysis of progeny from a prior genetic cross identified a genomic region on chromosome XII that correlated with the observed strain-dependent phenotype. This QTL region contains two isoforms of the T. gondii enzyme nucleoside triphosphate hydrolase (NTPase) that were the prime candidates for mediating the observed strain-specific effect. Using reverse genetic analysis we show that deletion of NTPase I from a type I strain (RH) background restored the higher luciferase levels seen in the type II (ME49) strain. Rather than an effect on IFN-γ-dependent transcription, our data suggest that NTPase I was responsible for the strain-dependent difference in FLUC activity due to hydrolysis of ATP. We further show that NTPases I and II were not essential for tachyzoite growth in vitro or virulence in mice. Our study reveals that although T. gondii NTPases are not essential for immune evasion, they can affect ATP-dependent reporters. Importantly, this limitation was overcome using an ATP-independent Gaussia luciferase, which provides a more appropriate reporter for use with T. gondii infection studies.
INTRODUCTION
Toxoplasma gondii is an obligate intracellular protozoan parasite that can infect virtually all warm-blooded animals and humans (1). It is the cause of toxoplasmosis, a serious infectious disease in congenitally infected children and immunosuppressed individuals (2), and an important pathogen in livestock (3). Innate immunity provides a critical defense barrier against T. gondii infection and interferon gamma (IFN-γ) plays a key role in this defense (4). In IFN-γ-activated cells, the transcription factor signal transduction and activator of transcription factor 1 (STAT1) translocates to the nucleus and binds as a homodimer to gamma-activated sequence (GAS) sites in promoters of IFN-γ-responsive genes such as IFN regulatory factor 1 (IRF1), major histocompatibility complex type II (MHC-II), or immunity-related GTPases (5). IRF1 is an early response gene of the STAT1 pathway and a transcription coactivator of genes induced by IFN-γ (6). Over the past few years, multiple virulence factors of the parasite have been identified that work in concert to block effectors that have been preactivated by IFN-γ and which are upregulated by the STAT1 pathway (7). Central is the effector rhoptry protein 18, which has been identified as a major virulence determinant in the mouse by quantitative trait locus (QTL) analysis based on genetic crosses between T. gondii strains (8, 9). Virulent type I strains also contain a unique isoform of a nucleoside triphosphate hydrolase (NTPase) I, whereas NTPase II is universally present in all T. gondii strains (10). This difference has led to the speculation that NTPase I might also constitute an important virulence factor, and this possibility is supported by the fact that attempts to knock out NTPase I have been unsuccessful so far (11). The biological roles of NTPases I and II, however, still remain unknown. Interestingly, although these enzymes have only 16 amino acid differences between them (10), NTPase I preferentially hydrolyzes triphosphate nucleosides, while NTPase II hydrolyzes tri- and diphosphate nucleosides (10). Additionally, NTPase I cleaves ATP at 4.5 times the rate of NTPase II (10, 12). In vitro, it has been shown that dithiol compounds such as dithiothreitol (DTT) are essential for activation of the NTPases (13).
In cells that are infected prior to IFN-γ activation, T. gondii blocks IFN-γ-dependent gene transcription by an unknown effector mechanism (14, 15). This inhibition occurs independent of cell type in both mouse and human cells as shown by the inhibition of multiple genes that are upregulated by the STAT1 pathway such as IRF1, MHC-II, MHC transactivator (CIITA), and inducible nitric oxide synthase (i.e., the NOS2 gene) (14–16). Recently, a GAS element driven firefly luciferase (FLUC) reporter was used to measure strain-dependent inhibition in HEK293 cells treated with IFN-γ. Interestingly, no difference was detected between the three main clonal T. gondii lineage types I, II, and III (17), a finding consistent with previous transcriptional studies indicating that all three strain types suppress IFN-γ-dependent transcription (14, 15).
In the present study, we identified a strain-dependent difference using a similar GAS-driven FLUC reporter in RAW264.7 macrophages and HeLa cells. We identified the genomic locus responsible for the observed phenotype by QTL analysis of a previous genetic cross and generated disruptant mutants of the two most likely candidate genes of that locus: NTPase I and NTPase II. However, rather than being due to a transcriptional difference, our studies show that the FLUC assay is prone to artifacts due to the strong ATP hydrolysis activity of NTPases. This problem was circumvented by using a Gaussia luciferase (GLUC) that does not rely on ATP, thus confirming that all three strain types of T. gondii block IFN-γ-mediated transcription via STAT1.
MATERIALS AND METHODS
Parasite and host cell strains.
Toxoplasma gondii strains were maintained as tachyzoites by serial passage on human foreskin fibroblast (HFF) cell monolayers in regular growth media based on Dulbecco modified Eagle medium (Life Technologies, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA), 2 mM l-glutamine, 10 mM HEPES (pH 7.5), and gentamicin (20 μg/ml) at 37°C under 5% CO2. Parasites used in experiments were harvested after natural egress and separated from host cell debris by a 3-μm-pore-size nucleopore filter. T. gondii GT1 (ATCC 50853; type I), ME49 (ATCC 50611; type II), and CTG (ATCC 50842; type III) were used as representative type strains. A collection of progeny from a previous genetic cross between I (GT1SNFr) × II (ME49FUDRr) strains (18) was used for QTL mapping. RH (type I) strain expressing green fluorescent protein (GFP) (RHΔhxgprt/GFP; ATCC 50940) was used for the construction of CRISPR/Cas9 (19) guided NTPase I and II gene disruptant mutants. All parasite strains and cell lines were routinely diagnosed negative for Mycoplasma contamination using the e-Myco plus mycoplasma PCR detection kit (Boca Scientific, Boca Raton, FL).
Reporter cell line construction.
