Abstract
Although multifocal tumors and non-invasive/invasive components are commonly encountered in surgical pathology, their genetic relationship is often poorly characterized. We used next generation sequencing (NGS) to characterize somatic alterations in a patient with five spatially distinct, high grade papillary urothelial carcinomas (UC), with one tumor harboring an underlying invasive component. NGS of 409 cancer related genes was performed on DNA isolated from formalin fixed paraffin embedded (FFPE) blocks representing each papillary tumor (n=5), the invasive component of one tumor, and matched normal tissue. We identified 9 unique non-synonymous somatic mutations across the six UC samples, including five present in each carcinoma sample, consistent with clonal origin and limited intertumoral heterogeneity. Copy number and loss of heterogeneity (LOH) profiles were similar in all six carcinomas; however, the invasive carcinoma component uniquely showed focal CDKN2A loss and chromosome 9 LOH, and did not harbor gains of chromosomes 5p or X that were present in the other tumor samples. Phylogenetic analysis supported the invasive component arising from a shared progenitor prior to the outgrowth of cells in the non-invasive tumors. Results were extended to three additional cases of upper tract UC with paired non-invasive/ invasive components, which identified driving alterations exclusive to both non-invasive and invasive components. Lastly, we performed targeted RNAseq using a custom bladder cancer panel, which confirmed gene expression signature differences between paired non-invasive/invasive components. The results and approaches presented here may be useful in understanding the clonal relationships in multifocal cancers or paired non-invasive/invasive components from routine FFPE specimens.
Keywords: next generation sequencing, clonal evolution, bladder cancer, FFPE tissue
Introduction
Multifocal tumors are commonly encountered in surgical pathology practice. For example, patients with urothelial carcinoma (UC) frequently have spatially distinct tumors identified at the time of primary diagnosis or at tumor recurrence[2, 27]. Two non-mutually exclusive theories have been offered to explain this biological phenomenon. The field effect theory asserts the entire urothelium in patients with UC may be unstable, due to genetic hits and progressive outgrowth of genetically unstable cells throughout the entire urothelium. Findings consistent with possible field effects in multifocal UC include promoter methylation of cancer-associated genes in histologically normal urothelium adjacent to UC[16], such as E-cadherin promoter methylation in the urothelium of elderly patients[4], as well as widespread TP53 mutations in the urothelium of patients with high level radiation exposure[28]. The clonal expansion theory states a single neoplastic clone is responsible for multi-focal disease and recurrence. Evidence for the clonal expansion proposal includes data showing that identical genetic abnormalities are present in spatially distinct UCs from the same patient, such as identified by loss of heterozygosity (LOH) analysis, comparative genome hybridization (CGH), and TP53 mutational work [9, 17, 23, 25]. UCs may also manifest oligoclonally, with individual tumors showing two or more distinct sets of molecular hits, suggesting the lesions are derived from two or more neoplastic clones[24].
Similarly, many tumors demonstrate both invasive and non-invasive components, which may have similar or distinct morphology. For example, UCs typically arise as non-invasive tumors, which subsequently develop an invasive phenotype. This evolution to invasive carcinoma is not invariable. Many patients, particularly those with low grade carcinomas, never develop invasive disease. Studies evaluating genetic alterations associated with the deeply invasive phenotype have suggested a role for loss of the tumor suppressors PTEN, TP53, and RB1, as well as overexpression of mesenchymal markers such as MMP9[1, 5, 18]. Progression from superficial to advanced urothelial carcinoma has also been shown to be associated with 17p (TP53) and 18q losses in clonal recurrences by X chromosome inactivation studies[22]. However, the genetic alterations associated with invasive progression from non-invasive lesions are otherwise poorly understood.
Next generation sequencing (NGS) approaches have revolutionized our understanding of intra-/ and inter-tumoral heterogeneity/clonality and have recently been applied to metachronous UC[10, 11, 19]. However, such approaches generally use fresh frozen tissue, which is not routinely encountered in surgical pathology practice. We recently encountered a remarkable case of a man with five concurrent, spatially distinct UCs involving the bilateral renal pelvises, right distal ureter, urinary bladder, and prostatic urethra. With the exception of high stage invasive UC in one renal pelvis, the tumors were non-invasive or only focally invasive and the non-invasive component in all five tumors was histologically indistinguishable. Hence, we applied NGS approaches compatible with routine formalin fixed paraffin embedded (FFPE) tissue to evaluate clonality and directly compare transcriptional and genetic profiles between invasive and non-invasive components. Findings were extended by assessing paired invasive/non-invasive from three additional cases of upper tract UC.
Materials and Methods
Cohort
Diagnostic hematoxylin and eosin stained slides and FFPE blocks were retrieved from the University of Michigan Department of Pathology Tissue Archive. Sequencing of archived diagnostic FFPE tissue was performed with IRB approval (for patients without sequencing of normal tissue) and with patient consent (for patients where normal tissue was sequenced) through the University of Michigan Bladder Cancer Tissue Bank.
Case 1
We identified a case of a 70 year old man undergoing total urotheliectomy at the University of Michigan Health System for multifocal papillary UC. He had been diagnosed with UC of the bladder three years prior and had multiple documented recurrences (pTa and pT1) treated by TURBT. CT scan four months prior demonstrated marked right hydronephrosis with a soft tissue prominence in the posterior aspect of the bladder. No lymphadenopathy was observed. TURBT at that time demonstrated pT1 high grade papillary UC involving the urinary bladder and prostatic urethra. He was treated with 6 weeks of BCG with three instillations of mitomycin-C. Restaging TURBT two months prior demonstrated extensive high grade papillary pTa and pT1 UC involving the urinary bladder, prostatic urethra and penile urethra. A large left renal pelvis tumor was identified on panpyeloscopy one month prior, with biopsy demonstrating pTa papillary UC. At urotheliectomy, spatially distinct papillary tumors were grossly identified in the left renal pelvis (BL191), the right ureter (BL192), the prostatic urethra (BL194), the urinary bladder (BL195) and the right renal pelvis (BL196). By histology, all five locations demonstrated non-invasive, papillary UC; the right renal pelvis also showed underlying invasive UC (BL190) extending to the peripelvic adiopose tissue with areas suspicious for angiolymphatic invasion. He was followed after surgery at outside institutions and presented approximately one month after urotheliectomy with a 5 cm right flank mass diagnosed as poorly differentiated urothelial carcinoma with focal squamous differentiation. CT scan demonstrated multiple pulmonary nodules suspicious for metastatic disease. He received palliative radiation therapy to the flank mass without response and died approximately 6 months after urotheliectomy from metastatic UC. Metastatic foci were not available for histologic or molecular evaluation.
Cases 2-4
Three additional nephroureterectomy cases of upper tract UC, where non-invasive and invasive components could be separately isolated, were also identified. Case 2 was from an 87 year old man diagnosed with pT3 papillary (with focal inverted features) high-grade UC involving the renal pelvis (BL300 non-invasive/BL301 invasive). Case 3 was from a 70 year old man diagnosed with pT3 papillary high grade UC involving the distal ureter (BL302 non-invasive/BL303 invasive). Case 4 was from a 68 year old man diagnosed with pT4 high grade UC with inverted features involving the renal pelvis (BL305 non-invasive/BL306 invasive).
