Abstract
Salmonella enterica is among the most burdensome of foodborne disease agents. There are over 2,600 serovars that cause a range of disease manifestations ranging from enterocolitis to typhoid fever. While there are two vaccines in use in humans to protect against typhoid fever, there are none that prevent enterocolitis. If vaccines preventing enterocolitis were to be developed, they would likely protect against only one or a few serovars. In this report, we tested the hypothesis that probiotic organisms could compete for the preferred nutrient sources of Salmonella and thus prevent or treat infection. To this end, we added the fra locus, which encodes a utilization pathway for the Salmonella-specific nutrient source fructose-asparagine (F-Asn), to the probiotic bacterium Escherichia coli Nissle 1917 (Nissle) to increase its ability to compete with Salmonella in mouse models. We also tested a metabolically competent, but avirulent, Salmonella enterica serovar Typhimurium mutant for its ability to compete with wild-type Salmonella. The modified Nissle strain became more virulent and less able to protect against Salmonella in some instances. On the other hand, the modified Salmonella strain was safe and effective in preventing infection with wild-type Salmonella. While we tested for efficacy only against Salmonella Typhimurium, the modified Salmonella strain may be able to compete metabolically with most, if not all, Salmonella serovars, representing a novel approach to control of this pathogen.
INTRODUCTION
Salmonella infections are among the three most common foodborne infections in the United States and are the leading cause of hospitalization and death (1). Salmonella enterica subsp. enterica includes over 1,500 serovars that can be broadly classified into two pathovars, the gastrointestinal and the extraintestinal (2). The gastrointestinal pathovar consists of serovars that have a broad host range and robust pathways for anaerobic metabolism. The extraintestinal pathovar consists of more host-restricted serovars that cause systemic disease, i.e., typhoid fever (3, 4). The extraintestinal serovars are undergoing genome reduction as they lose host range determinants and the ability to respire anaerobically (2, 5). The gastrointestinal serovars Salmonella enterica serovar Typhimurium and Salmonella enterica serovar Enteritidis are the most medically significant serovars in the United States. They can infect a remarkably broad range of host species, including a large number of different animals and even plants (6–9). In humans, Salmonella serovar Typhimurium causes an acute enterocolitis characterized by an inflammatory diarrhea and fever (10–12). In rare cases, this is followed by reactive arthritis (13, 14). Another form of Salmonella disease, invasive nontyphoidal salmonellosis (iNTS), is emerging, especially in Africa (15, 16). This disease is associated with malaria infection in children and HIV infection in adults (17–20). Salmonella Typhimurium and Salmonella Enteritidis are the most common serovars associated with iNTS. Unfortunately, there are no vaccines for the gastrointestinal serovars (21, 22). Antibiotics are not recommended for uncomplicated cases of Salmonella-mediated enterocolitis but are used to treat the very young or elderly or when there are complications or invasive disease. However, multiple drug resistance is prevalent and increasing (15, 23). Novel therapeutic approaches are needed for the gastrointestinal salmonelloses.
Infection of slc11A1 mutant mice (formerly known as Nramp1), such as C57BL/6 or BALB/c, by Salmonella serovar Typhimurium is often used as a surrogate model for Salmonella serovar Typhi infection of humans. This is because the intestine shows little or no inflammation and there is no diarrhea but there is a systemic lethal infection. However, the lack of inflammation makes this a poor model for the natural disease caused by Salmonella Typhimurium, which is inflammatory diarrhea. It has been known for decades that the normal intestinal microbiota protects against systemic Salmonella infection, referred to as colonization resistance. For instance, the 50% lethal dose (LD50) for Salmonella Typhimurium in the BALB/c or C57BL/6 mouse decreases from 106 CFU for a mouse with normal microbiota to less than 10 CFU for a germfree mouse or a mouse pretreated with streptomycin (Strep treated [24, 25]). More recently, it was determined that the gastrointestinal tracts of germfree and Strep-treated mice become inflamed by Salmonella Typhimurium, mimicking the human disease (26). The germfree and Strep-treated murine models are now widely used to study Salmonella Typhimurium-induced inflammatory diarrhea (26–35). The Strep-treated model has the advantage that the mice have a normal immune system before disruption of the microbiota with streptomycin. The germfree mice have the advantage that defined microbial communities can be created, or they can accept transplants of microbiota from different animals or humans (35–37). Germfree mice are highly susceptible to intestinal infections, and we use them in this study to gauge the safety of our proposed probiotics. The newest inflammation model is the CBA/J mouse. These mice are Nramp1+/+ and are resistant to systemic Salmonella infection. However, these mice have the unusual attribute of allowing persistent intestinal colonization by Salmonella. It was recently discovered that the gastrointestinal tracts of these mice are becoming inflamed during these persistent infections, starting at 10 days postinfection (27, 38). Since this inflammation requires no streptomycin-mediated disruption of the microbiota, it has the most realistic microbial community composition of the Salmonella inflammation models.
In this report, we tested a probiotic approach to the prevention and treatment of salmonellosis. Probiotic microbial strains have long been used to prevent or treat illness. Probiotics could potentially replace antibiotics as growth promoters in agriculture or for prophylactic or therapeutic use in humans and animals. More research is needed to identify or design probiotic bacteria and to determine their mechanisms of action (39). With regard to specific infections, a collection of 11 Lactobacillus strains or a single Bacillus isolate has been found to be effective at reducing Salmonella colonization of poultry (40, 41). A probiotic Escherichia coli strain isolated from a healthy soldier in World War I, now called E. coli Nissle 1917 (here referred to as Nissle), has been shown to reduce Salmonella infection in a mouse model (42). This strain is closely related to uropathogenic isolates of E. coli but lacks virulence factors and has an abundance of fitness determinants, including at least six iron acquisition systems (43). Competition for iron is one mechanism by which Nissle inhibits Salmonella (42). Nissle is safe for use in animals and humans and has been shown to be effective in treating diarrhea, ulcerative colitis, and constipation and preventing necrotizing enterocolitis (NEC) in infants (44–47).
Here, we attempt to enhance the ability of Nissle to compete with Salmonella by adding the Salmonella fra locus to the Nissle genome. The fra locus contains five genes that confer the ability to utilize fructose-asparagine (F-Asn) as a carbon and nitrogen source (48). Mutants lacking this locus were identified in a genetic screen as highly attenuated in mouse models of inflammation (48). The fra locus is widely distributed among the gastrointestinal serovars of Salmonella but, like many loci involved with anaerobic metabolism, appears to be undergoing genome degradation in the extraintestinal serovars (2). Salmonella encodes two type 3 secretion systems (T3SS) encoded within Salmonella pathogenicity islands 1 and 2 (SPI1 and SPI2, respectively) that inject more than 40 effector proteins into host cells (10, 49–52). SPI1 elicits invasion of host cells, while SPI2 is required for survival within host cells. Loss of both renders Salmonella unable to cause inflammation and enterocolitis (33, 53). Consistent with a role in enterocolitis, the fra locus conferred a fitness benefit upon Salmonella only in mouse models that become inflamed from Salmonella infection and failed to confer a benefit in strain backgrounds lacking the ability to cause inflammation (lacking SPI1 and SPI2) (48). Therefore, we hypothesized that F-Asn is a significant nutrient source for Salmonella during inflammation and that adding the fra locus to Nissle would allow Nissle to compete with Salmonella for F-Asn and prevent or treat disease. Assuming that adding the fra locus to Nissle was going to increase the effectiveness of Nissle, we also pondered adding more Salmonella-specific nutrient acquisition systems to Nissle to increase effectiveness further. However, we realized that as we added these loci to Nissle, we would in effect be creating an avirulent Salmonella strain. To determine the effectiveness of this strategy, we included a Salmonella mutant lacking SPI1 and SPI2 as an example of an avirulent Salmonella strain that retains all of its nutrient acquisition loci.