All plasmids and primers used in this study are listed in Tables S1 to S3 in the supplemental material. RAW264.7 macrophages and HeLa cells were transduced with lentiviral particles containing four tandem GAS elements (5′-AGTTTCATATTACTCTAAATC-3′) fused to a murine cytomegalovirus (mCMV) promoter driving a FLUC (pGreenFire1-GAS-EF1-puro; System Biosciences, San Francisco, CA). Permanent cell lines were established under puromycin selection and single colonies were selected for activation by IFN-γ. Gibson assembly (Gibson assembly cloning kit; NEB, Ipswich, MA) was used to construct the GLUC expression vector. In short, five tandem GAS elements were fused to a mCMV promoter that is driving GLUC. A neomycin resistance cassette was added for stable cell transfection (pGLUC-5×GAS-mCMV-neoR). After transfection into RAW264.7 macrophages with Lipofectamine LTX (Thermo Fisher Scientific, Waltham, MA), stable colonies were selected under G418 (Sigma, St. Louis, MO) and tested for activation of GLUC by IFN-γ.
FLUC and GLUC reporter assays.
Reporter cells were plated into black 96-well plates with clear bottoms (BD Biosciences, Franklin Lakes, NJ) in regular growth media. Confluent monolayers were infected in triplicate with parasites at various multiplicities of infection (MOIs). At 2 h after infection, the cells were activated with 100 U of murine and human IFN-γ/ml, respectively, for 18 h. Cells containing FLUC were washed once with phosphate-buffered saline (PBS) and lysed with either cell culture lysis reagent (Promega, Madison, WI) or dithiothreitol (DTT)-free standard lysis buffer (100 mM potassium phosphate [pH 7.8], 1 mM EDTA, 7 mM 2-mercaptoethanol, 10% [vol/vol] glycerol) (20). Cells containing GLUC were lysed with luciferase cell lysis buffer (NEB). The luminescence intensity was measured after the addition of 100 μl of firefly luciferase assay substrate (Promega) after the addition of 50 μl of Gaussia luciferase assay buffer (NEB), according to the manufacturer's instructions. Luminescence was detected on a Cytation 3 Cell Imaging Multi-Mode plate reader (Biotek, Winooski, VT), and data were normalized to the uninfected, stimulated sample. For strain comparisons, we used the mean luminescence value generated at an MOI, at which the infection rate was ∼50% as determined by a quantitative invasion assay (21) that was performed in parallel. Samples were averaged over two to three biological replicates, each with three technical replicates.
QTL analysis.
For QTL association mapping, the ability of strains to interfere with the FLUC reporter system in RAW264.7 macrophages was expressed as a ratio of the uninfected, activated control. Each strain was measured at a range of five different MOIs in triplicate using a 96-well plate format. At 2 h after infection, the cells were activated with 100 U of IFN-γ/ml overnight for 18 h, after which the cells were lysed with lysis buffer (Promega), and the luciferase activity was measured. Log(1/P) values were plotted along the genome-wide coordinates of the 1,603 single nucleotide polymorphism markers described previously (18), with genome-wide significance thresholds determined from 1,000 permutations. For fine mapping the region in the identified QTL, we isolated and characterized new progeny from a previous genetic cross (18). These new progeny clones were typed with markers on chromosome XII using previously published markers (22) and at new genomic regions that were PCR amplified and sequenced with newly designed primers (see Table S2 in the supplemental material). Single nucleotide polymorphisms within these new markers were used to genotype the progeny with respect to the parental alleles.
CRISPR/Cas9-mediated generation of NTPase disruptant mutants.
For disruption of the NTPase locus, two CRISPR/Cas9 plasmids containing specific guide RNA sequences (see Table S3 in the supplemental material) matching either NTPase I or NTPase II were constructed by Q5 polymerase PCR mutagenesis (NEB), as described previously (19). After transfection of the plasmids, together with amplicons containing either hypoxanthine, xanthine, guanosine, phosphoribosyl transferase (HXGPRT), or dihydrofolate reductase (DHFR) by electroporation, transgenic parasites were selected in mycophenolic acid (25 μg/ml) and xanthine (50 μg/ml) and 3 mM pyrimethamine, respectively. Single-cell clones were obtained by limiting dilution, and gene disruption was verified by diagnostic PCR using primers listed in Table S3 in the supplemental material. PCRs were performed using Taq DNA polymerase (Sigma). Products were analyzed by electrophoresis in agarose gels with ethidium bromide.
Western blot analysis.
PCR-positive clones were further analyzed by Western blotting to confirm the absence of NTPase protein. In brief, T. gondii tachyzoite lysates were resolved on SDS-PAGE gels and subsequently transferred to nitrocellulose. Blotted proteins were blocked in LICOR blocking buffer (LICOR Biosciences, Lincoln, NE) and detected with rabbit anti-NTPase (23) and mouse anti-GRA7 (24) primary antibodies incubated in 5% milk in PBS for 1 h. Secondary antibodies (LICOR Biosciences) conjugated to IRDye 680CW (donkey anti-rabbit IgG) and IRDye 800CW (donkey anti-mouse IgG) were used to visualize signals on an Odyssey infrared imager (LICOR Biosciences).
Plaque and mouse virulence assay.
Mice were kept in an Association for Assessment and Accreditation of Laboratory Animal Care-approved facility. The Institutional Care Committee at Washington University School of Medicine approved all experiments. Groups of five female CD1 mice were injected intraperitoneally with 200 parasites. The survival of the animals was monitored over 30 days. The animal experiment was repeated once, and the cumulative mortality of both experiments (n = 10 mice per group) was plotted as Kaplan-Meyer curves in Prism (GraphPad Software, Inc., La Jolla, CA), as previously described (7). To test for parasite viability, parasites were inoculated onto HFF cell monolayers grown in six-well plates for 7 days without movement. Subsequently, cells were fixed with 70% ethanol and stained with 0.1% crystal violet. Plates were then scanned using an Epson scanner.