DNA/RNA isolation
From Cases 1-4, we cut 3-6×10um FFPE sections from all tumor components. Macrodissection was used to enrich for tumor content (estimated final tumor content ~70% from each site) and isolate non-invasive/invasive components. From Case 1, we isolated an area of normal kidney (BL193) and two areas of normal urothelium using microscopic evaluation of unstained slides (BL193A and BL193B) for macrodissection. From Case 3, we isolated an area of normal urothelium using microscopic evaluation of unstained slides (BL303MN). DNA and RNA were co-isoalted using the Qiagen Allprep FFPE DNA/RNA kit (Qiagen, Valencia, CA), according to the manufacturer’s instructions (except for adding a 2 minute room temperature incubation and extending centrifugation time to 5 minutes during the xylene deparaffinization and ethanol washing steps). DNA and RNA were quantified using the Qubit 2.0 fluorometer (Life Technologies, Foster City, CA).
Targeted Next Generation Sequencing
Targeted, multiplexed PCR-based next generation sequencing (NGS) was performed on isolated DNA from each component using the Ion Ampliseq Comprehensive Cancer Panel (CCP), which targets 1,688,650 bases from 15,992 amplicons representing the complete coding sequence of 409 cancer genes (http://tools.invitrogen.com/downloads/cms_103573.csv). Barcoded libraries were generated from 40ng of DNA per sample using the Ion AmpliSeq Comprehensive Cancer Panel (Life Technologies, Foster City, CA) and the Ion Ampliseq library kit 2.0 (Life Technologies, Foster City, CA) according to the manufacturer’s instructions with barcode incorporation. For Case 1, templates were prepared using the Ion PGM Template OT2 200 Kit (Life Technologies, Foster City, CA) on the Ion One Touch 2 according to the manufacturer’s instructions. Sequencing of multiplexed templates was performed using the Ion Torrent Personal Genome Machine (Life Technologies, Foster City, CA) on Ion 318 chips using the Ion PGM Sequencing 200 Kit v2 (200 base pair reads) according to the manufacturer’s instructions. Analysis was performed in Torrent Suite 3.6, with alignment by TMAP (version 3.6.39) using default parameters, and variant calling using the Torrent Suite Variant Caller plugin (version 3.6.63335) using default low-stringency somatic variant settings. Amplicon coverage summary files for copy number analysis were generated using the Torrent Suite Coverage Analysis plugin (v3.6.63324). Variants were annotated using Annovar [6, 26]. For BL193A and BL193B, samples were processed, sequenced and analyzed as just described, but were assessed using a custom Ion Torrent Ampliseq panel that targets ~130 cancer related genes.
For Cases 2-4, library preparation with the CCP was performed as for Case 1. Templates were prepared using the Ion PI Template OT2 200 Kit v3 on the Ion One Touch 2 according to the manufacturer’s instructions. Sequencing of multiplexed templates was performed using the Ion Torrent Proton Sequencer using Ion Proton P1 chips using the Ion PI Sequencing 200 Kit v3 (200 base pair reads) according to the manufacturer’s instructions. Analysis was performed in Torrent Suite 4.0.2, with alignment by TMAP using default parameters, and variant calling using the Torrent Suite Variant Caller plugin (version 4.0-r76860) using default low-stringency somatic variant settings. Amplicon coverage summary files for copy number analysis were generated using the Torrent Suite Coverage Analysis plugin (v4.0-r77897).
Somatic variant identification
To identify somatic variants for each tumor sample from Case 1, called variants were filtered to remove synonymous variants and those without adequate read support by removing variants with: flow corrected read depths (FDP) less than 20, flow corrected variant allele containing reads (FAO) less than 6, variant allele frequencies (FAO/FDP) less than 0.10 and skewed variant read support (>5-fold difference in number of forward (FSAF) vs. reverse (FSAR) reads containing the variant allele [FSAF/FSAR < 0.2 or FSAF/FSAR > 5]). Germline variants were removed by filtering variants from each tumor sample that were called in BL193. Lastly, all passing somatic variants were visually inspected in IGV to confirm sufficient coverage (FDP>20) and lack of variant read support in BL193 (FAO/FDP<1%).
For Cases 2-4, called variants in each tumor component were filtered to remove synonymous variants and those without adequate read support by removing variants with: FDP<20, FAO<6, FAO/FDP<0.10, indels with FSAF/FSAR <0.2 or FSAF/FSAR>5. Variants occurring exclusively in reads containing other variants (single nucleotide variants or indels) or those occurring in the last mapped base of a read were excluded. For Case 3, germline variants were removed by filtering variants from each tumor sample that were called in BL303MN. Passing somatic variants were visually inspected in IGV to confirm sufficient coverage (FDP>20) and lack of variant read support in BL303MN (FAO/FDP<1%).
For Cases 2 and 4, where matched normal tissue was not sequenced, germ line variants were removed by first filtering all variants with allele frequencies >0.5% in ESP6500 or 1000 genomes, or those reported in ESP6500 or 1000 genomes with observed variant allele frequencies between 0.35-0.65 or >0.9. Then, to prioritize potentially driving somatic variants and those informing on the relationship of paired invasive/invasive components, only variants that 1) differed between paired non-invasive/invasive samples, 2) have been reported in COSMIC, or 3) are deleterious variants in tumor suppressors, were retained. All variants passing the above criteria were then visualized in IGV.
Copy number analysis
To identify copy number alterations (CNAs) for Case 1, normalized, GC content corrected, total read counts per amplicon for each bladder tumor sample were divided by those from BL193, yielding a copy number ratio for each amplicon. Gene-level copy number estimates were determined by taking the coverage-weighted mean of the per-probe ratios, with expected error determined by the probe-to-probe variance (C.S.G. et al., manuscript submitted). For Case 3, the matched normal sample (BL303MN) was used as the reference. For cases 2&4, a composite “normal” sample consisting of multiple single and pooled normal DNA samples was used as the reference.
Loss of heterozygosity
To identify areas of loss of heterozygosity in Case 1, we identified all presumed heterozygous, adequately supported SNPs called in BL193 (0.40≤FAO/FDP≤0.60 and FAO>5) that were also called in at least one tumor sample (FAO>5). For each variant, the difference in variant allele frequencies (FAO/FDP) between BL193 and each tumor sample were calculated and plotted. Genes harboring at least two consecutive heterozygous SNPs where the absolute value of (BL193 variant frequency – tumor variant frequency) was >0.25 were considered to show LOH. For case 3, LOH was performed as just described except BL303MN was used in place of BL193.
Sanger sequencing to validate called somatic variants
Positions with candidate somatic mutations in at least one tumor sample in Case 1 were subjected to validation through Sanger sequencing of the paired normal tissue (BL193) and all tumor samples. Ten nanograms of DNA were used as template in PCR amplifications with Invitrogen Platinum PCR Supermix (Life Technologies, Foster City, CA) with the suggested initial denaturation and cycling conditions. Primer sequences were designed using Primer-BLAST software (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) to generate primers no closer than 20 base pairs to the target boundaries, producing products between 100 and 200 base pairs in length where possible, and were based on the hg19, February 2009 human genome assembly. The PCR products were subjected to Sanger sequencing by the University of Michigan DNA Sequencing Core after treatment with ExoSAP-IT (GE Healthcare) and sequences were analyzed using SeqMan Pro software (DNASTAR, Madison, WI).