MATERIALS AND METHODS
Strains and media.
Bacterial strains are listed in Table 1. Bacteria were grown in LB broth or on LB agar plates for routine culture (EM Science). XLD agar was used for recovery of Salmonella from mice (Becton, Dickinson). M9 minimal medium was made as described previously and contained either 5 mM glucose or 5 mM fructose-asparagine (F-Asn) as a carbon source (54). F-Asn was synthesized as previously described (48). When necessary, ampicillin (Amp) or kanamycin (Kan) was added to medium at 200 mg/liter or 60 mg/liter, respectively.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype | Reference or source |
|---|---|---|
| Escherichia coli strains | ||
| Nissle 1917 | E. coli Nissle, serotype O6:K5:H1 | 62 |
| 14028 | Wild-type Salmonella enterica serovar Typhimurium | American Type Culture Collection |
| ASD100 | 14028 Δ(ssrB-ssaU)1::Kan | Lambda Red mutation of SPI2 using primers BA2558 and BA2559 |
| ASD199 | 14028 Δ(avrA-invH)1 Δ(ssrB-ssaU)1::Kan | Δ(ssrB-ssaU)1::Kan mutation from ASD100 transduced into YD039 |
| ASD200 | 14028 Δ(avrA-invH)1 Δ(ssrB-ssaU)1 | Kan cassette in ASD199 was flipped out using pCP20 |
| ASD201 | 14028 Δ(avrA-invH)1 Δ(ssrB-ssaU)1 Δ(fraR-fraBDAE)4::Kan | Δ(fraR-fraBDAE)4::Kan mutation from CS1005 was transduced into ASD200 |
| ASD9000 | E. coli Nissle 1917 plus pWSK29 (Ampr) | E. coli Nissle 1917 electroporated with empty vector pWSK29 |
| ASD9010 | E. coli Nissle 1917 plus pASD5006 (Ampr) | E. coli Nissle 1917 electroporated with pASD5006 |
| CS1005 | 14028 Δ(fraR-fraBDAE)4::Kan | Lambda Red mutation of fra island using primers BA2515 and BA2538 |
| JLD1214 | 14028 IG(pagC-STM14_1502)::Cam | 48 |
| JM110 | rpsL thr leu thi-1 lacY galK galT ara tonA tsx dam dcm supE44 Δ(lac-proAB) | Stratagene |
| YD039 | 14028 Δ(avrA-invH)1 | 56 |
| Plasmids | ||
| pASD5006 | pWSK29 fraR-fraBDAE (Ampr) | 48 |
| pWSK29 | pSC101 cloning vector (Ampr) | 48 |
Addition of the Salmonella fra locus to E. coli Nissle 1917.
The low-copy-number plasmid pASD5006, containing the fra locus of Salmonella strain 14028, or the vector pWSK29 was electroporated into the E. coli dam dcm strain JM110 to decrease methylation and then purified and electroporated into E. coli Nissle 1917 with selection on LB-Amp. The ability of Nissle to grow on F-Asn was confirmed by growing Nissle plus pASD5006 (ASD9010) in M9 minimal medium with F-Asn as the sole carbon source compared to Nissle plus pWSK29 (ASD9000) (Fig. 1). This was done in a 96-well clear-bottom plate with the optical density at 600 nm recorded over an 18-h period at 37°C using a SpectraMax M5 microplate reader (Molecular Devices) and SoftMax Pro 6.1 software.
FIG 1.
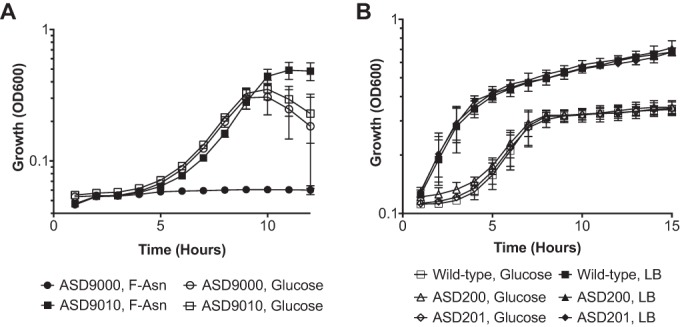
(A) Growth of Nissle plus vector (ASD9000) or Nissle plus fra (ASD9010) in M9 minimal medium containing either 5 mM glucose or 5 mM F-Asn as carbon source. (B) Growth of wild-type Salmonella (14028), the SPI1 SPI2 mutant (ASD200), and the SPI1 SPI2 fra mutant (ASD201) in either M9 glucose or LB.
Construction of a Salmonella SPI1 SPI2 mutant.
Lambda Red mutagenesis was used to construct the SPI2 mutant ASD100 (55). Oligonucleotides, including 40 nucleotides matching either ssrB or ssaU, including 30 nucleotides of the coding region of either target, were appended to sequences that bind pKD4, creating primers BA2558 and BA2559 (55). Oligonucleotides are listed in Table 2. These were used to amplify the Kan cassette from pKD4 using Taq DNA polymerase (NEB). The resulting PCR product, an FLP recombination target (FRT)-Kan-FRT cassette flanked by homology to ssrB and ssaU, was electroporated into strain 14028+pKD46, and transformants were selected on LB-Kan at 37°C. The insertion was verified by PCR using primers BA2582 and BA1922 (K1). This ΔSPI2::Kan mutation was transduced from ASD100 into the ΔSPI1 strain YD039 (56) using phage P22HTint, creating ASD199. The antibiotic resistance marker was deleted by electroporating ASD199 with pCP20 (55), which encodes the FLP recombinase, and transformants were selected on LB-Amp at 30°C. Deletion of the Kan cassette was verified using PCR with primers BA2582 and BA2583, as well as screened for loss of pCP20, creating ASD200.
TABLE 2.
Oligonucleotides
| Oligonucleotide | Sequence | Description |
|---|---|---|
| BA1922 | CAGTCATAGCCGAATAGCCT | Kanamycin cassette insertion verification primer |
| BA2515 | GCCTGCATGATTAATACGTACTGAAATAACTCTGGATCAGCATATGAATATCCTCCTTAG | Lambda Red mutagenic reverse primer for STM14_4328 with P2 priming site |
| BA2538 | ATGGATACAAATGATCGAGCAACCCGACAGTAAAAGCGCCGTGTAGGCTGGAGCTGCTTC | Lambda Red mutagenic forward primer for STM14_4332 with P1 priming site |
| BA2558 | ACGCCCCTGGTTAATACTCTATTAACCTCATTCTTCGGGCGTGTAGGCTGGAGCTGCTTC | Lambda Red mutagenic forward primer with homology to ssrB with P1 priming site |
| BA2559 | CCAAAAGCATTTATGGTGTTTCGGTAGAATGCGCATAATCCATATGAATATCCTCCTTAG | Lambda Red mutagenic reverse primer with homology to ssaU with P2 priming site |
| BA2582 | AAATAAGGGGATTCTACTATATCATGATCA | Reverse primer for confirmation of SPI2 deletion |
| BA2583 | GCCAGGCTAAAAGCGATTATTTTCAGTCTC | Forward primer for confirmation of SPI2 deletion |
| B2888 | GGATCCGCTTCGATACCTGAGTGGCAAAGTGTGCG | Forward primer for verification of fra island mutation with K1 |
Construction of SPI1 SPI2 fra triple mutant.