Immunofluorescence assay.
To assess the inhibition of the STAT1 pathway by the wild type and by NTPase disruptant mutants, HFF cells were infected for 16 h and subsequently activated with 100 U of IFN-γ/ml for 5 h. The cells were fixed with 4% formaldehyde in PBS for 15 min, permeabilized with 0.1% Triton X-100 for 15 min at room temperature, and blocked in 10% FBS for 20 min. Subsequently, samples were incubated with anti-IRF1 (Cell Signaling, Danvers, MA), washed in PBS, and incubated with secondary goat anti-rabbit IgG conjugated to Alexa Fluor 594 dye (Invitrogen, Carlsbad, CA). After staining, the slides were mounted with Prolong Gold plus DAPI (4′,6′-diamidino-2-phenylindole; Life Technologies) and examined with a Zeiss Axioskop 2 MOT Plus microscope equipped for epifluorescence and using a 63× PlanApochromat lens (numerical aperture [N.A.] of 1.40; Carl Zeiss, Inc., Thornwood, NY). Images were acquired with an AxioCam MRm camera (Carl Zeiss) using Axiovision and processed with Gimp 2.8. The mean red fluorescence of IRF1 in host cell nuclei was determined by the use of the ROI manager in ImageJ. A region of interest (ROI) was selected over DAPI-stained host cell nuclei and the unstained background. The corrected integrated density (CTCF) representing nuclear red fluorescence was calculated by using the following equation: CTCF = integrated density – (area of nucleus × mean fluorescence of background readings). Prism v6 (GraphPad) was used for blotting the results of three biological replicates, each with 15 technical replicates per condition.
Statistics.
Statistical analyses were conducted with Prism v6. The results were compared using the Kruskal-Wallis test or two-sample Student t tests, as noted (*, P < 0.05 was considered the minimum cutoff for significance).
RESULTS
Strain-dependent interference with GAS-FLUC assay.
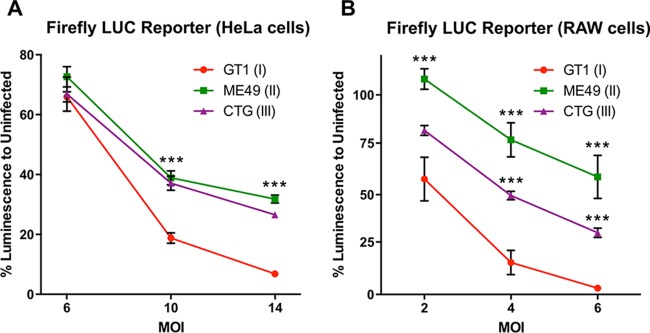
Clonal type strains of T. gondii (i.e., types I, II, and III) have previously been reported to equally interfere with the STAT1 signaling pathway and thereby block IFN-γ-dependent gene expression (15, 17). The exact mechanism of this block is still unknown despite several attempts to identify a T. gondii effector over the past 2 decades. To investigate the mechanism of STAT1 inhibition by T. gondii, we adopted a FLUC reporter system for use in HeLa cells and RAW267.4 macrophages. At a low MOI, we were unable to detect a significant strain-dependent difference in HeLa cells, a finding consistent with previous reports (17). However, at higher MOIs, the strain GT1 blocked the reporter system more efficiently than either ME49 or CTG (Fig. 1A). More dramatically, a strong strain-dependent effect was detected even at lower infection rates in RAW267.4 macrophages (Fig. 1B). These results led us to suspect that previous studies had failed to observed a strain-dependent effect either due to use of different reporter cell lines, lower MOIs, or a combination of effects.
FIG 1.
Toxoplasma gondii strain-dependent inhibition of luminescence in IFN-γ-inducible FLUC reporter lines. A tandem GAS element-driven FLUC reporter was stably expressed in HeLa cells (A) or RAW264.7 macrophages (B). Reporter cell lines were plated in 96-well plates and infected with different MOIs, as indicated. At 2 h after infection, the cells were stimulated with 100 U of IFN-γ/ml for 18 h. Subsequently, the cells were lysed, and the luciferase activity was measured. The data shown are means ± standard errors of the mean (SEM) of at least three independent experiments, each with three technical replicates. Kruskal-Wallis test with Dunn's correction (***, P < 0.0001) was performed for GT1 and each of the other T. gondii strains.
QTL analysis maps to a single locus on chromosome XII.
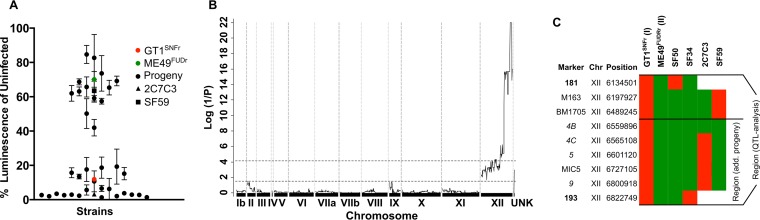
To investigate the observed strain-dependent effect further and to test whether the phenotype could be mapped to a genomic region, we took advantage of a previously established cross between GT1 and ME49 strains (18). We determined the ratio of IFN-γ-dependent FLUC activity from infected versus noninfected cells for each of 36 recombinant progeny (Fig. 2A). The parental GT1 strain strongly interfered with the FLUC signal, whereas the parental ME49 strain was only weakly inhibitory (Fig. 2A). Interestingly, almost all clones clustered close to one of the parental lineages, suggesting that a single genomic locus was responsible for the observed phenotype. We analyzed the interference of FLUC activation as a quantitative trait and mapped the locus to a region of ∼688 kb on the right side of chromosome XII (Fig. 2B). Initially, this region was bounded by recombination between markers 181 and 193, as typified by progeny SF50 and SF34 (Fig. 2B). In order to narrow down the region, we identified two new progeny from this cross (i.e., 2C7C3 and SF59) that were genotyped using a previous set of markers, as well as new genetic markers defined by direct sequencing of regions within this locus (Fig. 2C). Collectively, the crossovers defined by these progeny identify a region of ∼263 kb (TGME49_chrXII: 6559896.0.6822749) on chromosome XII containing 36 genes (http://ToxoDB.org) that was associated with the observed phenotype (Fig. 2C). Robust multiarray averaging (RMA) comparison for expression differences between GT1 and ME49 (25) across this region identified nucleoside triphosphate hydrolase (NTPase) as differentially expressed with higher levels being detected in the type I GT1 strain. Closer inspection revealed that this difference in expression occurred because of differences in gene copy number. Specially, NTPase I is only present in type I strains, while NTPase II is found in type I and other strains such as ME49 (10) (Fig. 3A). Additionally, the type I locus also contains a pseudogene that is not expressed (Fig. 3A).