Copy number quantitative PCR (qPCR)
To confirm CNAs identified by NGS in Case 1, we performed qPCR on genomic DNA from BL190-BL196. Copy number ratios of CDKN2A (9p21; lost in BL190) and FGFR1 (8p11; gained in all tumor samples) were determined using PIK3R1 (5q13; unaltered in all samples) as the reference gene. Primers and probes (5′ FAM; ZEN/Iowa Black FQ dual quenchers) were designed using PrimerQuest (http://www.idtdna.com/Primerquest/Home/Index) using genomic DNA sequences with repeat sequences masked. Assay specificity was confirmed using BLAST and BLAT and primers/probes in areas of SNPs were excluded. Primer/probes sequences are available upon request. qPCR reactions (15ul) were performed in triplicate using TaqMan Genotyping Master Mix (Applied Biosystems), 10ng genomic DNA per reaction and a final concentration of 0.9uM each primer and 0.25uM probe in 384 well plates on the Quantstudio 12K Flex (Applied Biosystems). Automatic baseline and CT thresholds of 0.2 were set using QuantStudio 12K Flex Real-Time PCR System Software. Log2 copy number of CDKN2A and FGFR1 were determined by the ΔΔCT method using PIK3R1 as the reference gene and BL193 (normal DNA) as the calibrator.
Phylogenetic analysis
For Case 1, no informative (present in more than one tumor but not in all) synonymous or non-coding somatic alterations were present. Hence, for each tumor sample, mutation status of the nine non-synonymous somatically mutated positions (representing each position mutated in at least one tumor), five regions of copy number gain/loss (chrs 5p, 8, CDKN2A locus, IGF2R locus and chr X), and copy neutral LOH at chromosome 9 and HRAS-NUP98 (chr 11) were considered as characters (present or absent) for evolutionary analysis using PHYLIP v 3.695. Evolutionary trees were constructed using Dollop (Dollo and polymorphism parsimony methods) using polymorphism parsimony with default parameters.
Targeted Multiplexed PCR based RNAseq
Multiplexed PCR based RNA sequencing was performed on all samples from Cases 1-4. For each sample, 20ng RNA was reverse transcribed, barcoded, and subjected to multiplexed PCR to generate libraries using a custom Ampliseq panel and the Ion Ampliseq RNA Library kit. The custom Ampliseq panel contained 8 housekeeping genes and 103 target genes assessing major transcriptional programs in bladder cancer identified from publically available data and the Oncomine database[1, 7, 8]. Template generation and sequencing was performed as for Case 1 on the Ion PGM. Data analysis was performed using Torrent Suite (4.0.2) and the Coverage Analysis RNA Plug-in (v4.0-r77897). For each amplicon, read counts were log2 transformed (read count + 1). Then, to determine normalized expression for each target gene, the log2 count was normalized to the median of the logx counts of the 8 houskeeping genes. Unsupervised centroid linkage hierarchical clustering with median centering of genes was performed using Cluster 3.0 and visualized using Java TreeView.
Results
We encountered a remarkable case of a patient presenting for urotheliectomy with multiple spatially distinct papillary UCs, including paired invasive/non-invasive components (Case 1, Fig 1A). All five spatially distinct non-invasive papillary urothelial carcinomas (BL191, 192, and 194-196) demonstrated virtually identical morphology (Fig 1B). Namely, at low power, the growth pattern in all was predominantly exophytic, with a smaller component demonstrating inverted growth features. Tumor cell nuclei were round, disordered, and hyperchromatic, and frequent mitotic figures were present, of sufficient extent to establish the diagnosis of high grade papillary urothelial carcinoma. All tumors lacked overt anaplasia. The invasive urothelial carcinoma in the right renal pelvis (BL190) was of the conventional type, consisting of irregular, infiltrative nests of tumor cells invading the renal parenchyma and inciting a desmoplastic response (Fig 1C&D). Tumor cell nuclei were high grade, showing nuclear enlargement, anisonucleosis, and hyperchromasia; mitotic figures were increased. No features of divergent histologic differentiation (e.g. nested, micropapillary, plasmacytoid) were present.
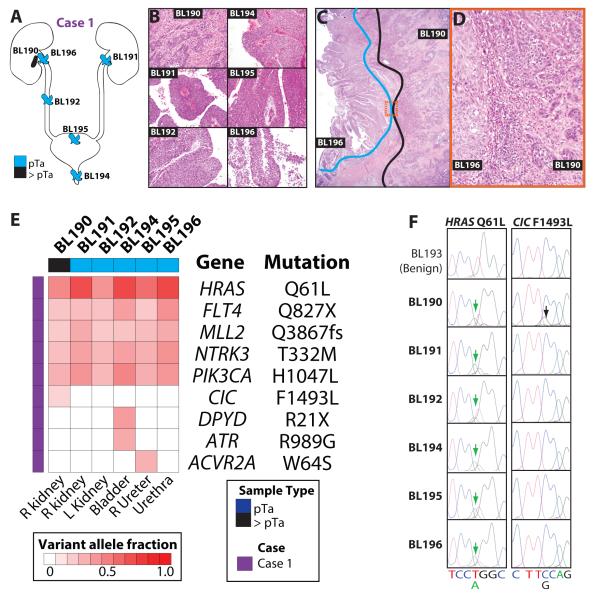
Figure 1. Next generation sequencing (NGS) of a case of urothelial carcinoma (UC) with extreme multifocality demonstrates monoclonality of spatially distinct tumors and non-invasive/invasive components.
A&B.A 70 yr old man underwent complete urotheliectomy (Case 1) for multifocal papillary tumors arising in the left (BL191) renal pelvis, right ureter (BL192), prostatic urethra (BL194), urinary bladder (BL195), and the right renal pelvis (BL196). Pathological assessment of representative sections demonstrated non-invasive, high grade papillary UC, with the exception of the right renal pelvis, which showed an underlying invasive component (BL190). Each component was macrodissected from unstained FFPE sections for NGS, in addition to matched benign renal tissue (BL193). C&D. Paired non-invasive (BL196) and invasive (BL190) UC components in the right renal pelvis were isolated separately by macrodissection. Areas isolated for BL190 (blue) and BL196 (black) are indicated, with the boxed region shown at high magnification in D. Original magnification 20x, 2x and 20x in B, C and D, respectively. E. NGS targeting the complete coding sequence of 409 cancer related genes by multiplexed PCR sequencing on the Ion Torrent PGM system was performed on each multifocal non-invasive/invasive tumor component. Assessment of matched benign renal tissue (BL193) was performed in parallel. All high confidence somatic variants passing Sanger sequencing validation are shown. Variant allele frequencies are indicated according to the color scale. F. Sanger sequencing traces for HRAS Q61L and CIC F1493L are shown. The position of the called variant is indicated by an arrow and the reference sequence and variant base are given.
To characterize the genetic relationship between the components, we performed comprehensive, targeted next generation sequencing using the Comprehensive Cancer Panel (CCP), which targets all coding exons of 409 cancer related genes. We generated an average of 5,003,158 mapped reads (304x targeted base coverage) per sample across the seven specimens: spatially distinct non-invasive high grade papillary urothelial carcinomas (BL191, BL192, BL194, BL195), paired invasive (BL190) and non-invasive (BL196) high grade papillary urothelial carcinoma components in the right renal pelvis and normal kidney (BL193) (Table S1).
Across the six tumor components, we identified a total of 36, high confidence, non-synonymous somatic mutations, representing 9 unique genomic positions (Fig 1E and Table S2). For each variant, Sanger sequencing was performed on the tumor and normal sample, with 34/36 (94%) confirmed as somatic variants (Fig 1F and data not shown). The two failed variants (MYH9 E1115K and E1244K in BL190) were not detected by Sanger sequencing in either the appropriate tumor or normal sample, and were called at low variant allele frequencies (0.14 and 0.17, respectively, Table S2).