Lambda Red mutagenesis was used to create an fraRBDAE island mutant (STM14_4332-STM14_4328), CS1005, using the protocol described above. Briefly, oligonucleotides BA2515 and BA2538 were used to amplify the Kan cassette from pKD4 using Taq DNA polymerase (NEB). The PCR product was electroporated into 14028+pKD46, and transformants were selected on LB-Kan at 37°C to create CS1005. The insertion of the Kan cassette was verified by PCR using BA1922 (K1) and BA2888. The resulting (fraR-fraBDAE)4::Kan island mutation was transduced into the ΔSPI1 ΔSPI2 strain ASD200 using the phage P22HTint, creating ASD201.
Animals.
Swiss Webster mice were obtained from Taconic Farms. CBA/J mice were obtained from Jackson Laboratories. Germfree C57BL/6 and Swiss Webster mice were bred at the Ohio State University (OSU) germfree animal facility. All mice were females between 6 and 10 weeks of age. All bacterial inocula were grown with shaking at 37°C overnight, resuspended in water, and administered by the intragastric route in a volume of 200 μl. For survival curves, mice were euthanized upon reaching the early-removal criteria of our approved animal protocol. For CFU determinations, ceca or feces were homogenized in 3 ml or 0.75 ml, respectively, of phosphate-buffered saline (PBS). One-hundred-microliter aliquots of serial dilutions were then plated on XLD agar plates containing the appropriate antibiotics, yielding a detection limit of 30 CFU for ceca and 8 CFU for feces.
Histopathology.
Cecal samples were removed from mice, and a portion was immersion fixed in 10% neutral buffered formalin, processed by routine methods, and embedded in paraffin wax by the Comparative Pathology and Mouse Phenotyping Shared Resource at the Ohio State University. Sections (4 μm) were stained with hematoxylin and eosin (H&E) and scored in a blinded fashion by a veterinary pathologist, board certified by the American College of Veterinary Pathologists (ACVP). The adapted, semiquantitative histopathologic scoring system (57) assessed enterocyte loss (none, 0; loss of single cell, 1; loss of groups of cells/erosion, 2; overt ulceration, 3), crypt inflammation (none, 0; 1 to 2 inflammatory cells, 1; cryptitis, 2; crypt abscess, 3), mononuclear cell inflammation (none, 0; mild, 1; moderate, 2; marked, 3), neutrophilic inflammation (none, 0; mild, 1; moderate, 2; marked, 3), epithelial hyperplasia (none, 0; mild, 1; moderate, 2; discrete nests of regenerated crypts delineated from adjacent mucosa with no obvious disruption from overlying mucosal surface, 3), and edema (none, 0; mild/focal/single layer affected, 1; moderate/multifocal/multiple layers affected, 2; marked/widespread/transmural involvement, 3).
Animal assurance.
All animal work was performed in accordance with the protocols approved by our Institutional Animal Care and Use Committee (OSU 2009A0035). The IACUC ensures compliance of this protocol with the U.S. Animal Welfare Act, the Guide for the Care and Use of Laboratory Animals (63), and the Public Health Service Policy on Humane Care and Use of Laboratory Animals.
RESULTS
A fra mutant of Salmonella is attenuated in several murine inflammation models, suggesting that F-Asn is a nutrient that is important to Salmonella fitness in the inflamed intestine (48). Therefore, we hypothesized that adding the fra locus to a probiotic organism would enhance the ability of that organism to compete with Salmonella for F-Asn and prevent or treat Salmonella infections. To test this hypothesis, we cloned the Salmonella fra locus on a low-copy-number plasmid and placed this plasmid in the well-characterized probiotic strain E. coli Nissle 1917 (Nissle). As expected, Nissle carrying the fra plasmid (ASD9010) was able to grow on F-Asn as the sole carbon source while Nissle carrying the vector alone (ASD9000) was not (Fig. 1A). We considered adding more Salmonella-specific nutrient acquisition systems to Nissle but realized that this was much like creating a nonpathogenic Salmonella strain. Therefore, instead of adding more nutrient acquisition systems to Nissle, we constructed a mutant of Salmonella lacking SPI1 and SPI2 (ASD200). This strain should compete with wild-type Salmonella for all nutrients without causing disease. In later experiments, we also constructed and tested an SPI1 SPI2 fra triple mutant (ASD201) to determine the fra dependence of any observed effects. Both ASD200 and ASD201 grow similarly to the wild type in M9 glucose and LB (Fig. 1B). We will refer to these four strains as the “probiotics” throughout this report.
To determine if the probiotics could protect mice from wild-type Salmonella, we started with germfree mice, which have no colonization resistance. We used both Swiss Webster and C57BL/6 mice (Nramp1+/+ and Nramp1G169D/G169D, respectively). A 109-CFU quantity of a probiotic strain or sham (water) was administered by oral gavage to groups of five mice. The following day, the mice were challenged with a lethal dose of 104 CFU of virulent Salmonella (strain JLD1214, which is a chloramphenicol-resistant derivative of ATCC 14028). In both germfree C57BL/6 mice and germfree Swiss Webster mice, all of the probiotics enhanced survival compared to sham (Fig. 2). Nissle plus fra protected slightly better than Nissle plus vector in germfree C57BL/6 mice, but this was not statistically significant (P = 0.075). Interestingly, Nissle plus vector was highly protective in germfree Swiss Webster mice (100% survival), but Nissle plus fra was less protective, with no survival (P = 0.004). The Salmonella SPI1 SPI2 mutant was the most protective in both types of mice. The Salmonella triple mutant (SPI1 SPI2 fra) was used only in the germfree Swiss Webster mice. While it appeared less protective than the double mutant (SPI1 SPI2), this was not statistically significant (P = 0.091).
FIG 2.
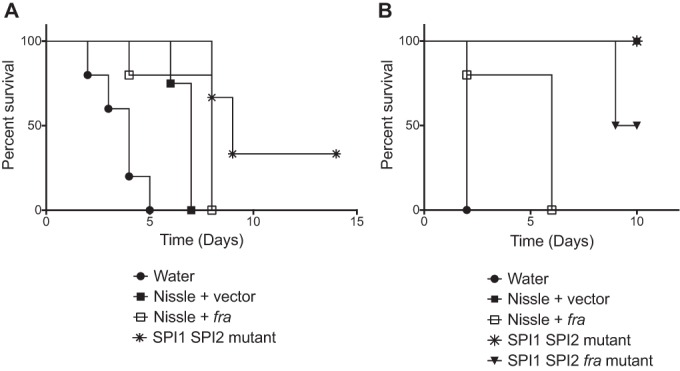
Evaluation of probiotics as prophylactics in germfree mice. On consecutive days, groups of five germfree C57BL/6 mice (A) or germfree Swiss Webster mice (B) were orally administered a probiotic strain (109 CFU of strains listed below the graph) or sham (water) and then virulent Salmonella (104 CFU of JLD1214). Survival was monitored over time. The statistical significance of each treatment being different from the others was determined with log rank (Mantel-Cox) tests, without correction for multiple comparisons, with a P value of <0.05 considered significant. In both panels, the sham was statistically significantly different from all of the treatments (P < 0.014 or better). In panel A, Nissle plus vector was not different from Nissle plus fra (P = 0.07), but it was different from the SPI1 SPI2 mutant (P = 0.02). In panel B, Nissle plus vector was different from Nissle plus fra (P = 0.003) but was not different from the SPI1 SPI2 mutant (P = 1.0) or the SPI1 SPI2 fra mutant (P = 0.09). In panel B, the SPI1 SPI2 mutant was not different from the SPI1 SPI2 fra mutant (P = 0.09).