FIG 2.
Mapping of a single target locus on chromosome XII. (A) GT1 (type I) and ME49 (type II) parental strains and 36 progeny were tested for their ability to interfere with the GAS-FLUC in RAW264.7 macrophages. The data shown are normalized averages of three technical replicates ± standard deviations (SD). (B and C) QTL scan of normalized percent inhibition identified a single locus of ∼688 kb (TGME49_chrXII: 6134501.0.6822749) on chromosome XII defined by the two markers 181 and 193 and the progeny SF50 and SF34. Fine mapping with additional PCR markers (see Table S2 in the supplemental material) within the QTL identified two additional informative progeny (2C7C3 and SF59) and narrowed the region responsible for the observed strain difference to ∼263 kb (TGME49_chrXII: 6559896.0.6822749) comprising 36 annotated genes.
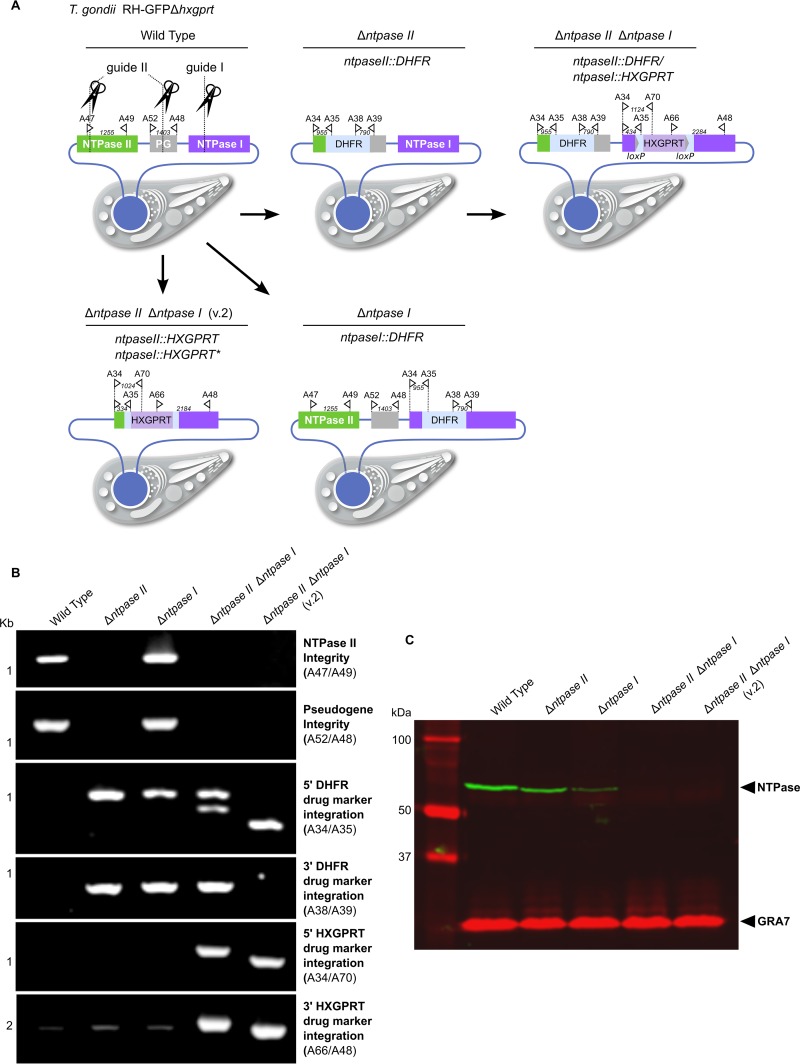
FIG 3.
Generation and verification of NTPase I and II single- and double-disruptant mutants. (A) Diagram of single NTPase II (Δntpase II; RHΔhxgprt/ntpaseII::DHFR) and NTPase I (Δntpase I; RHΔhxgprt/ntpaseI::DHFR) mutants and two versions of double-knockout mutants (Δntpase II Δntpase I, RHΔhxgprt/ntpaseII::DHFR/ntpaseI::HXGPRT; Δntpase II Δntpase I v.2, RHΔhxgprt/ntpaseII::HXGPRT/ntpaseI::HXGPRT) generated by CRISPR/Cas9 guided gene disruption. The pseudogene (PG) lacks a suitable start codon and therefore is not transcribed (30). However, it shares complete sequence homology in part with either NTPase II or I for which reason the guide RNA II targets both NTPase II and the pseudogene resulting in a double cut. The type I RH strain (Δhxgprt) was the background for all the mutants described. For the locations and sequences of guides, see Table S3 in the supplemental material. (B) Diagnostic PCR of disruptant mutants with primer pair locations as indicated in panel A. For primer locations and sequences, see Table S3 in the supplemental material. (C) Western blot analysis of protein loss in NTPase disruptant mutants. GRA7 served as loading control.