Among confirmed non-synonymous somatic mutations, five identical mutations were present in all six UC samples: HRAS (Q61L), FLT4 (Q827X), MLL2 (Q3867fs), NTRK3 (T332M), and PIK3CA (H1047L), consistent with a clonal relationship for all five non-muscle-invasive UCs present at distinct locations and the paired invasive component of one tumor (Fig 1E). Compared to the overlying non-invasive tumor (BL196), the paired invasive carcinoma component (BL190) also demonstrated a confirmed mutation in CIC (F1493L). Three unique mutations were seen in the remaining bladder (BL195) and prostatic urethra (BL194) tumors--ACVR2A W64S in BL195, and DPYD R21X and ATR R989G in BL194—consistent with continued, but very limited, clonal evolution in potential cancer drivers after divergence from the founding clone.
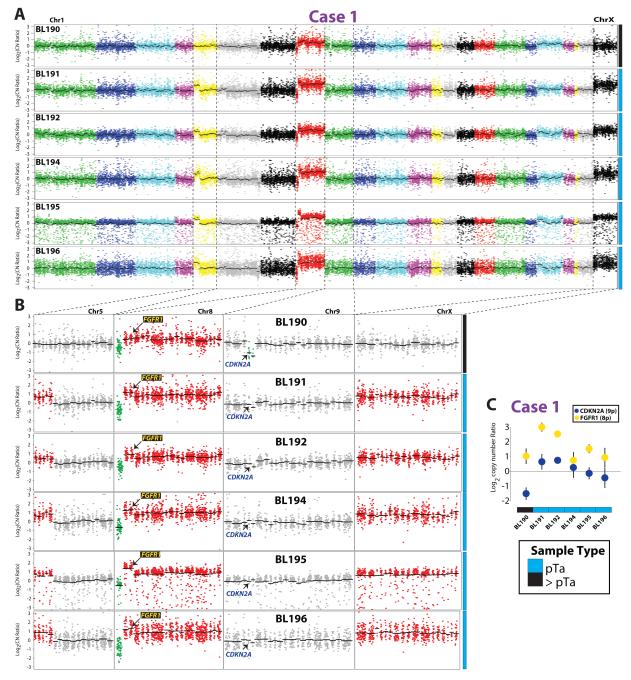
We next compared somatic CNAs between the 6 urothelial tumor samples using amplicon read counts normalized by counts from paired normal tissue (see Methods). Visualization of log2 copy number ratios demonstrated globally similar CNAs between the tumors (Fig 2A), consistent with clonal origin, with each tumor showing loss of 8p (WRN) and broad gain of the remainder of 8p and 8q (Fig 2B). Intriguingly, although the five non-invasive tumors all showed focal gain on chromosome 5p and broad gain of chromosome X, these CNAs were not seen in the invasive tumor (BL190) (Fig 2B). Conversely, BL190 harbored a focal homozygous deletion of the CDKN2A locus which was not present in any of the non-invasive components (Fig 2B). Loss of CDKN2A exclusively in BL190 and gain of FGFR1 in all tumor samples (located at 8p11) was confirmed by qPCR (Fig 2C).
Figure 2. Copy number analysis of multifocal urothelial carcinoma (UC) supports monoclonal origin and identifies private lesions in the invasive tumor component (BL190).
For each multifocal UC sample from Case 1, copy number ratios compared to matched normal tissue (BL193) were determined for each amplicon from next generation sequencing (NGS) data. A. Log2 copy number ratios per amplicon are plotted (chromosomes indicated by different colors), with gene-level copy number estimates (black bars) indicated. B. Indicated chromosomes are shown in higher resolution. Regions of gain and loss are shown in red and green, respectively. All tumor samples show the same loss of WRN on 8p and gain of the remainder of 8p and 8q. All tumor samples but the invasive component (BL190) show the same gain on chromosomes 5p and X. BL190 has a private high level loss of CDKN2A on chr 9. The location of genes validated by copy number qPCR (FGFR1 and CDKN2A) are indicated. C. qPCR confirmation of CNAs identified by next generation sequencing. qPCR on genomic DNA from BL190-196 was performed to determine copy number ratios of CDKN2A (9p21; lost in BL190) and FGFR1 (8p11; gained in all tumor samples) using PIK3R1 (5q13; unaltered in all samples) as the reference gene. Normalized mean CDKN2A (blue) and FGFR1 (yellow) log2 copy number ratios using BL193 (normal DNA) as the calibrator from triplicate qPCR +/- S.D. are plotted.
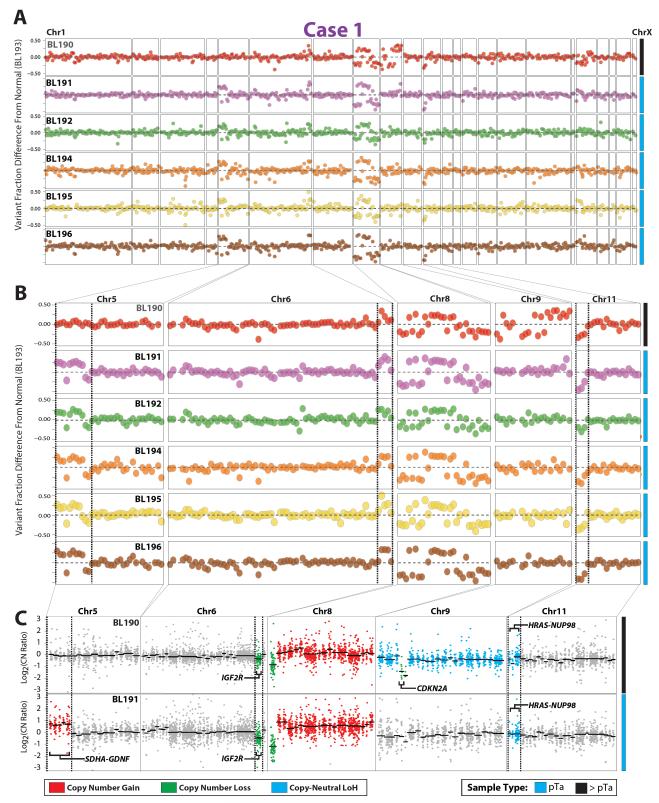
We similarly assessed loss of heterozygosity (LOH) in the tumors, which can be due to copy number alteration or copy number neutral, by comparing variant allele frequencies for heterozygous SNPs between the normal tissue sample (BL193) and each tumor sample. Concordant changes in the variant allele frequencies (towards 1 or 0) in tumor samples indicate either gain or loss of the same parental chromosome (in areas of copy number change) or clonal, copy number neutral LOH. We identified 629 well-supported, heterozygous SNPs present in BL193 that were also called in at least one tumor sample. As shown in Figure 3A&B, we observed concordant changes in SNP variant allele frequencies across all tumors on chromosome 8, consistent with the broad copy number loss/gain described above. We also identified concordant changes in variant allele frequencies for SNPs in IGF2R (chr 6q25) across all six tumors (Fig 3A&B), with focal low level IGF2R copy loss observed in all six tumors (Fig 3C). Likewise, all five non-invasive tumors showed concordant SNP variant allele frequency changes in the area of copy number gain on chromosome 5p, which was not present in BL190 (Fig 3A&B). These findings support our copy number analysis and the clonal origin of these tumors, through demonstration of gain/loss of the same parental chromosome in each shared alteration.
Figure 3. Loss of heterozygosity (LOH) analysis from next generation sequencing (NGS) data supports copy number alteration analysis and clonal origin of multifocal urothelial carcinoma (UC).