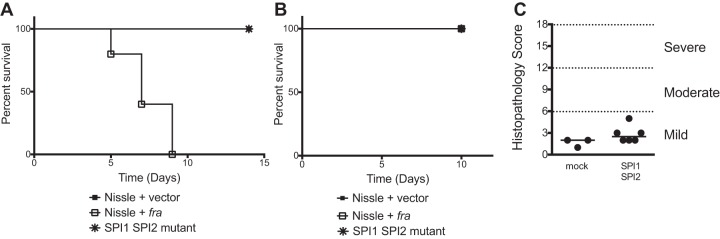
To test the safety of the probiotics, each strain was administered at a dose of 109 CFU to a group of germfree mice and mortality was monitored (Fig. 3). The Salmonella SPI1 SPI2 mutant and Nissle plus vector were completely safe in both types of mice (no mortality). Nissle plus fra caused no mortality in the Swiss Webster mice but caused 100% mortality in the C57BL/6 mice (Fig. 3). This indicates that the addition of the fra locus to Nissle increased its virulence in germfree C57BL/6 mice. In a separate experiment, we infected germfree Swiss Webster mice with a dose of 109 CFU of the Salmonella SPI1 SPI2 mutant and then quantitated inflammation of the ceca after 6 days of colonization using histopathology. The Salmonella SPI1 SPI2 mutant was safe with regard to inflammation (Fig. 3).
FIG 3.
(A and B) Safety of probiotics in germfree C57BL/6 mice (A) and germfree Swiss Webster mice (B). Groups of five mice were orally administered a probiotic strain (109 CFU of strains listed in the legend), and survival was monitored over time. (C) A group of six germfree Swiss Webster mice were orally administered 109 CFU of the Salmonella SPI1 SPI2 mutant, while three mice were inoculated with water (mock). After 6 days, the mice were euthanized and their ceca were scored for inflammation using histopathology. Bars indicate the median. In panels A and B, the statistical significance of each treatment being different from the others was determined with log rank (Mantel-Cox) tests, without correction for multiple comparisons, with a P value of <0.05 considered significant. In panel A, the Nissle plus fra was significantly different from Nissle plus vector and SPI1 SPI2 mutant (P = 0.0026). In panel B, the groups were not different. In panel C, the statistical significance of differences between groups was determined using a Mann-Whitney test and the groups were not different (P = 0.13).
The experiments in germfree mice revealed that the Nissle-plus-fra strain was less effective than Nissle plus vector at preventing death in germfree Swiss Webster mice (Fig. 2B), and it gained the ability to kill germfree C57BL/6 mice (Fig. 3A). Thus, the ability to utilize F-Asn enhanced the virulence of Nissle. In contrast, the Salmonella SPI1 SPI2 mutant was safe and effective in protecting both germfree C57BL/6 and germfree Swiss Webster mice from wild-type Salmonella.
To further test the ability of these strains to protect against a lethal Salmonella infection, we moved to a Strep-treated Swiss Webster mouse model. Mice with a normal microbiota are highly resistant to Salmonella-mediated inflammation, but treatment with streptomycin disrupts the microbiota and allows Salmonella-mediated inflammation to occur within a day of infection. Thus, in this experiment the mice were treated with streptomycin. One day later, they were treated with a dose of 109 CFU of a probiotic strain or sham; 1 day after that, they were challenged with a lethal dose of Salmonella (107 CFU of JLD1214). All of the probiotic strains except Nissle plus vector provided statistically significant protection compared to sham (Fig. 4). The protection provided by Nissle plus fra was statistically significantly different from that of sham but was not different from that of Nissle plus vector (P = 0.523), making it difficult to conclude that the ability to utilize F-Asn improved the ability of Nissle to protect against Salmonella (Fig. 4). The Salmonella SPI1 SPI2 mutant and the SPI1 SPI2 fra triple mutant both provided protection statistically significantly different from that provided by sham, but they were not different from each other (P = 0.684), indicating that protection is not dependent upon the ability to utilize F-Asn (Fig. 4).
FIG 4.
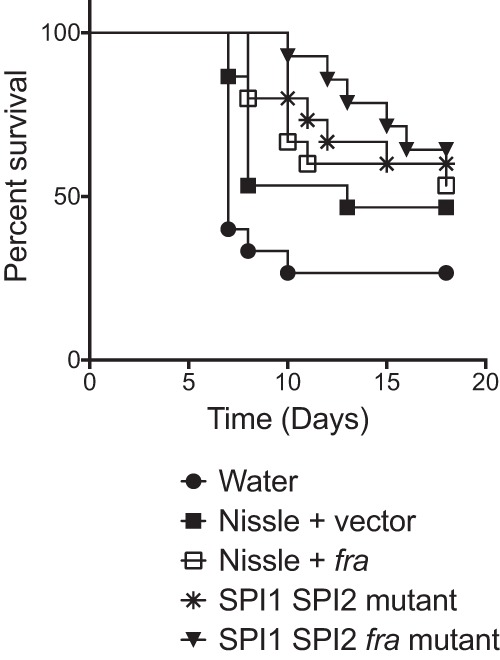
Evaluation of probiotics as prophylactics in Strep-treated Swiss Webster mice. On consecutive days, groups of 15 mice were administered streptomycin, then a probiotic strain (109 CFU of strains listed below the graph) or sham (water), and then virulent Salmonella (107 CFU of JLD1214). Survival was monitored over time. The statistical significance of each treatment being different from the others was determined with log rank (Mantel-Cox) tests, without correction for multiple comparisons, with a P value of <0.05 considered significant. The sham is statistically significantly different from all treatments (P < 0.03 or better) except Nissle plus vector (P = 0.10). Nissle plus vector is not different from Nissle plus fra (P = 0.52), SPI1 SPI2 mutant (P = 0.27), or the SPI1 SPI2 fra mutant (P = 0.40). The SPI1 SPI2 mutant is not different from the SPI1 SPI2 fra mutant (P = 0.68).