Generation of NTPase single- and double-disruptant mutants.
To verify that the NTPase locus was responsible for the mapped phenotypic difference, we used CRISPR/Cas9 technology (19) to generate single- and double-gene disruptant mutants in the type I RH strain. Initially, we used a DHFR cassette that confers resistance to pyrimethamine to isolate a gene disruptant mutant lacking either NTPase I (Δntpase I; RHΔhxgprt/ntpaseI::DHFR) or NTPase II (Δntpase II; RHΔhxgprt/ntpaseII::DHFR) (Fig. 3A). Subsequently, a double-disruptant mutant was generated using an HXGPRT resistance cassette inserted into the NTPase I gene in the Δntpase II background (Δntpase II Δntpase I; RHΔhxgprt/ntpaseII::DHFR/ntpaseI::HXGPRT). All mutants were confirmed by diagnostic PCR (Fig. 3B). To confirm the absence of protein products in the single or double gene knockouts, we used a polyclonal rabbit antibody that has been raised previously against the full-length NTPase II protein (23). Western blot data indicate that in the RH strain NTPase I is more abundant than NTPase II (Fig. 3C). We independently generated a second version (v2) double-knockout clone using HXGPRT resistance cassette as an insert (Δntpase II Δntpase I v.2, RHΔhxgprt/ntpaseII::HXGPRT/ntpaseI::HXGPRT) (Fig. 3A).
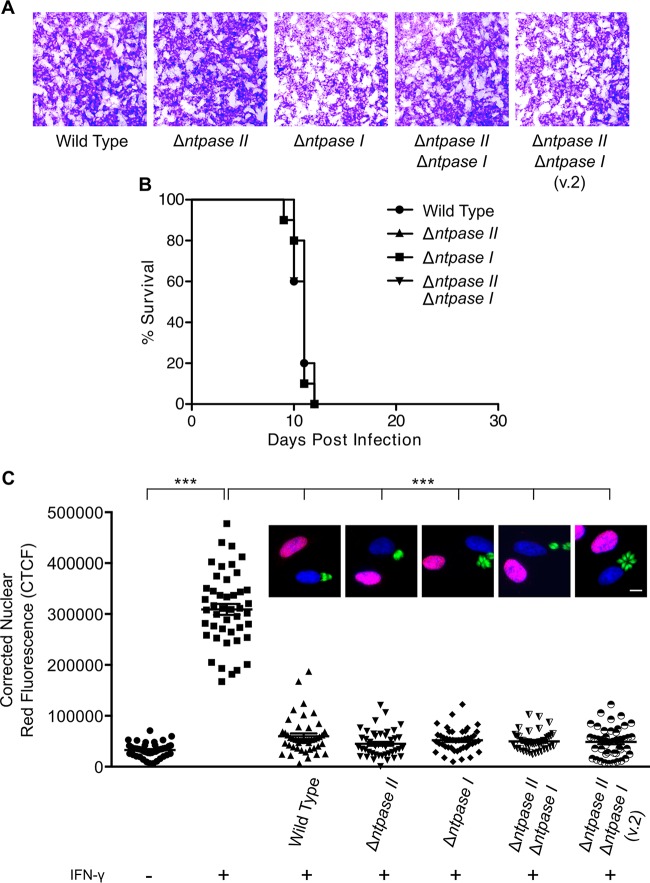
Knockouts of NTPase II and I are not essential for growth or virulence in CD1 mice and do not affect STAT1 transcription.
Since NTPase I is present in type I strains (10), it has been speculated that NTPase I is a primary virulence factor (11). Notably, we were not able to identify any growth defect in either of the single or double knockouts in vitro (Fig. 4A). To test whether the absence of NTPase I or NTPase II, as well as the disruption of both genes, would have a virulence defect in vivo, we infected CD1 mice. No significant difference in survival was identified between single and double knockouts compared to wild-type RH (Fig. 4B). To test whether NTPase I and NTPase II interfere with the STAT1 pathway, we used IRF1 as IFN-γ-inducible protein marker. Normally, IRF1 is upregulated by STAT1-mediated transcription, and it accumulates in the nucleus after treatment with IFN-γ; this pathway is blocked by prior infection with T. gondii (15). Unexpectedly, we could not identify any difference in the ability to block the IRF1 induction between wild-type RH compared to single and double NTPase disruptant mutants (Fig. 4C). This finding suggested that any potential effects of NTPases on the FLUC assay were not due to disruption of STAT1-mediated transcription but rather to some interference in the assay.
FIG 4.
Growth, virulence, and interference with STAT1 pathway. (A) Plaque assay on HFF monolayers examining growth of wild-type and NTPase knockout parasites. (B) Virulence of NTPase knockout strains in CD-1 mice infected intraperitoneally with 200 tachyzoites per mouse. Groups of five mice per strain were infected, and the survival was monitored. The experiment was repeated once, and the results of both experiments were plotted together. (C) A fluorescence assay performed to examine the interference of wild-type T. gondii with IRF1 protein expression as a readout for STAT1-activated transcription compared to NTPase knockout strains showed no NTPase-dependent effect. HFF monolayers were infected for 2 h and subsequently activated with 100 U of IFN-γ/ml for 5 h. Quantification of the IRF1 fluorescence of infected cells versus controls. Data points represent the mean red fluorescence in a ROI overlaying the host cell nucleus. IRF1, red; DAPI, blue; T. gondii-GFP, green. Scale bar, 10 μm. ***, P < 0.0001 (Kruskal-Wallis with Dunn's correction).
NTPases I and II were responsible for the ATP-dependent artifact in firefly luciferase assay.