LOH analysis was performed through assessment of the change in variant allele frequency of heterozygous germline SNPs (see Methods) between normal tissue (BL193) and each tumor sample in Case 1. Positive and negative values in areas of copy number gain or loss support LOH secondary to chromosomal gain or loss and those in areas without copy number gain support copy number neutral LOH. Tumor samples with concordant positive or negative values indicate gain/loss of the same chromosome. A&B. Genome wide (A) and high resolution (B) LOH plots, with each point representing the difference in variant frequency for a heterozygous SNP between each tumor sample and the normal tissue (BL193). Colors indicate different tumor samples. Focal LOH in B is indicated by dashed lines. C. Copy number plots (as in Fig 3) for the invasive component (BL190) and a representative non-invasive tumor (BL191 shown) in areas of LOH. Colors indicate genes/chromosomes with copy number gain (red), loss (green), or copy-neutral LOH (cyan), with genes in areas of focal copy number neutral or LOH in areas of copy number gain/loss are indicated.
We also observed focal concordant copy number neutral LOH in contiguous SNPs extending from HRAS to NUP98 on chromosome 11p15 (rs41258054, rs35404087, rs11029446) in all six tumors (Fig 3C). In addition, we observed broad copy neutral LOH across chromosome 9 in BL190, which was not present in the overlying non-invasive component (BL196) or any of the other non-invasive UCs (Fig 3A-C). Although the CCP is not designed to assess LOH (few amplicons target common heterozygous SNPs), our results demonstrate that this approach can be used to support copy number alteration analysis and identify copy neutral LOH.
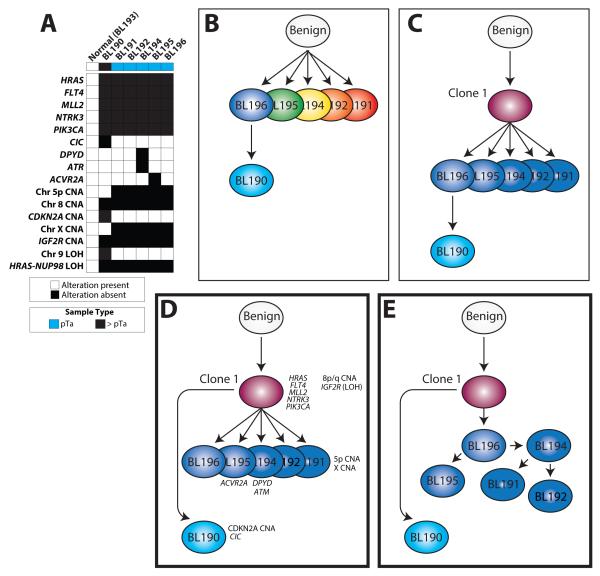
Phylogenetic analysis (see Methods) was performed to characterize the evolutionary relationship between these six tumor samples; however the limited number of informative somatic mutations/CNAs/LOH events (Fig 4A) resulted in multiple equally-plausible phylogenetic tree models for tumor evolution. Critically, none of these models supported multiclonal disease in the papillary urothelial tumors (Fig 4B) or derivation of the invasive clone from the overlying clonal papillary urothelial tumors (Fig 4C). All equally-plausible models demonstrated that BL190 diverged from the clone that gave rise to all five non-invasive components, consistent with the CDKN2A homozygous deletion and chromosome 9 LOH present only in BL190, and the chromosome 5p and X CNAs found exclusively in the five non-invasive lesions (Fig 4D&E).
Figure 4. Evolution and progression of multifocal urothelial carcinoma (UC) as demonstrated by targeted next generation sequencing (NGS).
A. Phylogenetic analysis was performed using indicated non-synonymous mutations, CNAs and LOH events from Case 1. Multiple equally likely trees were identified (modeled in D&E). B-E. Multiple evolutionary mechanisms could account for the progression from a benign cell to the multifocal non-invasive papillary UC tumors with an invasive component underlying one tumor. B. Separate tumor clones could have seeded each papillary tumor, with evolution of the invasive component from the overlying non-invasive component. C. A single tumor clone could have seeded each papillary tumor, with evolution of the invasive component from the overlying non-invasive component. D-E. Phylogentic analysis of next generation sequencing data from Case 1 tumors supports a model where a single clone seeded each papillary tumor (which may represent detachment and implantation from the first tumor, as modeled in E), with the invasive component (BL190) derived from the original clone, rather than the overlying non-invasive tumor. Alterations present at each stage and sample are indicated. Genes harboring somatic mutations (italicized), chromosomal regions with copy number gain/loss, and copy neutral loss of heterozygosity (LOH) are indicated. In D&E, subclonal relationships of the papillary non-invasive tumors could not be further characterized due to the lack of informative mutations/alterations.
One potential mechanism for the similar genetic findings in the spatially distinct tumors is transformation of the “normal” appearing intervening urothelium, the so called “field effect.” Similarly, the normal urothelium may have been infiltrated with rare transformed cells that seeded each papillary tumor. However no carcinoma in situ (including involvement by individual cancerous cells) was appreciated microscopically in this case. Hence, we performed careful macrodissection of two sections of histologically normal urothelium (Fig S1), one through the left ureteral orifice, and performed targeted NGS using a custom Ion Torrent Ampliseq panel that targets ~130 cancer related genes, including the observed HRAS and PIK3CA mutations. At high coverage over the HRAS (375x in BL193A and 381x in BL193B) and PIK3CA mutation positions (381x in BL193A and 459x in BL193B) in these normal urothelial samples, we observed no evidence of the clonal mutations found in all tumor samples (0 mutant allele containing reads), as shown in Fig S1. Sequencing of BL195 with this panel confirmed the presence of the HRAS and PIK3CA mutations at 63% and 31%, respectively, consistent with results from the CCP (data not shown).
To extend the findings from this unique case, we identified three additional cases (Cases 2-4) of upper tract UC where we could isolate separate non-invasive/invasive components by macrodissection (Table S1). All cases represented high grade, invasive (at least pT3) UC, with histology shown in Fig 5A. All samples were assessed using the CCP as for Case 1, except sequencing was performed using the Ion Torrent Proton Sequencer. Across the seven samples (matched normal benign urothelium was also sequenced for Case 3), we generated an average of 9,376,584 mapped reads (575x targeted base coverage) per sample (Table S1).
Figure 5. Next generation sequencing (NGS) of paired non-invasive/invasive urothelial carcinoma (UC) supports clonality and identifies heterogeneity in potential cancer driver alterations.