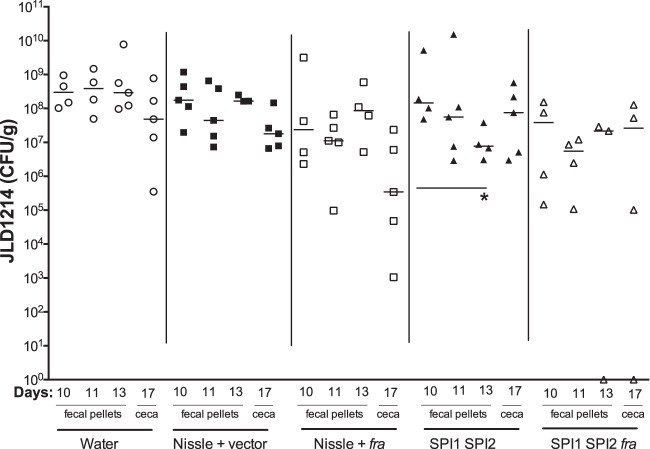
A more recent mouse model of Salmonella-mediated inflammation is the CBA/J model. These mice are Nramp1+/+, they tend to carry Salmonella for long periods in their intestinal tract, and their intestinal tract becomes inflamed by day 10 postinfection (27, 38). With no need for disruption of the microbiota with antibiotics, this model may have the most realistic microbiota composition during inflammation. To test the ability of our probiotic strains to treat a Salmonella infection, we inoculated the CBA/J mice with 109 CFU of Salmonella, waited 10 days for inflammation to begin, and then treated the mice with 109 CFU of probiotic or sham. Thus, this is a therapeutic rather than a prophylactic model. Salmonella shedding in feces was measured on days 10 (just before probiotic inoculation), 11, and 13, and ceca were harvested on day 17 (Fig. 5). Nissle plus fra appeared to reduce Salmonella shedding in ceca by day 17, but this just missed statistical significance, with a P value of 0.055. The only probiotic strain to cause a statistically significant decrease in fecal counts of virulent Salmonella over time was the Salmonella SPI1 SPI2 mutant (day 13 compared to day 10). The SPI1 SPI2 fra triple mutant was not different over time, which might suggest that protection is fra dependent; however, it was not different from the SPI1 SPI2 mutant either (P = 0.999), leaving the fra dependence unlikely.
FIG 5.
CBA/J mice were orally inoculated with 109 CFU of virulent Salmonella strain JLD1214. Ten days postinfection, groups of five mice were treated with 109 CFU of probiotic or sham. Salmonella (JLD1214) shedding in feces was measured on days 10 (just before probiotic inoculation), 11, and 13. Salmonella (JLD1214) in the ceca was measured on day 17. The limit of detection was 30 CFU for ceca and 8 CFU for feces. Statistical significance between groups was determined using a Mann-Whitney test. *, P < 0.05.
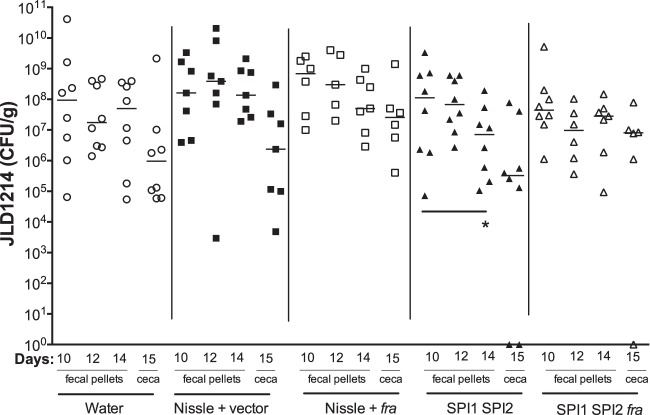
We used the CBA/J model a second time, in which we increased the number of mice per group from 5 to 8 and increased the number of probiotic doses from one to three, administered on days 10, 12, and 14 postinfection (Fig. 6). As in the previous experiment, only the SPI1 SPI2 mutant reduced the counts of virulent Salmonella over time (day 14 compared to day 10). Again, the SPI1 SPI2 fra triple mutant was not different over time, suggesting that there is fra dependence to the protection. However, the triple mutant was not different from the double mutant (P = 0.527), again leaving the fra dependence in question. For this experiment, we also performed histopathology on ceca harvested on day 15 to determine if inflammation was reduced by the probiotics. This showed that there were no statistically significant differences between the treatment and sham groups (Fig. 7). However, the mice treated with the Salmonella SPI1 SPI2 mutant appeared to fall into two categories, with half having little or no inflammation while the other half were highly inflamed. As a group, there may be no statistically significant improvement, but for some individuals, the treatment may be effective. Consistent with this, the only mice that were completely cleared of wild-type Salmonella from their cecum were two mice that had been treated with the Salmonella SPI1 SPI2 mutant and one mouse that had been treated with the Salmonella SPI1 SPI2 fra triple mutant (Fig. 6). These three mice also had the lowest inflammation scores in their respective groups.
FIG 6.
CBA/J mice were orally inoculated with 109 CFU of virulent Salmonella strain JLD1214. Groups of eight mice were treated with 109 CFU of probiotic or sham three times, on days 10, 12, and 14 postinfection. Salmonella (JLD1214) shedding in feces was measured on the same days just before probiotic inoculation. Salmonella (JLD1214) in the ceca was measured on day 15. The limit of detection was 30 CFU for ceca and 8 CFU for feces. Statistical significance between groups was determined using a Mann-Whitney test. *, P < 0.05.
FIG 7.
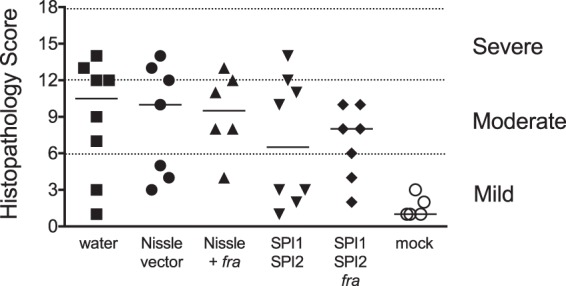
Histopathology scores of ceca harvested from the mice in Fig. 6 on day 15 postinfection. The bar represents the median.
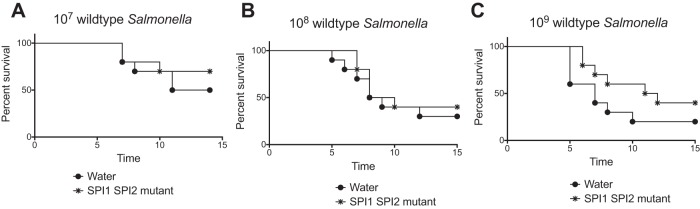
The CBA/J model demonstrated that the Salmonella SPI1 SPI2 mutant can reduce the CFU counts of wild-type Salmonella in fecal samples, but this was not a dramatic effect. This may be because the wild type had a 10-day head start before the probiotic was administered. We decided to return to the Strep-treated Swiss Webster model, but rather than testing the ability of the Salmonella SPI1 SPI2 mutant to prevent an infection, as in Fig. 4, we would use the mutant to treat an existing infection. In this experiment, the mice were treated with streptomycin; the following day, they received either 107, 108, or 109 CFU of wild-type Salmonella (JLD1214). Then, 24 h later, they received 109 CFU of probiotic or sham (water). Survival was monitored over time (Fig. 8). At 107 and 108 CFU of wild-type Salmonella, administration of the SPI1 SPI2 mutant had no effect on the survival curve (Fig. 8A and B). At 109 CFU of wild-type Salmonella, administration of the SPI1 SPI2 mutant appeared to improve survival of the mice, but this was not statistically significant (Fig. 8C).
FIG 8.
Evaluation of the Salmonella SPI1 SPI2 mutant as a therapeutic in the Strep-treated Swiss Webster model. On consecutive days, groups of 10 mice were administered streptomycin, then wild-type Salmonella JLD1214 (107 CFU in panel A, 108 CFU in panel B, or 109 CFU in panel C), and then 109 CFU of the probiotic (Salmonella SPI1 SPI2 mutant, ASD200) or sham (water). Survival was monitored over time. The statistical significance of each treatment being different from the others was determined with log rank (Mantel-Cox) tests, without correction for multiple comparisons, with a P value of <0.05 considered significant. The sham was not statistically significantly different from the treatment in panel A (P = 0.44), panel B (P = 0.58), or panel C (P = 0.17).