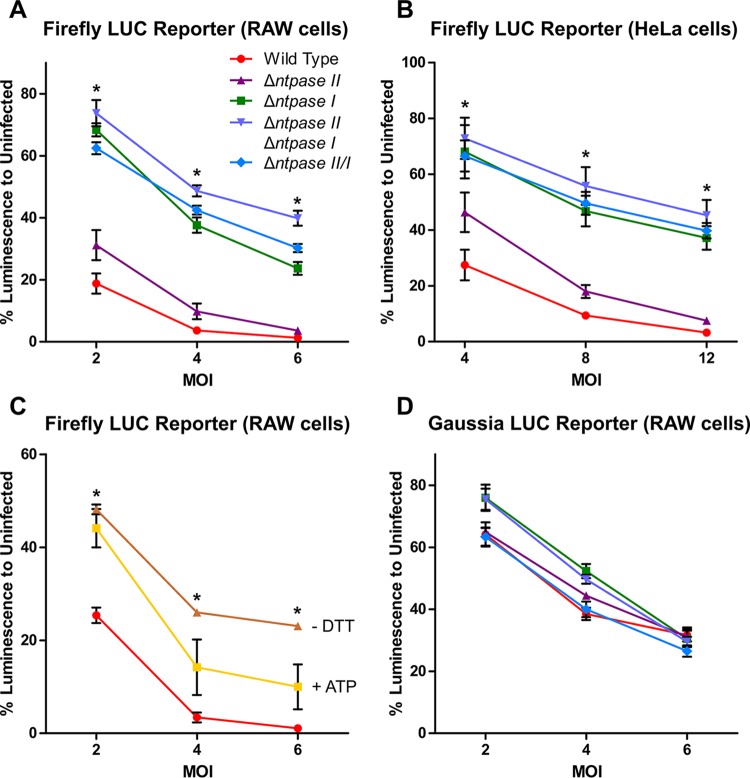
To examine the role of NTPase enzymes in luciferase assays, we compared wild-type and mutant lines in the reporter cell assays. We observed a strong negative influence of NTPase I on the FLUC activity in both HeLa cells and RAW264.7 macrophages (Fig. 5A and B). Notably, the NTPase I knockout in the type I strain mirrors the less inhibitory phenotype of type II (ME49) and III (CTG) strains, which naturally lack NTPase I (Fig. 5A and B). In contrast, deletion of NTPase II showed only a mild change in inhibition of the FLUC assay (Fig. 5A and B). Furthermore, the inhibitor effect of NTPases on the FLUC assay was partially rescued by adding 20 mM ATP during host cell lysis and by the absence of dithiol compounds, respectively (Fig. 5C). These findings suggest that the inhibition of FLUC activity is due to consumption of ATP by the NTPases.
FIG 5.
Influence of NTPases I and II on firefly and Gaussia luciferase reporter lines. IFN-γ-inducible tandem GAS element-driven FLUC reporter lines were infected with different MOIs for 3 h and subsequently stimulated with 100 U of IFN-γ/ml for 18 h. NTPase I and double-knockout mutants were significantly less capable at suppressing FLUC activity in both RAW264.7 macrophages (A) and HeLa cells (B) than the wild-type RH strain. Asterisks (*) indicate a P value of <0.05 between the wild type and NTPase I and double-knockout mutants. (C) The addition of ATP during cell lysis or the absence of DTT in the lysis buffer partially reversed the interference with the firefly luciferase reporter. Asterisks (*) indicate a P value of <0.05 between cell lysis with or without DTT. (D) An IFN-γ-inducible tandem GAS element-driven ATP-independent GLUC RAW264.7 macrophage reporter line showed no NTPase-dependent effect. The data shown are averages ± SEM of at least three independent experiments, each performed with three technical replicates (A, C, and D), and averages ± SD of two independent experiments with three technical replicates (B).
To overcome the limitation of FLUC assays, we implemented the ATP-independent GLUC as an alternative reporter. Using this ATP-independent system, the original strain-dependent difference in luciferase activity was no longer apparent. Moreover, when we compared the NTPase disruptant mutants, they showed no difference from wild-type T. gondii parasites (Fig. 5D).
DISCUSSION
In this study, we aimed to identify T. gondii effectors responsible for blocking the IFN-γ immune response in host cells. Previously, it has been shown that T. gondii inhibits STAT1-mediated transcription in IFN-γ-activated cells and that different clonal parasite strain types do so with similar efficiency (15, 17). We developed multiple GAS-FLUC reporter lines in human and mouse cells and contrary to previous studies uncovered a strain-dependent inhibition of IFN-γ signaling. Initially, we attributed this difference to different reporter cell lines or to the higher MOIs used in our studies. Based on observing a strong strain-specific difference in the inhibition of FLUC activity in RAW264.7 macrophages, we reasoned that this trait was amenable to QTL analysis. We took advantage of a previous genetic cross (18) and analyzed recombinant progeny based on their ability to inhibit IFN-γ-driven GAS-FLUC activity in infected macrophages.
Using QTL analysis, we mapped the strain-dependent phenotype to a single locus containing the NTPase I and NTPase II genes, which have previously been implicated in virulence (10, 11). After we identified the QTL on chromosome XII, we narrowed the locus to a region containing 36 genes. Based on detecting a higher level of gene expression in the type I GT1 lineage, we reasoned that the NTPase I and NTPase II genes might indeed be involved in this phenotype. NTPase I is found only in virulent type I strains (10), and previous studies have suggested it is refractory to deletion (11), suggesting that it might play a role in acute virulence. NTPases I and II are located in dense granules and constitute 2 to 8% of the total tachyzoite protein content (12, 13, 26). It has previously been reported that NTPases might be involved in tachyzoite egress or purine salvage (27). However, it is unclear how the NTPases might affect STAT1 signaling, since both enzymes are not thought to be released into the host cell cytosol but rather occupy the parasitophorous vacuole (23).