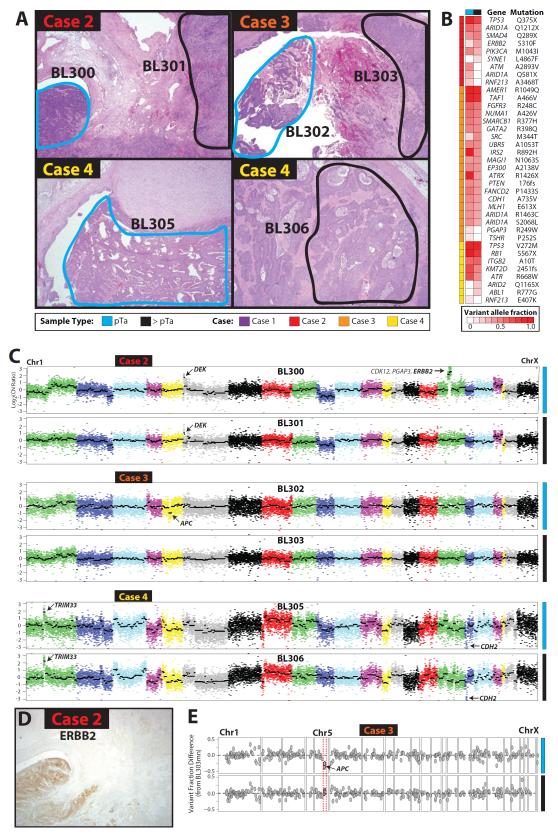
Non-invasive and invasive components were macrodissected from three cases of upper tract, high grade, invasive UC (>pT2) and subjected to targeted NGS as for Case 1 (Figs 1-4). A. Histology of each case according to the legend. Original magnification 2x for all panels. B. Shared somatic mutations demonstrate clonality but identify heterogeneity of potential driving mutations in paired non-invasive/invasive components. Heatmap of all high confidence somatic mutations from each case are shown. Variant allele frequencies are indicated according to the color scale. C. Copy number analysis from NGS data supports clonality but identify potentially driving alterations exclusively in paired non-invasive components. Focal high level CNAs are indicated. D. Validation of ERBB2 amplification by IHC exclusively in the non-invasive component of Case 2 (BL300). Compare to H&E stained section in A. 2x original magnification. E. LOH support of APC loss exclusively in the non-invasive component of Case 3 (BL302). LOH analysis was performed as in Fig 4. Variant allele fraction shifts in APC heterozygous SNPs were exclusively present in BL302 (top) and not the paired invasive component (BL303), consistent with LOH due to copy number loss in BL302.
Across the paired non-invasive tumor components in Cases 2, 3 and 4, we identified a total of 14, 38 and 13 high confidence, non-synonymous somatic mutations, representing 9, 20 and 8 unique genomic positions, respectively (Fig 5B and Table S2). In each case, known driving alterations were present in both non-invasive and invasive components, including ARID1A Q1212X, ERBB2 S310F and TP53 Q375X in Case 2, FGFR3 R248C, ARID1A R1463C/S2068L and MLH1 E613X in Case 3, and KMT2D 2451fs, RB1 567X and TP53 V272M in Case 4, consistent with clonality. Case 2 harbored a low variant frequency SYNE1 L4867F mutation exclusively in the invasive component (BL301), but three mutations exclusively in the non-invasive component (BL300, including ARID1A Q581X). Case 3 harbored no somatic mutations exclusively in the invasive component (BL303) and two low frequency mutations exclusively in the non-invasive component (BL302). Lastly, Case 4 harbored two somatic mutations in the invasive component (BL306, including ARID2 Q1165X) and one low frequency mutation exclusively in the non-invasive component (BL305).
Copy number analysis of Cases 2-4 supported clonality between each non-invasive/invasive pair, consistent with mutation data (Fig 5C). Focal, high level CNAs were identified in both components of Cases 2 (DEK amplification) and 4 (TRIM33 amplification and CDH2 deletion). Intriguingly, however, focal, high level CNAs involving known cancer genes were identified exclusively in the non-invasive components of Cases 2 (ERBB2 amplification) and 3 (APC deletion). To confirm these CNAs, we performed immunohistochemistry for ERBB2 on Case 2, which demonstrated strong (3+) staining exclusively in the non-invasive component (BL300), as shown in Fig 5D. Likewise, LOH analysis on Case 3 demonstrated changes in APC SNP variant allele frequencies (compared to BL303mn) exclusively in the non-invasive component (BL302), as shown in Fig 5E. Taken together, CNA analysis demonstrated surprising heterogeneity between paired non-invasive/invasive components, with alterations in key cancer drivers occurring exclusively in the non-invasive component in two cases.
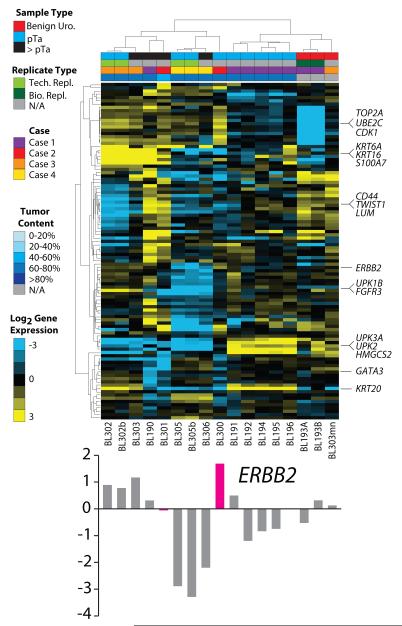
Lastly, we assessed transcriptional signatures in all tumors/components from Cases 1-4 using targeted, multiplexed PCR based RNAseq, which utilizes 20ng FFPE isolated RNA. We designed a custom Ampliseq RNA panel assessing 8 housekeeping genes and 103 target genes representing key transcriptional modules identified through analysis of publically available UC profiling studies and meta-analysis of UC studies in the Oncomine database[1, 7, 8]. In total, we sequenced three benign macrodissected urothelium samples (BL193A, BL193B and BL303mn), 12 UC samples from Cases 1-4, and two technical replicates with additional amplification cycles during library preparation (BL302b and BL205b). Across the 17 samples, through multiplexed sequencing on the PGM, we generated an average of 384,867 mapped reads (91% on target) resulting in an average of 104/111 amplicons with at least 10 mapped reads (Table S3). Normalized target gene expression was highly concordant for biological (BL193A/B) and technical (BL302/b, BL305/b) replicates ( Pearson’s r2 = 0.90, 0.97 and 0.89, respectively). Unsupervised hierarchical clustering identified four major clusters: 1) benign urothelium, 2) non-invasive UC from Case 1, and 3&4) non-invasive/invasive UC from Cases 2-4 and invasive UC from Case 1. Similarly, robust transcriptional signatures of genes representing proliferation, basal and luminal transcriptional programs were identified. Lastly, targeted RNAseq confirmed the over-expression of ERBB2 exclusively in the non-invasive component of Case 2 (BL300), consistent with the results from CNA analysis and IHC as shown in Fig 5.
Discussion
Here we used targeted, multiplexed PCR based NGS from routine FFPE tissue to assess clonality, DNA/RNA based alterations in multifocal and paired non-invasive/invasive tumor components in an extreme case of multifocal urothelial cancer (Case 1) and three additional cases of upper tract UC with distinct non-invasive/invasive components (Cases 2-4). In Case 1, amongst the five, spatially distinct papillary UCs, we identified five identical somatic point mutations in each tumor, demonstrating clonality. The shared HRAS Q61L and PIK3CA H1047L mutations are well described recurrent hotspot mutations in UC, and MLL2 was recently identified as one of the most significantly mutated genes (predominantly inactivating frameshift mutations) in UC by The Cancer Genome Atlas (TCGA) effort [1, 3, 13-15]. Through assessment of amplicon read counts, we identified clonal CNAs of chromosomes 5p, 8, X and IGF2R in all five spatially distinct tumors. Lastly, LOH analysis confirmed that these CNAs affected the same parental chromosome in each tumor, and further identified clonal copy number neutral LOH of HRAS-NUP98 (chr 11p15) in each spatially distinct tumor. Together, these results exclude multiclonal origin for the papillary urothelial tumors, and instead support a clonal origin and surprisingly minimal genetic differences in tumors arising tens of centimeters apart in the urinary tract. We sequenced only 400 cancer related genes (targeting over 50% of the Cancer Gene Census), precluding complete analysis of intertumoral heterogeneity. However, we identified only three private somatic non-synonymous mutations and no private high level CNAs amongst the five tumors, supporting very limited inter-tumoral heterogeneity amongst likely cancer driver genes.