DISCUSSION
The fra locus was identified in a genetic screen for Salmonella genes that are differentially required for fitness in germfree mice colonized, or not, with the commensal organism Enterobacter cloacae (48). Further experimentation revealed that a fraB mutation was severely attenuated in its ability to compete with wild-type Salmonella in four mouse models of inflammation: germfree, germfree colonized with human fecal microbiota, Strep treated, and interleukin-10 (IL-10) knockout. Interestingly, the fraB mutant was not attenuated in conventional mice that fail to become inflamed from Salmonella infection. It was also determined that a fraB mutant has no phenotype if the competition experiment is performed in a Salmonella genetic background lacking SPI1 and SPI2. These results were interpreted to mean that SPI1 and SPI2 are required for Salmonella to induce inflammation (in models that are permissive), and the inflammation may be killing microbes that would otherwise compete for F-Asn (48). This model gave rise to the idea that adding the fra locus to probiotic species, such as E. coli Nissle 1917, could give them the ability to compete with Salmonella for a critical nutrient source and thus prevent infection. However, since then we have learned that the fraB phenotype is primarily due to the accumulation of a toxic metabolite during growth on F-Asn rather than F-Asn being a critical nutrient source (B. M. M. Ahmer, unpublished data). Despite this, there seemed to be some fra dependence with regard to the ability of the Salmonella SPI1 SPI2 mutant to compete with wild-type Salmonella, especially in CBA/J mice. It appeared that protection was fra dependent because the SPI1 SPI2 double mutant, but not the SPI1 SPI2 fra triple mutant, was significantly different from sham. However, the double mutant is not statistically significantly different from the triple mutant, leaving the fra dependence in question. Furthermore, the Nissle strain modified to contain the fra locus was altered in its ability to kill germfree C57BL/6 mice and in its ability to protect germfree Swiss Webster mice against Salmonella infection, compared to the original Nissle strain. These results suggest that F-Asn is a significant nutrient source in some situations but definitely not the only nutrient source available to E. coli and Salmonella in the inflamed intestine.
The mechanism by which virulence of Nissle is enhanced by the fra locus is unknown. Virulence enhancement was observed only in germfree mice and resulted in the killing of C57BL/6 mice and a reduced ability to protect Swiss Webster mice against Salmonella, compared to wild-type Nissle (Nissle plus vector). We have unpublished results that indicate that F-Asn concentrations are quite high in the intestines of germfree mice. However, it is still surprising that simply providing another nutrient source to Nissle had these effects. The C57BL/6 mice are mutated at the Nramp1 locus, while the Swiss Webster mice are not, which makes the C57BL/6 mice more susceptible to systemic infections. It is possible that Nissle carrying the fra locus simply grew to higher numbers in the intestine, which allowed escape to a permissive systemic environment. In the Swiss Webster mice, wild-type Nissle was 100% effective in preventing killing of the mice by Salmonella, while Nissle plus fra delayed the killing compared to sham but still resulted in no survival. Why Nissle plus fra would have a reduced ability to protect against Salmonella is unclear. While adding Nissle to fra was not a successful strategy, this does not rule out the possibility that adding fra to a different probiotic organism, such as Lactobacillus or Bifidobacterium, might enhance the ability of these organisms to compete with Salmonella.
The Salmonella SPI1 SPI2 mutant looks promising. This strain was included in the study to determine what would happen if we continued adding Salmonella-specific nutrient acquisition loci to Nissle, essentially creating an avirulent Salmonella strain. Unlike Nissle, the Salmonella SPI1 SPI2 mutant has all of the same nutrient acquisition loci as does wild-type Salmonella. This strain was safe, and noninflammatory, even at doses of 109 CFU in highly susceptible germfree C57BL/6 mice (Fig. 3). This strain was also effective at prevention of Salmonella infection using the germfree and Strep-treated models (Fig. 2 and 4). A much more difficult task is to treat an existing infection. The Salmonella SPI1 SPI2 mutant was indeed modestly effective at treating an existing infection using the CBA/J model (Fig. 5 and 6) but not the Strep-treated Swiss Webster model (Fig. 8). In the CBA/J model, the wild-type Salmonella was administered 10 days before the probiotic. Despite being administered 10 days after the wild-type Salmonella, the probiotic was able to reproducibly reduce the CFU of wild-type Salmonella. Overall, however, the Salmonella SPI1 SPI2 mutant is much more effective as a preventative than as a therapeutic.
Currently, a cya crp mutant of Salmonella is used as a live attenuated vaccine strain in agriculture (58–60). This strain is metabolically attenuated so that it cannot compete metabolically with wild-type Salmonella but instead creates a lasting immune response against a single serovar. The use of a Salmonella SPI1 SPI2 mutant as a probiotic takes a different approach in which the strain is metabolically competent, so that it may be able to compete effectively against hundreds of serovars of Salmonella. There is precedent for this approach in the literature. A nontoxigenic Clostridium difficile strain can compete with wild-type C. difficile to resolve infection and prevent recurrence (61). Of the different mouse models, the Salmonella SPI1 SPI2 mutant was the most effective in protecting germfree mice from wild-type Salmonella. This suggests that this strain might be particularly effective in preventing Salmonella colonization of neonatal agricultural animals such as newly hatched poultry or swine. The probiotic approach could be used as an alternative, or in conjunction with vaccination, as the probiotic may protect during the first week or two of life while responses to vaccination are developing.
ACKNOWLEDGMENTS
We thank Edward Behrman for synthesis of fructose-asparagine. We thank Venkat Gopalan for critical reading of the manuscript. We thank Haley Steiner for care and assistance with the germfree mice. We thank the Comparative Pathology & Mouse Phenotyping Shared Resource, Department of Veterinary Biosciences and the Comprehensive Cancer Center, The Ohio State University, Columbus, OH, for slide preparation and histopathologic scoring of mouse ceca.
This work was supported by NIH grants 1R01AI116119 and R01AI097116 to B.M.M.A. and P30 CA016058 to the Comparative Pathology and Mouse Phenotyping Shared Resource Cancer Center. H.M.B. was partially supported by an undergraduate summer research fellowship from the OSU undergraduate research office.