Despite previous predictions, deletion of the genes encoding either or both of the NTPase enzymes had no effect on growth or virulence of T. gondii. Using the highly efficient CRISPR/Cas9 system (19), we were easily able to delete each of the NTPase genes, as well as generate the double mutant using several different strategies. Although these findings do not rule out another role in different life cycle stages or different hosts, it is apparent they are not essential under normal growth conditions or in the murine model. However, we did confirm that the presence of NTPase I was associated with greater inhibition of the FLUC assay, thus supporting the original strain-dependent difference we observed. Consistent with our QTL mapping result, deletion of the NTPase I gene in the type I RH strain restored levels of FLUC to levels similar to the ME49 strain, which contains only NTPase II. However, the inhibitory effect observed in the FLUC assay was not due to blockage of STAT1-induced transcription, as shown by the induction of IRF1 in an immunofluorescence assay. Transcription of IFN-γ-inducible genes such as IRF1 is initiated after phosphorylated STAT1 homodimers bind to gamma-activated sequence (GAS) elements in their promoter regions (28). Since IFN-γ-dependent transcription is globally blocked in T. gondii-infected cells (15, 29), we would have expected this block to be absent in NTPase knockout strains if these genes were responsible. However, when we examined the ability of NTPase mutants to inhibit STAT1-induced gene expression, we found that the loss of both enzymes had no effect. This result led us to suspect that the inhibition of FLUC activity was due to the potent activity of NTPase I in consuming ATP, which is necessary for initiation of the light reaction in several luciferase systems, such as FLUC and click beetle luciferase.
Taken together, the combination of QTL mapping and reverse genetic approaches using CRISPR/Cas9 confirmed that type I strains inhibit FLUC to a greater extent and that the NTPase genes were responsible for this trait. However, the failure of NTPase mutants to alter IRF1 induction suggested that the original FLUC assay was not a faithful readout for STAT1 activity. Instead, our findings strongly suggest that ATP hydrolysis by T. gondii NTPases was responsible for the observed interference with the FLUC assay. This conclusion is supported by the presence of NTPase I, which is more strongly expressed and has a higher ATP hydrolysis rate, in type I strains that show greater potential to inhibit the FLUC assay. Additional support is provided by the fact that NTPase activity was enhanced by dithiol compounds such as DTT (13), a component of our FLUC assay, and addition of ATP during cell lysis or DTT-free lysis buffer partially reversed the inhibition of FLUC activity by T. gondii.
Notably, the enzymatic effect of NTPases was independent of the species or cell type background of the reporter assay, as reflected by very similar results in both murine RAW264.7 macrophages and human HeLa cells. Hence, the artifact described here is likely to confound other ATP-dependent assays involving T. gondii infection of host cells. Although the use of T. gondii strains null for both NTPase I and NTPase II could be used to avoid interference artifacts in ATP-dependent assays, the use of a non-ATP-dependent luciferase such as Gaussia luciferase avoids this problem. Therefore, the Gaussia luciferase assay provides the preferred platform for future studies on gene expression inhibitor screening and growth assays involving T. gondii.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jennifer Barks for technical assistance and Michael Behnke for advice on QTL mapping.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00077-16.
REFERENCES
- 1.Sibley LD, Khan A, Ajioka JW, Rosenthal BM. 2009. Genetic diversity of Toxoplasma gondii in animals and humans. Philos Trans R Soc Lond B Biol Sci 364:2749–2761. doi: 10.1098/rstb.2009.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elmore SA, Jones JL, Conrad PA, Patton S, Lindsay DS, Dubey JP. 2010. Toxoplasma gondii: epidemiology, feline clinical aspects, and prevention. Trends Parasitol 26:190–196. doi: 10.1016/j.pt.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Dubey JP. 2010. Toxoplasmosis of animals and humans. CRC Press, Boca Raton, FL. [Google Scholar]
- 4.Hunter CA, Sibley LD. 2012. Modulation of innate immunity by Toxoplasma gondii virulence effectors. Nat Rev Microbiol 10:766–778. doi: 10.1038/nrmicro2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Platanias LC. 2005. Mechanisms of type-I- and type-II-interferon-mediated signaling. Nat Rev Immunol 5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 6.Kroger A, Koster M, Schroeder K, Hauser H, Mueller PP. 2002. Activities of IRF-1. J Interferon Cytokine Res 22:5–14. doi: 10.1089/10799900260289837. [DOI] [PubMed] [Google Scholar]
- 7.Etheridge RD, Alagan A, Tang K, Turk BE, Sibley LD. 2014. ROP18 and ROP17 kinase complexes synergize to control acute virulence of Toxoplasma in the mouse. Cell Host Microbe 15:537–550. doi: 10.1016/j.chom.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saeij JPJ, Boyle JP, Coller S, Taylor S, Sibley LD, Brooke-Powell ET, Ajioka JW, Boothroyd JC. 2006. Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science 314:1780–1783. doi: 10.1126/science.1133690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor S, Barragan A, Su C, Fux B, Fentress SJ, Tang K, Beatty WL, Haijj EL, Jerome M, Behnke MS, White M, Wootton JC, Sibley LD. 2006. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science 314:1776–1780. doi: 10.1126/science.1133643. [DOI] [PubMed] [Google Scholar]