In addition to the five spatially distinct papillary urothelial components in Case 1, we also sequenced the invasive UC component (BL190) underlying the non-invasive tumor in the right kidney (BL196). BL190 and all non-invasive components shared the same five somatic mutations, chromosome 8 and IGF2R CNAs (supported by LOH analysis), and copy number neutral LOH in HRAS-NUP98. A validated CIC F1493L somatic mutation, focal homozygous loss of CDKN2A, and chromosome 9 LOH were present exclusively in BL190, while BL190 lacked the chromosome 5p and X chromosome CNAs that were present in all five non-invasive tumors. Although loss of CDKN2A is the most common focal somatic event in urothelial cancer and has been shown to be more common in muscularis propria invasive (pT2) vs non-invasive UC (pTa)[1, 20], to our knowledge it has not previously been described as a driver of the invasive phenotype from non-invasive carcinoma. The role of the CIC in UC invasion is unknown. Likewise, results from sequencing three additional cases with paired non-invasive/invasive components identified an ARID2 Q1165X mutation exclusively in the invasive component of Case 4, nominating another potential mediator of invasion in UC.
Taken together, our results confirm clonality of the spatially distinct UC and the underlying invasive component in this case. However, phylogenetic analysis supports divergence of the invasive component from the clone that gave rise to all five papillary non-invasive tumors (Fig 5D&E), rather than the invasive component being derived from the overlying non-invasive tumor (Fig 5B&C). In sequencing fresh frozen metachronous urothelial carcinomas (initial non-invasive urothelial carcinomas and later invasive urothelial carcinomas), Nordendoft et al. similarly showed that while both non-invasive and invasive cancers showed multiple identical mutations, indicating clonal origin, mutational events unique to both non-invasive and invasive tumors were present[19]. Their findings similarly indicate divergence of non-invasive and invasive components from a common ancestor, although they were sequencing temporally separated tumors, rather than separate non-invasive/invasive components of the same tumor. In the present study, we cannot entirely exclude the possibility that the invasive clone was derived from the overlying non-invasive tumor (Fig 5C), which would require loss of the gained regions of chromosomes 5p and X seen in the non-invasive component; however, this explanation is less parsimonious. One possible explanation is that the invasive clone colonized the collecting duct system and then invaded into the kidney, rather than invading from the overlying papillary tumor. Similarly, the remarkable homogeneity amongst the papillary UCs (despite their spatial separation by tens of centimeters), without evidence of a field effect in histologically normal urothelium, raises the question of how these tumors arose. One possibility is that malignant clones detached from the surface of a primary tumor, “floated” to a distant portion of urothelium inside the urinary space, and implanted without invasion through the lamina propria. However, we are not aware of this mechanism, which would require contralateral (if from the upper tract) or bilateral dissemination (if from the bladder), and hence must be considered speculative. Nevertheless, the findings in this case are in contrast to the heterogeneity observed amongst driving alterations even at the intratumoral level in some cancers, most notably clear cell renal cell carcinomas[10, 11]. Given the remarkable presentation of this patient, whether our findings are applicable to other urothelial cancers requires further study and must be interpreted with caution.
Given the intriguing differences between the paired non-invasive/invasive components in Case 1, we similarly assessed three additional cases of >pT2 upper tract UC where we could separately isolate non-invasive/invasive components. In each case, somatic mutations and CNA profiles were highly concordant in each pair, consistent with clonal origin. Likewise, known driving alterations, such as DEK amplification and TP53 mutations were identified in each case. Intriguingly, in Cases 2, we identified focal amplification of ERBB2 and a deleterious ARID1A mutation (Q1212X) exclusively in the non-invasive component. Of note, a known somatic driving mutation in ERBB2 (S310F)[12, 21] and a second deleterious mutation in ARID1A (Q1212X) were present in both the non-invasive and invasive components from this case. Variant frequencies of other somatic alterations in both components of this case (including a homozygous TP53 Q375X mutation) support similar tumor content, yet the ERBB2 S310F variant was present at much lower frequency in the non-invasive vs. invasive component (0.13 vs. 0.37), consistent with amplification of non-mutant ERBB2 allele in the non-invasive component. Together, these results demonstrate heterogeneity in known driving alterations between paired non-invasive/invasive components and support continued selection for somatic alteration of these genes in the non-invasive tumor component after divergence of the invasive clone.
Lastly, we performed targeted RNAseq on all tumor components from Cases 1-4 as well as macrodissected urothelium using a custom panel designed to assess major transcriptional programs in urothelial cancer. Importantly, this approach is highly robust as demonstrated through biological and technical replicates across 5-6 orders of magnitude of target gene expression. This analysis supported heterogeneity seen at the DNA level and identified cases with both highly concordant and distinct transcriptional profiles between paired non-invasive components. For example, Case 3 (BL302/303), which showed highly similar mutational and CNA profiles (including FGFR3 R248C mutation), showed highly correlated transcriptional profiles in both components (basalhi/luminallow/p63low/CK20hi/proliferationhi/EMTlow/FGFR3hi). Likewise, Case 4 (BL305/306) showed highly correlated basallow/ luminallow/ p63hi/ CK20low/ proliferationhi/ EMTlow/ FGFR3low transcriptional programs in both components. On the other hand, in Case 1, the invasive component (BL190: basalhi/ luminallow/ p63hi/ CK20low/ proliferationmod/ EMThi/ FGFR3low), overlying non-invasive component (BL196: basalhi/ luminallow/ p63low/ CK20hi/ proliferationlow/ EMTlow/ FGFR3mod) and other non-invasive papillary tumors (BL191, BL192, BL194 and BL195: basallow/ luminalhi/ p63low/ CK20hi/ proliferationlow/ EMTlow/ FGFR3mod) showed marked differences in key transcriptional signatures. Likewise, Case 2 showed distinct profiles in the non-invasive (BL300: basallow/ luminalmod/ p63mod/ CK20low/ proliferationhi/ EMTlow/ FGFR3mod) and invasive (BL301: basalhi/ luminallow/ p63hi/ CK20low/ proliferationhi/ EMThi/ FGFR3mod) components, in addition to higher ERBB2 expression in BL300 as expected based on DNA and IHC data (Fig 5). Taken together, our results demonstrate the utlity of targeted RNAseq to assess key transcriptional signatures in FFPE UC tissue, and demonstrate unique profiles in distinct paired non-invasive/invasive components and cases.
Importantly, our approaches are compatible with small amounts of DNA (≤40ng) and RNA (≤20ng) obtained from routine FFPE surgical pathology specimens. Further, careful macrodissection, such as that required to isolated normal urothelium or non-invasive/invasive components, is not possible with fresh frozen tissue. Hence, in addition to informing on inter-tumor genetic relationships and non-invasive/invasive tumor components, this FFPE compatible approach may be useful for determining clonal relationships and driving alterations in precursor lesions and bona fide cancers, as well as assessing distinct areas of differentiation/morphology within a tumor.
Supplementary Material
Figure 6. Targeted, multiplexed PCR based next generation RNA sequencing (RNAseq) identifies distinct transcriptional signatures in multifocal and paired non-invasive/invasive urothelial carcinoma (UC).