REFERENCES
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerging Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nuccio SP, Bäumler AJ. 2014. Comparative analysis of Salmonella genomes identifies a metabolic network for escalating growth in the inflamed gut. mBio 5:e00929-14. doi: 10.1128/mBio.00929-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong VK, Baker S, Pickard DJ, Parkhill J, Page AJ, Feasey NA, Kingsley RA, Thomson NR, Keane JA, Weill F-X, Edwards DJ, Hawkey J, Harris SR, Mather AE, Cain AK, Hadfield J, Hart PJ, Thieu NTV, Klemm EJ, Glinos DA, Breiman RF, Watson CH, Kariuki S, Gordon MA, Heyderman RS, Okoro C, Jacobs J, Lunguya O, Edmunds WJ, Msefula C, Chabalgoity JA, Kama M, Jenkins K, Dutta S, Marks F, Campos J, Thompson C, Obaro S, Maclennan CA, Dolecek C, Keddy KH, Smith AM, Parry CM, Karkey A, Mulholland EK, Campbell JI, Dongol S, Basnyat B, Dufour M, Bandaranayake D, Naseri TT, Singh SP, Hatta M, Newton P, Onsare RS, Isaia L, Dance D, Davong V, Thwaites G, Wijedoru L, Crump JA, de Pinna E, Nair S, Nilles EJ, Thanh DP, Turner P, Soeng S, Valcanis M, Powling J, Dimovski K, Hogg G, Farrar J, Holt KE, Dougan G. 2015. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat Genet 47:632–639. doi: 10.1038/ng.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gunn JS, Marshall JM, Baker S, Dongol S, Charles RC, Ryan ET. 2014. Salmonella chronic carriage: epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol 22:648–655. doi: 10.1016/j.tim.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker S, Dougan G. 2007. The genome of Salmonella enterica serovar Typhi. Clin Infect Dis 45(Suppl 1):S29–S33. doi: 10.1086/518143. [DOI] [PubMed] [Google Scholar]
- 6.Swearingen MC, Porwollik S, Desai PT, McClelland M, Ahmer BMM. 2012. Virulence of 32 Salmonella strains in mice. PLoS One 7:e36043. doi: 10.1371/journal.pone.0036043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iniguez AL, Dong Y, Carter HD, Ahmer BMM, Stone JM, Triplett EW. 2005. Regulation of enteric endophytic bacterial colonization by plant defenses. Mol Plant Microbe Interact 18:169–178. doi: 10.1094/MPMI-18-0169. [DOI] [PubMed] [Google Scholar]
- 8.Dong Y, Iniguez AL, Ahmer BMM, Triplett EW. 2003. Kinetics and strain specificity of rhizosphere and endophytic colonization by enteric bacteria on seedlings of Medicago sativa and Medicago truncatula. Appl Environ Microbiol 69:1783–1790. doi: 10.1128/AEM.69.3.1783-1790.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith JN, Dyszel JL, Soares JA, Ellermeier CD, Altier C, Lawhon SD, Adams LG, Konjufca V, Curtiss R, Slauch JM, Ahmer BMM. 2008. SdiA, an N-acylhomoserine lactone receptor, becomes active during the transit of Salmonella enterica through the gastrointestinal tract of turtles. PLoS One 3:e2826. doi: 10.1371/journal.pone.0002826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Heijden J, Finlay BB. 2012. Type III effector-mediated processes in Salmonella infection. Future Microbiol 7:685–703. doi: 10.2217/fmb.12.49. [DOI] [PubMed] [Google Scholar]
- 11.Sánchez-Vargas FM, Abu-El-Haija MA, Gómez-Duarte OG. 2011. Salmonella infections: an update on epidemiology, management, and prevention. Travel Med Infect Dis 9:263–277. doi: 10.1016/j.tmaid.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Godinez I, Keestra AM, Spees A, Bäumler AJ. 2011. The IL-23 axis in Salmonella gastroenteritis. Cell Microbiol 13:1639–1647. doi: 10.1111/j.1462-5822.2011.01637.x. [DOI] [PubMed] [Google Scholar]
- 13.Tuuminen T, Lounamo K, Leirisalo-Repo M. 2013. A review of serological tests to assist diagnosis of reactive arthritis: critical appraisal on methodologies. Front Immunol 4:418. doi: 10.3389/fimmu.2013.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ajene AN, Fischer Walker CL, Black RE. 2013. Enteric pathogens and reactive arthritis: a systematic review of Campylobacter, Salmonella and Shigella-associated reactive arthritis. J Health Popul Nutr 31:299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okoro CK, Kingsley RA, Connor TR, Harris SR, Parry CM, Al-Mashhadani MN, Kariuki S, Msefula CL, Gordon MA, de Pinna E, Wain J, Heyderman RS, Obaro S, Alonso PL, Mandomando I, Maclennan CA, Tapia MD, Levine MM, Tennant SM, Parkhill J, Dougan G. 2012. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat Genet 44:1215–1221. doi: 10.1038/ng.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon MA. 2011. Invasive nontyphoidal Salmonella disease. Curr Opin Infect Dis 24:484–489. doi: 10.1097/QCO.0b013e32834a9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bronzan RN, Taylor TE, Mwenechanya J, Tembo M, Kayira K, Bwanaisa L, Njobvu A, Kondowe W, Chalira C, Walsh AL, Phiri A, Wilson LK, Molyneux ME, Graham SM. 2007. Bacteremia in Malawian children with severe malaria: prevalence, etiology, HIV coinfection, and outcome. J Infect Dis 195:895–904. doi: 10.1086/511437. [DOI] [PubMed] [Google Scholar]
- 18.Reddy EA, Shaw AV, Crump JA. 2010. Community-acquired bloodstream infections in Africa: a systematic review and meta-analysis. Lancet Infect Dis 10:417–432. doi: 10.1016/S1473-3099(10)70072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mooney JP, Lokken KL, Byndloss MX, George MD, Velazquez EM, Faber F, Butler BP, Walker GT, Ali MM, Potts R, Tiffany C, Ahmer BMM, Luckhart S, Tsolis RM. 2015. Inflammation-associated alterations to the intestinal microbiota reduce colonization resistance against non-typhoidal Salmonella during concurrent malaria parasite infection. Sci Rep 5:14603. doi: 10.1038/srep14603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takem EN, Roca A, Cunnington A. 2014. The association between malaria and non-typhoid Salmonella bacteraemia in children in sub-Saharan Africa: a literature review. Malar J 13:400. doi: 10.1186/1475-2875-13-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strugnell RA, Scott TA, Wang N, Yang C, Peres N, Bedoui S, Kupz A. 2014. Salmonella vaccines: lessons from the mouse model or bad teaching? Curr Opin Microbiol 17:99–105. doi: 10.1016/j.mib.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Martin LB. 2012. Vaccines for typhoid fever and other salmonelloses. Curr Opin Infect Dis 25:489–499. doi: 10.1097/QCO.0b013e328356ffeb. [DOI] [PubMed] [Google Scholar]
- 23.Chen H-M, Wang Y, Su L-H, Chiu C-H. 2013. Nontyphoid Salmonella infection: microbiology, clinical features, and antimicrobial therapy. Pediatr Neonatol 54:147–152. doi: 10.1016/j.pedneo.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Bohnhoff M, Drake BL, Miller CP. 1954. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc Soc Exp Biol Med 86:132–139. doi: 10.3181/00379727-86-21030. [DOI] [PubMed] [Google Scholar]
- 25.Collins FM, Carter PB. 1978. Growth of salmonellae in orally infected germfree mice. Infect Immun 21:41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez CA, Winter SE, Rivera-Chávez F, Xavier MN, Poon V, Nuccio S-P, Tsolis RM, Bäumler AJ. 2012. Phage-mediated acquisition of a type III secreted effector protein boosts growth of Salmonella by nitrate respiration. mBio 3:e00143-12. doi: 10.1128/mBio.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Bäumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sekirov I, Gill N, Jogova M, Tam N, Robertson M, de Llanos R, Li Y, Finlay BB. 2010. Salmonella SPI-1-mediated neutrophil recruitment during enteric colitis is associated with reduction and alteration in intestinal microbiota. Gut Microbes 1:30–41. doi: 10.4161/gmic.1.1.10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valdez Y, Grassl GA, Guttman JA, Coburn B, Gros P, Vallance BA, Finlay BB. 2009. Nramp1 drives an accelerated inflammatory response during Salmonella-induced colitis in mice. Cell Microbiol 11:351–362. doi: 10.1111/j.1462-5822.2008.01258.x. [DOI] [PubMed] [Google Scholar]