- 10.Asai T, Miura S, Sibley LD, Okabayashi H, Takeuchi T. 1995. Biochemical and molecular characterization of nucleoside triphosphate hydrolase isozymes from the parasitic protozoan Toxoplasma gondii. J Biol Chem 270:11391–11397. doi: 10.1074/jbc.270.19.11391. [DOI] [PubMed] [Google Scholar]
- 11.Nakaar V, Samuel BU, Ngo EO, Joiner KA. 1999. Targeted reduction of nucleoside triphosphate hydrolase by antisense RNA inhibits Toxoplasma gondii proliferation. J Biol Chem 274:5083–5087. doi: 10.1074/jbc.274.8.5083. [DOI] [PubMed] [Google Scholar]
- 12.Nakaar V, Beckers CJM, Polotsky V, Joiner KA. 1998. Basis for substrate specificity of the Toxoplasma gondii nucleoside triphosphate hydrolase. Mol Biochem Parasitol 97:209–220. doi: 10.1016/S0166-6851(98)00153-4. [DOI] [PubMed] [Google Scholar]
- 13.Asai T, O'Sullivan WJ, Tatibana M. 1983. A potent nucleoside triphosphate hydrolase from the parasitic protozoan Toxoplasma gondii. J Biol Chem 258:6816–6822. [PubMed] [Google Scholar]
- 14.Lüder CGK, Lang T, Beuerle B, Gross U. 1998. Down-regulation of MHC class II molecules and inability to up-regulate class I molecules in murine macrophages after infection with Toxoplasma gondii. Clin Exp Immunol 112:308–316. doi: 10.1046/j.1365-2249.1998.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SK, Fouts AE, Boothroyd JC. 2007. Toxoplasma gondii dysregulates IFN-γ inducible gene expression in human fibroblasts: insights from a genome-wide transcriptional profiling. J Immunol 178:5154–5165. doi: 10.4049/jimmunol.178.8.5154. [DOI] [PubMed] [Google Scholar]
- 16.Luder CG, Algner M, Lang C, Bleicher N, Gross U. 2003. Reduced expression of the inducible nitric oxide synthase after infection with Toxoplasma gondii facilitates parasite replication in activated murine macrophages. Int J Parasitol 33:833–844. doi: 10.1016/S0020-7519(03)00092-4. [DOI] [PubMed] [Google Scholar]
- 17.Rosowski EE, Saeij JP. 2012. Toxoplasma gondii clonal strains all inhibit STAT1 transcriptional activity but polymorphic effectors differentially modulate IFNγ-induced gene expression and STAT1 phosphorylation. PLoS One 7:e51448. doi: 10.1371/journal.pone.0051448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Behnke MS, Khan A, Wootton JC, Dubey JP, Tang K, Sibley LD. 2011. Virulence differences in Toxoplasma mediated by amplification of a family of polymorphic pseudokinases. Proc Natl Acad Sci U S A 108:9631–9636. doi: 10.1073/pnas.1015338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen B, Brown KM, Lee TD, Sibley LD. 2014. Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. mBio 5:e01114-14. doi: 10.1128/mBio.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanley PE, Kricka J (ed). 1991. Bioluminescence and chemiluminescence: current status. Wiley, Chichester, United Kingdom. [Google Scholar]
- 21.Buguliskis JS, Brossier F, Shuman J, Sibley LD. 2010. Rhomboid 4 (ROM4) affects the processing of surface adhesins and facilitates host cell invasion by Toxoplasma gondii. PLoS Pathog 6:e1000858. doi: 10.1371/journal.ppat.1000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su C, Howe DK, Dubey JP, Ajioka JW, Sibley LD. 2002. Identification of quantitative trait loci controlling acute virulence in Toxoplasma gondii. Proc Natl Acad Sci U S A 99:10753–10758. doi: 10.1073/pnas.172117099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sibley LD, Niesman IR, Asai T, Takeuchi T. 1994. Toxoplasma gondii: Secretion of a potent nucleoside triphosphate hydrolase into the parasitophorous vacuole. Exp Parasitol 79:301–311. doi: 10.1006/expr.1994.1093. [DOI] [PubMed] [Google Scholar]
- 24.Alaganan A, Fentress SJ, Tang K, Wang Q, Sibley LD. 2014. Toxoplasma GRA7 effector increases turnover of immunity-related GTPases and contributes to acute virulence in the mouse. Proc Natl Acad Sci U S A 111:1126–1131. doi: 10.1073/pnas.1313501111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bahl A, Davis PH, Behnke M, Dzierszinski F, Jagalur M, Chen F, Shanmugam D, White MW, Kulp D, Roos DS. 2010. A novel multifunctional oligonucleotide microarray for Toxoplasma gondii. BMC Genomics 11:603. doi: 10.1186/1471-2164-11-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asai T, Kim T, Kobayashi M, Kojima S. 1987. Detection of nucleoside triphosphate hydrolase as a circulating antigen in sera of mice infected with Toxoplasma gondii. Infect Immun 55:1332–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferguson DJ, Cesbron-Delauw MF, Dubremetz JF, Sibley LD, Joiner KA, Wright S. 1999. The expression and distribution of dense granule proteins in the enteric (coccidian) forms of Toxoplasma gondii in the small intestine of the cat. Exp Parasitol 91:203–211. doi: 10.1006/expr.1998.4384. [DOI] [PubMed] [Google Scholar]
- 28.Boehm U, Klamp T, Groot M, Howard JC. 1997. Cellular responses to interferon gamma. Annu Rev Immunol 15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 29.Lang C, Hildebrandt A, Brand F, Opitz L, Dihazi H, Luder CG. 2012. Impaired chromatin remodelling at STAT1-regulated promoters leads to global unresponsiveness of Toxoplasma gondii-infected macrophages to IFN-γ. PLoS Pathog 8:e1002483. doi: 10.1371/journal.ppat.1002483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakaar V, Bermudes D, Peck KR, Joiner KA. 1998. Upstream elements required for expression of nucleoside triphosphate hydrolase genes of Toxoplasma gondii. Mol Biochem Parasitol 92:229–239. doi: 10.1016/S0166-6851(97)00220-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.