Benign urothelium (BL193A, BL193B and BL303mn) and all tumor components from Cases 1-4 were subjected to targeted RNAseq using 20ng FFPE isolated RNA per sample and a custom Ampilseq panel assessing 8 housekeeping genes and 103 target genes. Biological and technical replicates (BL302b and BL305b, subjected to 3 extra amplification cycles during library construction to assess for over-amplification effects) are indicated in the legend. Normalized, median centered, log2 target gene expression is shown in the heatmap (according to the color scale), with centroid linkage clustering of genes and samples. From top to bottom, genes representing proliferation, basal, EMT, ERBB2, FGFR3, luminal, GATA3 and CK20 (KRT20) modules are indicated. ERBB2 expression to support amplification and over-expression by IHC exclusively in BL300 compared to BL301 (see Fig 5) is shown below the heatmap. Values for all genes are given in Table S4.
Acknowledgments
The authors thank Mandy Davis and Angela Fullen for technical assistance and Arul Chinnaiyan and Eric Fearon for critical review of the manuscript. This work was supported by the University of Michigan Department of Anatomic Pathology Operations Committee (to J.I.W. and SAT) and generous gifts from the Teeter and MacLeod families for bladder cancer research at the University of Michigan.
Footnotes
Disclosures/conflicts of interest: S.A.T has a separate sponsored research agreement with Compendia Bioscience/Life Technologies. None of the study described herein was supported by Compendia Bioscience/Life Technologies and they had no role in the data collection, interpretation, or analysis, and did not participate in the study design or the decision to submit for publication. All authors have no disclosures or conflicts of interest otherwise.
References
- 1.Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014 doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babjuk M, Oosterlinck W, Sylvester R, et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder, the 2011 update. Eur Urol. 2011;59:997–1008. doi: 10.1016/j.eururo.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 3.Balbas-Martinez C, Sagrera A, Carrillo-de-Santa-Pau E, et al. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat Genet. 2013;45:1464–1469. doi: 10.1038/ng.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bornman DM, Mathew S, Alsruhe J, Herman JG, Gabrielson E. Methylation of the E-cadherin gene in bladder neoplasia and in normal urothelial epithelium from elderly individuals. Am J Pathol. 2001;159:831–835. doi: 10.1016/S0002-9440(10)61758-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castillo-Martin M, Domingo-Domenech J, Karni-Schmidt O, Matos T, Cordon-Cardo C. Molecular pathways of urothelial development and bladder tumorigenesis. Urologic Oncology. 2010;28:401–408. doi: 10.1016/j.urolonc.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 6.Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet. 2012;49:433–436. doi: 10.1136/jmedgenet-2012-100918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi W, Porten S, Kim S, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152–165. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damrauer JS, Hoadley KA, Chism DD, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci U S A. 2014;111:3110–3115. doi: 10.1073/pnas.1318376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denzinger S, Mohren K, Knuechel R, et al. Improved clonality analysis of multifocal bladder tumors by combination of histopathologic organ mapping, loss of heterozygosity, fluorescence in situ hybridization, and p53 analyses. Hum Pathol. 2006;37:143–151. doi: 10.1016/j.humpath.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 10.Gerlinger M, Horswell S, Larkin J, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014 doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England Journal of Medicine. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greulich H, Kaplan B, Mertins P, et al. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc Natl Acad Sci U S A. 2012;109:14476–14481. doi: 10.1073/pnas.1203201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo G, Sun X, Chen C, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. 2013;45:1459–1463. doi: 10.1038/ng.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyer G, Al-Ahmadie H, Schultz N, et al. Prevalence and co-occurrence of actionable genomic alterations in high-grade bladder cancer. J Clin Oncol. 2013;31:3133–3140. doi: 10.1200/JCO.2012.46.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kompier LC, Lurkin I, van der Aa MN, van Rhijn BW, van der Kwast TH, Zwarthoff EC. FGFR3, HRAS, KRAS, NRAS and PIK3CA mutations in bladder cancer and their potential as biomarkers for surveillance and therapy. PLoS One. 2010;5:e13821. doi: 10.1371/journal.pone.0013821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin HH, Ke HL, Wu WJ, Lee YH, Chang LL. Hypermethylation of E-cadherin, p16, p14, and RASSF1A genes in pathologically normal urothelium predict bladder recurrence of bladder cancer after transurethral resection. Urologic oncology. 2012;30:177–181. doi: 10.1016/j.urolonc.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Louhelainen J, Wijkstrom H, Hemminki K. Allelic losses demonstrate monoclonality of multifocal bladder tumors. Int J Cancer. 2000;87:522–527. [PubMed] [Google Scholar]
- 18.Netto GJ. Molecular biomarkers in urothelial carcinoma of the bladder: are we there yet? Nat Rev Urol. 2012;9:41–51. doi: 10.1038/nrurol.2011.193. [DOI] [PubMed] [Google Scholar]
- 19.Nordentoft IL,P, Birkenkamp-Demtroder K, Shumansky K, Vang S, Hornshoj H, Juul M, Villesen P, Hedegaard J, Roth A, Thorsen K, Hoyer S, Borre M, Reinert T, Fristrup N, Dyrskjot L, Shah S, Pedersen JS, Orntoft TF. Mutational context and diverse clonal developments in early and late bladder cancer. Cell Rep. 2014 doi: 10.1016/j.celrep.2014.04.038. [DOI] [PubMed] [Google Scholar]
- 20.Rebouissou S, Herault A, Letouze E, et al. CDKN2A homozygous deletion is associated with muscle invasion in FGFR3-mutated urothelial bladder carcinoma. J Pathol. 2012;227:315–324. doi: 10.1002/path.4017. [DOI] [PubMed] [Google Scholar]
- 21.Ross JS, Wang K, Gay LM, et al. A high frequency of activating extracellular domain ERBB2 (HER2) mutation in micropapillary urothelial carcinoma. Clin Cancer Res. 2014;20:68–75. doi: 10.1158/1078-0432.CCR-13-1992. [DOI] [PubMed] [Google Scholar]
- 22.Sidrandsky DF,P, Von Eschenbach A, Oyasu R, Preisinger AC, Vogelstein B. Clonal origin of bladder cancer. The New England journal of medicine. 1992;326:737–740. doi: 10.1056/NEJM199203123261104. [DOI] [PubMed] [Google Scholar]
- 23.Simon R, Eltze E, Schafer KL, et al. Cytogenetic analysis of multifocal bladder cancer supports a monoclonal origin and intraepithelial spread of tumor cells. Cancer Res. 2001;61:355–362. [PubMed] [Google Scholar]
- 24.Stoehr R, Hartmann A, Hiendlmeyer E, Murle K, Wieland W, Knuechel R. Oligoclonality of early lesions of the urothelium as determined by microdissection-supported genetic analysis. Pathobiology : journal of immunopathology, molecular and cellular biology. 2000;68:165–172. doi: 10.1159/000055919. [DOI] [PubMed] [Google Scholar]
- 25.Stoehr R, Knuechel R, Boecker J, et al. Histologic-genetic mapping by allele-specific PCR reveals intraurothelial spread of p53 mutant tumor clones. Lab Invest. 2002;82:1553–1561. doi: 10.1097/01.lab.0000035022.29742.85. [DOI] [PubMed] [Google Scholar]
- 26.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:3. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xylinas E, Rink M, Margulis V, et al. Multifocal carcinoma in situ of the upper tract is associated with high risk of bladder cancer recurrence. Eur Urol. 2012;61:1069–1070. doi: 10.1016/j.eururo.2012.02.042. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto S, Romanenko A, Wei M, et al. Specific p53 gene mutations in urinary bladder epithelium after the Chernobyl accident. Cancer Res. 1999;59:3606–3609. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.