- 32.Woo H, Okamoto S, Guiney D, Gunn JS, Fierer J. 2008. A model of Salmonella colitis with features of diarrhea in SLC11A1 wild-type mice. PLoS One 3:e1603. doi: 10.1371/journal.pone.0001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, Mering von C, Hardt WD. 2007. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5:2177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hapfelmeier S, Hardt W-D. 2005. A mouse model for S. typhimurium-induced enterocolitis. Trends Microbiol 13:497–503. doi: 10.1016/j.tim.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 35.Stecher B, Macpherson AJ, Hapfelmeier S, Kremer M, Stallmach T, Hardt WD. 2005. Comparison of Salmonella enterica serovar Typhimurium colitis in germfree mice and mice pretreated with streptomycin. Infect Immun 73:3228–3241. doi: 10.1128/IAI.73.6.3228-3241.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL. 2012. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rivera-Chávez F, Winter SE, Lopez CA, Xavier MN, Winter MG, Nuccio S-P, Russell JM, Laughlin RC, Lawhon SD, Sterzenbach T, Bevins CL, Tsolis RM, Harshey R, Adams LG, Bäumler AJ. 2013. Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Pathog 9:e1003267. doi: 10.1371/journal.ppat.1003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Behnsen J, Deriu E, Sassone-Corsi M, Raffatellu M. 2013. Probiotics: properties, examples, and specific applications. Cold Spring Harb Perspect Med 3:a010074. doi: 10.1101/cshperspect.a010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higgins JP, Higgins SE, Vicente JL, Wolfenden AD, Tellez G, Hargis BM. 2007. Temporal effects of lactic acid bacteria probiotic culture on Salmonella in neonatal broilers. Poult Sci 86:1662–1666. doi: 10.1093/ps/86.8.1662. [DOI] [PubMed] [Google Scholar]
- 41.Tellez G, Pixley C, Wolfenden RE, Layton SL, Hargis BM. 2012. Probiotics/direct fed microbials for Salmonella control in poultry. Food Res Int 45:628–633. doi: 10.1016/j.foodres.2011.03.047. [DOI] [Google Scholar]
- 42.Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, Libby SJ, Fang FC, Raffatellu M. 2013. Probiotic bacteria reduce Salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, Dobrindt U. 2004. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J Bacteriol 186:5432–5441. doi: 10.1128/JB.186.16.5432-5441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cukrowska B, LodInova-ZadnIkova R, Enders C, Sonnenborn U, Schulze J, Tlaskalová-Hogenová H. 2002. Specific proliferative and antibody responses of premature infants to intestinal colonization with nonpathogenic probiotic E. coli strain Nissle 1917. Scand J Immunol 55:204–209. doi: 10.1046/j.1365-3083.2002.01005.x. [DOI] [PubMed] [Google Scholar]
- 45.Kruis W, Fric P, Pokrotnieks J, Lukás M, Fixa B, Kascák M, Kamm MA, Weismueller J, Beglinger C, Stolte M, Wolff C, Schulze J. 2004. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 53:1617–1623. doi: 10.1136/gut.2003.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Buenau R, Jaekel L, Schubotz E, Schwarz S, Stroff T, Krueger M. 2005. Escherichia coli strain Nissle 1917: significant reduction of neonatal calf diarrhea. J Dairy Sci 88:317–323. doi: 10.3168/jds.S0022-0302(05)72690-4. [DOI] [PubMed] [Google Scholar]
- 47.Schroeder B, Duncker S, Barth S, Bauerfeind R, Gruber AD, Deppenmeier S, Breves G. 2006. Preventive effects of the probiotic Escherichia coli strain Nissle 1917 on acute secretory diarrhea in a pig model of intestinal infection. Dig Dis Sci 51:724–731. doi: 10.1007/s10620-006-3198-8. [DOI] [PubMed] [Google Scholar]
- 48.Ali MM, Newsom DL, Gonzalez JF, Sabag-Daigle A, Stahl C, Steidley B, Dubena J, Dyszel JL, Smith JN, Dieye Y, Arsenescu R, Boyaka PN, Krakowka S, Romeo T, Behrman EJ, White P, Ahmer BMM. 2014. Fructose-asparagine is a primary nutrient during growth of Salmonella in the inflamed intestine. PLoS Pathog 10:e1004209. doi: 10.1371/journal.ppat.1004209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V. 2009. Salmonella takes control: effector-driven manipulation of the host. Curr Opin Microbiol 12:117–124. doi: 10.1016/j.mib.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller AJ, Kaiser P, Dittmar KEJ, Weber TC, Haueter S, Endt K, Songhet P, Zellweger C, Kremer M, Fehling H-J, Hardt W-D. 2012. Salmonella gut invasion involves TTSS-2-dependent epithelial traversal, basolateral exit, and uptake by epithelium-sampling lamina propria phagocytes. Cell Host Microbe 11:19–32. doi: 10.1016/j.chom.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 51.Agbor TA, McCormick BA. 2011. Salmonella effectors: important players modulating host cell function during infection. Cell Microbiol 13:1858–1869. doi: 10.1111/j.1462-5822.2011.01701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ibarra JA, Steele-Mortimer O. 2009. Salmonella—the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell Microbiol 11:1579–1586. doi: 10.1111/j.1462-5822.2009.01368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hapfelmeier S, Stecher B, Barthel M, Kremer M, Müller AJ, Heikenwalder M, Stallmach T, Hensel M, Pfeffer K, Akira S, Hardt W-D. 2005. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J Immunol 174:1675–1685. doi: 10.4049/jimmunol.174.3.1675. [DOI] [PubMed] [Google Scholar]
- 54.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 55.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teplitski M, Al-Agely A, Ahmer BMM. 2006. Contribution of the SirA regulon to biofilm formation in Salmonella enterica serovar Typhimurium. Microbiology (Reading, Engl) 152:3411–3424. doi: 10.1099/mic.0.29118-0. [DOI] [PubMed] [Google Scholar]
- 57.Kennedy RJ, Hoper M, Deodhar K, Erwin PJ, Kirk SJ, Gardiner KR. 2000. Interleukin 10-deficient colitis: new similarities to human inflammatory bowel disease. Br J Surg 87:1346–1351. doi: 10.1046/j.1365-2168.2000.01615.x. [DOI] [PubMed] [Google Scholar]
- 58.Curtiss R III, Kelly SM. 1987. Salmonella typhimurium deletion mutants lacking adenylate cyclase and cyclic AMP receptor protein are avirulent and immunogenic. Infect Immun 55:3035–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hassan JO, Curtiss R III. 1991. Control of colonization by virulent Salmonella typhimurium by oral immunization of chickens with avirulent delta cya delta crp S. typhimurium. Res Microbiol 142:109. doi: 10.1016/0923-2508(91)90103-H. [DOI] [PubMed] [Google Scholar]
- 60.Kelly SM, Bosecker BA, Curtiss R III. 1992. Characterization and protective properties of attenuated mutants of Salmonella choleraesuis. Infect Immun 60:4881–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gerding DN, Meyer T, Lee C, Cohen SH, Murthy UK, Poirier A, Van Schooneveld TC, Pardi DS, Ramos A, Barron MA, Chen H, Villano S. 2015. Administration of spores of nontoxigenic Clostridium difficile strain M3 for prevention of recurrent C. difficile infection: a randomized clinical trial. JAMA 313:1719–1727. doi: 10.1001/jama.2015.3725. [DOI] [PubMed] [Google Scholar]
- 62.Nissle A. 1959. Explanations of the significance of colonic dysbacteria & the mechanism of action of E. coli therapy (mutaflor). Medizinische 4:1017–1022. [PubMed] [Google Scholar]
- 63.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]